ABSTRACT
Objective: To investigate and define the underlying molecular mechanism of tripartite motif-containing 58 (TRIM58) in regulating the tumor growth of gastric cancer (GC).
Methods: TRIM58 expression in GC tissues and cells was detected by real-time PCR and Western blot, followed by lentiviral-induced overexpression or knockdown of TRIM58. Subsequently, CCK8, BrdU-ELISA, flow cytometry, immunoprecipitation, in vitro animal experiments and immunochemistry were performed to explore the function of TRIM58. Western blotting was used to detect β-catenin, C-myc, Cyclin D1, and survivin expression.
Results: TRIM58 expression was significantly reduced in tumor tissues of GC patients and GC cell lines, whereas β-catenin, C-myc, Cyclin D1, and survivin were highly expressed. Overexpression of TRIM58 in GC cells resulted in decreases in β-catenin, C-myc, Cyclin D1, and survivin protein expression and significantly suppressed proliferation by preventing cell-cycle progression and promoting cell apoptosis. Conversely, TRIM58 knockdown resulted in the opposite effects. Furthermore, the effect of TRIM58 knockdown on GC cells was potently reversed by a β-catenin inhibitor, XAV939. Immunoprecipitations showed the interaction between TRIM58 and β-catenin, and TRIM58 overexpression significantly enhanced β-catenin degradation. In addition, we found a significant decrease in the growth and weight of tumors and an increase in tumor cell apoptosis in TRIM58-overexpression nude mice, which were also accompanied by reduced β-catenin expression.
Conclusions: These data suggest that TRIM58 may function as a tumor suppressor in GC and potentially suppress the tumor growth of gastric cancer by inactivation of β-catenin signaling via ubiquitination.
KEYWORDS: Tripartite motif-containing protein 58 (TRIM58), β-catenin signaling, ubiquitination, proliferation, apoptosis, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common malignancies and is the second leading cause of cancer deaths worldwide, with prominent occurrence in Eastern Asia, South America, and Eastern Europe.1–3 In Japan, gastric cancer accounts for 18% of all cancer deaths, ranking second in cancer-related fatalities.4 In China, the incidence of gastric cancer in the northwest and eastern coastal areas is significantly higher than that in the southern regions. Despite great progress in diagnosis and treatment, the five-year survival rate of GC remains low, at only 20%, due to high recurrence rates and tumor metastases.5,6 Thus, GC remains a huge health burden and there is an urgent need to identify new tumor biomarkers that can predict the risk of GC progression.
Deregulation of ubiquitin ligases is reported to be associated with various biological processes in diseases.7–9 Tripartite motif-containing (TRIM) proteins, comprising a RING-finger domain, a B box, and a coiled-coil domain, possess E3 ubiquitin ligase activities in diverse processes and diseases.10–12 A study revealed that TRIM proteins are involved in many cancers and regulate transcription factor activity.13 Aberrant gene methylation of TRIM58, a member of the TRIM protein family, has been associated with poor prognosis in liver and lung cancer patients.14,15 In addition, TRIM58 is a tumor suppressor in colorectal cancer, and downregulation of TRIM58 is associated with poor patient outcome due to the modulation of EMT via the Wnt/β-catenin pathway.16 Studies have revealed that activated Wnt signaling promotes nuclear translocation of β-catenin, which, in turn, drives expression of two important Wnt target genes that serve as oncogenes, Cyclin D1 and C-myc.17,18 Aberrant activation of Wnt/β-catenin signaling has been reported to play an important role in the initiation and development of cancer, and an increase of Wnt/β-catenin signaling can promote the metastasis of lung cancer cells.19 Furthermore, TRIM29 is found to act as an oncogene in GC, and also is involved in Wnt/β-catenin signaling activity.20,21 However, the effect of TRIM58 in GC and its potential mechanisms are still little known.
In this study, reduced TRIM58 expression was observed in GC tissues and cells, whereas β-catenin, C-myc, Cyclin D1, and survivin were highly expressed. Overexpression of TRIM58 in GC cells resulted in decreases in β-catenin, C-myc, Cyclin D1, and survivin protein expression and significantly suppressed proliferation by preventing cell-cycle progression and promoting cell apoptosis. Conversely, TRIM58 knockdown resulted in the opposite effects, which was reversed by a β-catenin inhibitor. Immunoprecipitations showed an interaction between TRIM58 and β-catenin, and TRIM58 overexpression significantly enhanced β-catenin degradation. In addition, we found a significant decrease in tumor growth and tumor weight and an increase in tumor cell apoptosis in TRIM58-overexpression nude mice, accompanied by reduced β-catenin expression. These data suggest that TRIM58 may act as a tumor suppressor in GC, potentially by modulating β-catenin signaling.
Materials and methods
GC patient tissue
Twenty-three GC patients, with a mean age of 55 years (ranging from 50 to 70 years old), treated at Fudan University Cancer Hospital (Shanghai, China) were enrolled in this study. After a written informed consent was obtained, cancer and normal tissue samples from the patients were collected between May 25 and November 30, 2017, and stored in liquid nitrogen prior to use. The expression of TRIM58 in tissues was detected by real-time PCR and immunohistochemistry (IHC). All experiments performed in this study were approved by the Ethics Committee of Shanghai Medical College, Fudan University (Shanghai, China).
Cell culture
Five human GC cell lines, MKN45, BGC823, HGC27, AGS and SNU719, and a gastric mucosa cell line, GES-1, were purchased from Type Culture Collection of the Chinese Academy of Science (Shanghai, China). Cells were cultured with RPMI-1640 medium (SH30809.01B, HyClone, GE Healthcare Life Sciences, Logan, UT, USA), containing 10% fetal bovine serum (FBS; 16000–044, GIBCO, Thermo Fisher Scientific, Inc.) and 1% antibiotic (100×, penicillin and streptomycin; P1400-100, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at 37°C in a 5% CO2 humidified-incubator (Thermo Forma 3111, Thermo Fisher Scientific, Waltham, MA, USA). The medium was replaced every 2 days. GC cell lines (MKN45, BGC823, HGC27, AGS, and SNU719) were previously utilized in studies of other biological mechanisms that potentially regulate the development and progression of GC through regulation of β-catenin signaling.22–26
Plasmid constructs and lentiviral infection
After synthesis of shRNA sequences targeting three sites of the TRIM58 gene (NM_015431.3; shown in Table 1), the shRNA construct was formed by double-strand annealing and inserted into the Agel I/EcoR I restriction sites in the pLKO.1-puro vector (Addgene, Inc., Cambridge, MA, USA). The full-length, 1461 bp, coding DNA sequence (CDS) region of TRIM58 was synthesized (GENEWIZ, Suzhou, Jiangsu, China), and inserted into the pLVX-Puro vector (Addgene, Inc.) at the EcoRI/BamHI restriction sites. After confirmation of successful insertion using DNA sequencing (Shanghai Majorbio Pharmaceutical Technology Co., Ltd., Shanghai, China), pLKO.1-shTRIM58 (1,000 ng) or pLVX-Puro-TRIM58 (1,000 ng) plasmids were co-transfected into 293T cells with psPAX2 and pMD2G, viral packaging plasmids (Addgene, Inc.) using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.). After 48 h of transfection, viral particles were collected by ultracentrifugation.
Table 1.
TRIM58 interference target design results.
| Name | Sequences |
|---|---|
| TRIM58 target site 1 (893–911) | GGAGGGAGCTCTTAAGGAA |
| TRIM58 target site 2 (1293–1311) | GGACTATGAAGCCGGTGAA |
| TRIM58 target site 3 (1454–1472) | GGGCATCCAGGGATCATTT |
Experimental grouping
In vitro, AGS and HGC27 cells were infected with the lentiviruses containing vector or TRIM58 overexpression (TRIM58). In addition, SNU719 cells were infected with negative control (shNC) or TRIM58 interference (shTRIM58-1/shTRIM58-2/shTRIM58-3). Medium-treated cells were used as a control. The efficiency of shTRIM58 and TRIM58 overexpression in GC cells was determined. Furthermore, cell proliferation, cell-cycle progression, and apoptosis as well as the expression of several related proteins were examined.
After treatment of shNC + DMSO, shTRIM58 + DMSO, shNC + XAV939 (Wnt/β-catenin inhibitor, 10 µmol/l; S1180, Selleck), and TRIM58 + XAV939 (10 µmol/l) in SNU719 cells, cell proliferation and the expression of several related proteins were examined.
Twelve nude mice were randomly subcutaneously inoculated with AGS cells with TRIM58 (Six) or vector (Six), after the formation of a tumor, the tumor sizes in the mice were measured by vernier caliper every 3 days. HE and TUNEL stainings were used to analyze tumor cell apoptosis and TRIM58 and β-catenin expression was detected by Western blot.
Real-time polymerase chain reaction (RT-PCR) assay
Total RNA of GC tissues or cells was isolated using Trizol reagent (1596–026, Invitrogen, Thermo Fisher Scientific, Inc.). After quantification and confirmation of RNA integrity, 1 µg of RNA was reverse transcribed into cDNA by a reverse transcription kit (#K1622, Fermentas, Thermo Fisher Scientific, Inc.). RT-PCR using the cDNA templates was carried out on an ABI 7300 Real-Time PCR system (ABI-7300, Applied Biosystems, Thermo Fisher Scientific, Inc.), with the following program: 95°C, 10 min (95°C, 15 Sec; 60°C, 45 Sec) × 40; 95°C, 15 Sec; 60°C, 1 min; 95°C, 15 Sec; 60°C, 15 Sec.27 Subsequently, using the 2−ΔΔCq method,28 the TRIM58 mRNA, relative to GAPDH, was calculated. Primers are listed as follows: TRIM58, Forward (F): 5ʹ-GCTGCTGAAAGGGAATGAG-3ʹ, Reverse (R): 5ʹ-GTTGTGGGTGGCAAGATAAG-3ʹ; C-myc, F: 5ʹ-AGAGTTTCATCTGCGACCCG-3ʹ, R: 5ʹ-GGCTGCCGCTGTCTTTGC-3ʹ; Cyclin D, 5ʹ-TACCCCGATGCCAACCTC-3ʹ, R: 5ʹ-TGTTCCTCGCAGACCTCC-3ʹ; survivin: 5ʹ-CAAGGAGCTGGAAGGGTG-3ʹ, R: 5ʹ-CGGATAAAACTGCGTGGC-3ʹ; GAPDH, F: 5ʹ-AATCCCATCACCATCTTC-3ʹ, R: 5ʹ-AGGCTGTTGTCATACTTC-3ʹ.
Western blot analysis
RIPA buffer (R0010, Beijing Solarbio Science & Technology Co., Ltd.), containing protease and phosphatase inhibitors, was used to extract total protein. After quantification by a BCA kit (PICPI23223, Thermo Fisher Scientific, Inc.), the proteins were separated on a 10% or 12% SDS-PAGE gel and transferred to polyvinylidene fluoride (PVDF) membranes (HATF00010, EMD Millipore, Billerica, MA, USA). Membranes were blocked in 5% skimmed milk (BYL40422, BD Biosciences, Franklin Lakes, NJ, USA) at room temperature for 1 h, and then incubated overnight at 4°C with primary antibodies against TRIM58 (1: 500, Ab90362, Abcam, Cambridge, UK), β-catenin (1: 1000; #8480, Cell Signaling Technology [CST], Inc., Danvers, MA, USA), survivin (1: 1000, #2803, CST), C-myc (1: 1000; Ab32072, Abcam), Cyclin D1 (1: 1000; #2922, CST) and GAPDH (1: 2000; #5174, CST) with gentle shaking. Following 5–6 washes with TBST, the membranes were incubated in goat anti-rabbit (A0208) secondary antibodies labeled with HRP (1: 1000; Beyotime Institute of Biotechnology, Haimen, China) for 2 hours at room temperature. Membranes were washed 5–6 times with TBST and developed with a chemiluminescent reagent (WBKLS0100, EMD Millipore) and exposed on ECL imaging system (Tanon-5200, Tanon Science and Technology Co., Ltd., Shanghai, China). Using Image J version 1.47v (National Institutes of Health, Bethesda, MD, USA), the expression of proteins, relative to GAPDH, was analyzed and calculated.
Histopathology
After embedding, fixing and sectioning, the tissue slices were subjected to a 30-min incubation at 65°C in an oven. Later, the slices were soaked in xylene I and xylene II (Shanghai Sinopharm) in order for 15 min then soaked for 5 min in 100%, 95%, 85%, and 75% ethanol sequentially, followed by a 10-min rinse with tap water. A brief procedure of IHC was described as follows: Slides were submerged in 0.01 M sodium citrate buffer (pH 6.0) for 10–15 min for antigen retrieval followed by a 10-min incubation with 0.3% H2O2 in a wet-box. Next, the slides were washed and incubated for 1 h with TRIM58 Rb-antibody, followed by a 20–30 min incubation with HRP-labeled secondary antibodies (Changdao, D-3004, Shanghai, China). TUNEL staining was conducted as follows: The slide was immersed in trypsin for 40 min and then washed with PBS. The specimen was incubated with 50 μl of TUNEL reaction mixture for 1 h in a dark-wet-box at 37°C, followed by a 30-min reaction with 50 µl of POD in a dark-wet-box at 37°C (covered with a coverslip). Subsequently, the slides were subjected to DAB staining, hematoxylin staining and alcohol differentiation with 1% hydrochloric acid (for TUNEL staining: 75% alcohol, 85% alcohol, 95% alcohol, 100% alcohol, for step by step dehydration, 3 min each). Hematoxylin-eosin (HE) staining was performed as follows: The slices were stained for 5 min in hematoxylin solution and color-separated for several seconds in ammonia water. After rinsing for 15 min with running water, the slides were dehydrated sequentially in 70% and 90% alcohol for 10 min, followed by 1–2 min staining with eosin solution and complete dehydration by alcohol. The images of the slides were taken on a microscope (NIKON, ECLIPSE Ni) and were analyzed using the IMS image analysis system (NIKON, DS-Ri2).
Cell proliferation assay
GC cells in a logarithmic growth phase were trypsinized and re-suspended at 30,000 cells/mL cell, and then plated in 96-well plates (TR4001, TRUELINE) at 3,000 cells/ml for overnight culture in the incubator. After treatment for 0, 24, 48, and 72 h, the cells were incubated at 37°C for 1 h with 100 µl of Cell Counting Kit-8 (CCK-8; CP002, SAB, USA) solution (CCK-8: serum-free medium = 1:10). The absorbance value (OD) at 450 nm, indicating cell proliferation, was measured using a microplate reader (DNM-9602, Perlong, Beijing, China). Cell proliferation was assessed using the BrdU Cell Proliferation ELISA Kit (Ab126556, Abcam) per manufacturer’s instructions. Cell proliferation was also assessed by BrdU-ELISA as follows: After incorporation of BrdU into the DNA of dividing cells, the cells were subjected to immobilization, permeabilization, and DNA denaturation. Next, the cells were incubated with anti-BrdU monoclonal antibody for 1 h and washed to remove unbound antibody. Cells were then incubated with horseradish peroxidase-conjugated goat anti-mouse antibody. Finally, the colored reaction that indicates cell proliferation was quantified using a spectrophotometer.
Cell-cycle detection
When in a logarithmic growth phase, GC cells were trypsinized and seeded at 300,000 cells/well in six-well plates for 24 h of adherent growth, and then treated as indicated. The cells were collected and washed with 1 ml of pre-cooled PBS. After 5 min of centrifugation at 1,000 g, the cells were resuspended in 300 µl of PBS contained 10% FBS (16000–044, GIBCO), followed by 24 h of fixation at 4°C in 700 µl of absolute ethanol (−20°C pre-cooled). After centrifugation and washing, the cells were incubated in 100 µl of RNase A solution (1 mg/ml; R8020-25, Solarbio) for 30 min in the dark at 37°C, followed by incubation in 400 µl of propidium iodide (PI) solutions (50 µg/ml; C001-200, 7Seabiotech, Shanghai, China) for 10 min. The red fluorescence at 488 nm (excitation wavelength) was determined by flow cytometry (Accuri C6, BD Biosciences), and the cell-cycle progression was analyzed using the FLOWJO software.
Cell apoptosis assay
After treatment, GC cells were collected and subjected to an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double stain (Beyotime, C1063) according to the manufacturer’s instructions. Briefly, 195 µl of Annexin V-FITC binding buffer was used to re-suspend 500,000–1,000,000 cells, followed by 15 min incubation in the dark at 4°C in 5 µl of Annexin V-FITC. After that, the cells were incubated in 5 µl of PI for 5 min in the dark at 4°C. A tube without Annexin V-FITC and PI was used as a control. The percentage of apoptotic GC cells was evaluated by a Flow cytometer using the BD AccuriTM C6 Software (Version 1.0.264.21, BD Biosciences).
Immunoprecipitation (IP) and ubiquitination detection
After treatment and extraction, the proteins were incubated at 4°C with rabbit-IgG (1 µg; Sc-2027, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or IP-indicated antibody (1 µg) overnight, untreated proteins as an input control. After the formation of the immune complex in Protein A/G PLUS-Agarose for 2 h, samples were centrifuged for 4 min at 3000 rpm at 4°C, washed with 1 ml of lysis buffer 4 times, and then boiled for 5 min in the appropriate protein loading buffer. Samples were centrifuged again and the supernatant was collected in a new tube for western blot analysis. Anti-TRIM58 antibody (Ab186357) and Anti-β-catenin antibody (#8480) were applied for IP detection, and Anti-TRIM58 antibody (Ab90362) and Anti-β-catenin antibody (Ab6301) were used in western blot analysis. Following the 1 h of incubation in Protein A/G PLUS-Agarose (Sc-2003, Santa Cruz Biotechnology, Inc.), the proteins were incubated overnight at 4°C in 1 µg rabbit-IgG (Sc-2027, Santa Cruz Biotechnology) or rabbit-Anti-β-catenin (#8480) antibody. After the formation of the immune complex, the proteins were centrifuged, washed with lysis buffer, and boiled in protein loading buffer. The proteins were then subjected to SDS-PAGE and Western blot was performed using the Anti-Ubiquitin antibody (Ab7780).
Tumors in nude mice
After the preparation of 5,000,000 cells/ml suspensions, 12 nude mice were randomly grouped for subcutaneous injection with 100 µl of vector-AGS cells (Six) or TRIM58-AGS cells (Six). After injection, tumors formed after 1–2 weeks. After tumor formation, tumor sizes were measured by vernier caliper every 3 days. After the tumors reached maximum size, the nude mice were sacrificed and photographed, and then the tumors were harvested for weighing and recording. Tumor cell apoptosis was analyzed by H&E and TUNEL staining and western blot was used to detect the expression of TRIM58 and β-catenin.
Statistical analysis
Statistical significance in this study was analyzed using GraphPad prism 7.0 software (GraphPad Software, Inc., La Jolla, CA, USA). One-way analysis of variance (ANOVA) with Tukey’s multiple comparison was used to assess the significance among multiple comparisons. A paired or non-paired Student’s t-test was used for two groups. Experimental values were presented as mean ± standard deviation (SD) of three individual repeats and a P value of less than 0.05 was indicated as statistically significant.
Results
TRIM58 expression is significantly reduced in GC tissues and cell lines
TRIM58 expression was detected in paired cancer and normal tissues from 23 GC patients. We observed that TRIM58 expression was significantly lower in the tumor tissues of GC patients compared to that in paired normal tissue (Figure 1A). IHC staining of 10 pairs of cancer and normal samples has shown that TRIM58 expression in 7 of the cancer tissues was lower than that in normal tissue (Figure 1B), which further supported the hypothesis that GC tumors have low expression of TRIM58. RT-PCR and Western blot analysis also demonstrated that TRIM58 expression in GC cells (MKN45, BGC823, HGC27, AGS, and SNU719) was significantly reduced compared to normal gastric mucosa cells (GES-1), with TRIM58 expression relatively lower in HGC27 and AGS, and relatively higher in SNU719 (Figure 1C). On the contrary, β-catenin was highly expressed in cancer samples (Figure 1D) and GC cells (Figure 1E). Moreover, compared to normal tissue, the expression of C-myc, Cyclin D1 and survivin in GC tumors was significantly increased (Figure 1F). Consistent with this, C-myc, Cyclin D1 and survivin were also highly expressed in GC cell lines (Figure 1G). These findings suggested that TRIM58 may function as a tumor suppressor in GC progression, and β-catenin signaling may mediate the effects of TRIM58. HGC27, AGS, and SNU719 cell lines were selected for the following experiments.
Figure 1.
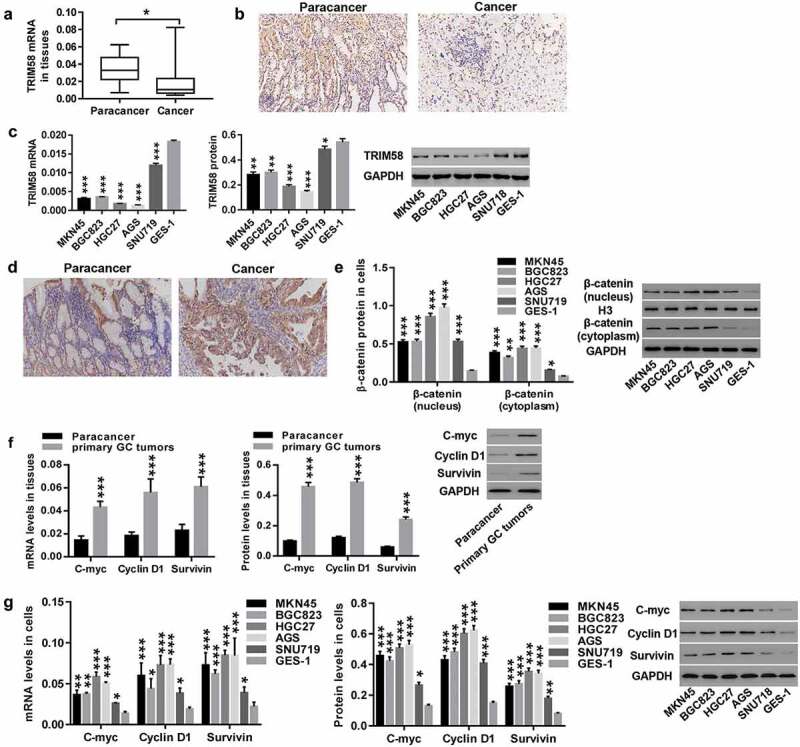
TRIM58 expression is significantly reduced in GC tissues and cell lines Paired cancer and normal tissues of 23 patients were collected. (A) TRIM58 mRNA expression was examined by RT-PCR. (B) TRIM58 in malignant and normal tissues was detected by IHC. (C) mRNA and protein expression of TRIM58 in GC cell lines were examined. (D) β-catenin in cancer and normal tissues was detected by IHC. (E) β-catenin expression (nuclear and cytoplasmic) in GC cell lines was detected. (F-G) mRNA and protein expression of C-myc, Cyclin D1, and survivin in GC tumors and cell lines were detected. *P < .05, **P < .01 and ***P < .001 were compared to Paracancer or GES-1.
Overexpression and knockdown of TRIM58 in GC cells by lentiviral infection
In vitro, AGS and HGC27 cells were infected with TRIM58 lentivirus, and SNU719 cells were infected with shTRIM58-1, shTRIM58-2, and shTRIM58-3 lentiviruses. Figure 2 shows that infection with TRIM58 lentivirus significantly upregulated both mRNA and protein expression of TRIM58 in AGS (Figure 2A) and HGC27 (Figure 2B) cells, while all three shTRIM58 lentiviruses significantly downregulated TRIM58 expression in SNU719 cells (Figure 2C). shTRIM58-1 and shTRIM58-2 showed the best effects; therefore, shTRIM58-1 and shTRIM58-2 were used for further study.
Figure 2.
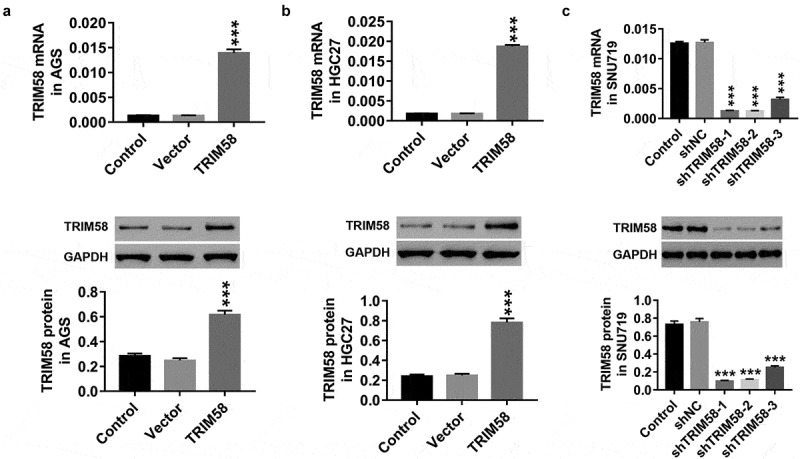
Overexpression and knockdown of TRIM58 in GC cells by lentiviral infection After lentiviral infection with vector or TRIM58 and shNC or shTRIM58 (shTRIM58-1, shTRIM58-2, and shTRIM58-3) (A-B) the overexpression efficiency of TRIM58 in AGS and HGC27 cells was detected. (C) The knockdown efficiency of shTRIM58-1, shTRIM58-2, and shTRIM58-3 in SNU719 cells was also detected. ***P < .001 was compared to vector or shNC.
Overexpression of TRIM58 inhibits GC cell proliferation via promoting cell-cycle arrest and cell apoptosis
A previous study demonstrated that the promotion of TRIM58 expression downregulated colorectal cancer cell invasion.16 In our study, as shown in Figure 3, we found that overexpression of TRIM58 in AGS and HGC27 cells significantly inhibited cell proliferation (Figure 3A), stalled cell-cycle progression in the G1-phase, thus reducing S/G2-phase cells (Figure 3B), and promoted cell apoptosis (Figure 3C), whereas TRIM58 knockdown in SNU719 cells showed opposite effects. Furthermore, the expression of β-catenin, C-myc, Cyclin D1, and survivin was significantly decreased by TRIM58 overexpression and increased by TRIM58 knockdown (Figure 3D). Based on these findings, we conjectured that overexpression of TRIM58 in GC cells could inhibit cell proliferation by arresting cell-cycle progression and promoting cell apoptosis, which is potentially mediated by β-catenin signaling.
Figure 3.
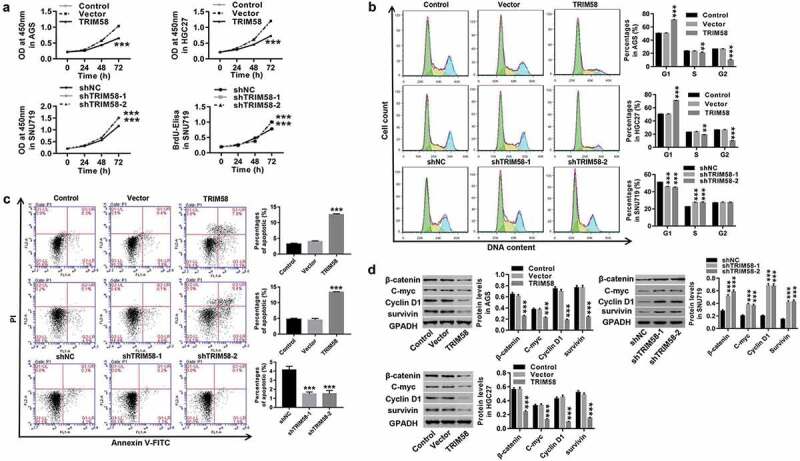
Overexpression of TRIM58 inhibits GC cell proliferation via cell-cycle arrest and increase in cell apoptosis AGS, HGC27, or SNU719 cells were infected with shTRIM58 (shTRIM58-1 and TRIM58-2) or TRIM58 lentiviruses. (A) Cell proliferation was assessed by CCK-8 or BrdU-ELISA at 0, 24, 48, and 72 h. (B-C) After 48 h of infection, cell-cycle phase proportions and apoptosis were assessed by flow cytometry. (D) The levels of related proteins (β-catenin, C-myc, Cyclin D1, and survivin) were analyzed by Western blot. **P < .01 and ***P < .001 were compared to vector or shNC.
TRIM58 inhibition of GC cell proliferation may be due to β-catenin ubiquitination and inactivation
We further investigated the underlying mechanisms of TRIM65 in regulating GC cells. A previous study revealed that Wnt/β-catenin signaling activation was a critical oncogenic event in tumor initiation and development.29 C-myc and Cyclin D1 are two important proto-oncogenes that are associated with cell proliferation and cell-cycle progression and are downstream target genes of Wnt/β-catenin signaling.29–31 Survivin is a regulator of cell-cycle progression and is critically required for inhibition of apoptosis.32,33 Here, as shown in Figure 4A, TRIM58 knockdown-induced GC cell proliferation was potently rescued by treatment with the Wnt-β-catenin inhibitor, XAV939. Likewise, TRIM58 knockdown correlated with significantly decreased expression of β-catenin, C-myc, Cyclin D1 and survivin (Figure 4B). These are in agreement with a previous report that activation of Wnt/β-catenin promotes cell proliferation in cancer.34 In addition, Co-IP demonstrated that the interaction between TRIM58 and β-catenin (Figure 4C). Furthermore, overexpression of TRIM58 enhanced β-catenin ubiquitination in GC cells (Figure 4D). Altogether, the data implicate that TRIM58 inhibition of GC cell proliferation may be mediated by ubiquitination and inactivation of β-catenin signaling.
Figure 4.
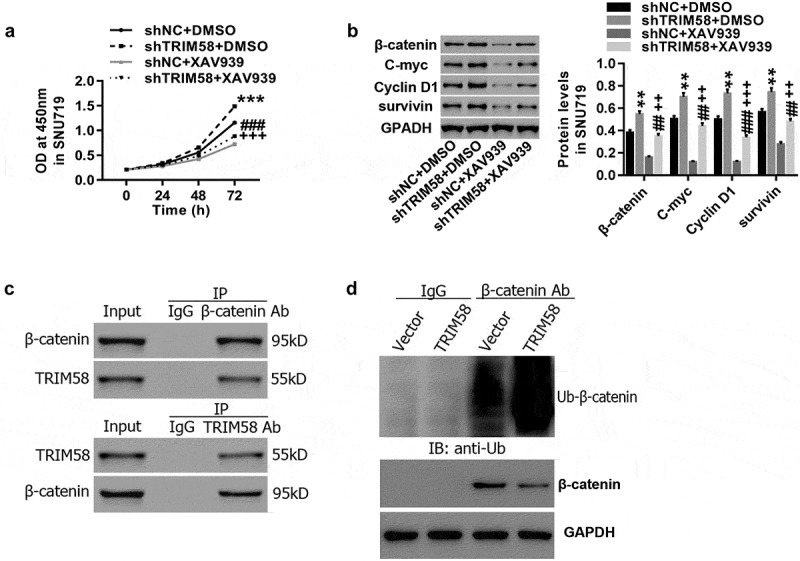
TRIM58 inhibition of GC cell proliferation may be due to β-catenin ubiquitination and inactivation After treatment with shTRIM58 lentivirus and XAV939 in SNU719 cells, (A) Cell proliferation was assessed at 0, 24, 48, and 72 h, and (B) the levels of related proteins (β-catenin, C-myc, Cyclin D1 and survivin) were analyzed by Western blot. (C) A Co-IP shows the co-existence of TRIM58 and β-catenin in the precipitant. (D) β-catenin ubiquitination was examined after infection with TRIM58 lentivirus, and β-catenin expression was detected by western blot. **P < .01 and ***P < .001 compared to shNC + DMSO, ##P < .01 and P < .001 compared to shTRIM58 + DMSO, and ++P < .01 and +++P < .001 compared to shNC + XAV939.
TRIM58 inhibited tumor growth and promoted apoptosis in nude mice through inactivation of β-catenin signaling
After subcutaneous injection of vector-AGS cells (Six) or TRIM58-AGS cells (Six) in nude mice, tumor growth and apoptosis were measured. As shown in Figure 5A–B, the growth and weight of tumors in TRIM58-overexpression nude mice was significantly inhibited, concurrent with a significant increase in apoptosis. Furthermore, the expression of β-catenin protein in TRIM58-overexpression nude mice was significantly decreased (Figure 5C), which was consistent with IHC analysis of β-catenin expression (Figure 5D). These results further prove that TRIM58 may potentially inhibit GC tumor growth and apoptosis through the inactivation of β-catenin signaling.
Figure 5.
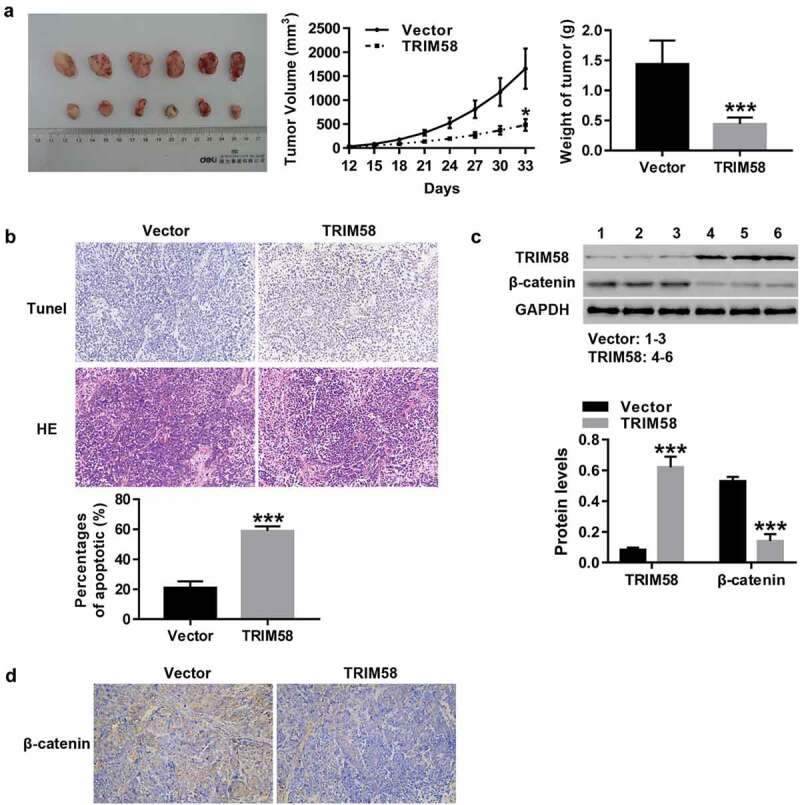
TRIM58 inhibited tumor growth and promoted apoptosis in nude mice through inactivation of β-catenin signaling Twelve nude mice were randomly injected with vector-AGS cells (Six) or TRIM58-AGS cells (Six). (A) The volume and weight of tumor in the mice were measured and a tumor growth curve was drawn. (B) The percentages of apoptosis were examined by Tunel and H&E staining. (C) The protein levels of TRIM58 and β-catenin were determined. (D) β-catenin expression in tissues of nude mice as detected by IHC. *P < .05 and ***P < .001 compared to vector.
Discussion
Cumulative studies have demonstrated that TRIM proteins are involved in various human cancers. For instance, the expression of TRIM26 and TRIM44 is suggested to be correlated with the overall survival of hepatocellular carcinoma patients,35,36 and TRIM29 overexpression in invasive bladder cancer indicates a poor prognosis.37 Additionally, knockdown of TRIM37 and TRIM46 inhibits breast cancer cell proliferation and tumor growth.38,39 In the present study, significantly reduced TRIM58 expression was observed in GC tissues and cell lines, and overexpression of TRIM58 in GC cells significantly inhibited cell proliferation by arresting cell-cycle progression and promoting apoptosis, while TRIM58 knockdown had opposite effects. These findings suggested that TRIM58 may function as a tumor suppressor in GC and TRIM58 overexpression may be associated with good prognostic outcomes of GC, which may guide decisions for GC treatment.
We also investigated the underlying mechanisms by which TRIM58 plays a role in GC. Studies have implicated β-catenin activation, as well as C-myc, Cyclin D1 and survivin expression, in the progression of cancer, and overexpression of β-catenin and Cyclin D1 is reported to be a marker of poor prognosis in cancer.40–43 These were consistent with the findings in our study that β-catenin, C-myc, Cyclin D1, and survivin were highly expressed in GC tumors and cell lines. Furthermore, knockdown of TRIM58-induced cell proliferation and expression of β-catenin, C-myc, Cyclin D1, and survivin was strongly reversed by treatment with the β-catenin inhibitor, XAV939. In line with previous reports,44,45 these results indicate that the decrease of Cyclin D1 could inhibit the progression of the cell cycle by arresting cells in the G1-phase, thereby inhibiting cell proliferation. We inferred that TRIM58 inhibited GC cell proliferation by inactivation of β-catenin signaling. This is consistent with previous studies, which has shown that activation of β-catenin signaling is a frequent cause in GC and correlates with patient survival in GC.46,47 It is also reported that TRIM24 and TRIM31 are up-regulated in human GC and promote the growth of GC cells, possibly by regulating the ubiquitin-proteasome system.48,49 Likewise, our data showed the interaction between TRIM58 and β-catenin, and overexpression of TRIM58 significantly promoted β-catenin degradation. Importantly, IHC staining showed that β-catenin expression in the tissues of TRIM58-overexpression nude mice was significantly decreased. Altogether, our results demonstrate that TRIM58 inhibition of GC cell proliferation is possibly due to ubiquitination and inactivation of β-catenin. This view was further supported by our in vivo experiments of nude mice. In nude mice, the growth and weight of tumors in TRIM58-overexpression cells was significantly inhibited, concurrent with a significant increase in apoptosis and increased β-catenin protein.
In summary, the current study demonstrates that TRIM58 may function as a tumor suppressor in GC progression. Overexpression of TRIM58 can significantly inhibit GC cell proliferation via inhibition of cell-cycle progression and the promotion of apoptosis. These effects are mediated by ubiquitination and inactivation of β-catenin signaling. Therefore, high TRIM58 expression may be a good prognostic marker for GC and targeting TRIM58 may be an effective therapeutic strategy in GC.
Funding Statement
This study is supported by the National Natural Science Foundation of China [81502027] and the Shanghai Committee of Science and Technology Funds [17411963200].
References
- 1.Siegel R, Ma J, Zou Z, Jemal A.. Cancer statistics, 2014. Ca A Cancer J Clin. 2014;67:7. doi: 10.3322/caac.21387. [DOI] [Google Scholar]
- 2.Roder DM. The epidemiology of gastric cancer. Gastric Cancer. 2002;5:5–11. [DOI] [PubMed] [Google Scholar]
- 3.Herszényi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci. 2010;14:249–258. [PubMed] [Google Scholar]
- 4.Tominaga S. Epidemiologic trends of stomach cancer in Japan and world. Nihon Rinsho. 2001;59(Suppl 4):5. [PubMed] [Google Scholar]
- 5.Kagawa S, Shigeyasu K, Ishida M, Watanabe M, Tazawa H, Nagasaka T, Shirakawa Y, Fujiwara T. Molecular diagnosis and therapy for occult peritoneal metastasis in gastric cancer patients. World J Gastroenterol. 2014;20:17796. doi: 10.3748/wjg.v20.i47.17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 7.Jamal A, Swarnalatha M, Sultana S, Joshi P, Panda SK, Kumar V. The G1 phase E3 ubiquitin ligase TRUSS that gets deregulated in human cancers is a novel substrate of the S-phase E3 ubiquitin ligase Skp2. Cell Cycle. 2015;14:2688–2700. doi: 10.1080/15384101.2015.1056946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyungsoo K, Frederick DT, Levesque MP, Cooper ZA, Feng Y, Clemens K, Brill L, Samuels Y, Hayward NK, Perlina A, et al. Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep. 2015;11:1458–1473. doi: 10.1016/j.celrep.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snoek BC, De Wilt LH, Jansen G, Peters GJ. Role of E3 ubiquitin ligases in lung cancer. World J Clin Oncol. 2013;4:58–69. doi: 10.5306/wjco.v4.i3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. Embo J. 2014;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozato K, Shin DM, Chang TH, Iii HCM. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napolitano LM, Meroni G. TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- 13.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;770:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 14.Qiu X, Huang Y, Zhou Y, Zheng F. Aberrant methylation of TRIM58 in hepatocellular carcinoma and its potential clinical implication. Oncol Rep. 2016;36:811–818. [DOI] [PubMed] [Google Scholar]
- 15.Kajiura K, Masuda K, Naruto T, Kohmoto T, Watabnabe M, Tsuboi M, Takizawa H, Kondo K, Tangoku A, Imoto I. Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget. 2016;8:2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Zhang X, Cai J, Li Y, Luo Q, Wu H, Yang Z, Wang L, Chen D. Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncol Rep. 2018;40:1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He T, Sparks AB, Rago C, Hermeking H, Zawel L, Costa L, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a Target of the APC Pathway. Science. 1998;281:1509. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 18.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen DX, Chiang AC, Zhang XHF, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massagué J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K, Tanaka F, Watanabe M, Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for Lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi: 10.1245/s10434-007-9461-1. [DOI] [PubMed] [Google Scholar]
- 21.Feng Q, Jian-Ping X, Jun D, Xiao-Jun X. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol. 2015;8:5053–5061. [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Park CH, Jung KC, Rhee HS, Yang CH. Negative regulation of β-catenin/Tcf signaling by naringenin in AGS gastric cancer cell. Biochem Biophys Res Commun. 2005;335:771–776. doi: 10.1016/j.bbrc.2005.07.146. [DOI] [PubMed] [Google Scholar]
- 23.Ikenoue T, Ijichi H, Kato N, Kanai F, Masaki T, Rengifo W, Okamoto M, Matsumura M, Kawabe T, Shiratori Y, et al. Analysis of the β‐catenin/T cell factor signaling pathway in 36 gastrointestinal and liver cancer cells. Jpn J Cancer Res. 2002;93:1213–1220. doi: 10.1111/j.1349-7006.2002.tb01226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X-Z, Cheng -T-T, He Q-J, Lei Z-Y, Chi J, Tang Z, Liao Q-X, Zhang H, Zeng L-S, Cui S-Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Guo J, Wang Q, Zhou J, Xu C, Teng R, Chen Y, Wei Q, Liu Z-P. LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin. J Cancer Res Clin Oncol. 2014;140:1997–2008. doi: 10.1007/s00432-014-1753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Xu J, Fu H, Zhang Y, Zhang X, Yang D, Zhu Z, Wei Z, Hu Z, Yan R, et al. TRIM32 promotes cell proliferation and invasion by activating β-catenin signalling in gastric cancer. J Cell Mol Med. 2018;22:5020–5028. doi: 10.1111/jcmm.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong JY, Kang B, Kim A, Hwang S, Ahn J, Lee S, Kim J, Park J-H, Cheon D-S. Development of a highly sensitive real-time one step RT-PCR combined complementary locked primer technology and conjugated minor groove binder probe. Virol J. 2011;8:330. doi: 10.1186/1743-422X-8-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 30.Klopfleisch R, Gruber AD. Differential expression of cell cycle regulators p21, p27 and p53 in metastasizing canine mammary adenocarcinomas versus normal mammary glands. Res Vet Sci. 2009;87:91–96. doi: 10.1016/j.rvsc.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 32.Shin S, Sung B-J, Cho Y-S, Kim H-J, Ha N-C, Hwang J-I, Chung CW, Jung YK, Oh BH. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and −7. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 33.Granziero L, Ghia P, Circosta P, Gottardi D, Strola G, Geuna M, Montagna L, Piccoli P, Chilosi M, Caligaris-Cappio F. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- 34.Lim JH, Park JW, Chun YS. Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677. doi: 10.1158/0008-5472.CAN-06-3171. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, He D, Yang L, Wen B, Dai J, Zhang Q, Kang J, He W, Ding Q, He D. TRIM26 functions as a novel tumor suppressor of hepatocellular carcinoma and its downregulation contributes to worse prognosis. Biochem Biophys Res Commun. 2015;463:458–465. doi: 10.1016/j.bbrc.2015.05.117. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Wu Y, Miao X, Li C, Yin H, Yang S, Lu X, Liu Y, Chen Y, Shen R, et al. High expression of TRIM44 is associated with enhanced cell proliferation, migration, invasion, and resistance to doxorubicin in hepatocellular carcinoma. Tumor Biol. 2016;37:1–14. doi: 10.1007/s13277-016-5316-3. [DOI] [PubMed] [Google Scholar]
- 37.Palmbos PL, Lidong W, Huibin Y, Yin W, Jacob L, Ahmet MKL, Wilkinson JE, Kumar-Sinha C, Ney GM, Tomlins SA, et al. ATDC/TRIM29 drives invasive bladder cancer formation through miRNA-mediated and epigenetic mechanisms. Cancer Res. 2015;75:5155–5166. doi: 10.1158/0008-5472.CAN-15-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchita B, Claude G, Lynn C, Jianhong O, Xiaochun Z, Tushir JS, Virbasius C-M, Lin L, Zhu LJ, Wajapeyee N, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–120. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Li X, Wei D, Sun C, Guo D, Zhang L. Mmu-miR-1894-3p inhibits cell proliferation and migration of breast cancer cells by targeting Trim46. Int J Mol Sci. 2016;17:609. doi: 10.3390/ijms17040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L, Wu YX, Yu JH, Chen X, Luo XJ, Yin YR. Research of the relationship between β-catenin and c-myc-mediated Wnt pathway and laterally spreading tumors occurrence. Eur Rev Med Pharmacol Sci. 2017;21:252. [PubMed] [Google Scholar]
- 41.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen ZH, Zhou XJ, Pathology DO. The expression and correlation of β-catenin and survivin in nasopharyngeal carcinoma. Jilin Med J. 2018; 39:608-609. [Google Scholar]
- 43.Hai W, Haiyan W, Mohammad Shahidul M, Juanjuan W, Yingqing D, Qunli S, Liu Q, Zhou X, Wang J. Overexpression of β-catenin and cyclinD1 predicts a poor prognosis in ovarian serous carcinomas. Int J Clin Exp Pathol. 2013;7:264–271. [PMC free article] [PubMed] [Google Scholar]
- 44.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. [DOI] [PubMed] [Google Scholar]
- 45.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clements WM, Wang J, Sarnaik A, Kim OJ, Macdonald J, Fenogliopreiser C, Groden J, Lowy AM. beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–3506. [PubMed] [Google Scholar]
- 47.Zhou YN, Xu CP, Han B, Li M, Qiao L, Fang DC, Yang J-M. Expression of E-cadherin and β-catenin in gastric carcinoma and its correlation with the clinicopathological features and patient survival. World J Gastroenterol. 2002;8:987–993. doi: 10.3748/wjg.v8.i6.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao ZF, Wang ZN, Zhao TT, Xu YY, Wu JH, Liu XY, Xu H, You Y, Xu H-M. TRIM24 is upregulated in human gastric cancer and promotes gastric cancer cell growth and chemoresistance. Virchows Archiv. 2015;466:525–532. doi: 10.1007/s00428-015-1737-4. [DOI] [PubMed] [Google Scholar]
- 49.Takeyuki S. The cellular level of TRIM31, an RBCC protein overexpressed in gastric cancer, is regulated by multiple mechanisms including the ubiquitin-proteasome system. Cell Biol Int. 2013;35:657–661. [DOI] [PubMed] [Google Scholar]


