ABSTRACT
Plant leaf margins produce small outgrowths or teeth causing serration in a regular arrangement, which is specified by auxin maxima. In Arabidopsis, the spatiotemporal pattern of auxin dependents on both, the transcription factor CUC2 and the signal peptide EPFL2, a ligand of the growth-promoting receptor kinase ERECTA (ER). Ectopic expression of CUC2 can have contrary effects on leaf growth. Ubiquitous expressed CUC2 suppresses growth in the whole leaf, whereas cuc2-1D mutants have enlarged leaves, through ER-dependent cell proliferation in the teeth. Here we investigated the growth dynamics of cuc2-1D leaves and the growth restricting the function of CUC2 using the ubiquitous inducible CUC2-GR transgene. In time courses, we dissected the serration promoting the function of CUC2 in the leaf margin and ectopic growth inhibition by CUC2 in the leaf plate. We found that CUC2 limits growth rather by cell cycle inhibition than by cell size control. Furthermore, endogenous CUC2 was rapidly induced by CUC2-GR indicating a possible auto-inducible feedback. In contrast, EPFL2 was quickly decreased by transient CUC2 induction but increased in cuc2-3 mutant leaves suggesting that CUC2 can also counteract the EPFL2-ER pathway. Therefore, tooth growth promotion and growth inhibition by CUC2 involve partially the same mechanism but in contrary ways.
KEYWORDS: Arabidopsis thaliana, leaf development, leaf growth, leaf margin, cell division, signal transduction, transcriptional regulation
1. Introduction
Plant leaves evolved to capture light and carbon dioxide for photosynthesis, while at the same time managing water loss and tissue temperatures within an optimum range. The leaves of Arabidopsis thaliana (Arabidopsis) are typical for dicotyledonous plants, their leaf blades are broad and thin and are held in a horizontal plane by the leaf petiole.1 Based on the results of more than three decades of research in Arabidopsis, several pathways are known controlling leaf growth and shape but we are far from understanding all interconnections of the components or the complexity of the gene networks shaping the leaf. Leaves are established at the flank of the shoot apical meristem (SAM) rising from a few founder cells that are distinguished by high auxin concentrations and downregulation of the KNOTTED-like homeodomain class I (KNOXI) transcription factors KNAT1, KNAT2 (for knotted-like from Arabidopsis thaliana) and SHOOT MERISTEMLESS (STM); whereas KNAT1 misexpression induces lobed leaves.2-5 In the simple leaves of Arabidopsis, KNOXI genes are widely transcriptionally and epigenetically repressed involving transcription factors such as ASYMMETRIC LEAVES 1 (AS1) and AS26-9 and SAWTOOTH 1 (SAW1) and SAW2.10 In young leaf primordia, three axes are established very quickly, and subsequent growth, expansion and differentiation follow along the proximodistal, the dorsoventral and the mediolateral axis.2 In contrary to shoots that grow from the SAM, cells that build the leaf blade come from the meristematic zone of the leaf primordia, which is located at the junction between the leaf blade and the leaf petiole in Arabidopsis.11 The meristematic zone can be distinguished in two types of dividing tissues producing the leaf plate and the leaf margin, respectively.12
Current research of leaf shape concentrates on the role of pattern formation in the leaf margin that controls the number and the size of leaf teeth (serration) or lobes. The process of leaf serration formation is related to the leaf primordia formation in the SAM, both are dependent on auxin maxima established by auxin efflux carrier PIN-FORMED 1 (PIN1).13 The transcription factor CUP-SHAPED COTYLEDON 2 (CUC2), expressed in the leaf sinuses, promotes the generation of PIN1-dependent auxin activity maxima while auxin represses CUC2 expression.14 In leaves, CUC2 is specifically targeted by MIR164A, which triggers the cleavage of the CUC2 mRNA.15 Expression of MIR164A-resistant CUC2 phenocopies the strong-serrated mir164a mutant leaves suggesting that higher CUC2 expression is linked to enhance leaf serration.16 Consistently, cuc2-1D mutant which carries a point mutation in the MIR164A-targeting site shows both, high CUC2 expression and stronger outgrowth leaf serrations, which is independent of KNAT1.17,18 On the other hand, the constitutive misexpression of CUC2 and its homologue CUC1 causes strong inhibition of overall leaf growth.15,19-21 In cuc2-3 loss of function mutants, the auxin maxima is abolished and accompanied by less cell proliferation around the remaining tips suggesting that CUC2 supports the outgrowth of leaf teeth by promoting cell division.22 Additionally, CUC2 represses growth cell-autonomously to enhance leaf serration by inhibition of growth in the sinus.14,16 Recent findings showed that the first morphological visible event is the repression of growth at the area of CUC2 expression, whereas the CUC2-dependent outgrowth of the tooth occurs later.23
The EPIDERMAL PATTERNING FACTOR (LIKE) (EPF and EPFL) gene family encodes plant-specific secretory peptides, which play important roles in leaf development including control of stomata density and patterning in the epidermis.24,25 Several of the EPF peptides, including EPFL2, are ligands of the three ERECTA-family leucine-rich repeat receptor-like kinases ERECTA (ER), ERECTA-LIKE1 (ERL1) and ERL2.26,27 er mutant plants display round leaves with short petioles,28,29 whereas the phenotype of er erl1 erl2 triple mutants is more severe with a small rosette with small, round leaves that lack petiole elongation caused by substantially reduced cell proliferation.30,31 The binding of EPFL2 peptide to the ER family receptors is required for leaf tooth growth that is accompanied by repression of auxin response in growing leaf margins.26
The opinion, how CUC2 promotes leaf serration, changes over the years several times.23 CUC2 has been proposed to either locally repress growth to form the leaf sinuses16 to promote tooth outgrowth in an auxin-dependent manner22 or in a combination of both.14 In the latter variant, the CUC2-dependent growth repression occurs initially forming the leaf sinuses, whereas, during a secondary phase, CUC2 stabilizes non-cell autonomously PIN1 locations and so indirectly auxin maxima that are required for tooth formation.14,23 Recently, showed that EPFL2 signaling promotes leaf tooth growth via repression of auxin response in the growing leaf margin.26 Our study shows a more precise analysis of cuc2-1D and 35S::CUC2-GR (CUC2-GR)17,19 focused on the role of CUC2 in leaf growth and leaf margin development revealing new aspects of these Arabidopsis plant lines and growth control by CUC2. Here, we provide evidences that CUC2 inhibits growth in the sinus by controlling rather cell division than cell size. We demonstrate that growth inhibition by CUC2 is a consequence of reduced cell division and that the spatiotemporal pattern of ectopic CUC2 in the whole leaf blade or the leaf sinus affects leaf size and shape. Transient ubiquitous induction revealed that CUC2 can repress EPFL2 and therefore counteract the cell proliferation promoting ER signaling. Furthermore, we discuss how our findings expand the common view of CUC2-dependent growth regulation in leaves by including cell cycle control in the leaf plate to the margin-centered growth model.
2. Results
2.1. CUC2misexpression can have contrary effects to the growth of Arabidopsis leaves
While studying leaf serration in mutants defective in the epigenetic machinery, we used loss-of-function cuc2-3 mutants and dominant cuc2-1D mutants carrying a MIR164-resistant CUC2 allele, as controls, which either loses all or has enhanced leaf serration, respectively (Figure 1a–c, 32, 16, 17). We were wondering why cuc2-1D mutants have a significant increased total area of rosette leaves (Figure 1a,c; 18), whereas constitutive misexpression of CUC2 is described as reducing leaf size.32 In order to get a better understanding of the growth dynamics, we measured the leaf surface area of cuc2-1D in comparison with Col-0 and cuc2-3 in a time course (Figure 1d). Interestingly, the leaf surface area of cuc2-1D was not distinguishable before all three leaf types, Col-0, cuc2-3, and cuc2-1D, stopped to grow (Figure 1d, 36 DAG). Using toothless cuc2-3 leaves, it was shown that wild-type teeth emerge rather by the outgrowth of the leaf tooth tip than by growth repression in the sinus.22 To prove whether the increased leaf area of cuc2-1D leaves is primarily caused by enhanced tooth growth, we also measured the width and the length in different areas of the leaves (Figure 1e and S1). The width between the first tooth tips and length of leaf blades and petioles was slightly increased in cuc2-1D mutants verifying the results from 17, which also mentioned that leaf width at the sinus of cuc2-1D were significantly wider than Col-0 suggesting that the increased medial-lateral expansion of cuc2-1D leaves was not only due to an outgrowth of leaf teeth but a general enhancement of leaf expansion. In contrast, we found a significant reduction of the leaf wide at some sinuses indicating growth inhibition inside of the leaf blade at least in the area of these sinuses (Figure 1e). This growth suppression occurred early (before 24 DAG) underneath the sinus in cuc2-1D leaves and was also later not covered by tooth outgrowth, which became significant 36 DAG at the very moment when total leaf growth stopped in all three leaf types (Figure 1d,e).
Figure 1.
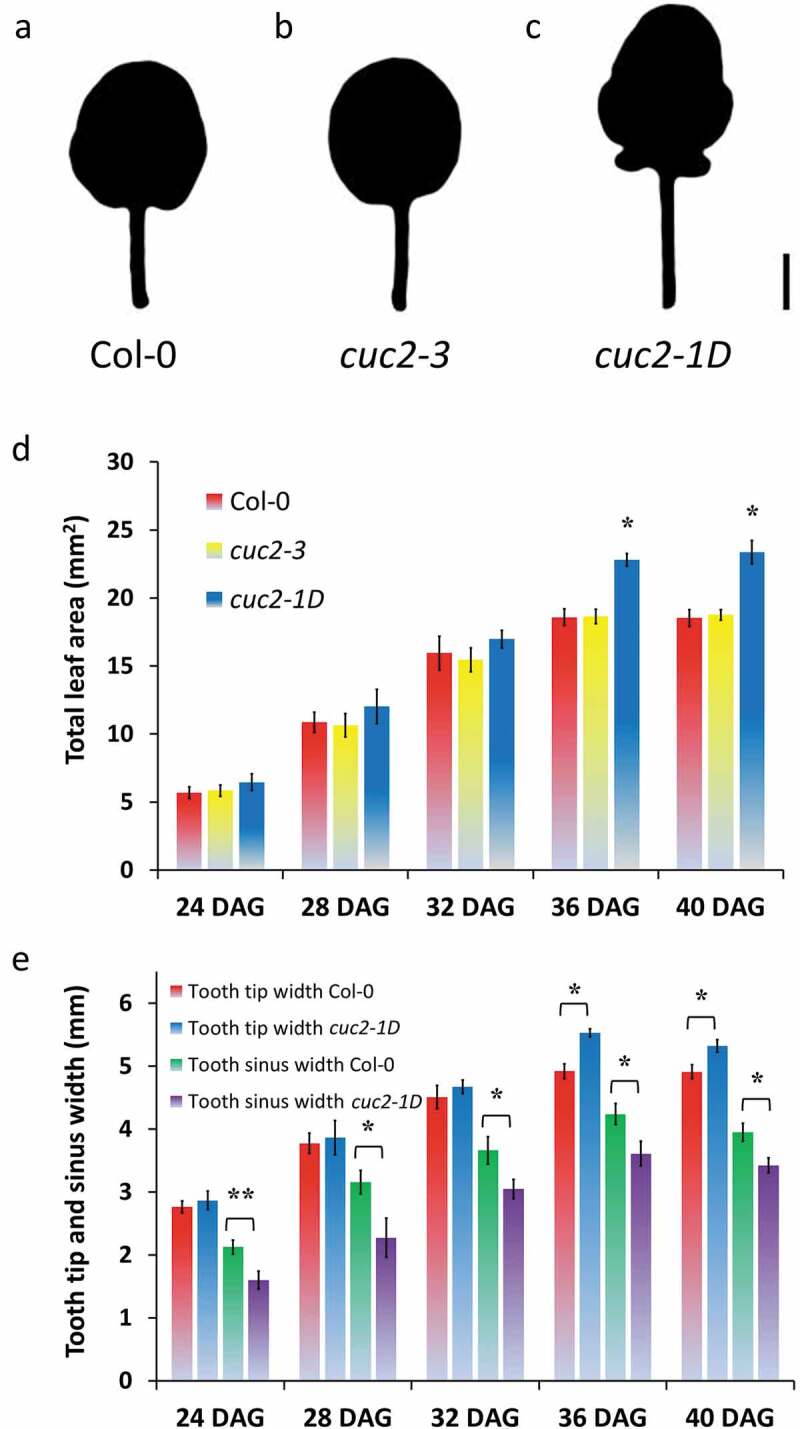
Contrary effects on leaf growth by gaining of CUC2 function in cuc2-1D mutants. A-C, Silhouette of fourth rosette leaf of wild-type (Col-0) (a), cuc2-3 (b) and cuc2-1D mutants (c), 40 DAG. Scale Bars = 2 mm. D, Time course of the dynamics of the total leaf area. *, Significant enlarged (Student’s t-test p < .05) in comparison with Col-0 and cuc2-3 mutant leaves of the same age. Note that each genotype did not show any significant difference between 36 and 40 DAG indicating the leaf growth of all three genotypes terminated around 36 DAG. E, Antagonistic dynamics of the leaf width at the 1st tooth tips and at the sinuses between the 1st and 2nd tooth of the fourth rosette leaf of Col-0 and cuc2-1D mutants, 40 DAG. D-E, N = 10 (24–36 DAG), N = 20 (40 DAG); ± SE. Significant changes (Student’s t-test, *, p < .05, **, p < .01) between Col-0 and cuc2-1D mutant leaves of the same age.
Our results suggest that in contrast to earlier reports misexpression of CUC2 inhibits growth at least in some areas of cuc2-1D leaves. However, our findings may apply only to a subset of rosette leaves as we analyzed mainly juvenile leaves. Leaves, which rise after vegetative phase change, display stronger serrated margins.33 That might increase medial-lateral expansion at the leaf tooth margin, which could cover the growth inhibition in the sinus but we found rather stronger growth suppression in the sinus of later rosette leaves (Figure S1D). Hence, leaf growth inhibition seems a shared feature of plants constitutively over-expressing CUC2 and cuc2-1D mutants.
2.2. Cuc2-dependent growth suppression is not limited to early leaf stages
During early leaf development, CUC2 is expressed along the whole margin of wild-type leaf primordia. Later, CUC2 expression is ceased in the developing teeth and restricted to the sinus area.14,16 When the leaf growth has stopped, CUC2 expression is harder to observe in the sinus regions and then finally vanish [16, Xiaoyu Li and Ralf Müller-Xing, unpublished data]. This raises the question whether leaf cells are competent to react to ectopic CUC2 long after endogenous CUC2 expression is terminated and earlier studies with constitutive expressed CUC2 did not examine that matter in detail.15,19-21 Considering our data about growth dynamics in cuc2-1D leaves, it seems that the spatiotemporal pattern of CUC2 could influence whether CUC2 promotes or suppresses growth in leaves. In order to investigate the temporal aspect, we decided to use an inducible system with an unmodified CUC2 cDNA for our induced overexpression experiments [CUC2-GR friendly provided by Ben Scheres].19 Using that genetic tool insured that the natural regulation of CUC2 by MIR164A was not compromised, in contrast to earlier used microRNA resistance versions [CUC2m].16,34,35,32
We investigated carefully the consequences of starting or withdrawing continued CUC2 overexpression at different time points (Figure 2a). First, we compared the rosette and leaf phenotype of continuously dexamethasone (DEX) treated CUC2-GR and Col-0 plants with untreated controls (Figures 2b–g and 3a,c). In the presence of DEX, we observed no changes in Col-0 plants but strong growth inhibition in all areal parts of CUC2-GR plants resembling constitutive CUC2 expression (Figures 2b–f and 3a,c), whereas the total leaf area of induced CUC2-GR was reduced to 25% (Figure 3a). Interestingly, we found strong enhanced teeth in continuously induced CUC2-GR leaves (Figures 2f,g and 3c, S2A). We measured further growth parameters of the continuously induced CUC2-GR leaves and found overall inhibition of growth along the proximo-distal and the medio-lateral axes (Figure S3).
Figure 2.

Early continued induction of CUC2-GR can promote growth in leaf teeth but decrease total leaf size. A, Scheme of induction time courses of DEX treatments. Red lines indicating DEX induction, black one non-treatment. B-X, All leaf phenotypes were analyzed at 22 DAG. B-E, Leaf rosettes of Col-0 after no DEX (b) and continuous DEX treatment (c), CUC2-GR with no DEX (d) and continuous DEX (e). F, Leaf silhouettes of the fourth rosette leaf; +, continuous DEX treatment, -, No DEX control. G, Fourth leaf continuously treated with DEX (Cont Ind). H-L, The fourth leaves were initially not treated with DEX, but continuous DEX-induced (ind) starting from 11 DAG (h), 12 DAG (i), 13 DAG (j), 14 DAG (k), No DEX control (l). M-R, Plants were initially not treated with DEX but continuous DEX-induced (ind) starting from 12 DAG (m), 14 DAG (n), 16 DAG (o), 18 DAG (p), 20 DAG (q) and 22 DAG (r) on. S-X, From the germination on, plants were treated continuously with DEX but DEX was withdrew (WD) from 12 DAG (s), 14 DAG (t), 16 DAG (u), 18 DAG (v), 20 DAG (w) and 22 DAG (x) on. Scale bars (b-x) = 1000 μm.
Figure 3.
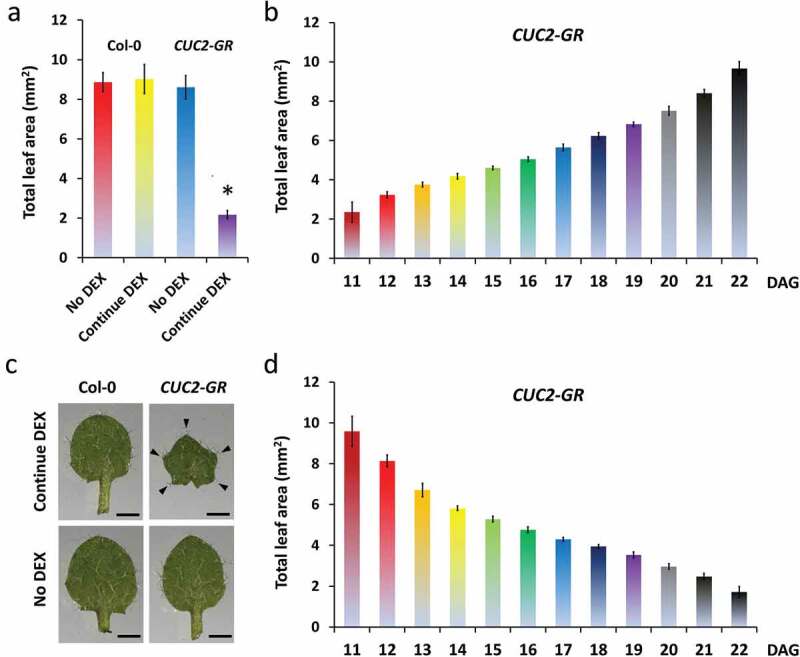
The effects of CUC2-GR on leaf growth correlate with the length of DEX treatment. A, C-D, Total leaf area of the fourth leaf of non-treated, temporary and continuous DEX-treated Arabidopsis plants; all leaves were measured at 22 DAG; N = 10, ± SE. A, Continuous DEX-treatment Col-0 and CUC2-GR leaves in comparison to non-treated. * Significant difference compared to the others (Student’s t-test p < .05) B, DEX-induction time course: The continuous DEX-treatment started between 11 DAG and 22 DAG. C, Fourth leaves of continuous DEX-treated (started 0 DAG) and non-treated Col-0 and CUC2-GR plants, 16 DAG. D, DEX-withdrew time course: The DEX-treatment was stopped between 11 DAG and 22 DAG. Note that there was no significant difference of the leaf areas between continuous DEX (a), continued DEX induction started at 11 DAG (b) and DEX withdrew at 22 DAG (d) in CUC2-GR plants.
In a second approach, we started or withdraw continued CUC2 overexpression at serial days between 10–22 DAG and found consistent gradients of increasing or decreasing the size of rosettes or leaves, respectively (Figures 2h–x and 3b,d, S2). Even starting induction or withdrawing of DEX only one or 2 days before measurement (22 DAG) had a clear impact on the total leaf area indicating that at least the same region of the leaf can react to ectopic CUC2 in very late developmental stages.
2.3. CUC2-GR can initiate ectopic tooth growth only in early leaf stages
Leaves, which were exposed to high CUC2 expression during their formation as in cuc2-1D or continuously induced CUC2-GR, show much stronger leaf serration than wild-type (Figures 1a,c, 2f–h, 3c). More in detail, we observed that an early start of continued CUC2-GR induction can promote growth in leaf teeth thereby the decrease of the total leaf size reached its maximum (Figures 2f–h, 3c). The teeth size of continuously DEX-treated CUC2-GR leaves was significant enlarged compared to the controls (Figure 3c, S2A). CUC2-GR leaves, with continuous DEX treatment started later than 12 DAG, were clearly smaller but did not longer display increased teeth (Figure 2i–l, S2A-B). Consistently, tooth size was also increased in CUC2-GR leaves initiated during DEX treatment although DEX was then early withdrawn (≥11 DAG, Figure 2s–x). Taken together, it seems that ectopic expressed CUC2 can enhance leaf serration only in very early leaf stages, whereas can inhibit the overall growth of the leaf blade also in later stages.
2.4. CUC2 represses several key regulators of leaf development but induces its own expression
Leaf growth and patterning involve coordinated regulation among transcription factors36 that build several partially interconnected networks. To test which down-stream transcription factors react rapidly to induced CUC2-GR, we performed quantitative RT-PCR analyses (qRT-PCRs; Figure 4) within 3 h after DEX induction (3 HAI) focusing on transcription factors that are involved in both growth regulation and formation of leaf serration and are predicted to work upstream or downstream of CUC2.
Figure 4.
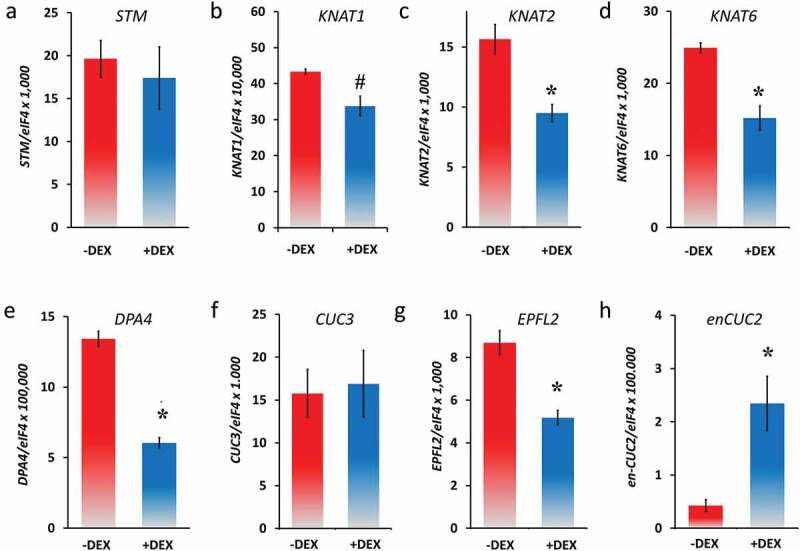
Expression analysis of genes related to leaf development in none and DEX-induced CUC2-GR leaves (qRT-PCR, N = 3, ± SE), 26 DAG, 3 HAI. A, STM, B, KNAT1 (BP), C, KNAT2, D, KNAT6, E, DPA4, F, CUC3, G, EPFL2 and H, endogenous CUC2 (en-CUC2). Asterisks indicate a significant change of expression (Student’s t-test: *, P < .05; #, P = .07) compared with the non-treated control.
In many plant species with strong serrated or compound leaves, KNOXI genes are expressed in the leaf sinus that plays a fundamental role in the serration process.37,38 Furthermore, the KNOX gene STM is misexpressed in the sinus of older leaves of plants carrying the CUC2g-m4 transgene, which is MIR164A-resistant resembling cuc2-1D mutants.22 Although the ectopic STM expression correlates rather with ectopic meristem formation than early CUC2 misexpression.22 To test whether the KNOXI genes response to temporary increased CUC2 activity, we checked their mRNA abundance in CUC2-GR leaves by qRT-PCR 3 HAI with DEX. Interestingly, the mRNA levels of KNAT1, KNAT2, and KNAT6 were slightly decreased, whereas the expression of STM was not significantly changed (Figure 4b–d) confirming a rather indirect correlation of high CUC2 activity and ectopic STM expression such as in CUC2g-m4 leaves. KNOXI are widely repressed in leaves, through the activities of transcription factors such as AS1, AS2, SAW1, and SAW2.6,7,9,10 Nevertheless, we found increased levels of all four KNOXI genes in cuc2-3 mutants (Figure S3A-D) suggesting that CUC2 could be part of the group of transcription factors, which suppresses KNOXI gene expression in wild-type leaves.
Next, we checked an up-stream component of the CUC2 signaling network controlling leaf serration. DEVELOPMENT-RELATED PcG TARGET IN THE APEX4 (DPA4/NGAL3) negatively regulates CUC2 expression independently of MIR164A preventing strong serration.39 The mRNA levels of DPA4 were slightly decreased at 3 HAI of CUC2-GR (Figure 4e), whereas DPA4 was up-regulated in cuc2-3 mutants (Figure S4) indicating a double-negative feedback loop between CUC2 and DPA4.
In contrast to CUC1,16 CUC3 is involved in leaf serration.40 Genetic analyses showed that CUC2 promotes leaf serration via two different pathways, one early independent of CUC3 promoting teeth emergence and outgrowth, and one latter requiring both CUC2 and CUC3, which sustain teeth formation.40 Although this genetic interactions and similar expression domains suggest CUC2 might be upstream of CUC3,40 we did not find any significant changes of CUC3 in response to temporary induced CUC2-GR (Figure 4f).
DEX-treated CUC2-GR leaves (Figures 2 and 3, S2) phenocopied either the smaller leaves of er mutants [Figure S5]28,29,41 or the much smaller leaves of er erl1 erl2 triple mutants.30 The petiole length of continuously induced CUC2-GR can be dramatically reduced to a certain degree of almost distinction (Figures 2f,g and 3c) that remarkably resembles the leaf phenotype of er erl1 erl2 triple mutants that lack petiole elongation.30 However, in average the petiole length of continuously induced CUC2-GR plants was reduced to 40.7% (Figure S3B) that reminds of the shorter petiole in Ler plants (Figure S5D). EPFL2 encodes one of the ligands of the ER-like receptor kinases ER ERL1 and ERL2.26 Three hours after DEX induction of CUC2-GR plants, EPFL2 mRNA levels had been significantly reduced compared with the non-DEX treated plants (Figure 4g), whereas EPFL2 was up-regulated in the cuc2-3 loss-of-function mutants and might be slightly reduced in cuc2-1D mutants (Figures 5 and 6a). cuc2-1D epfl2 mutants have teeth in mature leaves even if the teeth are smaller than those of cuc2-1D indicating that CUC2 can promote tooth growth in an EPFL2-independent manner.26 Nevertheless, the reduction of the tooth size of cuc2-1D leaves by loss of EPFL2 function suggests that the ectopic tooth growth is partially dependent of EPFL2-ER ligand-receptor module. Alternatively, ectopic CUC2 promotes serration also by cell autonomous repression of both growth and EPFL2 expression in the sinus and other areas of the leaf plate.
Figure 5.

Expression analysis of EPFL2 and endogenous CUC2 (en-CUC2) in seedlings (qRT-PCR, N = 3, ± SE). A, Col-0, cuc2-3 and cuc2-1D seedlings, 26 DAG. B, CUC2-GR seedlings were mock-treated (mock) or treated with cycloheximide (CHX), dexamethasone (DEX), or dexamethasone plus cycloheximide (CD). Asterisks indicate significant change of expression (Student’s t-test: *, P < .05; **, P < .01) compared with Col-0 or mock-treated CUC2-GR plants, respectively.
Figure 6.
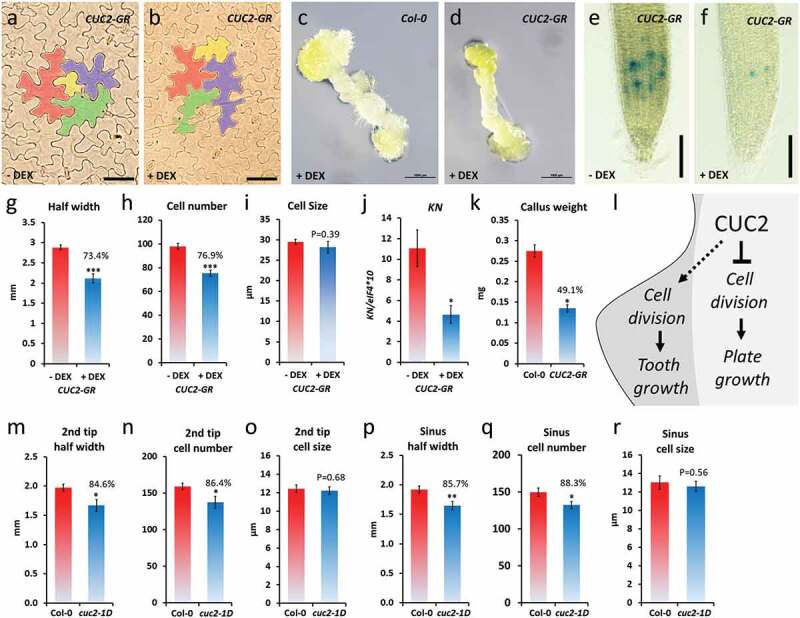
Growth repression by ectopic CUC2 based on reduced cell numbers. A-K, continuously DEX-treated CUC2-GR plants (+ DEX) in comparison to non-treated (- DEX) or Col-0. A-B, pavement epidermis cells in the fourth leave of CUC2-GR plants, 30 DAG, four cells of representative cell size range were marked by false colors. C-D, Callus of hypocotyl explants of DEX-treated CUC2-GR and Col-0 plants, 16 days on callus-inducing media (CIM). E-F, CYCB1;1::GUS expression in CUC2-GR roots. G-I, Half leaf width, cell number and cell size (diameter) was measured from the midrib to the tooth tip of the fourth leaf, 30 DAG, N ≥ 10, ± SE. J, expression of the cell cycle marker KN in CUC2-GR seedlings, 7 DAG (qRT-PCR, N = 3, ± SE), 7 DAG. E, Callus weight of hypocotyl explants of DEX-treated CUC2-GR and Col-0 plants, after 7 weeks on CIM, N = 20, ± SE. L, Conceptual model of leaf growth control by CUC2. M-R, Fourth leaf of Col-0 and cuc2-1D plants. Half leaf width, cell number and cell size (diameter) were measured from the midrib to the 2nd tooth tip (2nd tip) or the sinus between 1st and 2nd tooth (sinus), respectively, 20 DAG long day, N = 10, ± SE. Asterisks indicate significant differences (Student’s t-test: *, P < .05; **, p < .01) compared with the controls.
In order to test the response of the CUC2 promoter to transient higher CUC2 levels, we designed gene-specific amplification primers derived from untranslated region sequences that specifically amplified the endogenous CUC2 (en-CUC2) but not the transgenic CUC2-GR mRNA. Interestingly, en-CUC2 was significantly induced by CUC2-GR in the qRT-PCR analysis 3 HAI with DEX (Figure 4h). That indicates that CUC2 could induce its own expression by direct transcriptional activation or by binding to fast-reacting target genes encoding transcription factors, which activate CUC2 in turn. Nevertheless, as the transcription factor DPA4 is meant to be an up-stream repressor of CUC2,39 the repression of DPA4 could also play a role in indirect up-regulation of en-CUC2 in induced CUC2-GR plants. To test this hypotheses, we used cycloheximide (CHX) as a translation inhibitor42 and found even higher en-CUC2 expression in the samples treated with both, DEX and CHX suggesting that the activation of en-CUC2 by CUC2-GR is rather directly (Figure 5b).
To summarize, we found that some key regulators of leaf development, supposed to be genetically downstream of CUC2, do not react to temporary increased CUC2, such as CUC3 and STM, or are surprisingly downregulated such as KNAT1,2,6, DPA4, and EPFL2.
2.5. Ectopic CUC2 limits growth rather by cell cycle inhibition than by cell size control
Previous studies about the role of CUC2 in leaf serration focused on either promotion of tooth growth or growth repression in the sinus.14,16,22,23,40,43CUC2 contributes to the outgrowth of leaf teeth by promoting non-cell autonomously cell proliferation.22 Nevertheless, this does not answer how the cell autonomous function of CUC2 causes the growth inhibition in the leaf sinus of wild-type or plants with ectopic CUC2 expression. To determine the effects of ectopic CUC2 expression on cell size and cell number, we cleared rosette leaves of continuously induced CUC2-GR and control plants, 30 DAG. Like in the epidermis of wild-type leaves, the size of pavement cells varies strongly in CUC2-GR but we did not find any obvious changes in the overall cell size between induced and non-induced plants (Figure 6a,b). As a more direct test of the hypothesis that CUC2 controls rather numbers of cell than cell size, we examined the distance (half leaf width), cell number and cell diameters along a line between midrib to the tooth tip of the cleared leaves (Figure 6g–i). We found a strong correlation between the reduction of the half leaf wide, and the cell number (26.6% or 23.1%, respectively; Figure 6g–h) in continuously induced CUC2-GR indicating that ectopic CUC2 inhibits cell proliferation (Figure 6l). To test this hypothesis, we measured the expression of KNOLLE (KN), which encodes an M phase-specific syntaxin, involved in vesicle fusion during cytokinesis,44 by qRT-PCR. In continuously induced CUC2-GR seedlings, the abundance of KN transcripts was reduced more than half in comparison to the control (Figure 6j).
In order to test the general capacity of ectopic CUC2 in cell cycle inhibition, we examined the effects in callus and roots. Continuously induction of CUC2-GR significant reduced callus size and weight on callus inducing media (CIM) (Figure 6c–d,k). In continuously induced CUC2-GR root tips, expression of the cell cycle marker CYCB1;1::GUS was strongly reduced (Figure 6e–f, S6) suggesting that ectopic CUC2 can partially inhibit cell cycle in various tissues.
Next, we examined the half leaf width, cell number, and cell diameters also in cuc2-1D leaves (Figure 6m–r). As shown before, we found slightly deeper serration in the sinus between the first and second tooth (14.3%), which correlated with the slightly reduced cell number (11.7%) between midrib and sinus (Figure 6p–q). Interestingly, also the distance, and accordantly the cell number, between the second tooth tip and midrib was significant reduced (15.3% and 13.6%, respectively; Figure 6m–n). Hence, we did not find any significant changes in the average size of pavement cells in cuc2-1D leaves (Figure 6o,r).
To summarize, it was reported before that CUC2 promotes non-cell autonomously cell proliferation in tooth formation that is even enhanced by ectopic CUC2 expression.14,17,22 Here we show that ectopic CUC2 can repress cell division but did not alter the average epidermis cell size in CUC2-GR or cuc2-1D leaves suggesting that the cell autonomous growth inhibition by CUC2 in the sinus and other regions of the leaf plate is mainly caused by reduced cell proliferation.
3. Discussion
Leaf shape is controlled by a combination of factors either promote or inhibit growth.2,45 The transcription factor CUC2 shapes the leaf margin by both growth inhibition in the sinus and promoting non-cell autonomously tooth outgrowth, which involves the growth-promoting phytohormone auxin.14,23,2,16,developed a computer model of leaf serration basing on the interactions between auxin transport, PIN1 location, and CUC2 expression during leaf margin development. In the simulation, auxin locally promotes and CUC2 locally inhibits the propagation of a single cell layer representing the leaf margin.14 The computer model can successfully simulate the consequences of loss of CUC2 or PIN1, which both cause smooth leaf margins in the simulation as well as in planta, whereas simulating increased CUC2 expression in the leaf margin produces narrower leaves with deeper serrations.14 The latter-simulated morphology comes very close to phenotype of cuc2-1D and ubiquitous expressed CUC2-GR but cuc2-1D leaves significant wider and CUC2-GR leaves are rather shorter than only narrower (Figure S3A, C). In addition, the computer model does not include the control of cell division in the inner leaf plate,46 which also influences leaf shape and size. This limitation does not affect the simulation of wild-type leaves, because CUC2 expression is here limited to the sinus leaf margin, but it matters in leaves with ectopic CUC2 expression.
Our results suggest that dependent of the spatiotemporal pattern, ectopic CUC2 expression can enhance either tooth outgrowth or growth inhibition or both. Furthermore, ectopic CUC2 can repress cell division in the leaf plate causing reduced growth independently whether the plate area is underneath of sinuses or teeth. Temporary induction of CUC2 represses the KNOX1 genes KNAT1, KNAT2 and KNAT6 (Figure 4b–d). In as1 mutants, ectopic expression of KNAT1, KNAT2 and KNAT6 cause overproliferation in epidermal cells of the leaf petioles which is suppressed in as1 knat1 knat2 knat6 quadruple mutants.7 Nevertheless, the expression of KNOX1 genes is widely silenced in wild-type leaves8,10,47,48 making it unlikely that cell cycle inhibition by ectopic CUC2 is widely dependent on further downregulation of KNAT1, KNAT2, and KNAT6.
Our simplified model in Figure 6l suggests that the sinus area belongs rather to the leaf plate, in which CUC2 expression can inhibit cell autonomously cell division. As in wild-type leaves CUC2 expression is limited to a few cells in the sinus,16 the growth repression is rather weak. Loss of CUC2 function prevents tooth growth as well as growth inhibition in the sinus but both processes balanced each other so that wild-type and cuc2-3 leaves have the same total leaf area (Figure 1d). In leaves with ubiquitous expressed CUC2, growth repression can affect the whole leave. Furthermore, higher CUC2 activity, such as in cuc2-1D, promotes non-cell autonomously cell division during the ectopic tooth outgrowth (Figure 6l), which depends on the negative feedback loop of auxin and CUC2 expression.14
The increased leaf size in cuc2-1D dependents on increased cell proliferation, mediated through an ER-dependent pathway.17 Interestingly, transient induced CUC2 can repress EPFL2 (Figure 4g), which encodes a ligand of the receptors of the ER family promoting cell division in leaves and in general.26,30,31CUC2 can promote growth in leaf teeth both, dependently and independently of EPFL2,26 but repression of EPFL2 by CUC2 could still reduce cell proliferation in the leaf plate. Although EPFL2 is expressed in the whole leaf blade with the exception of vascular and tooth tips, loss of EPFL2 causes clearly less severe leave size reduction26 than ubiquitous expressed CUC2. This indicates that the repression of EPFL2 does not play more than a minor role in cell cycle repression by CUC2.
It will be a challenge to dissect both CUC2-dependent pathways, either promoting cell division in teeth or repressing cell division in the sinus. Our approach, using inducible CUC2-GR in time courses, was a first step to understand the developmental timing of both processes. In future studies, temporal induction and reduction of CUC2 expression under control of the CUC2 promoter in combination with separation of the cells of teeth and sinuses will provide more specific and accurate data of the downstream events and might identify different down-stream targets of CUC2 in both tissues.
4. Materials and methods
4.1. Plant materials and growth conditions
Arabidopsis thaliana (L.) plants were grown at 21°C under short day (10 h light/14 h dark) conditions. Columbia (Col-0, N70000) and Landsberg-0 (La-0, N6765), which were used as wild-type controls, and Landsberg erecta-0 (Ler-0, NW20) and the dominant cuc2-1D mutant (N16485) plant lines were obtained from the Nottingham Arabidopsis Stock Center. cuc2-316,32 and 35S::CUC2-GR [CUC2-GR]19 were kindly provided by Patrick Laufs and Ben Scheres, respectively. Peter Doerner kindly provided seeds of CYCB1;1::GUS.49 With the exception of La-0 and Ler-0, all genotypes were in Col-0 background. Surface-sterilized seeds were sowed on half-strength Murashige and Skoog salt mix (½ MS; Tian Da Chemical, China) media plates under aseptic conditions. After 10 days, the plants, which had approximately the same size, were transferred to soil in pots.
4.2. Dexamethasone treatments
Dexamethasone (DEX) (WAKO, Japan) 10 mmolL−1 was dissolved in 70% ethanol and kept at −20°C as a stock solution. In depletion and continues DEX-treatment experiments, CUC2-GR plants and Col-0 controls were grown on ½ MS tissue culture plates containing 10 µmolL1 (10 µM) DEX for 10 days and then transferred to soil. Plants, growing on soil, were directly sprayed one time per day with a 10 µM DEX solution containing 0.02% Silwet. The CUC2-GR and Col-0 seedlings were DEX treated and non-treated either continuously or with specific time courses, starting continuous treatment or withdrawn to different time points, as shown in Figure 2a. Hence, the phenotype of seedlings and dissected leaves were documented only at the last time point.
4.3. Phenotypic characterization and microscopy
The seedlings were photographed with a Nikon digital camera (D3200, AF-S Micro NIKKOR 60 mm1:2.8 G ED) in a different time course. The leaves were dissected at specific time points and scanned with a scanner (LIDE220, Cannon, Japan). The smaller leaves image was taken under an SMZ25 Microscope (Nikon, Japan). Leaf silhouettes were generated from scanned leaves by using the following steps in the computer program PhotoshopTM (Acrobat Systems incorporated): (I) Extraction of the black background by the magic wand tool (Tolerance, 50), (II) Removing overhanging trichomes and interfering artifacts, (III) Gaussian Blur (Radius, 2.0 pixels), (IV) Unsharp Mask (Amount, 250%; Radius, 500 pixels; Tolerance, 0 levels), (V) Magic wand tool (Tolerance, 50) and Rotate (Transform) for final arrangement of the leaves. All phenotype data were analyzed by using Student’s t-test (Excel, Microsoft).
4.4. Expression analysis
For RNA extraction, whole seedlings (without roots) or leaves (1st to the 4th) were collected and snap frozen in liquid nitrogen. The total RNA was purified using TRIZOL (Invitrogen) procedure. The extracted RNA was treated with DNase (Thermo Scientific) to remove DNA contamination. cDNA was synthesized using first-strand cDNA synthesis kit (Thermo Scientific) analyzed by qRT-PCR using the SYBER green supermix (Roche) at the Roche Lightcycler480 II. The DEX plus cycloheximide experiment was described before.42 The primers used in the qRT-PCR analysis are listed in Table S1.
4.5. GUS histochemical assay
CUC2-GR ♀ was crossed with CYCB1;1::GUS ♂.49 In the F1 generation, seedlings were grown on ½ MS tissue culture plates containing 10 µmolL-1 DEX or no DEX for 8 days. Detection of GUS activity in the roots was performed with whole seedlings as described with minor modifications.50
4.6. Callus induction
Col-0 and CUC2-GR seeds were sterilized with 70% ethanol (3 times) and germinated on half-strength MS medium (2.165 g/L MS basal medium with vitamin powder, 10 g/L sucrose, and 9 g/L agar, pH 5.8) for 7 days in short-day condition (3 days on fridge before transfer to short day). Hypocotyls were excised and transferred to callus inducing medium (CIM) (4.4 g/L MS basal medium, 10 g/L sucrose, and 9 g/L agar, 2.2 μM 2,4-D, 0.2 μM kinetin, pH 5.8) with 10 µM DEX. Callus weight was measured at 7 weeks after callus induction. Two independent biological replicates were performed.
4.7. Tissue clearing and cell counting
For cell counting and visualizing of cell size, rosette leaves were mounted in clearing solution (chloral hydrate:glycerol:water, 8:1:2 [w/w/v]) as described with minor modifications.52 The measurement of half leaf wide and cell number followed a line between midrib and leaf margin of tooth tip or sinus. The measurement lines ran at a right angle to the midrib. The average cell diameters were calculated by half leaf width/cell number (µm/N).
Funding Statement
This work was supported by the Fundamental Research Funds for the Central Universities, China [Grant No. 2572016DA03] and Natural Science Foundation of Heilongjiang Province of China, General Program [Grant No. C2016007] to Q.X., X.L., Z.H., and R.M.-X.
Acknowledgement
Seeds of cuc2-3 were kindly provided by Patrick Laufs, and seeds of 35S::CUC2-GR generously provided by Ben Scheres. Peter Doerner kindly provided CYCB1;1::GUS seeds.
Disclosure of potential conflicts of interest
The authors declare that they have no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Thomas B. Leaf development. Encyclopedia of applied plant sciences, 2nd. Netherlands: Elsevier Amsterdam. Vol. 1. 2016; 1–11. [Google Scholar]
- 2.Byrne ME. Making leaves. Curr Opin Plant Biol. 2012;15(1):24–30. doi: 10.1016/j.pbi.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6(12):1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379(6560):66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 5.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12(4):507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408(6815):967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 7.Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, Machida Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J Cell Mol Biol. 2010;61(1):70–82. doi: 10.1111/j.1365-313X.2009.04033.x. [DOI] [PubMed] [Google Scholar]
- 8.Lodha M, Marco CF, Timmermans MCP. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 2013;27(6):596–601. doi: 10.1101/gad.211425.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. [DOI] [PubMed] [Google Scholar]
- 10.Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell. 2007;19(9):2719–2735. doi: 10.1105/tpc.106.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichihashi Y, Kawade K, Usami T, Horiguchi G, Takahashi T, Tsukaya H. Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol. 2011;157(3):1151–1162. doi: 10.1104/pp.111.185066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukaya H. Leaf shape diversity with an emphasis on leaf contour variation, developmental background, and adaptation. Semin Cell Dev Biol. 2018:48-57. [DOI] [PubMed] [Google Scholar]
- 13.Scarpella E, Barkoulas M, Tsiantis M. Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol. 2010;2(1):a001511. doi: 10.1101/cshperspect.a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci U S A. 2011;108(8):3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131(17):4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 16.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18(11):2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larue CT, Wen J, Walker JC. A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J Cell Mol Biol. 2009a;58(3):450–463. doi: 10.1111/tpj.2009.58.issue-3. [DOI] [PubMed] [Google Scholar]
- 18.Larue CT, Wen J, Walker JC. Genetic interactions between the miRNA164-CUC2 regulatory module and BREVIPEDICELLUS in Arabidopsis developmental patterning. Plant Signal Behav. 2009b;4(7):666–668. doi: 10.4161/psb.4.7.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell. 2010;22(3):640–654. doi: 10.1105/tpc.109.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Current Biology CB. 2004;14(12):1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 21.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. [DOI] [PubMed] [Google Scholar]
- 22.Kawamura E, Horiguchi G, Tsukaya H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J Cell Mol Biol. 2010;62(3):429–441. doi: 10.1111/tpj.2010.62.issue-3. [DOI] [PubMed] [Google Scholar]
- 23.Biot E, Cortizo M, Burguet J, Kiss A, Oughou M, Maugarny-Calès A, Gonçalves B, Adroher B, Andrey P, Boudaoud A, et al. Multiscale quantification of morphodynamics: morphoLeaf software for 2D shape analysis. Development. 2016;143(18):3417–3428. doi: 10.1242/dev.134619. [DOI] [PubMed] [Google Scholar]
- 24.Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50(6):1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 25.Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One. 2013;8:e65183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tameshige T, Okamoto S, Lee JS, Aida M, Tasaka M, Torii KU, Uchida N. A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Current Biology CB. 2016;26(18):2478–2485. doi: 10.1016/j.cub.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Torii KU. Mix-and-match: ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012;17(12):711–719. doi: 10.1016/j.tplants.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Bowman J. Arabidopsis: an atlas of morphology and development. In: Bowman J, editor. New York (London): Springer-Verlag; 1994. [Google Scholar]
- 29.Rédei GP. A heuristic glance at the past of Arabidopsis genetics: a note on Columbia wild type and landsberg erecta. In: Koncz C, Chua N-H, Schell JS, editors. Methods in Arabidopsis research. Singapore (River Edge N.J.): WORLD SCIENTIFIC; 1992. p. 1–15. [Google Scholar]
- 30.Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131(7):1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- 31.Shpak ED, Lakeman MB, Torii KU. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell. 2003;15(5):1095–1110. doi: 10.1105/tpc.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134(6):1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 33.Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011;138(19):4117–4129. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- 34.Peaucelle A, Morin H, Traas J, Laufs P. Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development. 2007;134(6):1045–1050. doi: 10.1242/dev.02774. [DOI] [PubMed] [Google Scholar]
- 35.Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J Cell Mol Biol. 2008;55(1):65–76. doi: 10.1111/j.1365-313X.2008.03483.x. [DOI] [PubMed] [Google Scholar]
- 36.Moon J, Hake S. How a leaf gets its shape. Curr Opin Plant Biol. 2011;14(1):24–30. doi: 10.1016/j.pbi.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Bar M, Ori N. Leaf development and morphogenesis. Development. 2014;141(22):4219–4230. doi: 10.1242/dev.106195. [DOI] [PubMed] [Google Scholar]
- 38.Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. [DOI] [PubMed] [Google Scholar]
- 39.Engelhorn J, Reimer JJ, Leuz I, Göbel U, Huettel B, Farrona S, Turck F. Development-related PcG target in the apex 4 controls leaf margin architecture in Arabidopsis thaliana. Development. 2012;139(14):2566–2575. doi: 10.1242/dev.078618. [DOI] [PubMed] [Google Scholar]
- 40.Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 2011;23(1):54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tisné S, Barbier F, Granier C. The ERECTA gene controls spatial and temporal patterns of epidermal cell number and size in successive developing leaves of Arabidopsis thaliana. Ann Bot. 2011;108(1):159–168. doi: 10.1093/aob/mcr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner D, Sablowski RW, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285(5427):582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- 43.Maugarny-Calès A, Cortizo M, Adroher B, Borrega N, Gonçalves B, Brunoud G, Vernoux T, Arnaud N, Laufs P. Dissecting the pathways coordinating patterning and growth by plant boundary domains. PLoS Genet. 2019;15(1):e1007913. doi: 10.1371/journal.pgen.1007913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139(6):1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukaya H. Leaf development. The Arabidopsis Book. 2013;11:e0163. doi: 10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215(2):407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- 47.Guo M, Thomas J, Collins G, Timmermans MCP. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell. 2008;20(1):48–58. doi: 10.1105/tpc.107.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Li B, Liu J, Guo Z, Liu Y, Li Y, Shen W-H, Huang Y, Huang H, Zhang Y, et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J Integr Plant Biol. 2016;58(12):959–970. doi: 10.1111/jipb.v58.12. [DOI] [PubMed] [Google Scholar]
- 49.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J Cell Mol Biol. 1999;20(4):503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 50.Müller-Xing R, Clarenz O, Pokorny L, Goodrich J, Schubert D. Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell. 2014;26(6):2457–2471. doi: 10.1105/tpc.114.123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berleth T, Jurgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


