 Herein, by directly introducing mismatched reactant DNA, high-reactivity and high-threshold enzyme-free target recycling amplification (EFTRA) is explored.
Herein, by directly introducing mismatched reactant DNA, high-reactivity and high-threshold enzyme-free target recycling amplification (EFTRA) is explored.
Abstract
Herein, by directly introducing mismatched reactant DNA, high-reactivity and high-threshold enzyme-free target recycling amplification (EFTRA) is explored. The developed high-efficiency EFTRA (HEEFTRA) was applied as a programmable DNA signal converter, possessing higher conversion efficiency than the traditional one with perfect complement owing to the more negative reaction standard free energy (ΔG). Once traces of input target miRNA interact with the mismatched reactant DNA, amounts of ferrocene (Fc)-labeled output DNA could be converted via the EFTRA. Impressively, the Fc-labeled output DNA could be easily captured by the DNA tetrahedron nanoprobes (DTNPs) on the electrode surface to form triplex-forming oligonucleotide (TFO) at pH = 7.0 for sensitive electrochemical signal generation and the DTNPs could be regenerated at pH = 10.0, from which the conversion efficiency (N) will be accurately obtained, benefiting the selection of suitable mismatched bases to obtain high-efficiency EFTRA (HEEFTRA). As a proof of concept, the HEEFTRA as an evolved DNA signal converter is successfully applied for the ultrasensitive detection of miRNA-21, which gives impetus to the design of other signal converters with excellent efficiency for ultimate applications in sensing analysis, clinical diagnosis, and other areas.
Introduction
Enzyme-free target recycling amplification (EFTRA) based on the toehold strand displacement reaction (TSDR) could convert traces of input targets into amounts of output products to construct autocatalytic circuits for exponential signal amplification as a signal converter, which has the advantages of high specificity, low cost, and simple operation with less environmental interference.1–5 Nevertheless, with the lack of an accurate and sensitive measurement method, exactly measuring the conversion efficiency (N) of EFTRA remains a serious challenge, which further limits the exploitation of its inherent properties and expanded applications. Thus it is of significant importance to carve out an effective method for accurately monitoring the conversion efficiency of EFTRA. Herein, we utilize a tetrahedral DNA nanostructure with mechanical rigidity, chemical and structural stability, and highly ordered upright orientation6,7 to design a simple DNA tetrahedral nanoprobe (DTNP), which was immobilized on the electrode surface to effectively capture the Fc-labeled output DNA, the product converted by the quantified input target via EFTRA, to generate a sensitively monitorable electrochemical signal8–13 at pH = 7.0. When this signal value is the same as that generated by the independent and quantified Fc-labeled DNA (identical to the Fc-labeled output DNA), the experimental conversion efficiency of EFTRA (N) can be accurately obtained through the ratio of the concentration of Fc-labeled output DNA from the input target miRNA via EFTRA and the concentration of the input target miRNA. Significantly, the DTNP could be regenerated at pH = 10.0, achieving the continuous long-term usage of this sensing platform to accurately monitor the conversion efficiency of EFTRA and providing a new insight for DNA nanoprobe regeneration.
Promoting the conversion efficiency of EFTRA to develop a high-efficiency DNA signal converter is another significant goal for advancing the superiority and applicability of nucleic acid amplification in diagnostic applications, biological research, nanobiotechnology, and bioengineering. Since the rate-limiting step of TSDR is the branch migration in the displacement domain,14,15 the conversion efficiency of EFTRA could be affected by the reaction equilibrium of the branch migration process. Thus, suitable mismatched bases in the displacement domain of the resultant DNA can make the EFTRA reaction standard free energy (ΔG) more negative when compared with the completely matched reactant DNA in traditional EFTRA,16–23 which could enhance the driving force of EFTRA with increased reactivity and threshold for improving the conversion efficiency. Herein, we adopt the theoretical conversion efficiency of EFTRA (N′) obtained by the well-studied thermodynamic parameters of the nucleic acid hybridization reaction24–27 from NUPACK28 to provide some referential mismatched bases in the displacement domain of the reactant DNA, and then contrast the accurate experimental N to screen out the suitable mismatched bases for high-efficiency EFTRA (HEEFTRA). As a result, we carve out the HEEFTRA and apply it as a programmable DNA signal converter for biomarker assay, which gives impetus to exploit a new generation of nucleic acid amplification techniques for biosensing analysis and early disease diagnosis.
Results and discussion
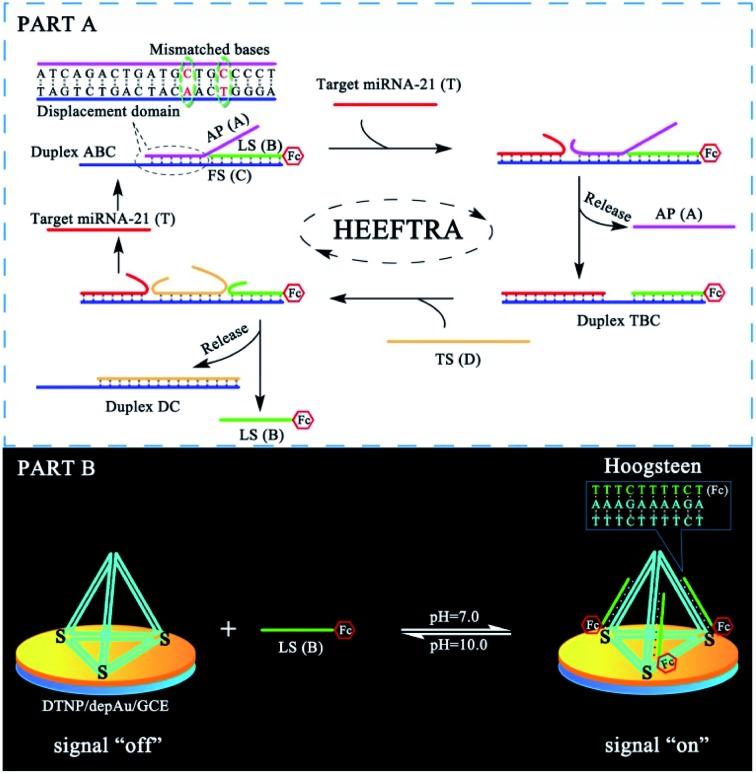
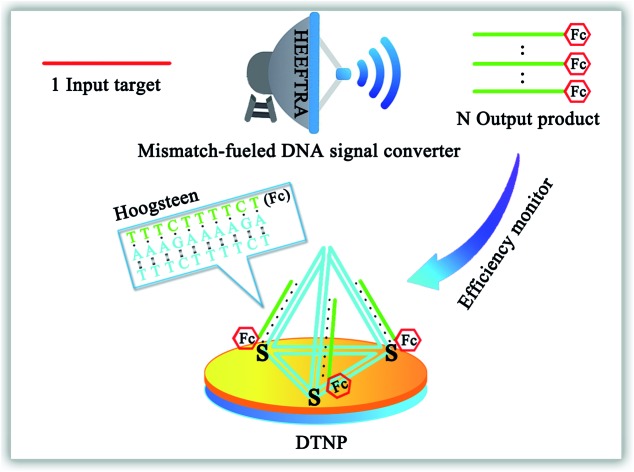
The reaction mechanism and conversion efficiency monitoring of the HEEFTRA are shown in Scheme 1. The target miRNA could trigger the first TSDR to hybridize with the toehold of the duplex (double helix) ABC (FS-AP-LS), then AP (A) will be displaced and released away, from which the next toehold in the middle of FS (C) will be exposed and then initiate the next TSDR in the presence of TS (D), accompanied by the simultaneous release of Fc-labeled output DNA LS (B) and target miRNA (T). Next, the released target miRNA can be reused to release more Fc-labeled DNA LS (B). As a result, one input of target miRNA can induce multiple outputs of Fc-labeled DNA LS (B) via the HEEFTRA (Part A). Then, the Fc-labeled output DNA LS (B) could be captured by the DTNP on the electrode surface to form triplex-forming oligonucleotide (TFO)29–31 at pH = 7.0 for generating the sensitive electrochemical signal response and the TFO could be dissociated in pH = 10.0 realizing the regeneration of DTNP, from which the experimental conversion efficiency of the EFTRA (N) can be accurately obtained (Part B). Ultimately, with the help of the theoretical N′ and experimental N, the suitable mismatched bases in the displacement domain of duplex ABC (two mismatched bases as shown in Part A) are selected for achieving HEEFTRA. Significantly, the evolved HEEFTRA is used to construct an ultrasensitive electrochemical biosensor for the detection of miRNA-21 with a detection limit about an order of magnitude beyond that of the wild-type EFTRA down to 0.25 fM, which could be further applied in miRNA-21 assay from breast cancer cell lysates.
Scheme 1. (Part A) Schematic Illustration of the high-efficiency enzyme-free target recycling amplification (HEEFTRA). (Part B) Illustration of the electrochemical monitoring of the EFTRA conversion efficiency by the DNA tetrahedral nanoprobe (DTNP) on the electrode surface.
Firstly, we carried out a series of polyacrylamide gel electrophoresis (PAGE) characterizations to prove the reaction mechanisms of the EFTRA (Fig. S1A†), DTNP (Fig. S1B†), and TFO (Fig. S2A†) and additionally harnessed the UV-vis absorption spectra to further verify the successful formation of TFO (Fig. S2B†), and the results were as expected. Next, cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS) for the self-assembly of the elaborated biosensor (Fig. S3†) and the characterization of the electrode surface (Fig. S4–S6†) all indicated the successful construction of this biosensor. Then TFO was formed under the optimal reaction time (90 min, Fig. S4A†) and pH (7.0, Fig. S4B†), and we also measured the binding constant (Ka) of TFO with a value of 1.56 × 107 M–1 (ESI, pages S15–S17†), which demonstrates that the designed DTNP has a high binding affinity to the Fc-labeled DNA LS (B) and can be used to sensitively capture the LS (B). We also adopted SWV to characterize the biosensor under different conditions, and the results (Fig. S9†) further displayed that this biosensing platform could effectively recognize the LS (B) and be successfully operated for accurately and sensitively measuring the EFTRA conversion efficiency (N).
To obtain the experimental N, we firstly determined the relationship of the current responses of the biosensor to different concentrations of the quantified independent Fc-labeled LS (B) (Fig. S10†) and different concentrations of the quantified target miRNA (T) (Fig. S11†) via EFTRA. When these two current response values corresponding to the quantified LS (B) and the quantified target miRNA respectively are the same, the concentration of the independent Fc-labeled LS (B) is identical to that of the output LS (B) from the input miRNA-21 via EFTRA. Thus the accurate experimental N could be obtained by proportioning the concentration of the quantified independent Fc-labeled LS (B) and the concentration of the quantified target miRNA under the same current response (ESI, pages S18–S21†), and the results of the experimental wild-type EFTRA conversion efficiency (N) are shown in Table S2.†
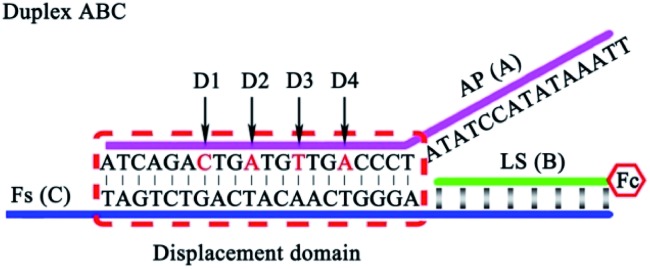
It is very important to choose suitable mismatches in the duplex ABC for achieving HEEFTRA since suitable mismatches can not only make the ΔG of the EFTRA more negative to induce a higher conversion efficiency but could also ensure the sufficient stability of the duplex ABC for the successful operation of the EFTRA. When the mismatches were introduced in FS (C) or LS (B), weak affinity of FS (C) for LS (B), target miRNA, or TS (D) was obtained, resulting in low EFTRA conversion efficiency (Scheme 1). Remarkably, the AP (A) with suitable mismatches not only ensured the stability of duplex ABC but could also be displaced by target miRNA more easily owing to the decreased interaction between AP (A) and FS (C), accompanied by the improved EFTRA conversion efficiency. According to the ΔG calculated from NUPACK as a duplex ABC stability evaluation index, we concluded that mismatched domains with number less than or equal to four at the middle instead of the tail of the displacement region in AP (A) (Fig. 1) could be permitted to introduce suitable mismatches.
Fig. 1. The four possible mismatched domains in the middle of the displacement region that can ensure the stability of duplex ABC and improve the EFTRA conversion efficiency simultaneously.
In view of the wide variety of mismatch types, we harnessed the theoretical computation of the EFTRA conversion efficiency to narrow their range to select the optimal mismatch type. Firstly, we simulated the EFTRA using a two-step reaction model for calculation of the theoretical N′:T + ABC ⇌ TBC + A; ΔG1D + TBC ⇌ DC + T + B; ΔG2where ΔG1 = ΔGTBC + ΔGA – ΔGABC – ΔGT and ΔG2 = ΔGB + ΔGT + ΔGDC – ΔGTBC – ΔGD, and T, A, B, C, and D represent target, AP, LS, FS, and TS, respectively. The equilibrium concentrations of all DNA species can then be derived by solving a set of equations: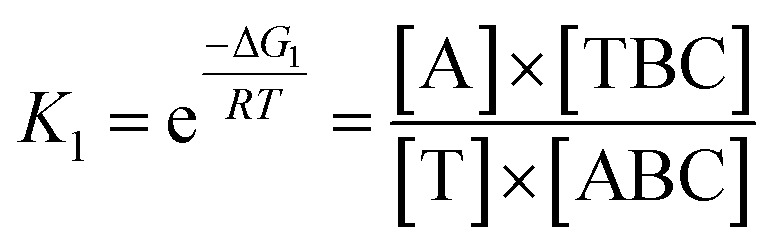
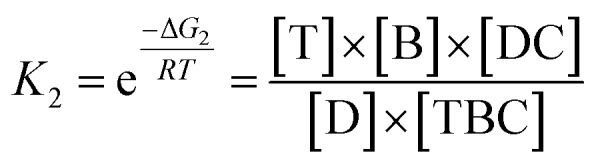
Then through the computation of the ratio of the balanced concentration of output Fc-labeled LS ([B]) and the initial concentration of the input target miRNA ([T]0) (ESI, page S21†), the theoretical N′ of the wild-type EFTRA ([B]/[T]0) was also obtained (Table S2†).
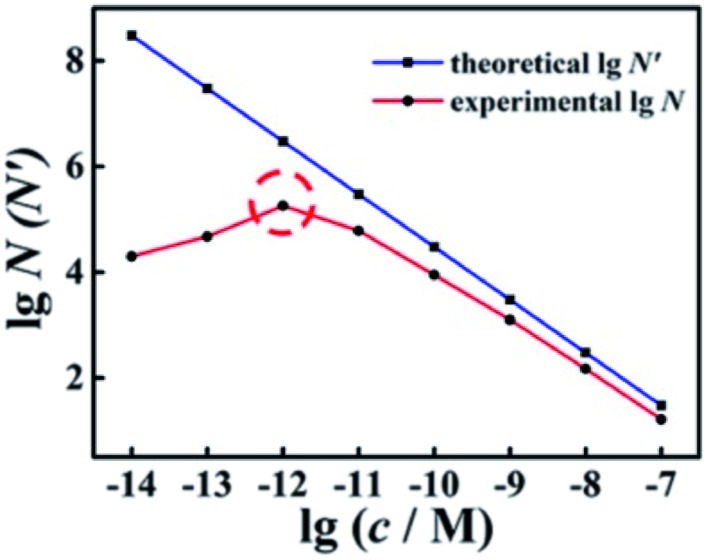
With different quantified target miRNA, the experimental N and theoretical N′ of the wild-type EFTRA were obtained (Table S2†). By comparing these results, we found that the trends of N and N′ to increase or decrease were almost the same. As shown in Fig. 2, with the target concentration increasing from 10 fM to 100 nM (1 × 10–14 to 1 × 10–7 M), the theoretical N′ decreased. In contrast, the trend of the experimental N was consistent with the theoretical N′ (target concentration, 1 pM to 100 nM (1 × 10–12 to 1 × 10–7 M)), while their trends were exactly opposite with the target concentration in the range of 10 fM to 1 pM (1 × 10–14 to 1 × 10–12 M), which can be ascribed to the TSDR, which is weakly thermodynamically favorable or even thermodynamically unfavorable within the limits of a sufficiently low concentration of the trigger of EFTRA (target)15 and the difference between the real ΔG values and the predicted values.32 Thus we chose a target concentration of 1.0 pM to deeply study the EFTRA conversion efficiency, at which the experimental N was practically maximum (Table S2†).
Fig. 2. Comparison of experimental and theoretical conversion efficiency of wild-type EFTRA with different concentrations of target from 10 fM to 100 nM: 10 fM, 100 fM, 1 pM, 10 pM, 100 pM, 1 nM, 10 nM, and 100 nM.
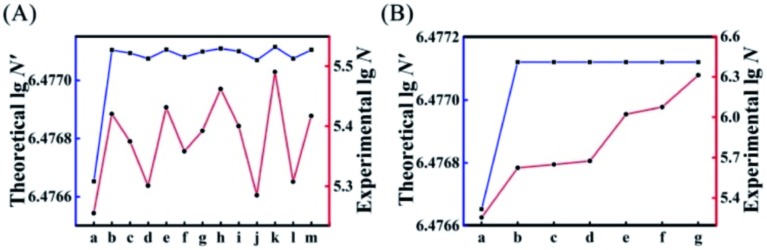
Next, we introduced all possible kinds of single-mismatched bases (Table S3†) in AP to obtain the corresponding theoretical N′ and experimental N. As shown in Fig. 3A, the experimental N changed in accordance with the trend of the theoretically predicted N′. Fig. 3B illustrates the corresponding experimental N (red curve) of some specific multiple-mismatched bases in AP (A) (Table S3†) with the high theoretical N′ (blue curve). The experimental N increased with the elevated number of mismatches, which was consistent with the trend of theoretical N′ to increase or decrease, further indicating that the theoretical N′ could indeed be used as a reference to choose suitable mismatches for achieving HEEFTRA.
Fig. 3. Comparison of experimental and theoretical EFTRA conversion efficiency with different (A) single-mismatched sequences: (a) AP, (b) AP-D1M1t, (c) AP-D1M1a, (d) AP-D1M1g, (e) AP-D2M1c, (f) AP-D2M1g, (g) AP-D2M1t, (h) AP-D3M1c, (i) AP-D3M1a, (j) AP-D3M1g, (k) AP-D4M1c, (l) AP-D4M1g, and (m) AP-D4M1t, and (B) multiple-mismatched sequences: (a) AP, (b) AP-D1D4M2, (c) AP-D2D4M2, (d) AP-D3D4M2, (e) AP-D1D3D4M3, (f) AP-D2D3D4M3, and (g) AP-D1D2D3D4M4 in duplex ABC (1.0 pM target miRNA).
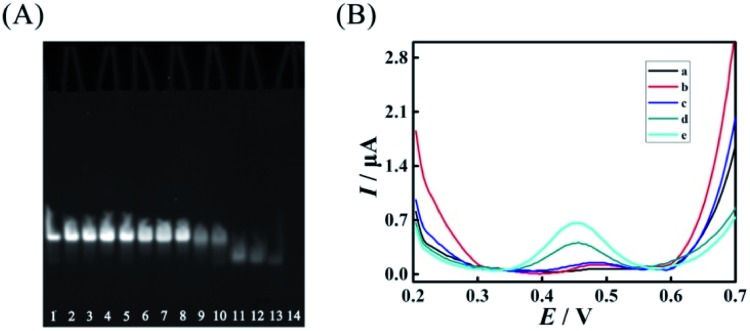
However, excess mismatches of AP (A) would destroy the stability of duplex ABC, bringing false-positive EFTRA operation and a high current background signal in the DNA signal converter in the absence of target miRNA. Thus, PAGE was firstly used to monitor the stability of duplex ABC with different mismatches. As shown in Fig. 4A, lanes 1–11 correspond to duplex ABC with sequences of AP, AP-D1M1t, AP-D2M1c, AP-D3M1c, AP-D4M1c, AP-D1D4M2, AP-D2D4M2, AP-D3D4M2, AP-D1D3D4M3, AP-D2D3D4M3, and AP-D1D2D3D4M4, respectively (Table S3,† D and M represent “domain” and “mismatch” separately) and the obvious bands directly indicate that the stability of duplex ABC was almost unaffected after the introduction of single (lanes 2–5) and double (lanes 6–8) mismatches and decreased slightly after the introduction of triple mismatches (lanes 9 and 10). Compared with lanes 12–14 which respectively correspond to single strand FS, AP and LS, the band in lane 11 representing duplex ABC vanished, displaying that duplex ABC could not be constructed steadily after the introduction of quadruple mismatches in AP. Secondly, in the absence of target miRNA, we also employed SWV to study the current background signal of the biosensor after different multiple mismatches in AP (A) were introduced to further verify the stability of duplex ABC. As displayed in Fig. 4B, the SWV current responses of the biosensing platform with matched duplex ABC (curve a), single-mismatched duplex ABC (curve b), or double-mismatched duplex ABC (curve c) were hardly noticeable; however, after the introduction of triple-mismatched bases (curve d) or quadruple-mismatched bases (curve e) in AP, the SWV signal responses all increased significantly. The high background signals of the biosensor with triple or quadruple mismatches further certified that excess mismatches would obviously destroy the duplex ABC stability and be out of the DNA signal converter development. Integrating the above results of PAGE and SWV with the experimental N (red curve) shown in Fig. 3B, the most suitable mismatches in duplex ABC that endow the EFTRA with excellent experimental N and low background signal simultaneously were double mismatches (AP-D3D4M2) instead of the triple or quadruple mismatches from the theoretical prediction, which might be ascribed to the effects of the cation concentration, pH, etc.33 Impressively, we introduced it in the EFTRA and achieved the HEEFTRA. The HEEFTRA was further harnessed for developing an ultrasensitive biosensor for miRNA analysis.
Fig. 4. (A) Nondenaturing PAGE characterization of duplex ABC with different mismatched sequences (8% PAGE, 90 min): lane 1, AP (2 μM); lane 2, AP-D1M1t (2 μM); lane 3, AP-D2M1c (2 μM); lane 4, AP-D3M1c (2 μM); lane 5, AP-D4M1c (2 μM); lane 6, AP-D1D4M2 (2 μM); lane 7, AP-D2D4M2 (2 μM); lane 8, AP-D3D4M2 (2 μM); lane 9, AP-D1D3D4M3 (2 μM); lane 10, AP-D2D3D4M3 (2 μM); lane 11, AP-D1D2D3D4M4 (2 μM) and single strands: lane 12, FS (2 μM); lane 13, AP (2 μM); lane 14, LS (2 μM). (B) Comparison of SWV current responses of the biosensing platform with different mismatches: (a) AP; (b) AP-D4M1c; (c) AP-D3D4M2; (d) AP-D2D3D4M3; (e) AP-D1D2D3D4M4 in the absence of target miRNA.
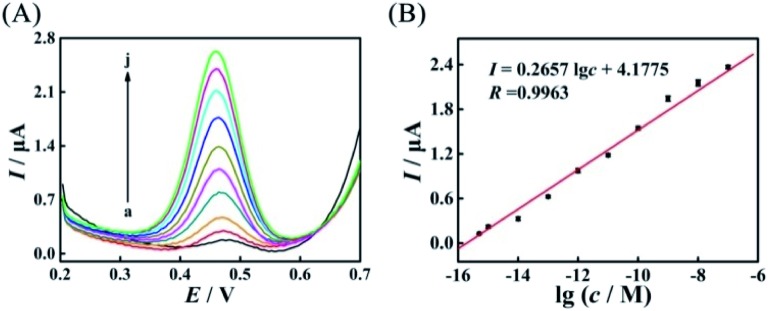
After the suitable mismatches (AP-D3D4M2) were introduced in duplex ABC, based on the evolved HEEFTRA and the elaborated DTNP, the current response of this proposed biosensor dramatically increased with the increase of the target concentration from 0.5 fM to 100 nM (5 × 10–16 to 1 × 10–7 M) (Fig. 5A) and showed a good linear relationship with the logarithm of the miRNA-21 concentration (Fig. 5B), and the regression equation was expressed as I = 0.2657 lg c + 4.1775 (R = 0.9963) with detection limit down to 0.25 fM for miRNA-21 analysis (Table S6†), exhibiting relatively desirable performance compared to the biosensor without mismatches (Fig. S11†) and other methods (Table 1).
Fig. 5. (A) SWV current responses of the biosensors to different concentrations of the target miRNA-21 with mismatches (AP-D3D4M2) in HEEFTRA: (a) 0.5 fM, (b) 1.0 fM, (c) 10 fM, (d) 100 fM, (e) 1.0 pM, (f) 10 pM, (g) 100 pM, (h) 1 nM, (i) 10 nM, and (j) 100 nM, and (B) corresponding calibration plot for the SWV peak current vs. lg c.
Table 1. Comparison of the developed biosensor with other miRNA detection methods.
| Analytical methods | Linear range | Detection limit | Ref. |
| Fluorescence | 100 fM to 10 nM | 58 fM | 41 |
| Electrochemiluminescence | 5.0 fM to 500 pM | 1.51 fM | 42 |
| Electrochemiluminescence | 10 fM to 0.1 nM | 6.6 fM | 43 |
| Chronocoulometry | 2.0 fM to 1.0 nM | 2.0 fM | 44 |
| Electrochemical | 20 fM to 10 pM | 8.2 fM | 45 |
| Electrochemical | 5 fM to 5.0 pM | 1.92 fM | 46 |
| Electrochemical | 10 fM to 100 nM | 1.22 fM | This work (without mismatches) |
| Electrochemical | 0.5 fM to 100 nM | 0.25 fM | This work (with mismatches) |
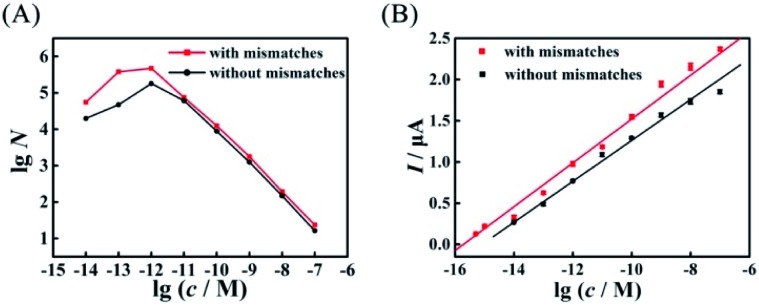
Compared with the biosensor based on wild-type EFTRA, the proposed biosensor with mismatches in HEEFTRA exhibited a higher conversion efficiency (Fig. 6A), a wider range of detection concentration (Fig. 6B) and a lower detection limit for target assay (Table 1), demonstrating that the introduction of suitable mismatches in the displacement domain of the reactant DNA successfully achieved the HEEFTRA. Moreover, the detection of tumour-specific circulating miRNA at ultrahigh sensitivity is of utmost significance for the early diagnosis and monitoring of cancer;34 thus, there is a great need to develop new approaches or sensing media with improved miRNA detection limits owing to its low abundance in total RNA samples and the susceptibility to degradation.35–37 In view of this, we indeed explored a practically valuable HEEFTRA.
Fig. 6. Comparison of (A) the experimental EFTRA conversion efficiency of the proposed biosensor with and without mismatches under different concentrations of target miRNA (accurate data are shown in Tables S2 and S5,† respectively) and (B) the corresponding calibration plots for the SWV peak current vs. lg c with and without mismatches.
In addition, the prepared biosensor based on the evolved HEEFTRA also exhibits excellent reproducibility, selectivity, and stability (Fig. S12†). As displayed in Fig. S13,† the capacity of the elaborated biosensor for miRNA-21 detection was investigated with total RNA extraction solutions from human cancer cell lines MCF-7 and HeLa (ESI, pages S6 and S7†) and the results (Fig. S13†) were consistent with previous reports.38–40 Finally, the reversible pH switching of the biosensor has also been investigated (ESI, pages S29 and S30†) and the regenerability of the proposed biosensor was excellent with more than seven pH switching cycles (Fig. S14†).
Conclusions
In summary, we proposed a mismatch-fueled high-efficiency DNA signal converter named HEEFTRA and applied it in the construction of ultrasensitive biosensor. Overall, first, we carved out an effective method to accurately and sensitively measure the conversion efficiency of EFTRA addressing the challenge of exact monitoring of the EFTRA conversion efficiency based on a DNA tetrahedral nanoprobe (DTNP) with multiple recognition sites at the lateral edges, high stability, and low surface-induced perturbation on the electrode surface. Second, through the introduction of suitable mismatches in reactant DNA, the conversion efficiency of EFTRA is obviously improved, providing a new idea for promoting the exploitation of the inherent properties and expanded applications of HEEFTRA. Third, as a practical application, the evolved HEEFTRA was applied to develop a biosensor with excellent specificity, stability, reproducibility, and regenerability for the ultrasensitive detection of miRNA-21, achieving the assay of miRNA from cancer cell lysates. Given these advantages, the programmable mismatch-fueled HEEFTRA shows great potential as a new generation of DNA signal converter to construct biosensors for sensing analysis and clinical diagnosis, like the detection of nucleic acids,47 ATP,48 and proteins,49 and even for applications in other areas50 after some small adjustments of the specific nucleic sequence in it.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This paper was financially supported by the National Natural Science Foundation of China (21775124, 21675129 and 21575116) and the Fundamental Research Funds for the Central Universities (XDJK2018AA003), China.
Footnotes
†Electronic supplementary information (ESI) available: Experimental section and related experimental data. See DOI: 10.1039/c9sc05084a
References
- Yin P., Choi H. M. T., Calvert C. R., Pierce N. A. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Hu J., Sheng Y., Kwak K. J., Shi J., Yu B., Lee L. J. Nat. Commun. 2017;8:1683. doi: 10.1038/s41467-017-01942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Z., Xu J., Zheng J., Yang S., Tan W., Yang R. H. Angew. Chem., Int. Ed. 2019;58:11574–11585. doi: 10.1002/anie.201812449. [DOI] [PubMed] [Google Scholar]
- Srinivas N., Parkin J., Seelig G., Winfree E., Soloveichik D. Science. 2017;358:eaal2052. doi: 10.1126/science.aal2052. [DOI] [PubMed] [Google Scholar]
- Oesinghaus L., Simmel F. C. Nat. Commun. 2019;10:2092. doi: 10.1038/s41467-017-01942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell N., Schlapak R., Kastner M., Armitage D., Chrzanowski W., Riener J., Hinterdorfer P., Ebner A., Howorka S. Angew. Chem., Int. Ed. 2009;48:525–527. doi: 10.1002/anie.200804264. [DOI] [PubMed] [Google Scholar]
- Pei H., Lu N., Wen Y., Song S., Liu Y., Yan H., Fan C. Adv. Mater. 2010;22:4754–4758. doi: 10.1002/adma.201002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne A., Bouchardon A., Moiroux J. J. Am. Chem. Soc. 2003;125:1112–1113. doi: 10.1021/ja028640k. [DOI] [PubMed] [Google Scholar]
- Fan C., Plaxco K. W., Heeger A. J. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Staveren D. R., Metzler-Nolte N. Chem. Rev. 2004;104:5931–5986. doi: 10.1021/cr0101510. [DOI] [PubMed] [Google Scholar]
- Xuan F., Fan T. W., Hsing I. ACS Nano. 2015;9:5027–5033. doi: 10.1021/nn507282f. [DOI] [PubMed] [Google Scholar]
- Zhu J., Gan H., Wu J., Ju H. Anal. Chem. 2018;90:5503–5508. doi: 10.1021/acs.analchem.8b01217. [DOI] [PubMed] [Google Scholar]
- Liu S., Fang L., Wang Y., Wang L. Anal. Chem. 2017;89:3108–3115. doi: 10.1021/acs.analchem.6b04871. [DOI] [PubMed] [Google Scholar]
- Genot A. J., Zhang D. Y., Bath J., Turberfield A. J. J. Am. Chem. Soc. 2011;133:2177–2182. doi: 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y., Winfree E. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- Jiang Y. S., Bhadra S., Li B., Ellington A. D. Angew. Chem., Int. Ed. 2014;53:1845–1848. doi: 10.1002/anie.201307418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. S., Zhang D. Y. Nat. Chem. 2015;7:545–553. doi: 10.1038/nchem.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. R., Wang J. S., Fang J. Z., R Evans E., Pinto A., Pekker I., Boykin R., Ngouenet C., Webster P. J., Beechem J., Zhang D. Y. Nat. Methods. 2015;12:1191–1196. doi: 10.1038/nmeth.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lei P., Ding S., Zhang Y., Yang J., Cheng Q., Yan Y. Biosens. Bioelectron. 2016;77:435–441. doi: 10.1016/j.bios.2015.09.069. [DOI] [PubMed] [Google Scholar]
- Wu C., Cansiz S., Zhang L., Teng I., Qiu L., Li J., Liu Y., Zhou C., Hu R., Zhang T., Cui C., Cui L., Tan W. J. Am. Chem. Soc. 2015;137:4900–4903. doi: 10.1021/jacs.5b00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Fan H., Satyavolu N. S. R., Wang W., Lake R., Jiang J., Lu Y. Angew. Chem., Int. Ed. 2017;56:8721–8725. doi: 10.1002/anie.201703540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yang Z., Chang Y., Qing M., Yuan R., Chai Y. Anal. Chem. 2018;90:9538–9544. doi: 10.1021/acs.analchem.8b02251. [DOI] [PubMed] [Google Scholar]
- Dai W. H., Dong H. F., Guo K. K., Zhang X. J. Chem. Sci. 2018;9:1753–1759. doi: 10.1039/c7sc04243d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Craig S. L. J. Am. Chem. Soc. 2005;127:13227–13231. doi: 10.1021/ja052352h. [DOI] [PubMed] [Google Scholar]
- Chen C., Wang W., Ge J., Zhao X. S. Nucleic Acids Res. 2009;37:3756–3765. doi: 10.1093/nar/gkp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. X., Fang J. Z., Duan W., Wu L. R., Zhang A. W., Dalchau N., Yordanov B., Petersen R., Phillips A., Zhang D. Y. Nat. Chem. 2017;10:91–98. doi: 10.1038/nchem.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang G. A., Mason S. D., Yang X., Yu Z., Tang Y., Li F. Chem. Sci. 2018;9:6434–6439. doi: 10.1039/c8sc02761g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Cecconello A., Idili A., Ricci F., Willner I. Angew. Chem., Int. Ed. 2017;56:15210–15233. doi: 10.1002/anie.201701868. [DOI] [PubMed] [Google Scholar]
- Wu N., Willner I. Nano Lett. 2016;16:6650–6655. doi: 10.1021/acs.nanolett.6b03418. [DOI] [PubMed] [Google Scholar]
- Walsh S., El-Sagheer A. H., Brown T. Chem. Sci. 2018;9:7681–7687. doi: 10.1039/c8sc02418a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Hicks D. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y., Chen S. X., Yin P. Nat. Chem. 2012;4:208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Gregory R. I. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T., Liang W., Li Y., Sun Y., Xiang Y., Zhang Y., Dai Z., Duo Y., Wu L., Qi K., Shivananju B. N., Zhang L., Cui X., Zhang H., Bao Q. Nat. Commun. 2019;10:28. doi: 10.1038/s41467-018-07947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Lin H., Guo Z., Lu J., Jia Y., Ye M., Su F., Niu L., Kang W., Wang S., Hu Y., Huang Y. Biosens. Bioelectron. 2019;143:111616. doi: 10.1016/j.bios.2019.111616. [DOI] [PubMed] [Google Scholar]
- Qu A., Sun M., Xu L., Hao C., Wu X., Xu C., Kotov N. A., Kuang H. Proc. Natl. Acad. Sci. U. S. A. 2019;116:3391–3400. doi: 10.1073/pnas.1810764116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Han J., Cui Y., Zhou X., Fan K. Biochem. Biophys. Res. Commun. 2013;439:384–389. doi: 10.1016/j.bbrc.2013.08.072. [DOI] [PubMed] [Google Scholar]
- Lu J., Tsourkas A. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Xu L., Ma W., Wu X., Sun M., Kuang H., Wang L., Kotov N. A., Xu C. J. Am. Chem. Soc. 2016;138:306–312. doi: 10.1021/jacs.5b10309. [DOI] [PubMed] [Google Scholar]
- Wang L., Deng R., Li J. Chem. Sci. 2015;6:6777–6782. doi: 10.1039/c5sc02784e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Zhang P., Chai Y., Yuan R. Anal. Chem. 2017;89:5036–5042. doi: 10.1021/acs.analchem.7b00418. [DOI] [PubMed] [Google Scholar]
- Zhang P., Jiang J., Yuan R., Zhuo Y., Chai Y. J. Am. Chem. Soc. 2018;140:9361–9364. doi: 10.1021/jacs.8b04648. [DOI] [PubMed] [Google Scholar]
- Fang C. S., Kim K., Yu B., Jon S., Kim M., Yang H. Anal. Chem. 2017;89:2024–2031. doi: 10.1021/acs.analchem.6b04609. [DOI] [PubMed] [Google Scholar]
- Chang J., Wang X., Wang J., Li H., Li F. Anal. Chem. 2019;91:3604–3610. doi: 10.1021/acs.analchem.8b05599. [DOI] [PubMed] [Google Scholar]
- Zhang K., Dong H., Dai W., Meng X., Lu H., Wu T., Zhang X. Anal. Chem. 2016;89:648–655. doi: 10.1021/acs.analchem.6b02858. [DOI] [PubMed] [Google Scholar]
- Wang F., Elbaz J., Orbach R., Magen N., Willner I. J. Am. Chem. Soc. 2011;133:17149–17151. doi: 10.1021/ja2076789. [DOI] [PubMed] [Google Scholar]
- Li X., Yang J., Xie J., Jiang B., Yuan R., Xiang Y. Biosens. Bioelectron. 2018;102:296–300. doi: 10.1016/j.bios.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Li F., Tang Y., Traynor S. M., Li X., Le X. C. Anal. Chem. 2016;88:8152–8157. doi: 10.1021/acs.analchem.6b01900. [DOI] [PubMed] [Google Scholar]
- Simmel F. C., Yurke B., Singh H. R. Chem. Rev. 2019;119:6326–6369. doi: 10.1021/acs.chemrev.8b00580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.