ABSTRACT
BACKGROUND:
Cystic fibrosis (CF) occurs in populations in Saudi Arabia and the Gulf area. Approximately 2000 known variants have been identified for the CF transmembrane conductance regulator (CTFR) gene. Screening for ten of the most common variants can detect 80% of alleles.
OBJECTIVE:
Determine the pattern of CFTR variants in the CF population of Saudi Arabia.
DESIGN:
A retrospective, descriptive.
SETTING:
Tertiary care center.
PATIENTS AND METHODS:
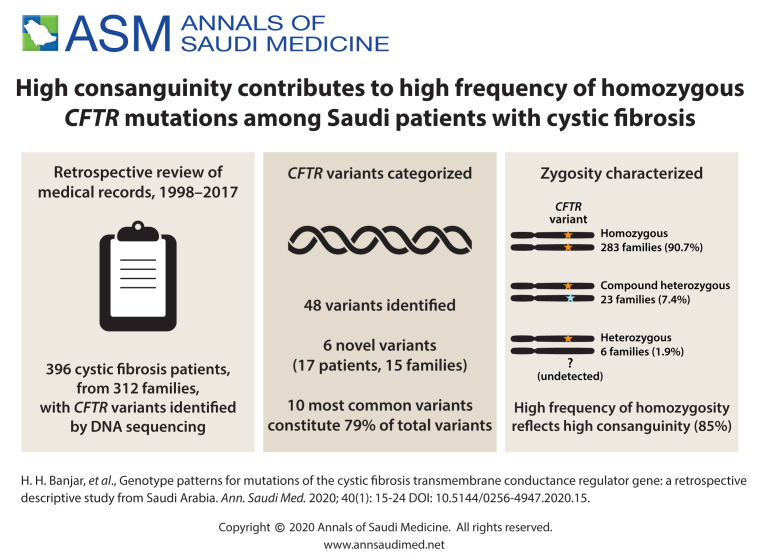
We examined the medical records of 396 confirmed CF patients of all age groups that were positive for a CFTR variant from the period of 1 January 1998 to 1 December 2017.
MAIN OUTCOME MEASURES:
Zygosity, morbidity and mortality patterns of different types of CFTR variants.
SAMPLE SIZE:
312 families that included 396 patients.
RESULTS:
Of 48 variants identified, 6 were novel, having not been described in the medical literature. A homozygous state was found in 283 families (90.7%) and compound heterozygosity in 23 (7.4%). Six families were heterozygous (1.9%). Median age (interquartile range) was 10.2 months (4.4 months to 5.7 years) at diagnosis and 9.7 (5.4-16.5) years at follow up. Of 396 patients, 378 patients (95.5%) survived and 18 (4.5%) died. The ten most common variants identified in descending frequency were: p.Gly473GlufsX54 in 98 alleles (16%), p.Ile1234Val in 66 alleles (11%), F508del in 64 alleles (11%), 711+1G>T in 62 alleles (10%), 3120+1G>A in 62 alleles (11%), p.His139Leuin 38 alleles (6.4%), p.Gln637Hisfs in 30 alleles (5.2%), p.Ser549Arg in 27 alleles (4.5%), p.Asn1303Lys in 14 alleles (2.3%), delExon19-21in 10 alleles (1.6%). This analysis identified 79.2% of our CFTR variants.
CONCLUSION:
CFTR mutational patterns in our CF population are characterized by a high allelic heterogeneity. The high prevalence of homozygous variants reflects the high level of consanguinity between parents.
LIMITATIONS:
Our CFTR screening reflected only about 80% of CF patients in Saudi Arabia.
CONFLICT OF INTEREST:
None.
INTRODUCTION
Cystic fibrosis (CF) is a monogenic and lethal disorder that affects multiple organ systems of the body.1,2 The Cystic Fibrosis Foundation reports that 60 000 to 70 000 people across the world suffer from CF.3 The prevalence of CF in the Middle East has been estimated to fall in the range of 1 in 2500-5000.4 Approximately 2000 known variants have been identified for the CF gene.5 Six different classes of variants are described depending on the fate of the protein coded by the cystic fibrosis transmembrane conductance regulator (CFTR) gene.2 Mutations in the gene lead to altered protein synthesis, which leads to insufficient active CFTR at the cell surface. In a previous study, we showed that screening for ten common CFTR gene mutations can detect 80% of cases positive for a CFTR variant.4
CFTR variants in the Saudi population have been described.1,2,5 In this report, we present up-to-date and comprehensive data on the allele frequency of CFTR mutations, zygosity, morbidity and mortality patterns of the largest cystic fibrosis center in the Gulf area and the only facility that performs whole genome sequencing for CFTR in Saudi Arabia.
PATIENTS AND METHODS
This study was a retrospective chart review of all CF patients referred to the CF clinic during the period from January 1998 to December 2017. CF was diagnosed based on a typical clinical picture of cough, sputum production and high sweat chloride test in two subsequent testing samples (>60 mmol/L) by the Wescor quantitative method.2 In the study, routine evaluation of all patients included a detailed medical and family history, physical examination, laboratory investigations and CFTR variant testing.
Pancreatic insufficiency was diagnosed based on stool elastase measurement of >100–200 ug/g in stool, or a 72-hour fecal fat estimation using the van de Kamer method;6 of which a positive result corresponded to a fecal fat content >7 g/24 hours. Bronchiectasis was identified as dilated bronchi through radiological studies like chest X-rays or computed tomography (CT). The percentage of each clinical presentation is counted in reference to the total CF population. The diagnosis of cystic fibrosis-related diabetes (CFRD) was based on fasting blood glucose (FBG) levels ≥126 mg/dL (7.0 mmol/L) or 2-hour postprandial plasma glucose levels ≥200 mg/dL (11.1 mmol/L) that persisted for more than 48 hours in duration, in the absence of a steroid effect. Respiratory cultures were taken from nasopharyngeal aspirates for patients younger than 5 years of age; and from induced sputum for patients older than 5 years of age. Culture of bronchoalveolar lavage specimens were taken from 10% of our CF population.
Definition of CFTR genotype
A detailed family history was taken to identify other family members affected with CF. A homozygous genotype was identified as two identical pathogenic CFTR variants. In a compound heterozygous genotype, two different pathogenic CFTR variants occurred in a trans configuration at two different alleles for the same chromosome. In a heterozygous genotype, one variant was identified in one allele, while the other pathogenic alleles were not identified due to the death of parents or unavailability of CFTR advanced detection technology at the time of CFTR screening. CFTR private mutations have only been described in 1 to 2 families.
CFTR allele counts for patients and families
Multiple siblings with a homozygous CFTR variant within the same family are counted as having two alleles. In a compound heterozygous CFTR variant, each variant is counted as one mutant allele separately (i.e., multiple CF-affected siblings within the same family will be counted with each CFTR variant separately). The percentage of alleles for the patients is calculated as the number of affected alleles for a certain CFTR variant divided by the total number of alleles for whole CF population (396 patients/792 alleles). The percentage of alleles for the families is calculated as the number of families with the affected alleles for a certain CFTR variant divided by the total number of alleles for whole CF population (312 families/624 alleles).
Pancreatic enzyme replacement
Patients with pancreatic insufficiency were placed on pancreatic enzyme according to the CF Foundation recommendation of 1000 units of lipase/kg per meal for children less than age of four and 500–2500 units of lipase/kg per meal, half of this with snacks for individuals older than 4 years of age. For dosing by fat content of meals, 500–4000 units of lipase per gram of fat ingested per day is considered to more closely mimic pancreatic enzyme secretion in response to a meal.
CFTR identification
The CFTR gene screen methodology involves DNA Isolation, polymerase chain reaction (PCR) amplification of genomic DNA, mutational analysis, and sequencing methods.7 Genomic DNA from the patient's lymphocytes was used to amplify the 27 exons and flanking sequences of the CFTR (NM_000492.3; NP_000483.3). The PCR products were analyzed by sequencing in both the forward and reverse directions were applied according to currently recommended methods.8-10 Variant detection was scored using publicly available variant databases for CF such as the Cystic Fibrosis Mutation Database (http://www.genet.sickkids.on.ca/CFTR/Home.html) and The Human Gene Mutation Database – Professional Edition. (http://www.hgmd.cf.ac.uk/ac/index.php). Both mutation databases provided an extensive repertoire of up-to-date sequence variants, deletions and insertions for the CFTR gene.
Ethical considerations and statistical methods
After obtaining ethical approval by the research advisory committee, the Declaration of Helsinki, good clinical practice guidelines were followed. Data collection and data entry were supervised by the principal investigator. All data were obtained by retrospective chart review and were stored in pediatrics research unit, accessed only by the principal investigator and the assigned clinical research coordinator. Patient information was kept strictly confidential. Each patient was given a study number, and all patient data were entered in to the designated data sheet without any patient identification. The department of Biostatistics Epidemiology and Scientific Computing (BESC) carried out statistical analysis of the data. Data is descriptive using median (interquartile range), mean (standard deviation) or number (percentage).
RESULTS
From 1 January 1998 to 1 December 2017, 396 patients had confirmed CF (312 families) and were positive for CFTR variants. Median age (interquartile range) was 10.2 months (4.4 months to 5.7 years) at diagnosis and 9.7 (5.4-16.5) years at follow up. Consanguinity between parents was 85%. Clinical presentations of the 396 CF patients were pancreatic insufficiency in 376 (95%), bronchiectasis in 76 (19%), meconium ileus in 39 (10%), persistent respiratory symptoms in 186 (47%), steatorrhea in 336 (85%), increased sweating in 181 (46%), abdominal pain in 114 (29%), clubbing in 51 (13%), cholestasis in 118 (30%), and CF-related diabetes in 47.4 (12%). Vitamin A, D, E and K deficiency was detected among 37% to 70% of patients, and 371 (94%) had mild to severe malnutrition. Respiratory cultures at presentation showed Pseudomonas aeruginosa in 237 (60%), Staphylococcus aureus in 122 (31%), Haemophilus influenzae in 83 (21%) and Streptococcus pneumoniae in 63 (16%).
Of the 48 CFTR variants identified, 283 of the 312 families (90.7%) were homozygous, 23 (7.4%) families were compound heterozygous, and 6 (1.9%) families were in heterozygous (only one allele identified; the other allele could not be detected with the present technology). Of the 48 variants identified, 42 were known variants (Tables 1 and 2) and 6 were novel variants (Table 3). Among the 42 CFTR variants, 10 were identified as the top most common variants which were present in 243 families/471 alleles (79.2%) (Table 1).11-25 The remaining 32 reported variants were found in 54 different families. Twenty-nine variants were detected at exonic locations,26-51 whereas only 3 variants were detected at intronic locations in the CFTR gene (Table 2).52,53 Thirty-two of 312 families (10.2%) had private mutation.
Table 1.
Ten most common CFTR mutations in the Saudi population (243 families/318 patients).
| Ref | Location | Nucleotide change | Protein change | Alive | Died | Patients (n) | Affected alleles (patient)a | Families (n) | Affected alleles (families)b | Homozygous | Heterozygous | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | Exon 11 | c.1418delG | p.Gly473GlufsX54 | 46 | 22 | 68 | 134 (17.0) | 50 | 98 (16.0) | 48 (95.8) Ho | 2 (4.2) comp.He* | rs397508205 |
| 15 | Exon 22 | c.3700A>G | p.Ile1234Val | 38 | 8 | 46 | 92 (12.0) | 33 | 66 (11.0) | 33 (100) Ho | 0.0 | rs75389940 |
| 16 | Exon 11 | c.1521_1523delCTT | p.Phe508del | 25 | 21 | 46 | 90 (11.4) | 34 | 64 (11.0) | 30 (88.2) Ho | 2 (5.9) Comp. He * and 2 He | rs113993960 |
| 26 | Intron 5 | c.579+1G>T | 711+1G>T | 26 | 10 | 36 | 72 (10.0) | 31 | 62 (10.0) | 31 (100) Ho | 0.0 | rs77188391 |
| 18 | Intron 18 | c.2988+1G>A | 3120+1G>A | 22 | 21 | 43 | 82 (10.2) | 33 | 62 (11.0) | 29 (7.9) Ho | 4 (12.1) Comp. He* | rs75096551 |
| 17 | Exon 4 | c.416A>T | p.His139Leu | 20 | 6 | 26 | 48 (6.1) | 20 | 38 (6.4) | 18 (90) Ho | 2 (10) Comp. He* | rs76371115 |
| 21 | Exon 14 | c.1911delG | p.Gln637Hisfs | 15 | 5 | 20 | 38 (5.0) | 16 | 30 (5.2) | 14 (87.5) Ho | 2 (12.5) Comp. He* | rs1554389296 |
| 22 | Exon 12 | c.1647T>G | p.Ser549Arg | 9 | 5 | 14 | 27 (3.5) | 14 | 27 (4.5) | 13 (92.9) Ho | 1 (7.1) Comp. He* | rs121909005 |
| 22 | Exon 24 | c.3909C>G | p.N1303K | 8 | 3 | 11 | 22 (3.0) | 7 | 14 (2.3) | 7 (100) Ho | - | |
| 23 | delExon 19-21 | 7 | 1 | 8 | 16 (2.1) | 5 | 10 (1.6) | 5 (100) Ho | - | rs80034486 | ||
| Total | 318 | 632 (80.3) | 243 | 471 (79) | 228 Ho | 13 Comp. He, 2 He |
Data are number or number (percent) unless noted otherwise.
Number of affected alleles for a certain CFTR mutation divided by total number of alleles for whole CF population (396 patients/792 alleles).
Number of families with the affected alleles for a certain CFTR mutation divided by total number of alleles for whole CF population (312 families / 624 alleles). Families (compound heterozygous are counted twice in Table 1 and 2). Ho: homozygous, He: heterozygous.
Compound heterozygous:
Reference 11: p.Gly473GlufsX54; 1 patient with p.Asp579Gly(c.1736A>G); Exon 13, missense mutation, and the other patient with p.K716RfsX6 (c.2147_2149delAGAinsGG); Exon 14, Indel frameshift mutation.
16-F508del; 2 patients with p.Arg709Ter(c.2125C>T); Exon 14, nonsense mutation. The other patient had no other mutation detected with the present technology.
18-3120+1G>A; the other associated mutations were: 2 patients with p.Gln637Hisfs (c.1911delG); Exon 14, single nucleotide deletion, 1 patient with p.H139L(c.416A>T); Exon 4, missense mutation, and 1 patient with p.Trp1098Ter (c.3294G>A) Exon 20, nonsense mutation.
19-p.H139L; 1 patient with p.Gly1249Glu (c.3746G>A); Exon 23, missense mutation, and the other patient with 3120+1G>A (c.2988+1G>A); Intron 18; splice donor site.
21-p.Gln637Hisfs; 1 patient with 3120+1G>A (c.2988+1G>A); Intron 18, splice donor site. The other patient with p.Tyr849Ter(c.2547C>A); Exon 15, nonsense mutation.
22-p.Ser549Arg; 1 patient with p.Gly542Ter(c.1624G>T); Exon 12, nonsense mutation.
Table 2.
Other CFTR mutations in the Saudi population (54 families/61 patients).
| Reference | Location | Nucleotide change | Protein change | Mutation effect | #Pts | Affected alleles (patient)a | Families | Affected alleles (families)b | Zygosity | Accession No. |
|---|---|---|---|---|---|---|---|---|---|---|
| 17 | Exon19 | c.3529A>T | p.Lys1177Ter | Nonsense | 5 | 10 (1.3) | 4 | 8 (1.3) | 4 (100) Ho | rs397508578 |
| 48 | Exon.21 | c.3419T>A | p.Met1140Lys | Missense | 4 | 8 (1.0) | 4 | 8 (1.3) | 4 (100) Ho | rs397508558 |
| 37 | Exon 12 | c.1657C>T | p.Arg553Ter | Missense | 3 | 6 (1.0) | 3 | 6 (1.0) | 3 (100) Ho | rs74597325 |
| 34 | Exon 11 | c.1399C>T | p.Leu467Phe | Missense | 3 | 8 (1.0) | 3 | 6 (1.0) | 3 (100) Ho | rs1800089 |
| 50 | Exon 10 | c.1375_1383del9 | pSer459_Gly461del | Deletion | 5 | 10 (1.3) | 3 | 6 (1.0) | 3 (100) Ho | rs786205658 |
| 38 | Exon 13 | c.1736A>G | p.Asp579Gly | Missense | 4 | 5 (0.6) | 4 | 5 (0.82) | 1 (25) Ho, 2 (50) He and 1 (25) Comp He* | rs397508288 |
| 31 | Exon 24 | c.3889dupT | p.Ser1297Phefs | SNDel | 3 | 6 (0.8) | 2 | 4 (0.7) | 2 (100) Ho | rs121908808 |
| 44 | exon15 | c.2551C>T | p.Arg851Ter | Nonsense | 2 | 4 (0.6) | 2 | 4 (0.7) | 2 (100) Ho | rs121909012 |
| 42 | Exon14 | c.2421A>G | p.Ile807Met | Missense | 3 | 6 (0.8) | 2 | 4 (0.7) | 2 (100) Ho | rs1800103 |
| 28 | Exon 4 | c.443T>C | p.Ile148Thr | Missense | 2 | 4 (0.6) | 2 | 4 (0.7) | 2 (100) Ho | rs35516286 |
| 52 | Intron 2 | IVS2+12T>C | c.164+12T>C | Splice Donor-Site | 2 | 4 (0.6) | 2 | 4 (0.7) | 2 (100) Ho | rs121908790 |
| 49 | Exon 23 | c.3746G>A | p.Gly1249Glu | Missense | 2 | 2 (0.3) | 2 | 2 (0.33) | 1 (50) He and 1 (50) Comp He* | rs121909040 |
| 45 | Exon22 | c.3472C>T | p.Arg1158Ter | Nonsense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs79850223 |
| 47 | exon 7-19 | Large deletionc exon 7-19 | - | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | ||
| 54 | Intron 16 | IVS16 + 5G>Ac | c.2657+5G>A | Consensus Splice Site (Exon skipping) | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs80224560 |
| 43 | Exon 15 | c.2547C>A | p.Tyr849Ter | Nonsense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs397508394 |
| 40 | Exon 14 | c.2125C>T | p.Arg709Ter | Nonsense | 3 | 3 (0.4) | 2 | 2 (0.33) | 2 (100) Comp He * | rs121908760 |
| 27 | exon14 | c.2215delG | p.Val739Tyrfs | Frameshift | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs397508353 |
| 39 | Exon 13 | c.1697C>A | p.Ala566Asp | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs1375786834 |
| 55 | Exon11 | c.1597T>C | p.Phe533Leu | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs397508238 |
| 35 | Exon 11 | c.1516A>G | p.Ile506Val | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs1800091 |
| 19 | Exon 8 | c.920G>A | p.Ser307Asn | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs397508817 |
| 31 | Exon 7 | c.853A>T | p.Ile285Phe | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs151073129 |
| 26 | Exon 5 | c.532G>A | p.Gly178Arg | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs80282562 |
| 30 | Exon 4 | c.422C>A | p.Ala141Asp | Missense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | rs397508700 |
| 27 | Exon 4 | G115Xc | Nonsense | 1 | 2 (0.3) | 1 | 2 (0.33) | 1 (100) Ho | ||
| 25 | Exon 3 | c.202A>G | p.Lys68Glu | Missense | 2 | 4 (0.6) | 1 | 2 (0.33) | 1 (100) Ho | rs397508332 |
| 47 | Exon 20 | c.3294G>A | p.Trp1098Ter | Nonsense | 1 | 1 (0.14) | 1 | 2 (0.33) | 1 (100) Comp He * | rs397508533 |
| 22 | Exon 12 | c.1624G>T | p.Gly542Ter | Nonsense | 1 | 1 (0.14) | 1 | 1 (0.2) | 1 (100) Comp He * | rs113993959 |
| 53 | Intron 5 | IVS5-1G>Tc | c.580-1G>T; | Splice Donor-Site | 1 | 1 (0.14) | 1 | 1 (0.2) | 1 (100) Comp He* | rs121908793 |
| 55 | Exon 4 | 425del 42 exon4 | 425del 42c exon4 | Inframe Deletion | 1 | 1 (0.14) | 1 | 1 (0.2) | 1 (100) He | |
| 26 | Exon 3 | c.254G>A | p.Gly85Glu | Inframe deletion | 1 | 1 (0.14) | 1 | 1 (0.2) | 1 (100) Comp He * | rs75961395 |
| 61 | 111 (15.4) | 54 | 96 (16.2) | 42 Ho 4 He 8 Comp He |
Data are number or number (percent) unless noted otherwise
Number of affected alleles for a certain CFTR mutation divided by total number of alleles for whole CF population (396 patients/792 alleles).
Number of families with the affected alleles for a certain CFTR mutation divided by total number of alleles for whole CF population (312 families / 624 alleles).
Private mutation. Families (compound heterozygous are counted twice in Table 1 and 2). Ho: Homozygous, He: heterozygous.
Compound heterozygous:
Reference 38: p.Asp579Gly; the other associated mutation is p.Gly473GlufsX54 (c.1418delG); Exon 11, frameshift deletion
Reference 48: p.Gly1249Glu; the other associated mutation is p.H139L (c.416A>T) Exon 4, Missense mutation 40
Reference 40: p.Arg709Ter; the other associated mutation is F508del (c.1521_1523delCTT); Exon 11, inframe deletion
Reference 47: p.Trp1098Ter; the other associated mutation is 3120+1G>A (c.2988+1G>A); Intron 18; splice donor site
Reference 22: p.Gly542Ter; the other associated mutation is p.Ser549Arg (c.1645A>C); Exon 12, missense mutation
Reference 26: c.254G>A; the other associated mutation is F508del (c.1521_1523delCTT); Exon 11, inframe deletion
Table 3.
Novel CFTR mutations in the Saudi population in this study (15 families/17 patients).
| Location | Mutation | Nucleotide change | Mutation effect | Alive | Died | Patients (n) | Affected alleles (patient)a | Families (n) | Affected alleles (families)b | Zygosity |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 10 | DS459_G461 | c.1375_1383del9 | Inframe mutation | 4 | 1 | 5 | 10 (1.3) | 4 | 8 (1.3) | 4 (100) Ho |
| Exon 17 | p.Y913X | c.2739T>G | Nonsense mutation | 3 | 0 | 3 | 6 (0.8) | 3 | 6 (1.0) | 3(100) Ho |
| Exon 14 | p.K716RfsX6 | c.2147_2149delAGAinsGG | Indel Framshift Mutation | 2 | 1 | 3 | 5 (0.7) | 3 | 5 (0.82) | 2 (66.6) Ho 1 (33.3)* |
| Intron 19 | IVS19-10T>G | c.3140-10T>G | Consensus Splice Acceptor-Site mutation | 3 | 0 | 3 | 6 (0.8) | 2 | 4 (0.7) | 2 (100) Ho |
| Exon 22 | p.P1181R | c.3542C>G | Missense mutation | 2 | 0 | 2 | 3 (0.4) | 2 | 3 (0.68) | 1 (50) Ho, 1 (50)* |
| Exon 15 | p.E873Dc | c.2619G>C | Missense mutation | 1 | 0 | 1 | 2 (0.3) | 1 | 2 (0.3) | 1 (100) Ho |
| Total | 17 | 32 (4.3) | 15 | 28 (4.8) | 13 Ho, 2* |
Data are number or number (percent) unless noted otherwise.
Number of affected alleles for a certain CFTR mutation divided by total number of alleles for whole CF population (396 patients/792 alleles).
Number of families with the affected alleles for a certain CFTR mutation divided by total number of alleles for whole CF population (312 families/624 alleles).
Private mutation. Families (compound heterozygous are counted twice in Table 1 and 2). Ho: Homozygous, He: heterozygous.
Compound heterozygous:
Raw #3, Exon 14: p.K716RfsX6; the other associated mutation is p.G473EfsX54 (c.1418delG); Exon 11, inframe deletion
Raw #5, Exon 22: p.P1181R; the other associated mutation is (p.R1162Q) (c.3485G>A); Exon 22, missense mutation
Novel Mutations
Six novel variants were found in 15/312 families (4.6%) that constituted 28 alleles (Table 3):
The variant allele, DS459_G461 (predicted deleterious due to in-frame deletion); Exon 10 was found in 4/312 families (8 alleles, 1.3%). The most common presentations included: bronchiectasis 3/4 (60%), P aeruginosa colonization (60%), and pancreatic insufficiency (40%).
The variant allele, c.2739T>G (p.Y913X) (predicted deleterious due to premature translation termination); Exon 17 was found in 3/312 families (6 alleles 1.0%). The clinical presentations included: echogenic liver parenchyma and mild cholestasis on ultrasonographic imaging study.
The variant allele, c.2147_2149delAGAinsGG (p.K716RfsX6) (predicted deleterious due to premature translation termination); Exon 144 was found in 3/312 families (5 alleles, 0.8%). The most common presentation included: respiratory colonization (100%) with P aeruginosa (100%) and S aureus/H influenza (50%).
The variant allele, c.3140-10T>G (IVS19-10T>G) (suspected consensus splice acceptor site mutation); Intron13 was found in 2/312 families (4 alleles 0.7%). One patient had CFRD (33%), and another patient had bilateral sequential lung transplant, chronic pancreatitis, Aspergillus niger pneumonia, acute rejection with severe bronchiolitis obliterans and died. The most common presentation included bronchiectasis (66.6%).
The variant allele, c.3542C>G (p.P1181R) (predicted deleterious in silico); Exon 22 was found in 2/312 families (3 alleles, 0.5%). Clinical presentations included lipodystrophy, high liver enzymes, speech and cognitive function delay, mild persistent asthma, night sweats, obstructive sleep apnea, large adenoids and tonsils, and nephromegaly.
T he variant allele, c.2619G>C (p.E873D) (predicted deleterious in silico); Exon 156 was found in 1/312 family (2 alleles 0.3%). The clinical presentations included recurrent chest infections and failure to thrive.
Survival and mortality rate
In our study, 378 (95.5%) survived, and 18 (4.5%) died. The median survival was 20 years (75% of those diagnosed in this study lived to a mean of 17 years. In comparison to the North American data, the median age of survival in Canada was 50.9 years (95% CI: 50.5-52.2) and in the United States the median age of sujrvival was 40.6 (95% CI: 39.1-41.8) years respectively).54 There was no significant difference between males and females (P=.245).
DISCUSSION
The most common CFTR variant in the Western world is F508del, which is found in approximately 66-75% of CF population.56 In the Saudi population there is no single common variant but 10 different variants constitute 79% of the total CFTR variants. We believe that the high rates of familial intermarriages among carriers of these variants have perpetuated certain CFTR variants. Consanguinity between parents in our CF population was 85% compared to 50% overall consanguinity in Saudi Arabia. We believe that CF has become a common disease in our population due to the consanguinity phenomenon despite being an orphan disease in other parts of the world.
The variant allele, c.1418delG (p.Gly473GlufsX54),13,55,57 c.416A>T (p.His139Leu) 2 and c.579+1G>A (711+1G>T) 2 were first described in Saudi Arabia as novel variants, and were of native Saudi descent, and all showed symptoms early in life with severe lung disease and pancreatic insufficiency.
The variant c.1521_1523delCTT (F508del) is the most common variant in North America and Western Europe, accounting for 60-75% of all CF patients.13,15,54 These variants only constituted 11% of variants in the Saudi CF population, and had a milder pulmonary disease compared to other Saudi-specific CF variants.
The variant c.3700A>G (p.Ile1234Val) is mainly described in an extended Saudi native tribe who resides around the Arabian Gulf area and travels frequently between all Gulf countries with high frequency of consanguinity. The variant has been reported in 35 patients from Qatar (which constituted 95% of their total CFTR mutations).58 The Qatari family is the same extended family tribe that has been reported from Saudi Arabia and carries a good prognosis. This variant was first described in Southern France in 1992.14,15
The variant c.1647T>G (p.Ser549Arg) was reported in native United Arab Emirates and Omani patients in more than 50% of the CF patients with severe lung disease, early Pseudomonas aeruginosa colonization and early death.59-62 whereas the variant c.1911delG (p.Gln637Hisfs) was reported in 31% 73 of CF patients in Bahrain. The proximity of Saudi Arabia, UAE and Bahrain allowed members of the same tribal origin to move freely between Gulf countries and intermarriages occur between similar tribes.
The variant allele, c.2988+1G>A (3120+1G>A) has been studied well by Dörk et al,63 who indicated a common origin of this variant. He analyzed DNA samples from 17 unrelated CF families from 4 different populations (Arabic, Greek, Native African, and African-American) and found a common origin of this variant. His impression was that a common ancestor had evolved separately in the respective population.
In our report, we have shown that the top 10 most common variants identified (79.2% of our CF population) could be used as a screening panel for newly diagnosed CF patients in our population that could also be applied to the rest of the Arab countries. We have also shown that a significant number of our CFTR variants are private mutations (20 variants in 20 families). Even though private mutations were only present in 20 families, they accounted for 41.7% of all variants encountered in the study. This is a clear reflection of the power of consanguinity to render homozygous many pathogenic alleles and is consistent with our previous experience of tremendous allelic heterogeneity that characterizes recessive disorders in our consanguineous population.64 For these reasons, special attention should be taken for CFTR screening of such patients in our community, and proper family counseling should be applied. Physicians in the Western countries who receive Arab patients traveling abroad for CFTR variant identification should consider using our screening panel rather than the Western prepared panel for CFTR variants. Novel variants in our population were mainly in a homozygous state and of severe clinical presentation. There was a big difference in median survival between our CF population (20 years) and North American population (from 50.9 to 40.6 years). This could be explained by the delay in diagnosis, early colonization with P aeruginosa, poor compliance to medications and poor nutritional status.54
In conclusion, there is a high allelic heterogeneity that characterizes CFTR mutational patterns in our CF population, with at least 10 common mutations that accounted for 79.2% of all CFTR mutations due to the spread of the variant in different tribes despite intermarriages between relatives of certain tribes. There is a high prevalence of homozygous variants, which reflects the power of consanguinity within the same tribe. Efforts must be made to improve the prognosis of this debilitating disease using preventative methods, early identification and referral to specialized centers.
ACKNOWLEDGMENTS
Fowzan S Alkuraya, MD (Hons) ABP ABMG, Maha Al-Eid PhD, Dhefaf AlAbdaly, Sara AlKaf, Abdelmoneim Eldali PhD, Edward Devol PhD, Manal Al Shike, Mohammed Alhamed, PhD. Khalid AlThubaiti, MD. AbdulAziz AlEnazi, MD.
Funding Statement
None.
REFERENCES
- 1.Banjar HH. Cystic fibrosis: presentation with other diseases, the experience in Saudi Arabia. Journal of Cystic Fibrosis 2003; 2:155–9. doi: 10.1016/s1569-1993(03)00058-4. [DOI] [PubMed] [Google Scholar]
- 2.De Boeck K. Cystic fibrosis in the year 2020: a disease with a new face. Acta Paediatr. 2020; [DOI] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Registry Reports - United States [Internet]. [cited 2020 Jan 11]. Available from: http://www.cysticfibrosisdata.org/ReportsUS.html
- 4.Banjar H, Angyalosi G.. The road for survival improvement of cystic fibrosis patients in Arab countries. Int J Pediatr Adolesc Med. 2015;2(2):47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cystic Fibrosis Mutation Database [Internet]. [cited 2020 Jan 11]. Available from: http://www.genet.sickkids.on.ca/
- 6.Singh VK, Schwarzenberg SJ.. Pancreatic insufficiency in Cystic Fibrosis. Journal of Cystic Fibrosis [Internet]. 2017. [cited 2020 Jan 11];16:S70–8. Available from: https://www.cysticfibrosisjournal.com/article/S1569-1993(17)30813-5/abstract [DOI] [PubMed] [Google Scholar]
- 7.Brunstein J. PCR: the basics of the polymerase chain reaction. MLO: medical laboratory observer; 2013; 45(4), 32–34. [PubMed] [Google Scholar]
- 8.Chatterjee N, Banerjee T, Datta S.. Accurate Estimation of Nucleic Acids by Amplification Efficiency Dependent PCR. PLoS ONE 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes C, Martinez-Puchol S, Pons MJ, Bazán J, Tinco C, Valle JD, et al. . Evaluation of PCR Approaches for Detection of Barton-ella bacilliformis in Blood Samples. PLOS Neglected Tropical Diseases 2016;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suppan M, Shamsuddine S, Ismail A, Balakrishnan V.. PCR-based Gene Synthesis, Cloning, Expression, Purification and Characterization of Bst DNA Polymerase in E. coli Cells. Current Synthetic and Systems Biology. 2015;3:10000126. [Google Scholar]
- 11.Zhang XXQ-Q. Diagnostic Direct DNA Sequencing and Systemic Treatment with Voriconazole in Scedosporium apiospermum Keratitis? A Case Report. Journal of Clinical & Experimental Ophthalmology 2013;04.. [Google Scholar]
- 12.ScDawson KP, Frossard PM.. Cystic Fibrosis in the United Arab Emirates: An Under-Recognized Condition? Tropical Doctor 1995; 25:110–1. [DOI] [PubMed] [Google Scholar]
- 13.El-Harith EA, Dork T, Stuhrmann M, Abu-Srair H, Al-Shahri A, Keller KM, et al. . Novel and characteristic CFTR mutations in Saudi Arab children with severe cystic fibrosis. Journal of Medical Genetics 1997; 34:996–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean M, Gerrard B, Stewart C, Krueger L, Holsclaw D, Quittell L, et al. . Identification of Cystic Fibrosis Mutations. Advances in Experimental Medicine and Biology The Identification of the CF (Cystic Fibrosis) Gene 1991:45–51.. [DOI] [PubMed] [Google Scholar]
- 15.Claustres M, Gerrard B, Kjellberg P, Des-georges M, Demaille J, Dean M.. Screening for cystic fibrosis mutations in Southern France: Identification of a frameshift mutation and two missense variations. Human Mutation 1992; 1:310–3. [DOI] [PubMed] [Google Scholar]
- 16.Riordan J. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Trends in Genetics 1989; 5:363. [DOI] [PubMed] [Google Scholar]
- 17.Banjar H, Kambouris M, Meyer BF, Al-Mehaidib A, Mogarri I.. Geographic distribution of cystic fibrosis transmembrane regulator gene mutations in Saudi Arabia. Annals of Tropical Paediatrics 1999; 19:69–73. [DOI] [PubMed] [Google Scholar]
- 18.Wilschanski M, Zielenski J, Markiewicz D, Tsui L-C, Corey M, Levison H, et al. . Correlation of sweat chloride concentration with classes of the cystic fibrosis transmembrane conductance regulator gene mutations. The Journal of Pediatrics 1995; 127:705–10. [DOI] [PubMed] [Google Scholar]
- 19.Macek M Jr, Mackova A, Hamosh A, Hilman BC, Selden RF, Lucotte G, et al. . Identification of common cystic fibrosis mutations in African-Americans with cystic fibrosis increases the detection rate to 75%. American Journal of Human Genetics 1997; 60(5): 1122. [PMC free article] [PubMed] [Google Scholar]
- 20.Banjar H, Mogarri I, Meyer BF, Al-Hamed M, Kambouris M.. Genetic and Clinical data of cystic fibrosis patients in a tertiary care center in Saudi Arabia. The Kuwait Medical Journal 1998; 30(4): 312-316. [Google Scholar]
- 21.Fanen P, Ghanem N, Vidaud M, Besmond C, Martin J, Costes B, et al. . Molecular characterization of cystic fibrosis: 16 Novel mutations identified by analysis of the whole cystic fibrosis conductance transmembrane regulator (CFTR) coding regions and splice site junctions. Genomics 1992; 13:770–6. [DOI] [PubMed] [Google Scholar]
- 22.Kerem BS, Zielenski J, Markiewicz D, Bozon D, Gazit E, Yahav J, et al. . Identification of mutations in regions corresponding to the two putative nucleotide (ATP)-binding folds of the cystic fibrosis gene. Proceedings of the National Academy of Sciences 1990; 87:8447–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborne L, Knight R, Santis G, Hodson M.. A mutation in the second nucleotide binding fold of the cystic fibrosis gene. American journal of human genetics 1991; 48(3): 608. [PMC free article] [PubMed] [Google Scholar]
- 24.Feuillet-Fieux M, Ferrec M, Gigarel N, Thuillier L, Sermet I, Steffann J, et al. . Novel CFTR mutations in black cystic fibrosis patients. Clinical Genetics 2004; 65:284–7. [DOI] [PubMed] [Google Scholar]
- 25.Kılınç MO, Ninis VN, Da?lı E, Demirkol M, Özkınay F, Arıkan Z, et al. . Highest heterogeneity for cystic fibrosis: 36 mutations account for 75% of all CF chromosomes in Turkish patients. American Journal of Medical Genetics 2002; 113:250–7. [DOI] [PubMed] [Google Scholar]
- 26.Zielenski J, Bozon D, Kerem B, Markiewicz D, Durie P, Rommens JM, et al. . Identification of mutations in exons 1 through 8 of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 1991; 10: 229-235. [DOI] [PubMed] [Google Scholar]
- 27.Bobadilla JL, Macek M, Fine JP, Farrell PM.. Cystic fibrosis: A worldwide analysis of-CFTR mutations?correlation with incidence data and application to screening. Human Mutation 2002; 19:575–606. [DOI] [PubMed] [Google Scholar]
- 28.Claustres M, Altiéri JP, Guittard C, Templin C, Chevalier-Porst F, Des Georges M.. Are P.I148T, P.R74W and P.D1270N Cystic Fibrosis Causing Mutations? BMC Medical Genetics 2004; 5 (19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kambouris M F, Meyer H, Banjar H, Hamed MH, Mogarri I, and Ozand P.. ldentification of two novel CFTR exonic deletions 425 del 42 and 1549 del G in CF patients by mutation detection enhancement (MDE) heteroduplex analysis: possible founder effect associated with the 1540-* G polymorphism. Arnerican Journal of Human Genetics 1996; 59. [Google Scholar]
- 30.Gouya L, Pascaud O, Munck A, Elion J, Denamur E.. Novel mutation (A141D) in exon 4 of the CFTR gene identified in an Algerian patient. Human Mutation 1997; 10:86–7. [DOI] [PubMed] [Google Scholar]
- 31.Schrijver I, Ramalingam S, Sankaran R, Swanson S, Dunlop CL, Keiles S, et al. . Diagnostic Testing by CFTR Gene Mutation Analysis in a Large Group of Hispanics. The Journal of Molecular Diagnostics 2005; 7:289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck S, Penque D, Garcia S, Gomes A, Farinha C, Mata L, et al. . Cystic fibrosis patients with the 3272-26A?G mutation have mild disease, leaky alternative mRNA splicing, and CFTR protein at the cell membrane. Human Mutation 1999;14:133. [DOI] [PubMed] [Google Scholar]
- 33.Onay T, Topaloglu O, Zielenski J, Gokgoz N, Kayserili H, Camcioglu Y, et al. . Analysis of the CFTR gene in Turkish cystic fibrosis patients: identification of three novel mutations (3172delAC, P1013L and M1028I). Human Genetics 1998; 102:224–30. [DOI] [PubMed] [Google Scholar]
- 34.Elahi E, Khodadad A, Kupershmidt I, Ghasemi F, Alinasab B, Naghizadeh R, et al. . A Haplotype Framework for Cystic Fibrosis Mutations in Iran. The Journal of Molecular Diagnostics 2006; 8:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi K, Knowles MR, Boucher RC, O'Brien WE, Beaudet AL.. Benign missense variations in the cystic fibrosis gene. American journal of human genetics 1990; 47(4): 611. [PMC free article] [PubMed] [Google Scholar]
- 36.The molecular genetic epidemiology of cystic fibrosis : report of a joint meeting of WHO/IECFTN/ICF(?M)?A/ECFS, Genoa, Italy, 19 June 2002. World Health Organization; 1970. http://www.who.int/iris/handle/10665/68702 (accessed August 2, 2018). [Google Scholar]
- 37.Cutting GR, Kasch LM, Rosenstein BJ, Zielenski J, Tsui L-C, Antonarakis SE, et al. . A cluster of cystic fibrosis mutations in the first nucleotide-binding fold of the cystic fibrosis conductance regulator protein. Nature 1990; 346:366–9. [DOI] [PubMed] [Google Scholar]
- 38.Brancolini V, Cremonesi L, Belloni E, Pappalardo E, Bordoni R, Seia M, et al. . Search for mutations in pancreatic sufficient cystic fibrosis Italian patients: detection of 90% of molecular defects and identification of three novel mutations. Human Genetics 1995;96. [DOI] [PubMed] [Google Scholar]
- 39.Alibakhshi R, Kianishirazi R, Cassiman J-J, Zamani M, Cuppens H.. Analysis of the CFTR gene in Iranian cystic fibrosis patients: Identification of eight novel mutations. Journal of Cystic Fibrosis 2008; 7:102–9. [DOI] [PubMed] [Google Scholar]
- 40.Bonizzato A, Nicolis E, Castellani C, Borgo G, Mastella G, Cabrini G, et al. . Analysis of the complete coding region of the CFTR gene in a cohort of CF patients from North-Eastern Italy: identification of 90% of the mutations. Human Genetics 1995;95. [DOI] [PubMed] [Google Scholar]
- 41.Bienvenu T, Cazeneuve C, Beldjord C, Dusser D, Kaplan JC, Hubert D.. A new missense mutation (G27E) in exon 2 of the CFTR gene in a mildly affected cystic fibrosis patient. Human Molecular Genetics 1994; 3:365–6. [DOI] [PubMed] [Google Scholar]
- 42.Vankeerberghen A, Wei L, Jaspers M, Cassiman J-J, Nilius B, Cuppens H.. Characterization of 19 Disease-Associated Mis-sense Mutations in the Regulatory Domain of the Cystic Fibrosis Transmembrane Conductance Regulator. Human Molecular Genetics 1998; 7:1761–9. [DOI] [PubMed] [Google Scholar]
- 43.Castaldo G, Fuccio A, Cazeneuve CC, Picci L, Salvatore D, Scarpa M, et al. . A noval nonsense mutation (Y849X) in the CFTR gene of a CF patient from southern Italy. Human Mutation 1999; 14:272–. [DOI] [PubMed] [Google Scholar]
- 44.White MB, Leppert M, Nielsen D, Zielenski J, Gerrard B, Stewart C, et al. . A de Novo cystic fibrosis mutation: CGA (Arg) to TGA (stop) at codon 851 of the CFTR gene. Genomics 1991; 11:778–9. [DOI] [PubMed] [Google Scholar]
- 45.Ronchetto P, Orriols JJT, Fanen P, Cremonesi L, Ferrari M, Magnani C, et al. . A nonsense mutation (R1158X) and a splicing mutation (3849 4A ? G) in exon 19 of the cystic fibrosis transmembrane conductance regulator gene. Genomics 1992; 12:417–8. [DOI] [PubMed] [Google Scholar]
- 46.Hantash FM, Rebuyon A, Peng M, Redman JB, Sun W, Strom CM.. Apparent Homo-zygosity of a Novel Frame Shift Mutation in the CFTR Gene Because of a Large Deletion. The Journal of Molecular Diagnostics 2009; 11:253–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chillon M, Casals T, Gimenez J, Ramos M, Palacio A, Morral N, et al. . Analysis of the CFTR gene confirms the high genetic heterogeneity of the Spanish population: 43 mutations account for only 78% of CF chromosomes. Human Genetics 1994;93. [DOI] [PubMed] [Google Scholar]
- 48.Clavel C, Pennaforte F, Pigeon F, Verlingue C, Birembaut P, Férec C.. Identification of four novel mutations in the cystic fibrosis transmembrane conductance regulator gene: E664X, 2113delA, 306delTAGA, and ?M1140. Human Mutation 1997; 9:368–9.. [DOI] [PubMed] [Google Scholar]
- 49.Greil I, Wagner K, Rosenkranz W.. A New Missense Mutation G1249E in Exon 20 of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Gene. Human Heredity 1994; 44:238–40. [DOI] [PubMed] [Google Scholar]
- 50.Cheadle JP, Goodchild MC, Meredith AL.. Direct sequencing of the complete CFTR gene: the molecular characterisation of 99.5% of CF chromosomes in Wales. Human Molecular Genetics 1993; 2:1551–6. [DOI] [PubMed] [Google Scholar]
- 51.Malone G, Haworth A, Schwarz MJ, Cup-pens H, Super M.. Detection of five novel mutations of the cystic fibrosis transmembrane regulator (CFTR) gene in Pakistani patients with cystic fibrosis: Y569D, Q98X, 296 12(T>C), 1161delC and 621 2(T>C). Human Mutation 1998; 11:152–7.. [DOI] [PubMed] [Google Scholar]
- 52.Casals T, Gimenez J, Larriba S, Estivill X, Ramos MD, Nunes V.. High heterogeneity for cystic fibrosis in Spanish families: 75 mutations account for 90% of chromosomes. Human Genetics 1997; 101:365–70. [DOI] [PubMed] [Google Scholar]
- 53.Capurso G, Sbrozzi-Vanni A, Piane M, Begini P, Panzuto F, Libi F, et al. . Phenotype Expression in a Case of Adult Cystic Fibrosis Caused by an Extremely Rare Compound Heterozygous Genotype (2183AA>G/2789 5G>A). Pancreas 2009; 38:599–601. [DOI] [PubMed] [Google Scholar]
- 54.Stephenson AL, Sykes J, Stanojevic S, et al. . Survival Comparison of Patients With Cystic Fibrosis in Canada and the United States: A Population-Based Cohort Study. Ann Intern Med. 2017;166(8):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kambouris M, Banjar H, Moggari I, Nazer H, Al-Hamed M, Meyer BF.. Identification of novel mutations in Arabs with cystic fibrosis and their impact on the cystic fibrosis transmembrane regulator mutation detection rate in Arab populations. Eur J Pediatr. 2000;159:303–9. [DOI] [PubMed] [Google Scholar]
- 56.Saleheen D., Frossard P.. The cradle of the deltaF508 mutation. Journal of Ayub Medical College 2007; 20(4): 157-60. [PubMed] [Google Scholar]
- 57.Fanen P, Ghanem N, Vidaud M, Besmond C, Martin J, Costes B, et al. . Molecular characterization of cystic fibrosis: 16 Novel mutations identified by analysis of the whole cystic fibrosis conductance transmembrane regulator (CFTR) coding regions and splice site junctions. Genomics 1992; 13:770–6. [DOI] [PubMed] [Google Scholar]
- 58.Abdul Wahab A, Al Thani G, Dawod ST, Kambouris M, Al Hamed M.. Heterogeneity of the cystic fibrosis phenotype in a large kindred family in Qatar with cystic fibrosis mutation (I1234V). Journal of Tropical Pediatrics 2001; 47(2): 110-112. [DOI] [PubMed] [Google Scholar]
- 59.Frossard PM, Lestringant G, Girodon E, Goossens M, Dawson KP.. Determination of the prevalence of cystic fibrosis in the United Arab Emirates by genetic carrier screening. Clinical Genetics 1999; 55:496–7. [DOI] [PubMed] [Google Scholar]
- 60.Frossard P, Hertecant J, Bossaert Y, Dawson K.. Genotypephenotype correlations in cystic fibrosis: clinical severity of mutation S549R(T?G). European Respiratory Journal 1999; 13:103–6. [DOI] [PubMed] [Google Scholar]
- 61.Frossard PM, Dawson KP, Das SJ, Alexander PC, Girodon E, Goossens M.. Identification of cystic fibrosis mutation in Oman. Clinical Genetics 2000; 57: 235-236. [DOI] [PubMed] [Google Scholar]
- 62.Eskandarani HA. Cystic Fibrosis Transmembrane Regulator Gene Mutations in Bahrain. Journal of Tropical Pediatrics 2002; 48:348–50. [DOI] [PubMed] [Google Scholar]
- 63.Dörk T, El-Harith E-HA, Stuhrmann M, Macek M, Egan M, Cutting GR, et al. . Evidence for a Common Ethnic Origin of Cystic Fibrosis Mutation 3120 1G?A in Diverse Populations. The American Journal of Human Genetics 1998; 63:656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alkuraya FS. Genetics and genomic medicine in Saudi Arabia. Molecular Genetics & Genomic Medicine 2014;2:369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]