ABSTRACT
Gastric cancer (GC) is the second most prevalent carcinoma resulting in cancer-related deaths in the world, with differences among geographic areas. Although the incidence and mortality rates of GC in Asia are decreasing, the search for diverse and effective therapies of GC is still needed to be fully inquired. The present research explored the expression pattern, functional role and underlying mechanism of DLX6-AS1 in GC. Firstly, we measured DLX6-AS1 expression in GC and then found the elevated level of DLX6-AS1. To further inspect the function role of DLX6-AS1 involved in GC, we performed lost-of-function assays. The silencing of DLX6-AS1 suppressed cell proliferation, migration and EMT process of GC cells. Subsequently, we uncovered that MAP4K1 was also up-regulated in GC and could be positively regulated by DLX6-AS1. Moreover, MAP4K1 down-regulation similarly inhibited GC progression. In addition, DLX6-AS1 stabilized MAP4K1 via modulating FUS. In summary, DLX6-AS1 modulated GC progression through FUS-regulated MAP4K1. Our paper exposed the role and regulatory mechanism of DLX6-AS1 in GC, which suggested a novel and valid therapy for GC patients.
KEYWORDS: Gastric cancer (GC), lncRNA DLX6-AS1, MAP4K1, FUS
Introduction
Gastric cancer (GC), with high morbidity and mortality rates,1 is one of the most common tumors in the world2 and a specific regional genetic disease3 in the digestive system with more than 50% of cases occur in Eastern Asia.2 Despite the slowly declining incidence and mortality rates in Asia,2 GC remains a momentous health problem with limited therapies. Therefore, a deep comprehension of the molecular mechanisms in GC is awfully required for GC prevention, diagnosis and therapy.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without open reading frames that encode proteins.4 They were previously discovered to regulate gene expression in human tumors.5-7 And an increasing number of studies suggested their crucial roles in a variety of cellular processes, such as cell proliferation,8 differentiation9 as well as apoptosis.10 For example, lncRNA MEG3 has been found to inhibit proliferation and induce apoptosis in prostate cancer.11 LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma.12 In addition, lncRNA H19 promotes tumorigenesis in head and neck squamous cell carcinoma.13
LncRNA DLX6 antisense RNA 1 (DLX6-AS1), located in chromosome 7q21.3, has been revealed to be overexpressed in multiple carcinomas, including glioma carcinoma,14 pancreatic cancer,15 lung adenocarcinoma,16 renal cell carcinoma17 and hepatocellular carcinoma.18 Furthermore, DLX6-AS1 participates in tumorigenesis and metastasis in a diverse range of carcinomas, which mainly involve cell proliferation,17 migration18 and invasion.14 However, the biological role of DLX6-AS1 in gastric cancer was yet to be researched.
MAP kinase kinase kinase kinases (MAP4Ks) belong to the mammalian Ste20-like family of serine/threonine kinases.19 Mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1), a member of MAP4K family, has been elucidated to boost cancer progression, containing bladder urothelial carcinoma20 and colon carcinoma.21 Nonetheless, the influence of MAP4K1 on gastric cancer was still unclear.
RNA-binding proteins (RBPs) are key players in post-transcriptional events through its specific and affinitive RNA-binding activities22 for the sake of the interplay with particular mRNAs to influence cellular activities of diverse tumors.23,24 Fused in sarcoma/translocated in liposarcoma (FUS), a nuclear RNA-binding protein, modulates different steps of gene expression, including transcription, splicing and even mRNA transport.25 FUS has been implicated as an oncogene in various human cancers,26,27 involving gastric cancer.28
In our research, the expression pattern, functional role and underlying mechanism of DLX6-AS1 affecting the carcinogenesis and progression of GC were probed.
Materials and methods
Cell culture
Human gastric cancer cell lines (AGS, HGC-27, SGC-7901 and BGC-823) and one normal gastric epithelial cell line (GES-1) were purchased from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). All cell lines were grown in Roswell Park Memorial Institute (RPMI)-1640 (Hyclone, South Logan, UT, USA) which was added with 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) and 2.5% penicillin and streptomycin (Beyotime Biotechnology, Beijing, China) in a humidified air with 5% CO2 at 37°C.
Cell transection
The oligonucleotides encoding short hairpin RNAs (shRNAs) targeting DLX6-AS1 (sh-DLX6-AS1#1, sh-DLX6-AS1#2 and sh-DLX6-AS1#3), MAP4K1 (sh-MAP4K1#1, sh-MAP4K1#2 and sh-MAP4K1#3) and FUS (sh-FUS) were separately designed and synthetized by Shanghai GenePharma Co., Ltd. (Shanghai, China), respectively. Human FUS and MAP4K1 cDNA were also separately ligated into the pcDNA3.1 vector. All vectors were gained from Shanghai Integrated Biotech Solutions Co., Ltd. (Shanghai, China). All shRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the provider’s protocols.
Quantitative real-time PCR (qRT-PCR)
Total RNA was harvested using Trizol reagent (Invitrogen, CA, USA), referring to the manufacturer’s guidance. Then cDNA was synthesized by M-MLV reverse transcriptase (Invitrogen) and Oligo (dT18) RT primer was applied for the reverse transcription of lncRNA and mRNA. qRT-PCR was performed on the Bio-rad CFX96 real-time PCR System (Bio-rad, CA, USA) using KAPA PROBE FAST qPCR Kits (Kapa Biosystems, MA, USA) and TaqMan probes (Invitrogen). Human GAPDH was used for lncRNA and mRNA normalization. The primers to amplify DLX6-AS1 were 5′-AGTTTCTCTCTAGATTGCCTT-3′ (forward) and 5′-ATTGACATGTTAGTGCCCTT-3′ (reserve); for MAP4K1 were 5′-GACCAGTGCAGGTGATCCAA-3′ (forward) and 5′-CAGAGCAGACGGAAGGTGAG-3′ (reserve); for FUS were 5′-GTGGAGGCAGAGGTGGCATGGGCGG-3′ (forward) and 5′-ACATTCTCACCCAGGCCTTGCACAA-3′ (reserve) and for GAPDH were 5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′ (reserve).
Cell Counting Kit-8 (CCK-8) assay
Exponentially growing cells were plated into 96-well dishes at the density of 2 × 103 cells/well with compete medium. Post 12 h of incubation, 10 ml Cell Counting Kit 8 (Dojindo Laboratories, Kumamoto, Japan) was supplemented into each well, and the cells were cultured for additional 2 hours at 37℃. After the incubation, the plates were washed with phosphate-buffered saline (PBS). The absorbance value of each well was read at the wavelength of 450 nm and a proliferation curve according to the data collected was constructed.
Colony formation assay
After being collected, stably transfected cells were inoculated into 6-pore culture plates at a concentration of 0.5 × 103 cells/well for nearly two weeks. AGS and HGC-27 cells were divided into different groups with or without paclitaxel. In subsequence to the incubation, the formative colonies were fixed by employing 4% paraformaldehyde for about 10 min and stained by employing 0.1% crystal violet for extra 10 min, followed by rinsing twice in PBS. The number of colonies (more than 50 cells) were calculated manually and recorded precisely.
Transwell assay
Transwell chambers (Corning Incorporated, Corning, NY, USA) without Matrigel (BD Biosciences, Bedford, MA, USA) were used to monitor the migration capacity of AGS and HGC-27 cells. With the density of 1 × 104 cells per well, cells were placed into the top chambers which were only replenished with 200 μl DMEM medium. The bottom chambers were supplemented with 800 μl DMEM medium including 10% FBS. One day later, cells were fastened using 4% paraformaldehyde for 30 minutes and further stained using 0.5% crystal violet (Amresco Co., Solon, OH, USA) for 20 min. A light microscope (model no. BX51; Olympus Corporation, Tokyo, Japan) was utilized to calculate the dyed cells.
RNA immunoprecipitation (RIP) assay
RIP assay was performed by using Magna RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA). GC cells were obtained and washed once by PBS, and the pellets were resuspended in freshly prepared RIP lysis buffer, maintained on ice for 20 min and centrifuged for 10 min at 10,000 g. Then cell lysate was incubated in RIP buffer together with magnetic beads precoated with anti-FUS antibody (Cell Signaling Technology) or mouse anti-IgG (Merck Millipore). Antibody anti-IgG was used as negative control. Proteinase K was used to extract the immunoprecipitated RNAs from total RNAs. The purified RNAs were determined by qRT-PCR.
Western blot
GC cell lysates were prepared by using RIPA Lysis buffer (Beyotime, China) including protease inhibitor cocktail (Roche). Thereafter, protein samples were loaded for sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. After a blockage of 5% skim milk, the membrane was probed with primary antibodies. Post washing, the membrane was grown for 1 h with horseradish peroxidase (HRP)-conjugated secondary antibody (1:2000, Santa Cruz Biotechnology). The ECL detection system (Thermo Fisher, USA) and Quantity One software (Bio-Rad, Hercules, CA, USA) were respectively employed to visualize and quantify the signals. The primary antibodies were antibodies against MAP4K1, MAP4K2, MAP4K3, MAP4K4 and MAP4K5 (Santa Cruz, CA); FUS (Cell Signaling Technology); E-cadherin, N-cadherin, Vimentin, K-Ras and N-Ras (Millipore); p-p38 (Tyr182), p38, p-JNK (Tyr183 and Tyr185), JNK, p-ERK (Tyr204), ERK, p-STAT3 (Tyr705), STAT3, NF-κB and IκB-α as well as YAP, pYAP (S127), c-Myc and Cyr61 (Cell Signaling Technology). Antibody anti-GAPDH (Proteintech) was a normalization control.
Statistical analysis
All statistical analyses were performed with the SPSS statistical software version 16.0 (SPSS UK, Ltd., Woking, UK). Student’s t test was applied to evaluate significant differences in data obtained from two independent groups and ANOVA followed by Student-Newman-Keuls Q test for multi group comparisons. P < .05 was considered to indicate significant statistics. All experiments were repeated at least three times independently.
Results
Up-regulated DLX6-AS1 promotes cell proliferation, migration and EMT in gastric cancer
For purpose of investigating whether lncRNA DLX6-AS1 participated in GC progression, we first examined the expression profile of DLX6-AS1 in four GC cell lines (AGS, HGC-27, SGC-7901 and BGC-823) and one normal gastric epithelium cell line GES-1. qRT-PCR assay revealed that DLX6-AS1 expression was comparatively higher in GC cell lines than that in GES-1 (Figure 1a), exhibiting its possible oncogenic role in GC. Then we performed lost-of-function assays to explore the function role of DLX6-AS1 in GC. As DLX6-AS1 was overexpressed in GC cells, sh-NC, sh-DLX6-AS1#1, sh-DLX6-AS1#2 and sh-DLX6-AS1#3 were separately transfected into AGS and HGC-27 cells, which presented the highest expression level of DLX6-AS1. DLX6-AS1 expression was markedly silenced in both AGS and HGC-27 cells in the treatment group, compared to that in the NC group (Figure 1b). Here, sh-DLX6-AS1#1 was chosen for subsequent function assays as it dramatically downregulated the expression level of DLX6-AS1. Then the effects of DLX6-AS1 knockdown on cell proliferation and migration in GC were detected by CCK-8 and transwell migration assays. Firstly, CCK-8 assay showed that DLX6-AS1 knockdown inhibited proliferative ability in comparison with the control groups (Figure 1c). Secondly, migratory capacity was dramatically repressed by the silencing of DLX6-AS1 as tested by transwell migration assay (Figure 1d). At last, western blot was utilized to evaluate the protein levels of EMT-associated proteins including E-cadherin, N-cadherin and Vimentin. The levels of E-cadherin were significantly increased in sh-DLX6-AS1#1-transfected AGS and HGC-27 cells while N-cadherin and Vimentin levels were decreased conversely (Figure 1e). Taken together, high expression level of DLX6-AS1 enhances cell proliferation, migration and EMT process of gastric cancer.
Figure 1.
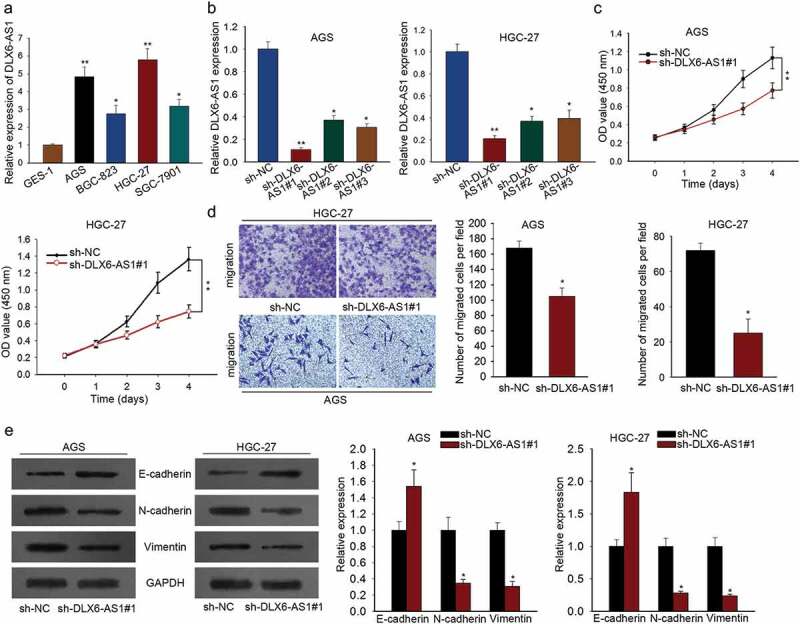
Up-regulated DLX6-AS1 promotes cell proliferation, migration and EMT in gastric cancer. (a) qRT-PCR analysis of the expression level of DLX6-AS1 in gastric cancer cell lines (AGS, HGC-27, BGC-823 and SGC-7901) and normal gastric epithelial cell GES-1. (b) DLX6-AS1 levels in AGS and HGC-27 cells with sh-DLX6-AS1#1, sh-DLX6-AS1#2 or sh-DLX6-AS1#3 treatment, compared with sh-NC group, were tested through qRT-PCR experiment. (c) CCK-8 assay was performed to detect the proliferative ability of DLX6-AS1-silenced AGS and HGC-27 cells. (d) Cell migration capacity of AGS and HGC-27 cells treated with sh-DLX6-AS1#1 was measured by transwell migration assay. (e) The expression levels of EMT-associated proteins, including E-cadherin, N-cadherin and Vimentin, in DLX6-AS1#1-transfected AGS and HGC-27 cells as respectively estimated by western blot. *P < .05, **P < .01.
MAP4K1 highly expressed in GC is regulated by DLX6-AS1 in a positive way
LncRNAs have been illustrated to regulate gene expression in abundant researches.29 DLX6-AS1 has been proved to modulate gene expression in several carcinomas8,18 and MAP4K family members are recognized as oncogenes in many carcinomas.21 Based on the information, we supposed that DLX6-AS1 might affect gastric cancer through regulating MAP4K family. Western blotting indicated that the levels of five MAP4K family members (MAP4K1, MAP4K2, MAP4K3, MAP4K4 and MAP4K5) were lowered by sh-DLX6-AS1#1 (Figure 2a). And MAP4K1 suffered the biggest change in response to DLX6-AS1 knockdown (Figure 2a). Hence, we tested MAP4K1 expression in four GC cells and normal GES-1 cells by qRT-PCR and found that the expression level of MAP4K1 was relatively high in GC cells (Figure 2b). Thereafter, qRT-PCR was employed to assess MAP4K1 expression in AGS and HGC-27 cells with DLX6-AS1 silence. The levels of MAP4K1 were obviously knocked down when sh-DLX6-AS1#1 was transfected into AGS and HGC-27 cells (Figure 2c). All data reveals that MAP4K1 is overexpressed in GC and DLX6-AS1 regulates MAP4K1 expression positively.
Figure 2.

MAP4K1 highly expressed in GC is regulated by DLX6-AS1 in a positive way. (a) Western blot results of the expression of five MAP4K family members: MAP4K1, MAP4K2, MAP4K3, MAP4K4 and MAP4K5, under the transfection of sh-DLX6-AS1#1. (b) The expression level of MAP4K1 in GC cell lines and normal gastric epithelial cell was individually assessed by qRT-PCR. (c) qRT-PCR analysis of the levels of MAP4K1 in AGS and HGC-27 cells with the silencing of DLX6-AS1. *P < .05, **P < .01, ***P < .001.
MAP4K1 enhances cell proliferation, migration and EMT in gastric cancer
To probe into the function role of MAP4K1 in GC cells, we performed lost-of-function experiments by knocking down the expression level of MAP4K1 using specific shRNAs. AGS and HGC-27 cells were both transfected with sh-NC, sh-MAP4K1#1, sh-MAP4K1#2 or sh-MAP4K1#3. MAP4K1 expression was strikingly low in AGS and HGC-27 cells with sh-MAP4K1#1/2/3 treatment as measured by qRT-PCR assay (Figure 3a). Since sh-MAP4K1#1 strikingly downregulated MAP4K1 expression than other two shRNAs (sh-MAP4K1#2 and sh-MAP4K1#3) in AGS and HGC-27 cells, we recruited sh-MAP4K1#1 for later assays. CCK-8 assay demonstrated that cell proliferation of AGS and HGC-27 cells was suppressed by the silencing of MAP4K1 (Figure 3b). And MAP4K1 knockdown evidently inhibited the migratory abilities of AGS and HGC-27 cells (Figure 3c). Furthermore, EMT-associated proteins were subjected to western blot, which elucidated that E-cadherin levels were increased while N-cadherin and Vimentin levels were decreased in MAP4K1 knockdown group (Figure 3d). In addition, colony formation assay was carried out for a therapeutic endpoint to the study of MAP4K1. The results displayed that the colony formation property was notably impaired by silencing of MAP4K1, and this effect was more apparent in paclitaxel group (Supplementary Figure 1A). These results exhibit the promotion of MAP4K1 on cell proliferation, migration and EMT in gastric cancer.
Figure 3.
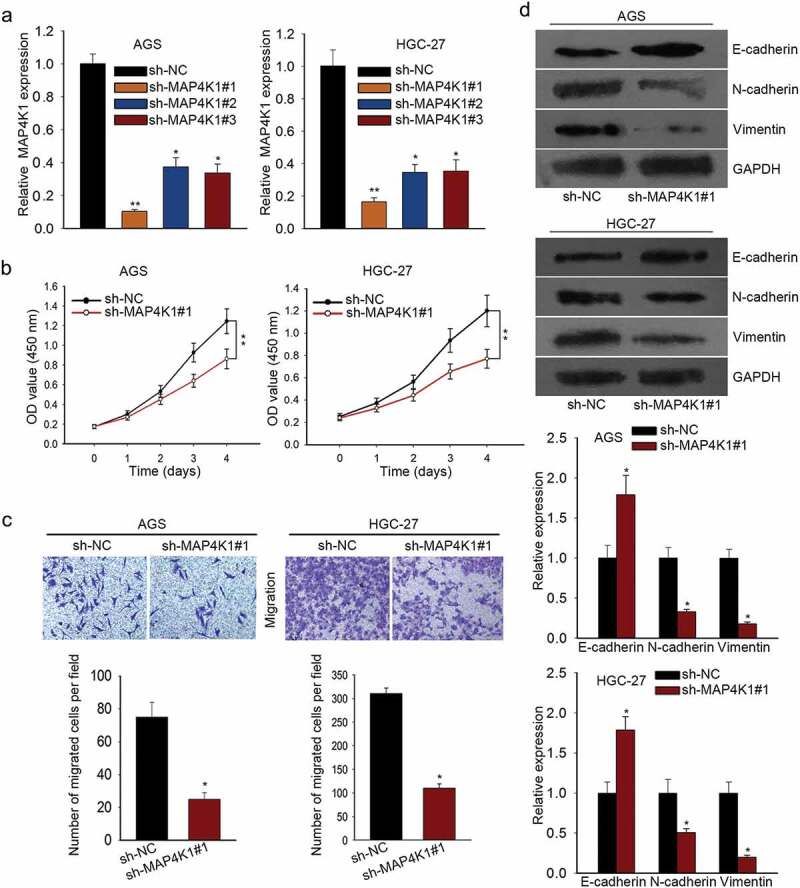
MAP4K1 enhances cell proliferation, migration and EMT in gastric cancer. (a) qRT-PCR result of MAP4K1 expression in AGS and HGC-27 cells with sh-MAP4K1#1/2/3 transfection. Here, sh-MAP4K1#1 transfection induced an obvious decrease in MAP4K1 expression. (b) CCK-8 experiment was carried out to evaluate the proliferation capacity of MAP4K1-silenced AGS and HGC-27 cells. (c) The migratory ability of AGS and HGC-27 cells with sh-MAP4K1#1 treatment was detected by transwell migration assay. (d) The levels of EMT-associated proteins, containing E-cadherin, N-cadherin and Vimentin, in sh-MAP4K1#1-transfected AGS and HGC-27 cells as tested by western blotting. *P < .05, **P < .01.
DLX6-AS1 stabilizes MAP4K1 by regulating FUS
Subsequently, we probed the modulation mechanism of MAP4K1 under DLX6-AS1 in GC. RBPs were selected for their function at post-transcriptional levels and critical roles in cellular activities of tumors, which could bind with specific mRNAs and stabilize their mRNA levels as molecular links in human tumors.26,30 From starbase v2.0, FUS was predicted as a binding protein of MAP4K1 (Figure 4a). FUS has been claimed to be an oncogene in cancers31 and stabilize gene expression specifically.26 And DLX6-AS1 was also predicted to interact with FUS according to starBase v2.0 (http://starbase.sysu.edu.cn/starbase2/browseRbpLncRNA.php). Hence we supposed that DLX6-AS1 might mediate MAP4K1 via FUS. To verify the interplay between FUS and MAP4K1, we resorted to RIP experiment. The immunoprecipitated complex by anti-FUS showed high enrichment of MAP4K1 as examined through qRT-PCR (Figure 4b). To further testify our hypothesis, we mechanistically tested FUS expression in GC cell lines and normal cell line GES-1. We detected the overexpression of FUS in GC cell lines (Figure 4c). According to qRT-PCR and western blot detection, the mRNA and protein levels of FUS were also downregulated by the silencing of DLX6-AS1 in AGS and HGC-27 cells (Figure 4d,e). Furthermore, we used actinomycin D, a RNA polymerase inhibitor causing RNA synthesis inhibition, to affirm the influence of DLX6-AS1 and FUS on MAP4K1 mRNA stability. The transfection efficiency of pcDNA3.1-FUS was assessed by qRT-PCR (Figure 4f). Unsurprisingly, after the addition of actinomycin D, the mRNA half-life of MAP4K1 was decreased by the down-regulation of DLX6-AS1 but partly reversed by the overexpression of FUS (Figure 4g), indicating that DLX6-AS1 regulated mRNA stability of MAP4K1 through FUS. What is known to all is that RAS mutations (K-, N-) are common in gastric cancer and can result in the expression variation of MAP4K family members. In western blot, K/N-Ras depletion lessened MAP4K1 levels, suggesting the involvement of MAP4K1 under RAS mutations in GC (Supplementary Figure 1B and C). Moreover, MAP4K family members have been linked to the alteration of signaling pathways via not only MAP kinase pathways but also Hippo pathway. Western blot demonstrated that MAP4K1 downregulation dramatically affected MAP4K pathways by increasing the expression of p-p38, p-JNK, IκB-α and decreasing that of p-ERK, p-STAT3 and NF-κB. It was also viewed that MAP4K1 downregulation activated Hippo pathway via lowering the levels of YAP, c-Myc and Cyr61 and augmenting the levels of pYAP (Supplementary Figure 1D and E). To sum up, DLX6-AS1 stabilizes MAP4K1 via modulating FUS for the cellular activities of GC cells.
Figure 4.
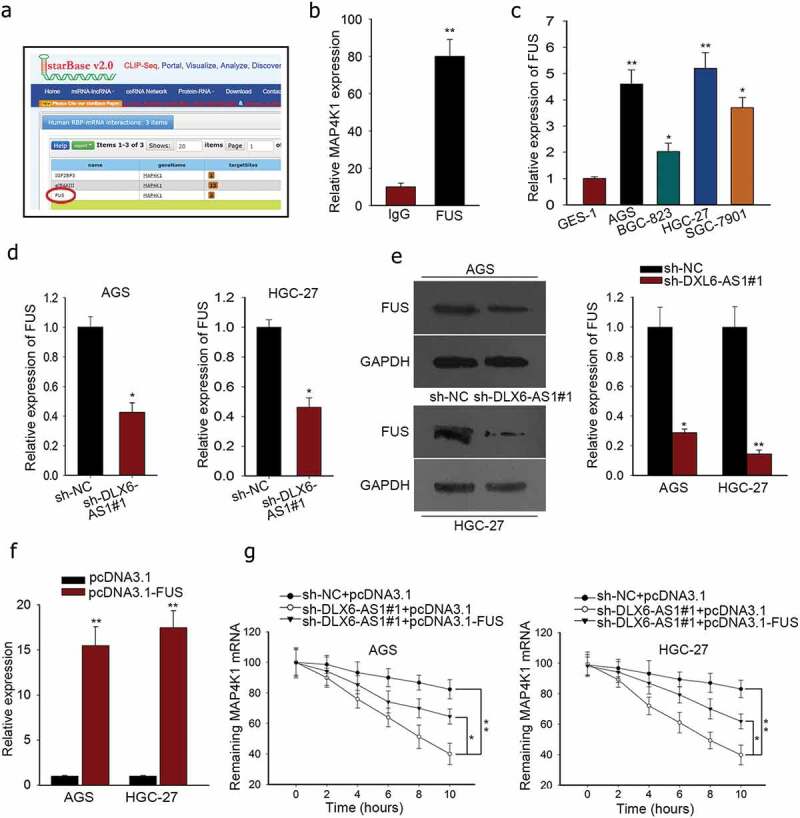
DLX6-AS1 stabilizes MAP4K1 by regulating FUS. (a) The binding of FUS and MAP4K1 from starbase v2.0. (b) RIP assay was conducted to confirm the combination of FUS with MAP4K1. (c) qRT-PCR analysis of the high FUS expression in GC cell lines. (d-e) The mRNA and protein levels of FUS in AGS and HGC-27 cells under sh-DLX6-AS1#1 treatment were evaluated by qRT-PCR. (f) AGS and HGC-27 cells were co-transfected with sh-DLX6-AS1#1 and pcDNA3.1 or sh-DLX6-AS1#1 and pcDNA3.1-FUS, with sh-NC and pcDNA3.1 as negative control. The efficiency of transfection was examined by qRT-PCR experiment. (g) Under actinomycin D treatment, the mRNA level of MAP4K1 with different treatments was dissected by qRT-PCR. *P < .05, **P < .01.
DLX6-AS1 improves cell proliferation, migration and EMT in GC by regulating MAP4K1
In order to validate the regulatory mechanism of DLX6-AS1 in GC, we conducted rescue experiments. First of all, sh-NC and pcDNA3.1, sh-DLX6-AS1#1 and pcDNA3.1 or sh-DLX6-AS1#1 and pcDNA3.1-MAP4K1 were respectively co-transfected into AGS and HGC-27 cells. Afterwards, the transfection efficacy of pcDNA3.1-MAP4K1 in AGS and HGC-27 cells was examined by qRT-PCR assay (Figure 5a). CCK-8 assay showed that the inhibited cell viability caused by DLX6-AS1 down-regulation was partly abolished by MAP4K1 overexpression in AGS and HGC-27 cells (Figure 5b). Cell migration of AGS and HGC-27 cells was repressed with the silencing of DLX6-AS1, which was partially reversed by the up-regulation of MAP4K1 as dissected by transwell migration assay (Figure 5c). At last, western blot was utilized to test EMT-associated proteins in AGS and HGC-27 cells. The increase of E-cadherin expression and the decrease of N-cadherin and Vimentin expression due to DLX6-AS1 knockdown were abrogated in part under pcDNA3.1-MAP4K1 treatment (Figure 5d). Totally, DLX6-AS1 boosts cell proliferation, migration and EMT in GC via regulating MAP4K1.
Figure 5.
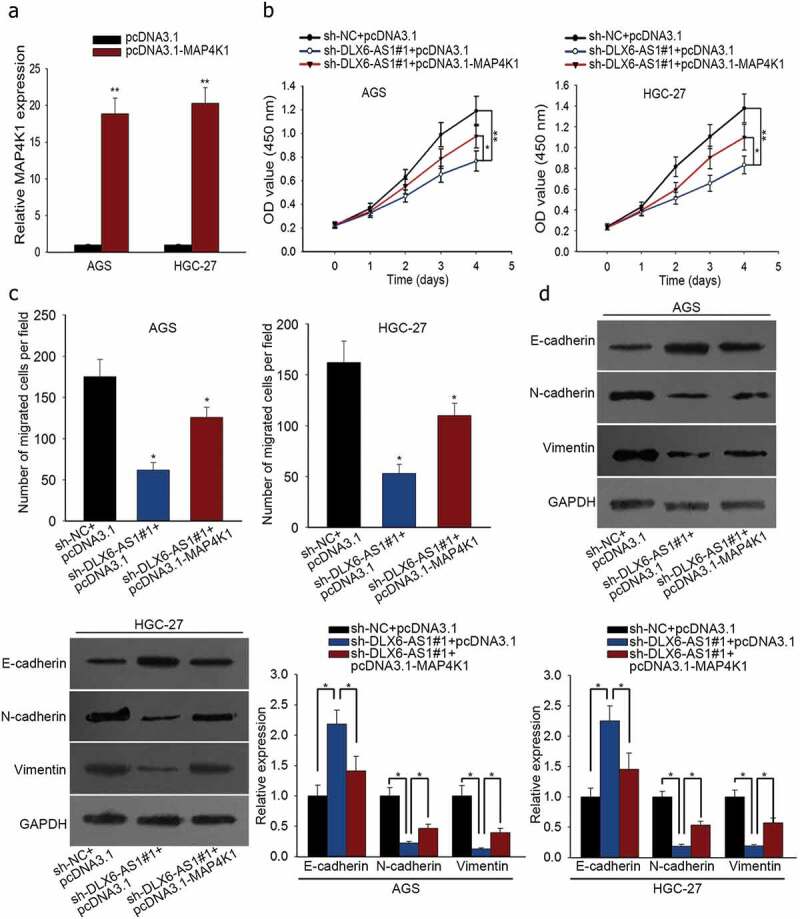
DLX6-AS1 improves cell proliferation, migration and EMT in GC by regulating MAP4K1. Cells were co-transfected with sh-DLX6-AS1#1 and pcDNA3.1 or sh-DLX6-AS1#1 and pcDNA3.1-MAP4K1, with sh-NC and pcDNA3.1 as normalization control. (a) MAP4K1 expression under pcDNA3.1-MAP4K1 transfection was confirmed by qRT-PCR. (b) CCK-8 experiment was applied to measure cell viability. (c) Cell migration was assessed by transwell migration assay. (d) Western blot analysis of EMT-associated proteins. *P < .05, **P < .01.
Discussion
As one of the most common malignant tumors in the digestive system, gastric cancer (GC) is a specific regional genetic disease3 and associated with high morbidity and mortality rates worldwide.1 Although more and more curative therapies have made progress for the treatment of GC patients, we still need to seek for novel therapies for the relapse of GC.
Increasing evidence has suggested that long non-coding RNAs (lncRNAs) with different levels of dysregulation act as oncogenes or tumor suppressors and influence biological functions in cancers.10,12 LncRNA DLX6 antisense RNA 1 (DLX6-AS1) is reported to be an oncogene and impact cellular activities of glioma,14 hepatocellular carcinoma,18 pancreatic cancer,15 renal cell carcinoma17 and lung adenocarcinoma.16 It was the first time that DLX6-AS1 was studied in GC. Consistently, the present study revealed that DLX6-AS1 was especially high in gastric cancer cell lines and the silencing of DLX6-AS1 could inhibit cell proliferation, migration and EMT of GC cells.
LncRNAs have been recognized to play vital roles in cancers via regulating gene expression. Here, carcinogenic MAP kinase kinase kinase kinases (MAP4Ks) were acknowledged as they were never discussed in gastric cancer. After DLX6-AS1 was silenced, only mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1) represented the biggest decline. Besides, MAP4K1 is known as an oncogene in bladder urothelial carcinoma20 and colon carcinoma.21 Our research uncovered that MAP4K1 with high expression in GC cells was positively regulated by DLX6-AS1. In addition, lost-of-function assays were conducted to determine the function of MAP4K1 in GC. In accordance with recent studies, MAP4K1 knockdown also repressed cell proliferation and migration as well as EMT in GC. And the promotion of MAP4K1 on the resistance of GC cells to paclitaxel was also figured out.
In order to inquire the regulatory mechanism underlying DLX6-AS1, we searched for RNA-binding proteins (RBPs) participating in tumor development and modulating genes at protein levels, intracellular localization and post-translation modifications.32 Recent researches implied their oncogenic or anticancer functions in diverse ranges of human cancers with mRNAs or lncRNAs.33,34 Here, the combination of fused in sarcoma/translocated in liposarcoma (FUS) and MAP4K1 or DLX6-AS1 was found from starBase v2.0. As a multifunctional DNA/RNA-binding protein, FUS is a key player in post-transcriptional events.22 Multiple studies have exposed the role of FUS as a regulator of mRNA stability in cancers,35-37 affecting cancer cell proliferation, apoptosis and metastasis etc. The current investigation documented that DLX6-AS1 stabilized MAP4K1 by regulating FUS expression. Furthermore, the participation of MAP4K1 under Ras mutations in gastric cancer and its effect on MAPK and Hippo pathways were confirmed.
Finally, rescue assays validated that DLX6-AS1 boosted cell proliferation, migration and EMT in GC via regulating MAP4K1 stabilized by FUS. Taken together, DLX6-AS1 contributes to cell proliferation, migration and EMT in GC via regulating FUS-stabilized MAP4K1. The current work implied the role of DLX6-AS1 as a biomarker in gastric cancer, and provided a new insight into GC pathogenesis.
Funding Statement
National Natural Science Foundation of China [No. 81570549; 81472242]; Shanghai Municipal Health Bureau Key Disciplines [Grant No. ZK2015A24]; Natural Science Foundation of the Science and Technology Commission of Shanghai Municipality [No. 15411971000; 16411972700]; The Research Fund of Shanghai Tongren Hospital, Shanghai Jiaotong University School of Medicine [No. TRYJ201511, TRYJ201613].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of our laboratories for their sincere advice and support.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Choi YJ, Kim N.. Gastric cancer and family history. Korean J Intern Med. 2016;31:1042–1053. doi: 10.3904/kjim.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20:4483–4490. doi: 10.3748/wjg.v20.i16.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coccolini F, Montori G, Ceresoli M, Cima S, Valli MC, Nita GE, Heyer A, Catena F, Ansaloni L. Advanced gastric cancer: what we know and what we still have to learn. World J Gastroenterol. 2016;22:1139–1159. doi: 10.3748/wjg.v22.i3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallardo M, Poltronieri P, D’Urso OF. Non-protein coding RNA biomarkers and differential expression in cancers: a review. J Exp Clin Cancer Res. 2008;27:19. doi: 10.1186/1756-9966-27-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y, Liu S, Fan J, Jin Y, Tian B, Zheng X, Fu H. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep. 2017;18:536–548. doi: 10.15252/embr.201643139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y, Zhang S, Zhong F, Liu C, Yin Y, et al. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat Commun. 2016;7:11730. doi: 10.1038/ncomms11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.She K, Huang J, Zhou H, Huang T, Chen G, He J. LncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673–2680. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- 9.Ponzio G, Rezzonico R, Bourget I, Allan R, Nottet N, Popa A, Magnone V, Rios G, Mari B, Barbry P. A new long noncoding RNA (lncRNA) is induced in cutaneous squamous cell carcinoma and down-regulates several anticancer and cell differentiation genes in mouse. J Biol Chem. 2017;292:12483–12495. doi: 10.1074/jbc.M117.776260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misawa A, Takayama K-I, Urano T, Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem. 2016;291:17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo G, Wang M, Wu X, Tao D, Xiao X, Wang L, Min F, Zeng F, Jiang G. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell Physiol Biochem. 2015;37:2209–2220. doi: 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 12.Ma DD, Yuan LL, Lin LQ. LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur Rev Med Pharmacol Sci. 2017;21:5143–5152. doi: 10.26355/eurrev_201711_13831. [DOI] [PubMed] [Google Scholar]
- 13.Guan G-F, Zhang D-J, Wen L-J, Xin D, Liu Y, Yu D-J, Su K, Zhu L, Guo -Y-Y, Wang K. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int J Med Sci. 2016;13:914–922. doi: 10.7150/ijms.16571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Zhang H, Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene. 2019;686:1–7. doi: 10.1016/j.gene.2018.10.065. [DOI] [PubMed] [Google Scholar]
- 15.An Y, Chen X-M, Yang Y, Mo F, Jiang Y, Sun D-L, Cai -H-H. LncRNA DLX6-AS1 promoted cancer cell proliferation and invasion by attenuating the endogenous function of miR-181b in pancreatic cancer. Cancer Cell Int. 2018;18:143. doi: 10.1186/s12935-018-0643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Li P, Zhao W, Yang R, Chen S, Bai Y, Dun S, Chen X, Du Y, Wang Y, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48. doi: 10.1186/s12935-015-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng X, Hu Z, Ke X, Tang H, Wu B, Wei X, Liu Z. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle. 2017;16:2212–2219. doi: 10.1080/15384101.2017.1361072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, He X, Jin T, Gang L, Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother. 2017;96:884–891. doi: 10.1016/j.biopha.2017.10.056. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Zhang H. Interactions between MAP4K1 and adaptor proteins. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2015;40:326–335. doi: 10.11817/j.issn.1672-7347.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Luo H, Li Y, Chen T, Wu S, Yang L. hsa-miR-96 up-regulates MAP4K1 and IRS1 and may function as a promising diagnostic marker in human bladder urothelial carcinomas. Mol Med Rep. 2012;5:260–265. doi: 10.3892/mmr.2011.621. [DOI] [PubMed] [Google Scholar]
- 21.Yang H-S, Matthews CP, Clair T, Wang Q, Baker AR, Li -C-CH, Tan T-H, Colburn NH. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Fagoonee S, Picco G, Orso F, Arrigoni A, Longo DL, Forni M, Scarfò I, Cassenti A, Piva R, Cassoni P , et al. The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget. 2016;8:10007–10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong H, Zhao W, Wang J, Seifer BJ, Ye C, Chen Y, Jia Y, Chen C, Shen J, Wang L, et al. Oncogenic mechanisms of Lin28 in breast cancer: new functions and therapeutic opportunities. Oncotarget. 2017;8:25721–25735. doi: 10.18632/oncotarget.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhar SK, Zhang J, Gal J, Xu Y, Miao L, Lynn BC, Zhu H, Kasarskis EJ, St. Clair DK. FUsed in sarcoma is a novel regulator of manganese superoxide dismutase gene transcription. Antioxid Redox Signal. 2014;20:1550–1566. doi: 10.1089/ars.2012.4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge Z, Cheng Z, Yang X, Huo X, Wang N, Wang H, Wang C, Gu D, Zhao F, Yao M, et al. Long noncoding RNA SchLAH suppresses metastasis of hepatocellular carcinoma through interacting with fused in sarcoma. Cancer Sci. 2017;108:653–662. doi: 10.1111/cas.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao L, Yuan L, Li P, Bu Q, Guo A, Zhang H, Cui N, Liu B. A FUS-LATS1/2 axis inhibits hepatocellular carcinoma progression via activating hippo pathway. Cell Physiol Biochem. 2018;50:437–451. doi: 10.1159/000494155. [DOI] [PubMed] [Google Scholar]
- 28.Sun H-D, Xu Z-P, Sun Z-Q, Zhu B, Wang Q, Zhou J, Jin H, Zhao A, Tang -W-W, Cao X-F. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8:10111. doi: 10.1038/s41598-018-27837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X-J, Wu J, Shi L, Li -X-X, Zhu L, Sun X, Qian J-Y, Wang Y, Wei J-F, Ding Q. PTEN expression is upregulated by a RNA-binding protein RBM38 via enhancing its mRNA stability in breast cancer. J Exp Clin Cancer Res. 2017;36:149. doi: 10.1186/s13046-017-0620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dormann D, Haass C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol Cell Neurosci. 2013;56:475–486. doi: 10.1016/j.mcn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Brinegar AE, Cooper TA. Roles for RNA-binding proteins in development and disease. Brain Res. 2016;1647:1–8. doi: 10.1016/j.brainres.2016.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, Dong N, He J, Sun Q, Lv G, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mang Y, Li L, Ran J, Zhang S, Liu J, Li L, Chen Y, Liu J, Gao Y, Ren G. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther. 2017;10:1003–1016. doi: 10.2147/OTT.S116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward CL, Boggio KJ, Johnson BN, Boyd JB, Douthwright S, Shaffer SA, Landers JE, Glicksman MA, Bosco DA. A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis. 2014;5:e1572–e. doi: 10.1038/cddis.2014.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong D, Wu Y-B, Jin C, Li -J-J, Gu J, Liao Y-F, Long X, Zhu S-Q, Wu H-B, Xu -J-J, et al. Elevated FUS/TLS expression is negatively associated with E-cadherin expression and prognosis of patients with non-small cell lung cancer. Oncol Lett. 2018;16:1791–1800. doi: 10.3892/ol.2018.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, Su X, Peng L, Jiao B. NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Bio. 2016;10:11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


