ABSTRACT
Oligosaccharins, which are biologically active oligosaccharide fragments of cell wall polysaccharides, may regulate the processes of growth and development as well as the response to stress factors. We characterized the effect of the oligosaccharin that stimulates rhizogenesis (OSRG) on the gene expression profile in the course of IAA-induced formation of adventitious roots in hypocotyl explants of buckwheat (Fagopyrum esculentum Moench.). The transcriptomes at two stages of IAA-induced root primordium formation (6 h and 24 h after induction) were compared after either treatment with auxin alone or joint treatment with auxin and OSRG. The set of differentially expressed genes indicated the special importance of oligosaccharin at the early stage of auxin-induced adventitious root formation. The list of genes with altered mRNA abundance in the presence of oligosaccharin included those, which Arabidopsis homologs encode proteins directly involved in the response to auxin as well as proteins that contribute to redox regulation, detoxification of various compounds, vesicle trafficking, and cell wall modification. The obtained results contribute to understanding the mechanism of adventitious root formation and demonstrate that OSRG is involved in fine-tuning of ROS and auxin regulatory modes involved in root development.
KEYWORDS: Adventitious roots, auxin, buckwheat, oligosaccharin, transcriptome
1. Introduction
Adventitious roots are plant roots that form after germination (postembryonic roots) from any non-root tissue (leaves, hypocotyl, stems, etc.). They are produced both during normal development and in response to stress conditions.1,2 The formation of adventitious roots is an excellent experimental system for studying important physiological and molecular events that occur during morphogenesis.
The development of adventitious roots involves several successive stages. Depending on the tasks and selected criteria, the number of stages of root formation varies from two3 to five.4 or even seven.5 stages. The most widely recognized phases of adventitious roots formation are induction, initiation, and expression.6 The induction stage is characterized by initial biochemical and molecular changes, which leads to reprogramming of target cells, but there are no visible changes. The initiation stage begins with the first visible cell divisions and ends with the formation of domed root primordia. The expression stage of adventitious roots formation includes the growth of root primordia through explant tissues, the establishment of vascular connections and the emergence of roots on the surface of the explant.6 The last stage does not require the use of exogenous auxin for the adventitious root formation,7 since the formed primordium becomes autonomous.3 Using auxin sensitivity as a criterion allows reducing the number of stages to two key stages: (i) when the exogenous auxin has a stimulating effect and (ii) when auxin is not required or even has an inhibitory effect.7,8
The importance of sensitivity to auxin as a criterion is determined by the fact that auxin is a key player in the root formation process and the other participants are considered to influence root formation through changes in its homeostasis, transport, or via signaling pathways mediated by this hormone.9 For example, signaling molecules such as NO,10 cGMP,11 Ca2+12 or H2O213 are involved in the formation of adventitious roots through an auxin-mediated signaling pathway.
Studies conducted over the past 20 years have shown that oligosaccharins – oligosaccharide fragments of cell wall polysaccharides that exhibit biological activity – can act as endogenous regulators of the process of root formation .14–17 The effects of the oligosaccharins produced from different cell wall polymers on the process of adventitious root formation have been shown in various model systems. Pectic cell wall fragments regulate the formation of roots in thin-layer explants of tobacco (Nicotiana tabacum L.)18 and buckwheat (Fagopyrum esculentum Moench) hypocotyls.19 Xyloglucan oligosaccharides stimulate the rooting of in vitro grown shoots of black pine (Pinus nigra J.F.Arnold) and birch (Betula paryrifera Marshall).20 Fragments of galactoglucomannan have been demonstrated to affect adventitious root formation in mung bean (Vigna radiata (L.) Wilczek) hypocotyls.16 Oligogalacturonides, the best-studied oligosaccharins, which are fragments of the linear pectic polymer homogalacturonan, show both inhibitory14,21 and activating22,23 effects on the root formation process.
Interactions with auxin have been demonstrated for several oligosaccharins, in addition to most effectors involved in the process of adventitious root formation. For example, oligogalacturonides with a polymerization degree of 6–18 inhibit auxin-induced root formation in tobacco explants.14 The inhibitory effect of oligogalacturonides is eliminated by increasing the concentration of auxin.24 Galactoglucomannan-derived oligosaccharins stimulate the formation of adventitious roots in the hypocotyls of mung beans without the addition of exogenous auxin and inhibit this process in its presence.16 Depending on the hormone concentration, fragments of the cell wall pectins of sycamore either induce or inhibit the formation of adventitious roots in thin-layer tobacco explants.18 The inhibitory effect of oligogalacturonides on auxin-induced adventitious root formation in Arabidopsis leaf explants is coupled to the changes in the expression of the genes involved in early auxin reactions (IAA5, SAUR16, and SAUR-AC1); changes in expression are detected within 30 minutes of treatment indicating that the oligogalacturonide-triggered cascade rapidly affects auxin signaling.21 The ability of oligogalacturonides at submicromolar concentrations to affect the activity of some (but not all) auxin-upregulated promoters was demonstrated in a set of transgenic plants harboring the GUS gene under the control of pNt114 of tobacco, prolB and prolD of Agrobacterium rhizogenes, or pGm-GH3 of soybean.24,25 All of these data indicate the existence of a relationship between the mechanisms of oligosaccharins and auxin action.
Most of the studies on the effects of oligosaccharins have been carried out using cell wall polysaccharide fragments obtained by enzymatic and chemical hydrolysis or chemical synthesis, raising the question of the presence of such compounds in vivo. Additionally, a method for obtaining biologically active oligosaccharides from plant homogenates without prior hydrolysis of cell wall polymers was developed,19 confirming the existence of such regulatory molecules in planta. Among the oligosaccharide fractions isolated from pea seedlings that exhibit biological activity in the test system of hypocotyl explants from buckwheat (Fagopyrum esculentum Moench.),19 one fraction presented the pronounced ability to stimulate IAA-induced formation of adventitious roots, and was named OSRG (oligosaccharine that stimulates rhizogenesis).26 Notably, treatment with OSRG alone (without IAA) did not stimulate root development in this test system compared to explants without both effectors.
The effects of oligosaccharins on the root formation in all studied plant systems have been characterized mainly by biochemical or histological approaches.16,27 Recently, the RNA sequencing method (RNA-Seq) has been successfully used to profile gene expression during the formation of adventitious roots in the presence or absence of auxin. Such studies have been performed for the hypocotyl segments of mung beans,28 carnation (Dianthus caryophyllus L.) stem cuttings,29 petunia (Petunia hybrida cv. Mitchell) cuttings,30 segments of tetraploid black locust (Robinia pseudoacacia L.)31 and tea (Camellia sinensis L.) nodal cuttings.32 Genes encoding members of the auxin signaling pathway involved in adventitious root formation have been identified, including auxin-induced the genes encoding some ARFs (AUXIN RESPONSE FACTOR) and GH3 (GRETCHEN HAGEN 3) and genes encoding the YUCCA flavin monooxygenases (enzymes involved in auxin biosynthesis), the auxin transporters PIN (PIN-FORMED) and ABCB/PGP (ATP BINDING B/P-GLYCOPROTEIN CASSETTE-TYPE), the high-affinity auxin influx carriers AUX1 and LIKE AUX1 (LAX1), and an auxin-sensitive transcription factor with a LOB domain (LATERAL ORGAN BOUNDARIES DOMAIN). A complete analysis of the changes in the transcriptome under the action of rhizogenic oligosaccharins has never been carried out before. The few transcriptome studies to characterize oligosaccharin effects have been performed for other auxin-dependent processes and only for oligogalacturonides.33,34
In the current work, we used RNA-Seq technology to reveal the effect of the oligosaccharin OSRG on gene expression patterns at two stages of auxin-induced adventitious root formation in buckwheat hypocotyls. The purpose of this study was to identify the genes and associated regulatory pathways responsible for the stimulating effect of this oligosaccharin on the development of adventitious root primordia, with the main focus on the interaction of the two effectors: auxin and oligosaccharin. This analysis sheds light on the mechanism of the formation of adventitious roots and the role of cell wall oligosaccharides in this process.
2. Materials and methods
2.1. Extraction, purification and compositional characterization of the oligosaccharin
The oligosaccharide extraction was carried out according to the method described by Zabotina with colleagues19 with minor modifications. A method for isolation and purification is based on the high hydrophilicity of neutral oligosaccharides and the absence of charges. The oligosaccharide fractions were isolated from pea seedlings (Pisum sativum L.) grown in tap water at 25°C under 100 μmol m−2 s−1 light with 12 h photoperiod for 8 days. The seedlings were fixed for 30 min at 100°C, dried at 60°C to a constant weight, and ground to a powder. Plant material was washed three times with chloroform:ethanol (1:2) mixture and the last extraction was carried out by boiling the solution for 30 min in a water bath. The pellet was washed four times with 70% ethanol and dried in air. The resulting alcohol insoluble residue was homogenized in phosphate buffer (50 mM KH2PO4, pH 7.0) and filtered.The pellet was additionally washed two times with the same buffer and the buffer extracts were combined.
Further purification of the fractions was performed by cation- and anion-exchange chromatography and gel filtration. The buffer extract was brought to pH 6.0 with 4 M HCl, applied to a Dowex 50W×2 column, and eluted with 50 mM phosphate buffer (pH 6.0). NaCl was added to the concentration of 1.5 M; the resulting solution was applied to a Dowex 50W×2 column and eluted with 1.5 M NaCl. The obtained solution was desalted on a Sephadex G-10 column. In cold conditions (2°C), the extract was brought to pH 2.2, passed through a Dowex 50W×2 column eluted with water (pH 2.2) and immediately neutralized to pH 7.0. The neutralized solution was concentrated, brought to pH 8.0 with ammonium hydroxide and applied to a DEAE-cellulose column eluted with 10 mM NH4Cl (pH 8.0). The solution was desalted on a Sephadex G-10 column and dried under vacuum. The obtained solution contained mainly neutral oligosaccharides, since a significant amount of proteins, phenols, and various acidic compounds, which included uronic acids, were absorbed on the resins.
The next step was gel-permeation chromatography on a Toyopearl HW-55F column eluted with 100 mM NaCl; the flow rate was 0.3 ml/min. The collected fractions were desalted on a Sephadex G-10 column and dried under vacuum. After this step, fractions (in equal amount of carbohydrate) were tested for the ability to stimulate IAA-induced formation of adventitious roots. The fraction with the highest activity was used for further purification. The last chromatography step was gel-permeation on a Biogel P-4 column. The elution was carried out with 0.02% NaN3; the flow rate was 0.3 ml/min. The fractions were collected and checked for the ability to stimulate IAA-induced formation of adventitious roots. The fraction with the maximal activity (fraction 3, OSRG) was characterized (Figure 1) and used in the experiments to analyze transcriptome changes.
Figure 1.
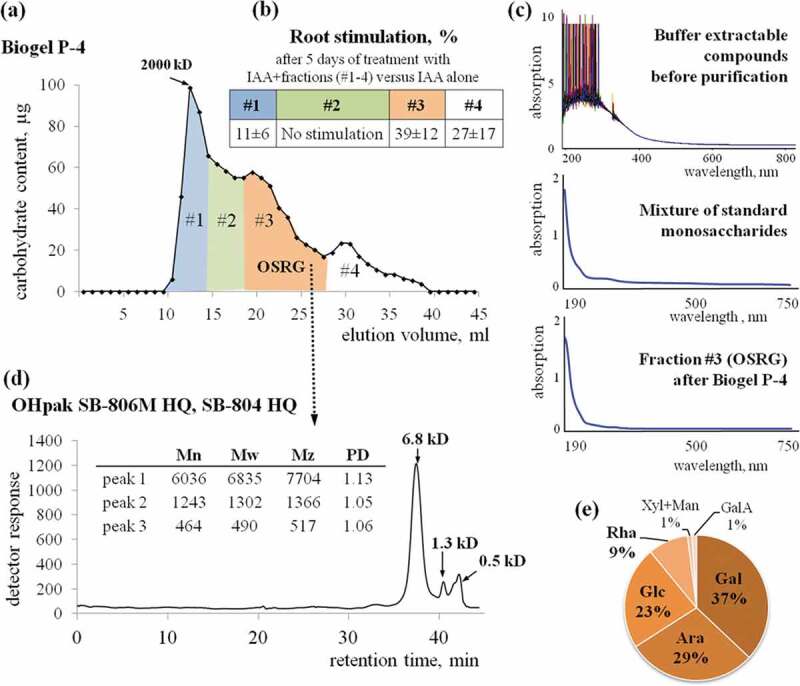
Characterization of the oligosaccharide fraction that stimulates the auxin-induced development of adventitious roots on the buckwheat hypocotyl. (a) Chromatography of the carbohydrate fraction obtained from the buffer-extractable compounds of pea seedlings on Biogel P-4, revealing a fraction of oligosaccharides that stimulates auxin-induced rhizogenesis (OSRG). (b) The increase in the number of auxin-induced adventitious roots per hypocotyl in the presence of OSRG (means ± standard error). (c) Absorption spectra obtained for buffer-extractable compounds before purification for the mixture of standard monosaccharides with a composition similar to that of OSRG and for the collected oligosaccharide fraction. (d) Determination of the molecular weight distribution (according to pullulan standards) using sequentially connected OHpak SB-806M HQ and OHpak SB-804 HQ columns. (e) Monosaccharide analysis of the collected oligosaccharide fraction performed by high-performance anion-exchange chromatography.
The molecular weight distribution of OSRG was determined by gel-filtrationon sequentially connected OHpak SB-G (6.0 × 50 mm, guard column), OHpak SB-806M HQ (8.0 × 300 mm) and OHpak SB-804 HQ (8.0 × 300 mm) columns (Shodex, Japan). Elution was carried out with 0.1 M NaCl, flow rate 0.5 ml/min, the temperature of the columns 40°C. Detection was carried out using an Agilent 1260 Infinity refractometric detector (Agilent, Germany). Dextran (2000 kDa; Sigma, USA), pullulans (380, 100, 23.7, and 5.8 kDa; Showa Denko, Japan) with low polydispersity indexes (1.09–1.19) and D-(+)-glucose (Merck, Germany) were used as standards for calibration. The number of analytical replicates was 2. The results were processed using Agilent GPC/SEC software.
To confirm the absence of non-carbohydrate components in the oligosaccharide fraction with biological activity, the types of chromophore groups were analyzed by wavelength scanning in the absorption range of 190–1100 nm. Scanning was performed on a UV/Lambda 25 spectrophotometer (Perkin Elmer, USA).
The monosaccharide composition of OSRG was characterized by anion-exchange chromatography. Samples were hydrolyzed with 2 M TFA at 120°C for 1 h, dried to remove TFA, dissolved in deionized water, and analyzed using a DX-500 system equipped with a CarboPac PA-1 column (4 × 250 mm) and an ED40 pulsed amperometric detector (Dionex, USA). The column temperature was 30°C, and the mobile phase (pumped at 1 ml/min) consisted of 100% A (0–20 min), 90% A (20–21 min), 50% A (22–41 min), 0% A (42–55 min), and, finally, 100% A (56–85 min), where A was 16.5 mM NaOH, and the other solvent (B) was 100 mM NaOH in 1 M sodium acetate. The results were analyzed using PeakNet software according to the calibrations obtained for monosaccharide standards treated in advance with 2 M TFA at 120°C for 1 h.
2.2. Treatment of buckwheat hypocotyl explants with oligosaccharide fractions
The testing of the oligosaccharide fractions for their ability to stimulate root formation was carried out in the previously developed test system of hypocotyl segments of buckwheat (Fagopyrum esculentum Moench.).19,35 The growth of buckwheat seedlings and the preparation and cultivation of explants were performed under sterile conditions. Seeds were sterilized for 10 min in a 2% sodium hypochlorite solution and then germinated in the dark at 25°C on hormone-free medium for 4–5 days at the half-stremgth Murashige-Skoog medium (MS/2)36 with the addition of 0.8% agar and 2% sucrose. Segments of 1 cm in length were cut from the middle part of the hypocotyls, additionally cut lengthwise into two parts, placed cut-side down in Petri dishes with MS/2 liquid medium supplemented with 2% sucrose, IAA alone or together with oligosaccharide fraction, and cultured in the dark at 25°C. The applied concentrations of IAA (3 μM) and OSRG (0.5 μg/ml) were determined as optimal in earlier experiments.26 The number of roots (at least 2 mm in length) that emerged on each explant was determined visually on the 5th day of incubation. Data on root numbers are expressed as the means ± standard error; the differences between the separate experimental groups were evaluated with the t-test, P˂0.05.
For the analysis of transcriptome changes, the isolated explants were collected in a liquid medium and divided into three groups. The first group was placed in medium without effectors (IAA or OSRG) and was sampled immediately after the preparation of all explants; this group was designated the control (CTR) and used as a reference. The second group was placed in medium with IAA, and the third group was placed in medium with IAA+OSRG. The samples for transcriptome analysis were collected 6 h and 24 h after the addition of effectors. For each treatment (IAA or IAA+OSRG) and each time point (6 and 24 h), 2 biological replicates, each consisting of 13 explants, were collected. The samples were fixed with liquid nitrogen and stored at −80°C until RNA extraction. Additionally, ten explants of each variant were left for long-term cultivation (5 days) to assess the effect of the treatments on root formation by counting the number of the emerged roots.
2.3. RNA extraction and sequencing
The total RNA from each collected sample was extracted using a TRIzol analogue combined with an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Polysaccharides and polyphenolics were removed from the RNA preparations by adding the Plant RNA Isolation Aid reagent (Ambion, USA) and β-mercaptoethanol to the lysis buffer RLT (according to37). The DNA residues from the obtained RNA samples were eliminated by DNase I treatment using a DNA-free kit (Ambion, USA). A total of 10 RNA samples (5 variants of two biological replicates in each) were obtained. Qualitative and quantitative evaluation of the RNA and cDNA libraries was performed with a Qubit fluorometer (Invitrogen, USA) and 2100 BioAnalyzer technology (Agilent Technologies) with the Agilent RNA 6000 Pico Kit (Agilent Technologies).
The preparation of cDNA libraries and subsequent sequencing were performed using the TruSeq Sample Prep Kit v3 (Illumina, USA) and the Illumina HiSeq 2500 platform according to the manufacturer’s instructions. Sequencing was carried out in single-end mode with a 60 nt read length and sequencing depth of 25 million mapped reads per sample.
2.4. RNA-Seq data analysis
Sequence trimming, mapping and expression level determination were carried out according to.38 For each gene, the total gene reads (TGR) were determined as the number of all reads that were mapped to this gene. Differentially expressed genes (DEGs) were identified for each pair of corresponding hypocotyl variants using the R package “DESeq2”.39 An FC (fold change)≥ |2| with an adjusted p-value (padj) <0.05 was considered the threshold for the identification of DEGs. The draft genome sequence of Fagopyrum esculentum and annotation available at NCBI under BioProject # PRJNA487881 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA487881) was used as a reference. The annotations of the mentioned Arabidopsis thaliana genes are provided according to the TAIR (https://www.arabidopsis.org/), UniProt (https://www.UniProt.org/UniProt) or SUBA4 (http://suba.live/) database. To validate RNA-Seq data, the expression level of 5 genes was assessed in all analyzed samples by qPCR. The results showed that qPCR data were in good agreement with those of RNA-Seq (Supplementary 1).
3. Results
3.1. Characterization of the obtained carbohydrate fraction and its effects on the formation of adventitious roots on buckwheat explants
The oligosaccharide fraction to be used as an effector for further transcriptome studies was determined based on the ability to stimulate root formation on the fragments of buckwheat hypocotyls. Fraction 3 (OSRG) obtained after separation on a column containing Biogel P-4 was the most effective in root induction and led to over 30% increase in root number (Figure 1a, b). For other fractions, the stimulating effect was completely absent or was below 10%. In the samples collected for transcriptome analysis, the number of adventitious roots on explants cultivated in the presence of OSRG constituted 13.4 ± 1.1 roots per explant compared to 10.1 ± 1.5 roots per explant for the samples treated only with auxin.
The separation of OSRG using high-performance liquid chromatography on series-connected OHpak columns revealed three peaks in the elution profile, whose retention times of corresponded to molecular weights of 6.8, 1.3 and 0.5 kDa according to calibration with pullulans (Figure 1c). The predominant components were those whose retention times corresponded to the highest molecular weight peak, with a degree of polymerization over 30 according to calibration with pullulans. The diversity of monosaccharides in the fraction and the potential presence of galactose and arabinose in cell wall-derived carbohydrates that are highly substituted and differ in their spatial structure from linear pullulans did not allow for strict correlation of the retention time in the column with the degree of polymerization.
Spectrophotometric analysis of the different types of chromophore groups in the sample obtained after chromatography purification of the buffer-extractable fraction of pea seedlings revealed one main absorption band with a maximum in the far UV region (190 nm) and a long shoulder of low intensity in the region of 220–340 nm (Figure 1c), both of which are characteristic of the absorption of sugars.40,41 The identical absorption spectrum of standard individual monosaccharides in the same proportions as the analyzed fraction confirmed that the absorption bands identified for OSRG were are carbohydrates and that there were no functional groups of non-carbohydrate compounds. The absorption spectrum of the non-purified fraction contained numerous bands corresponding to non-carbohydrate components (Figure 1c).
Monosaccharide analysis of the physiologically active carbohydrate fraction obtained after separation on Biogel P-4 column revealed the presence of mostly neutral components, among which galactose residues prevailed (37 mol%), and arabinose and glucose residues were present in approximately equal proportions (29 and 23 mol%), while rhamnose (9 mol%) and xylose together with mannose (1 mol% of each) were minor components, as was galacturonic acid, which was detected in a trace amount (Figure 1e). A mixture of individual monosaccharides with the same proportions found in OSRG and the same total concentration did not have any effect on IAA-induced root formation (data not shown).
3.2. General transcriptome characteristics of the analyzed samples
Transcriptome analysis was performed for buckwheat hypocotyls treated with IAA or IAA+OSRG for 6 or 24 h. The time intervals were chosen to take into account the stages of explant sensitivity to the applied effectors described in our previous study26 and the timing of events presented by other authors.13,28 For the appearance of adventitious roots to occur, buckwheat explants should be cultured in auxin solution for 24 hours, after which the hormone can be removed from the medium.26 This period of sensitivity to auxin, which some authors designate as a whole “root initiation”,7 is divided into two stages by other authors: the induction and initiation period.42 In the work on the hypocotyls of mung bean, the length of the first stage is determined as 6 hours, of the second – 24 hours.42
The samples for analysis were designated as IAA (6 h), IAA+OS (6 h), IAA (24 h), and IAA+OS (24 h). As a result of the global profiling of the transcriptome, approximately 300 million reads with a quality score above Q30 were obtained. For each treatment, two replicates were sequenced using the Illumina HiSeq 2500 platform. All raw data were uploaded onto NCBI SRA with the BioSample accession as PRJNA595876. Pearson’s correlation coefficients of the two replicates for each treatment were larger than 90%, suggesting an appreciable correlation between them (Table S1).
A total of 24,929 expressed genes with a normalized TGR> 5 were identified in two replicates of at least one sample (Table S2). Among these genes, 10,927 genes exhibited differential expression in various combinations of pairwise comparisons (Figure 2). IAA induced profound changes in the transcriptome of the buckwheat hypocotyl explants, with thousands of genes exhibiting a change in expression after either 6 or 24 h of explant cultivation (Figure 2). This finding is in accordance with many transcriptome studies on the effect of auxin on adventitious or lateral root formation.42,43
Figure 2.
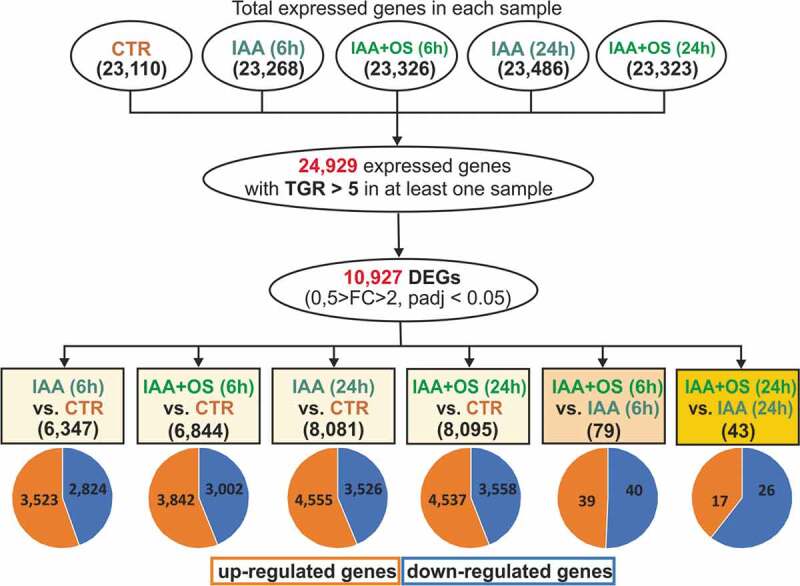
Quantitative characteristics of genes expressed in the analyzed samples.
The presence of the oligosaccharin stimulating the auxin-induced development of adventitious roots26 resulted in the differential expression of 122 genes identified with the applied thresholds (log2FC>1, padj<0.05) (Figure 2). These genes were within the focus of our current study. Two-thirds of these genes were detected after 6 h of sample incubation, indicating that the effect of oligosaccharin was more pronounced in the earlier stage of auxin-induced adventitious root formation.
3.3. Differentially expressed genes after 6 h of cultivation of buckwheat explants in iaa-containing medium with or without OSRG
In the samples collected after 6 h of incubation with both effectors compared to incubation with IAA alone, the abundance of the transcripts of 40 genes was increased, and that of 39 was decreased. Some of the genes that were grouped as being upregulated by OSRG addition (Figure 3) were actually downregulated by treatment with IAA alone, but their expression remained at the same level as in the 0 h sample without treatment with IAA or OSRG (control) or was even upregulated in the presence of OSRG (Figure 3). This was the case for the two genes with the highest fold-changes in mRNA abundance: tr_12709 and tr_18008. The closest Arabidopsis homologue (AT4G02340) of tr_12709 is annotated as a gene for hydrolase, an alpha/beta fold family domain-containing protein showing epoxide hydrolase activity (https://www.arabidopsis.org). The transcript tr_18008 is homologous to AT3G58640, a gene of the MAP3K family (mitogen-activated protein kinase kinase-like protein) that is involved in protein phosphorylation. The 6-h effect of IAA alone decreased the expression of this MAP3K, while the addition of OSRG maintained its expression at a similar level to that in all other analyzed samples (Figure 3). Similarly, the abundance of the tr_10564 (no homologs) and tr_17255 (homologue of AT3G08710) transcripts was decreased by IAA alone but remained at the initial level in the presence of OSRG. AT3G08710 encodes thioredoxin H-type 9, associated with the plasma membrane; it travels from cell to cell, suggesting a role in intercellular communication. The genes encoding the electron-transfer flavoprotein tr_24751 (homologue of AT5G43430) and the metallothionein tr_14620 (homologue of AT3G09390) were considerably upregulated compared to the samples treated with IAA alone but downregulated compared to the control (Figure 3).
Figure 3.
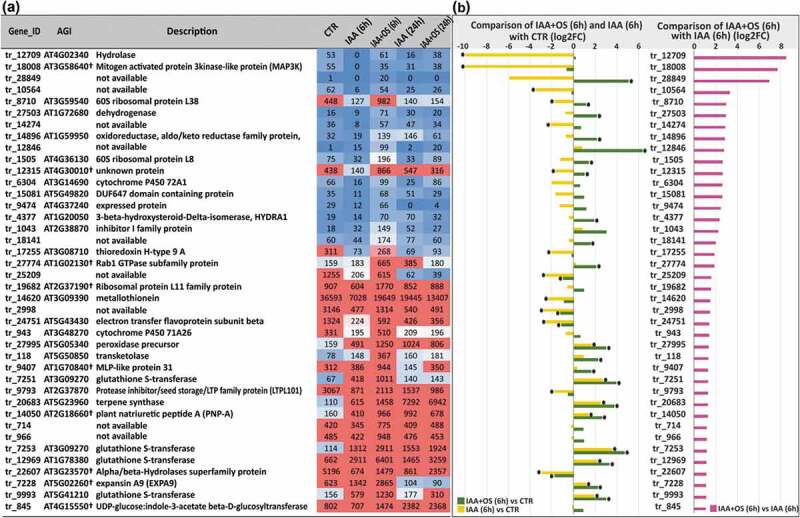
(a) List of upregulated genes in IAA+OS (6 h) samples compared with IAA (6 h) samples (cut off log2FC>1, padj<0.05) and the corresponding expression level indicators in the form of normalized TGR values. (b) Expression levels of upregulated genes (log2FC> 1, padj<0.05) in IAA+OS (6 h) samples compared to IAA (6 h) samples (right side) and in the comparisons of the IAA+OS (6 h) and IAA (6 h) groups with the control sample (CTR; 0 h) (left side): “*” indicates differential expression compared to control genes with a padj value <0.05.
The most extensive group of genes that were substantially upregulated in IAA+OS (6 h) group compared to both the control and IAA (6 h) samples consisted of genes encoding proteins that are potentially involved in redox regulation and the detoxification of various compounds. This group included four genes encoding glutathione S-transferases: tr_9993 (homologue of AT5G41210), tr_12969 (homologue of AT1G78380), tr_7253 and tr_7251 (both homologous to AT3G09270). After 6 h of incubation, the mRNAs of these genes were up to 25 times more abundant in the IAA+OS sample compared to the control and 2 times more abundant compared to the group treated with IAA alone (Figure 3). This group also included the oxidoreductase tr_14896 (homologue of AT1G59950), cinnamyl-alcohol dehydrogenase tr_27503 (homologue of AT1G72680), cell wall peroxidase tr_27995 (homologue of AT5G05340), and the cytochrome P450s tr_6304 (homologue of AT3G14690) and tr_943 (homologue of AT3G48270).
Three genes that were considerably upregulated after 6 h of incubation in the presence of OSRG compared to both the IAA-alone and control groups exhibited Arabidopsis homologs annotated as genes for putative ribosomal proteins: tr_8710 (homologue of AT3G59540 for 60S ribosomal protein L38), tr_1505 (homologue of AT4G36130 for 60S ribosomal protein L8) and tr_19682(homologue of AT2G37190 for 60S ribosomal protein L11). The transcripts of these genes were especially abundant under OSRG treatment in earlier stages of the IAA-induced formation of adventitious roots (Figure 3). The abundance of the transcript of tr_27774, which exhibits homology to AT1G02130, encoding a protein from the Rab1 GTPase subfamily, was also specifically increased in the presence of OSRG after 6 h of explant cultivation. This protein is required for membrane trafficking between the endoplasmic reticulum and the Golgi apparatus and for normal Golgi movement.44 Several genes encoding cell wall-related proteins were upregulated in IAA+OS (6 h) samples compared to IAA (6 h) samples: tr_7228 (homologue of AT5G02260, encoding expansin A9), tr_14050 (homologue of AT2G18660, encoding plant natriuretic peptide A, which belongs to a class of systemically mobile molecules distantly related to expansins), tr_9793 (homologue of AT2G37870, encoding lipid transfer protein-like), and tr_1043 (homologue of AT2G38870, encoding a serine protease inhibitor).
In IAA+OS (6 h) samples, the transcription of tr_845, which is homologous to AT4G15550, encoding UDP-glucose:indole-3-acetate beta-D-glucosyltransferase, was increased (Figure 3). This enzyme may be involved in the conjugation of IAA.45 The transcript of tr_118 showed increased abundance at the 6 h time point in the presence of OSRG, and this gene is similar to AT5G50850, encoding the E1β subunit of the mitochondrial pyruvate dehydrogenase complex, which oxidativelydecarboxylates pyruvate to form NADH and acetyl-CoA. This protein, designated MACCI-BOU 1 (MAB1), is a molecular player in auxin-regulated organ formation and has been suggested to contribute to PIN-dependent auxin polar transport via metabolic regulation.46 The Arabidopsis homologue of tr_4377, AT1G20050, which encodes a 3-beta-hydroxysteroid-delta-isomerase, the integral membrane protein HYDRA1, which has been demonstrated to play an important role in root patterning and influence the distribution of the PIN1 and PIN2 auxin efflux carriers;47 the abundance of tr_4377 transcripts was increased by OSRG.
Two upregulated genes were homologous to Arabidopsis genes that show Blast hits only to plant species and encode proteins of unknown function: tr_12315, homologous to AT4G30010, which encodes an unknown plant-specific protein located in mitochondria according to SUBA4 (http://suba.live/), and tr_9407, homologous to AT1G70840, encoding major latex protein (MLP)-like protein 31. For several differentially expressed genes, no functional annotations are available (Figure 3). TargetP (http://www.cbs.dtu.dk/services/TargetP/) indicated the localization of corresponding proteins for two of these genes (tr_14274 and tr_966) to the mitochondrion and one (tr_12846) to the chloroplast; no transmembrane domains were found in any of these genes according to TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) (Table S3).
The genes downregulated by OSRG at the 6 h time point included two genes, tr_6232 and tr_23526, whose Arabidopsis homologs encode proteins that are directly involved in the response to auxin: AT5G43700 (ATAUX2-11/IAA4) and AT1G04240 (SHORT HYPOCOTYL 2, SHY2/IAA3), respectively, both of which are AUX/IAA transcriptional regulator family proteins (Figure 4). The transcript abundance of both tr_6232 and tr_23526 was further decreased in the IAA+OS (24 h) samples (Figure 4). Two buckwheat genes whose Arabidopsis homologs encode transcription factors, tr_27441 (AT1G01060, encoding LATE ELONGATED HYPOCOTYL (LHY) from the MYB family involved in circadian rhythms) and tr_23365 (AT3G02550, encoding LOB domain-containing protein 41 (LBD41), were also downregulated in the IAA+OS (6 h) samples (Figure 4).
Figure 4.
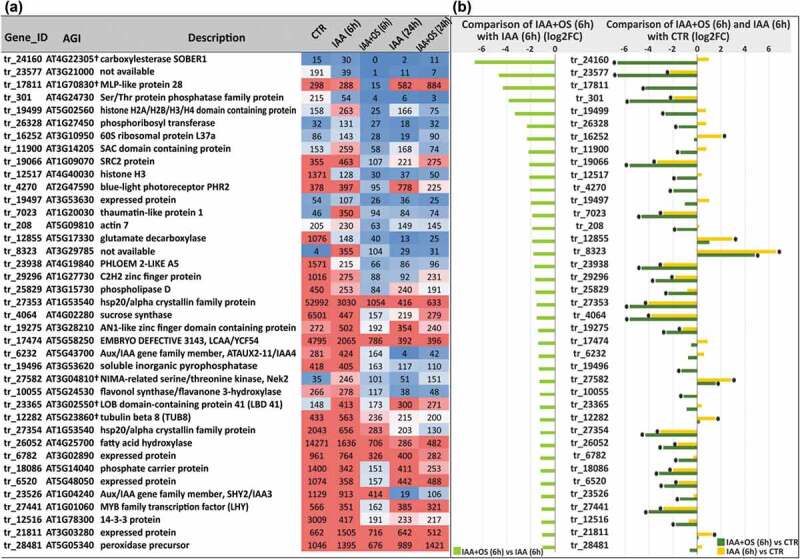
(a) List of downregulated genes in IAA+OS (6 h) samples compared with IAA (6 h) samples (cut off log2FC<1, padj<0.05) and the corresponding expression level indicators in the form of normalized TGR values. (b) Expression levels of downregulated genes (log2FC <1, padj<0.05) in IAA+OS (6 h) samples compared to IAA (6 h) samples (left) and in the comparison of IAA+OS (6 h) samples with the control sample (CTR; 0 h) (right): “*” denotes differentially expressed genes with a padj value <0.05.
Several small groups of functionally related downregulated genes were detected, indicating the importance of the regulation of the corresponding processes: tr_19499 and tr_12517, homologous to Arabidopsis genes encoding histones (AT5G02560 and AT4G40030, respectively); and tr_27353 and tr_27354, homologous to the same gene, AT1G53540, encoding the small ubiquitous chaperone hsp20 (Figure 4b). Three buckwheat genes (tr_23577, tr_8323, and tr_6520) whose Arabidopsis homologs (AT3G21000, AT3G29785, and AT5G48050, respectively) were annotated in TAIR (www.arabidopsis.org) as encoding retrotransposon family proteins, were also downregulated; however, both the expression levels of these buckwheat genes and their degree of similarity to the Arabidopsis genes (Table S2) were rather low.
The downregulation of the genes tr_208 (homologue of AT5G09810, encoding actin 7) and tr_12282 (homologue of AT5G23860, encoding tubulin beta 8) (Figure 4) in IAA+OS (6 h) samples indicated the possibility of an effect of OSRG on cytoskeletal rearrangement. tr_11900, homologous to AT3G14205, which encodes a SUPPRESSOR OF ACTIN domain protein 2 (SAC2), was also downregulated. SAC2 belongs to the phosphoinositide phosphatase family of proteins specific to land plants; such proteins localize to the tonoplast and are involved in vacuolar trafficking and turgor-driven cell expansion.48 Another down-regulated gene, tr_19066, is homologous to AT1G09070, encoding the SOYBEAN GENE REGULATED BY COLD-2 (SRC2) protein, which is also involved in vesicle trafficking to vacuoles.49
Several genes that were downregulated in the IAA+OS (6 h) samples exhibited homology to Arabidopsis genes encoding enzymes performing various post-translational modifications of proteins (Figure 4). One of these genes, tr_27502, is homologous to AT3G04810, encoding LSTK-1-like kinase, a member of the NIMA-related serine/threonine kinases (Neks), which have been linked to cell cycle regulation in fungi and mammals and might be involved in plant development.50 Other unknown proteins included a protein involved in N-terminal protein myristoylation (tr_21811 – AT3G03280) and a putative acyl-protein thioesterase (tr_24160 – AT4G22305), which is an enzyme that cleaves off lipid modifications on proteins.51 Myristoylation is a post-translational protein modification in which myristic acid is covalently attached to the N-terminal glycine residue; this modification influences the conformational stability of individual proteins and their ability to interact with membranes or the hydrophobic domains of other proteins.52 The considerably downregulated tr_17811 gene (Figure 4) is homologous to AT1G70830, encoding MLP-like protein 28, which can bind steroids (in vitro) and may also bind other types of hydrophobic ligands.53
3.4. Differentially expressed genes identified after 24-h of cultivation of buckwheat explants on iaa-containing medium with or without OSRG
After 24 h of effector action, several genes were found to be upregulated in the presence of OSRG to a higher level than in any other analyzed sample. These included tr_3050 (homologue of AT3G23790, encoding an AMP-binding enzyme with a poorly characterized function), tr_13010, homologue of AT5G08370, encoding α-galactosidase 2, which modifies the cell wall,54 tr_17004 (homologous to AT3G03280 (as was tr_21811, downregulated at 6 h)), encoding an unknown protein that is probably involved in N-terminal protein myristoylation (https://apps.araport.org), tr_22226 (homologue of AT4G38580, encoding a heavy metal-associated isoprenylated plant protein that may serve as a metallochaperone)55 and tr_19416 (homologue of AT1G26800, encoding the E3 ubiquitin-protein ligase MPSR1) (Figure 5). At the same time, the tr_26771 gene (homologue of AT5G02500, encoding heat shock protein 70 (a chaperone)) was also upregulated in the presence of OSRG (Figure 5).
Figure 5.
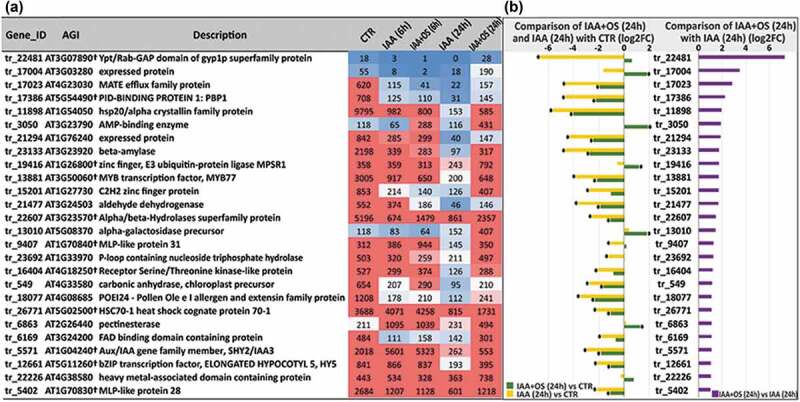
(a) List of upregulated genes in IAA+OS (24 h) samples compared with IAA (24 h) samples (cut off log2FC>1, padj<0.05) and the corresponding expression level indicators in the form of normalized TGR values. (b) Expression levels of upregulated genes (log2FC> 1, padj<0.05) in IAA+OS (24 h) samples compared to IAA (24 h) samples (right side) and in the comparisons of the IAA+OS (24 h) and IAA (24 h) groups with the control sample (CTR; 0 h) (left side): “*” indicates differential expression compared to control genes with a padj value<0.05.
Only two genes were upregulated in the presence of OSRG at both 6 h and 24 h; each of them encodes a protein with a poorly characterized function: tr_9407, homologous to AT1G70840, encoding major latex protein (MLP)-like protein 31, and tr_22607, homologous to AT3G23570, encoding an alpha/beta-hydrolase superfamily protein. The list of the upregulated genes identified at 24 h in the presence of OSRG also included tr_17023, a homologue of AT4G23030, encoding transporters from the multidrug and toxic compound extrusion (MATE) family, and tr_22481, a homologue of AT3G07890, encoding the Ypt/Rab-GAP domain of the gyp1p superfamily protein that modulates vesicle transport through interaction with RabGTPases by stimulating GTP hydrolysis.56 Additionally, the tr_23692 gene (encoding a putative GTP-hydrolase), homologous to AT1G33970 (encoding P-loop-containing nucleoside triphosphate hydrolase), was also upregulated in the presence of OSRG (Figure 5).
Genes encoding several transcription factors were differentially expressed in the presence of OSRG at the second studied stage of adventitious root formation. Among these transcription factors, MYB77 (tr_13881, homologous to AT3G50060) deserves special attention, since this transcription factor is involved in lateral root development; it interacts with ARF7 and regulates the expression of certain auxin-responsive genes.57 The genes encoding two other transcription factors, SHY2/IAA3 (tr_5571, homologue of AT1G04240) and HY5 (tr_12661, homologue of AT5G11260), were also upregulated in the presence of OSRG. HY5 is a shoot-to-root mobile transcription factor that is involved in light sensing and coordinates carbon and nitrogen metabolism.58 All three listed genes encoding transcription factors were actually downregulated by treatment with IAA alone at 24 h but remained at a similar level to that in the previous stage in the presence of oligosaccharin (Figure 5). Similar dynamics of mRNA abundance were detected for tr_16404, a homologue of AT4G18250, encoding a receptor serine/threonine-like protein located in the plasma membrane (Figure 5). The genes encoding two transcription factors, tr_14849 homologous to AT5G41315, encoding the basic helix loop helix domain protein GLABRA 3 (involved in the specification of root epidermal cell fate59), and tr_17421, homologous to AT2G35940, encoding the homeodomain factor BHL1, were downregulated in IAA+OS (24 h) samples compared to IAA (24 h) samples (Figure 6).
Figure 6.
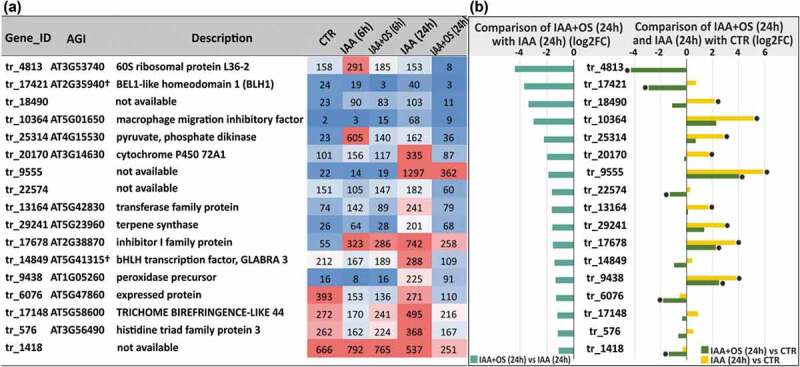
(a) List of downregulated genes in IAA+OS (24 h) samples compared with IAA (24 h) samples (cut off log2FC<1, padj<0.05) and the corresponding expression level indicators in the form of normalized TGR values. (b) Expression levels of downregulated genes (log2FC <1, padj<0.05) in IAA+OS (24 h) samples compared to IAA (24 h) samples (left) and in the comparison of IAA+OS (24 h) samples with the control sample (CTR; 0 h) (right): “*” denotes differentially expressed genes with a padj value <0.05.
In several cases, one orthologous gene in Arabidopsis had a number of buckwheat homologs that were differentially expressed in the presence of OSRG. Two buckwheat homologs (tr_29296 and tr_15201) of the Arabidopsis gene AT1G27730 were both downregulated at 6 h and upregulated at 24 h (Figure 4, 5). AT1G27730 encodes ZAT10, a zinc finger protein of the Cys2/His2 type that acts as a transcriptional repressor that is suggested to modulate or balance the response of plants to ROS and abiotic stress responses.60 Three buckwheat homologs of AT1G54050 were detected among the differentially expressed genes: two (tr_27353 and tr_27354) were downregulated at 6 h (Figure 4), and one (tr_11898) was upregulated at the later stage of root development (Figure 5). AT1G54050 encodes heat shock protein 20 (HSP20), which serves as a molecular chaperone. Two buckwheat homologs of AT2G38870, encoding a serine protease inhibitor, exhibited opposite expression dynamics, with tr_1043 being upregulated at 6 h (Figure 3) and tr_17678 being downregulated at 24 h (Figure 6).
The decrease in the gene expression of tr_17148 (homologous to AT5G58600, another name for the PMR5 gene, encoding the TRICHOME BIREFRINGENCE-LIKE 44 protein) observed after 24 h of cultivation in response to OSRG showed that oligosaccharin can also affect the properties of cell walls through the modification of cell wall polymers because trichome birefringence-like proteins are proposed to encode wall polysaccharide-specific O-acetyltransferases.61 Some genes that were differentially expressed at 24 h encoded other proteins that are able to modify the cell wall, including the upregulated tr_6863 gene (homologue of AT2G26440, encoding pectinesterase), the already mentioned tr_13010 gene (homologue of AT5G08370, encoding α-galactosidase 2), and tr_18077 (homologue of AT4G08685, encoding an extensin family protein) (Figure 5), and the downregulated tr_9438 gene (homologue of AT1G05260, encoding a cell wall peroxidase) (Figure 6).
4. Discussion
4.1. Modulation of the auxin response by OSRG
Comparative analysis of gene expression profiles between different time points in the process of root development and between the IAA and IAA+OSRG treatments revealed genes that were differentially expressed in the presence of the oligosaccharin OSRG, which stimulates IAA-induced root formation in hypocotyl explants of buckwheat, as shown previously.26 The addition of auxin to the excised hypocotyls cultivated in a liquid medium,induced adventitious root formation coupled with the differential expression of several thousand genes (Figure 2). Hundreds of these genes encode proteins directly involved in auxin signaling, including many transcription factors and regulators, as well as auxin transporters, auxin-binding proteins, and enzymes involved in auxin metabolism (Table S2). Despite the fact that OSRG affects IAA-induced adventitious root development and is weakly effective in the absence of the hormone,26 a few differentially expressed genes that are directly associated with auxinmetabolism or signaling were revealed in the presence of oligosaccharin. Among the most relevant auxin signaling genes, OSRG influenced the expression of Aux2-11/IAA4 and SHY2/IAA3, both of which belong to the family of Aux/IAA genes. SHY2/IAA3 is a known regulator of both lateral and adventitious root formation.62 Lateral root development is rather similar to adventitious root formation and is better characterized.63 This permits us to refer to studies on lateral root primordium development and emergence when discussing these processes for adventitious roots, while keeping in mind the differences between these root types, especially in the root induction stage.64 The SHY2/IAA3 gene encodes a protein that is a component of one of the four known AUX/IAA-ARF modules mediating auxin signaling at different stages of lateral root formation.65,66 It is also possible that a change in the expression of the SHY2/IAA3 gene reflects a change in the balance between the cytokinin- and auxin-mediated signaling pathways. Cytokinin and auxin antagonistically control the balance between cell division and differentiation in roots by influencing the expression of SHY2/IAA3.67 OSRG likely contributes to changes in the balance of one hormonal signaling pathway versus another at different stages in the formation of adventitious roots, suppressing SHY2/IAA3 gene expression in the first analyzed stage and increasing it in the second. Regulation of cytokinin activity can occur due to downregulation of the tr_26328, homologue of AT1G27450, encoding adenine phosphoribosyl transferase 1 (APT1), which catalyzes cytokinin inactivation through phosphoribosylation (Figure 4). Loss of APT1 activity leads to excess accumulation of active cytokinin, evoking cytokinin-regulated responses.68
AUX2-11/IAA4 also plays a certain role in root formation, although the corresponding AUX/IAA-ARF module has not been identified. Local expression of the AUX2-11 gene is observed at sites of the initiation of lateral roots.69 AUX2-11 is a unique member of the AUX/IAA family of proteins, since it exhibits a carboxyl terminal motif that acts as a substrate for geranylgeranyltransferaseI.70 The loss of activity of this enzyme increases the auxin-induced initiation of lateral roots, possibly through inactivation of AUX2-11 and an increase in the transcription of auxin-regulated genes.71
Notably, the expression of the gene encoding the transcription factor MYB77 was upregulated by OSRG (Figure 5). The roles of MYBs in the development and differentiation of roots have been proven.72 However, only MYB77 shows a direct interaction with the auxin response factor, which is usually associated only with transcriptional repressors from the AUX/IAA protein family. MYB77 binds ARF7, which is considered to be one of the key regulators of lateral root formation.57 Thus, upregulation of MYB77 by OSRG may lead to the modulation of auxin-inducible gene expression and further regulation of lateral root formation. Activation of MYB77 expression in the presence of OSRG occurred only at the second analyzed stage of root development. At the same time, HY5 expression was activated (Figure 5). HY5 is a transcription factor associated with light signaling; however, it is also involved in many aspects of root development, including lateral root formation and is thought to alter the balance of signaling through auxin and cytokinin.73 The known targets of auxin signaling through ARF7 and ARF19 are several members of the LBD gene family, which encodes plant-specific TFs.74 Members of the LBD family have been demonstrated to take part in adventitious and lateral root formation.74,75 Expression of LBD41 was downregulated by OSRGat early stage of adventitious root formation (Figure 4).
OSRG did not affect the expression of the genes encoding various transporters of auxin, including numerous pin-formed proteins (PIN), proteins of the AUXIN-RESISTANT1 and LIKE AUXIN-RESISTANT (AUX1/LAX) families, or WALLS ARE THIN 1 (WAT1), although many of these genes were significantly influenced by IAA and were differentially expressed at the two analyzed stages of adventitious root formation (Table S2). However, after 24 h of incubation with IAA and oligosaccharin, the tr_17386 gene, a homologue of AT5G54490, encoding PINOID (PID)-binding protein (PBP1), showed increased expression (Figure 5). PBP1 contains EF-hand calcium-binding motifs and acts as a positive regulator of auxin release from cells by regulating the activity of the protein kinase PINOID, which phosphorylates conserved serine residues in PIN auxin efflux carriers.76 The distribution of the auxin efflux carriers PIN1 and PIN2 and PIN-dependent auxin polar transport can also be influenced by the integral membrane protein HYDRA1,47 by MACCI-BOU 1 (MAB1),46 and by SAC2 and SRC2 proteins involved in vesicle trafficking to vacuoles48,49 since degradation of at least some PIN proteins may occur through this trafficking route.77 The transcription of the genes encoding listed proteins was affected by OSRG.Taken together, these results indicate that some elements of the auxin signaling pathway are influenced by the presence of oligosaccharin, as can be assumed from the differential expression of the corresponding genes. However, the changes are rather minor, especially compared with the effects of IAA itself.
4.2. The possible effect of OSRG through ROS metabolism
In a recent review of the morphogenesis of lateral root primordia in angiosperms,78 two major regulatory modes involved in lateral root formation in Arabidopsis were designated: the auxin mode and the reactive oxygen species (ROS) mode. The latter involves four peroxidases (PER7, PER9, PER57, and PER64), two transcription factors (MYB36 and the bHLH transcription factor UPBEAT1 (UPB1), which is expressed in the flanking region of the developing root primordium and regulates the expression of a subset PER genes),79 and RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) (which belongs to a family of NADPH oxidases that produce extracellular ROS).79,80 According to UniProt, all four designated peroxidases are cell-wall localized, while at least some members of the RBOH family, including the one encoded by the gene expressed in buckwheat explants at adventitious root induction, are plasma membrane proteins.
The ROS mode is considered to be especially important at the advanced stages of lateral root emergence, rather than at primordium specification.79,80 In our test system, the transcription of the genes encoding two ROS mode components, UPB1 (tr_4146, homologue of AT2G47270) and PER57 (tr_2938, homologue of AT5G17820), was increased after IAA addition at the early stage of adventitious root formation (Table S2). The transcription of the gene encoding RBOH (tr_6314, homologue of AT1G09090) was highly active in explanted hypocotyls even without the addition of IAA or OSRG and was maintained at a high level in the presence of these effectors. These findings indicate that the ROS regulatory mode may operate in buckwheat hypocotyl explants at the early stage of root development induction by auxin. Two genes encoding MYB36 (tr_20342 and tr_7854, homologs of AT5G57620) were activated at the second stage (Table S2). Such differences from lateral root development may occur because adventitious and lateral roots could have evolved from different cell types and may present differences in priming events.63,64 In addition, the involvement of the ROS mode in lateral root formation was studied in intact Arabidopsis roots, while in our test system, the wounding effect due to explant excision caused the formation of ROS at the very beginning of tissue reprogramming.
Among the known components of the ROS regulatory mode, the presence of oligosaccharin affects the transcription of two peroxidases (Figure 3). SRC2, which is encoded by a gene that is downregulated by OSRG (Figure 4), is known to affect at least one member of the RBOH family and to enhance its ROS-producing activity.81 Many other genes encoding proteins involved in redox regulation are also differentially expressed in the presence of OSRG. Significant activation of several genes encoding glutathione transferases was among the most obvious effects of OSRG, which occurred specifically at the earlier stage of adventitious root development. Glutathione transferases are soluble enzymes that are common in eukaryotic and prokaryotic species and are divided into several classes, designated as phi, tau, theta, etc.; in plant genomes, these enzymes are represented by large multigene families.82 Glutathione transferases conjugate the tripeptide glutathione (γ-Glu-Cys-Gly) to a broad range of molecules. OSRG stimulated the expression of four genes from this family (Figure 3), three of which have Arabidopsis homologs (AT1G78380 and AT3G09270) that belong to the tau group, which encodes a plant-specific class of glutathione transferases. Glutathione transferases are known to be involved in plant development under normal growth conditions, the reactions to various stresses and the detoxification of xenobiotics; they have also been shown to be auxin-binding proteins.83 Such binding may contribute to the temporary storage of auxin or its transport to its receptors. One of the functions of glutathione-S-transferase is to prevent cytotoxic events that may occur as a result of excessive accumulation of auxin molecules within cells.84 Thus, such enzymes can be involved in the intracellular modulation of both ROS and auxin action. As stated in a recent review,85 there are multiple functions of such enzymes in plant tissues, which are not well characterized and could still be underestimated. The expression of genes encoding other enzymes that may be involved in redox regulation (thioredoxin, metallothionein, cytochrome P450, dehydrogenase, oxidoreductase, electron transfer flavoprotein) is also upregulated by the action of oligosaccharin after 6 h of explant cultivation (Figure 3). ROS are known to act as signaling molecules in many cellular processes.86,87 Though neither all components of the ROS regulatory mode nor their interconnections have been established, it is clear that some of them localize to the cell wall (e.g., those affected by OSRG peroxidases) or the plasma membrane (the homologue of actively expressed RBOH has several cytoplasmic, transmembrane and extracellular domains). This is especially important when considering the possible mechanisms of oligosaccharin action. OSRG has a rather large size (Figure 1) and therefore cannot pass through the plasma membrane. The products of its possible hydrolysis (individual monosaccharide) do not exhibit an effect on IAA-induced adventitious root formation, at least at concentrations similar to the OSRG concentrations. Thus, the suggested site of OSRG action and the ROS regulatory mode partially overlap, giving rise to the possibility of their interaction. Most likely, oligosaccharin stimulates IAA-induced adventitious root formation in buckwheat hypocotyl explants through the action on the first stage of root development by changing the ROS level and regulating the operation of the ROS regulatory mode.
4.3. The sets of differentially expressed genes differed significantly at the two studied stages of adventitious root formation
The development of adventitious root involves the set of successive processes. The time points selected for the transcriptome analysis reflected different stages in the process of rhizogenesis occurring in excised hypocotyls: the first stage (6 h) encompasses the diverse processes involved in overcoming the consequences of explant cutting, founder cell specification, and maybe the first anticlinal/periclinal divisions, while at the second stage (24 h), the development of the dome-shaped primordium, which mainly involves cell division and growth inside the parental tissue before the appearance of a newly forming root on the hypocotyl surface, takes place.6,42 These stages are characterized by specific sets of genes that are differentially expressed in the presence of IAA and OSRG compared to the group treated with IAA alone, indicating the specificity of the processes that take place at each of the stages. For example, genes encoding several chaperones (heat shock proteins) are downregulated at 6 h (Figure 4) and upregulated at 24 h (Figure 5). The upregulation of the gene encoding the E3 ubiquitin-protein ligase MPSR1 at 24 h was notable (Figure 5). This enzyme recognizes emergent misfolded proteins independent of chaperones.88 The increased expression of the gene encoding this enzyme may indicate that in the second phase of the root formation process, activation of the protein degradation process through the 26S proteasome may be stimulated by OSRG, and this specific targeting of proteins for degradation may be an important mechanism of root formation.
The majority of differentially expressed genes identified in the presence of oligosaccharin were detected at the earlier stage of root development, in accordance with the data showing that OSRG has a maximum effect if added to the explants during early stages of IAA action or even in advance of IAA treatment.26 Rather unexpectedly, the genes encoding several ribosomal proteins were among these upregulated genes. There are not enough arguments to suggest the direct involvement of ribosomal proteins in adventitious root formation; however, our data are in accordance with an earlier report that a number of ribosomal protein-encoding gene mutants display a significant reduction in the number of adventitious roots that develop on the hypocotyl after whole root excision.89 Together, these data indicate the need for further research on the possible roles of ribosomal proteins in root development and plant morphogenesis in general.
Reprogramming of cell identities during development frequently requires changes in the chromatin state that need to be restricted to the correct cell populations, which has been well studied in animal cells.90 In plant cells, an auxin-regulated chromatin state switch that directs reprogramming from transit amplification to primordium founder cell fate in Arabidopsis inflorescences has been identified.91 The authors reporting this switch believe that this mechanism underlies additional auxin-controlled cell fate reprogramming events, for example, during embryo patterning and leaf morphogenesis. There may be a similar mechanism involved in reprogramming the fate of cells in the formation of the founder cells of the adventitious root primordium. A significant number of differentially expressed genes identified in the presence of oligosaccharin after 6 h of explant cultivation encoded proteins involved in chromatin reorganization and the remodeling of genetic material (histones, retrotransposons). Transposon-derived transcripts have been reported in several plant species, such as maize and rice; in rice roots, they are suggested to influence developmental processes through interactions with specific miRNAs.92
The emergence of adventitious roots requires the reorganization of cell walls, both in the primordium itself and in the tissues covering the primordia that favor its passage to the organ surface. Furthermore, lateral root induction is dependent on the mechanical properties of the overlying tissues that are modified by cell wall-modifying enzymes at the early stages of root development.93,94 The presence of OSRG resulted in the differential expression of several genes encoding cell wall proteins (according to the annotation of homologous genes in Arabidopsis): α-expansin, plant natriuretic peptide, peroxidase, lipid-transfer protein, α-galactosidase, and pectinesterase. The expression of tr_7228 (for α-expansin) was considerably activated by IAA and OSRG at the 6 h time point compared both to the control and the group treated with IAA alone; after 24 h, the expression of this gene decreased (Figure 4b), indicating particular importance at the early stage of primordium formation. The expression of tr_13010 (α-galactosidase) increased only after 24 h in auxin- and oligosaccharin-containing medium.
The tr_6863 gene (homologue of pectin esterase) was highly upregulated after 6 h in IAA-containing medium (as compared to the initial samples), irrespective of the presence of OSRG. At the later stage, oligosaccharin helped to maintain high expression of this gene, while its expression considerably decreased under treatment with IAA alone (Figure 6). Pectin demethylesterification performed by pectin esterases leads to the modification of pectin gel properties and often accompanies developmental properties.95 The degree of pectin methylation differs in the cell walls of the forming primordium and the cells of the tissues covering the primordium.96 Additional modification of cell wall polymers can occur due to the decrease in the expression of the tr_17148 gene, encoding the TRICHOME BIREFRINGENCE-LIKE (TBL44) protein, an acetyltransferase specific for the cell wall.61 O-acetylation affects rheological properties and may interfere with the enzymatic cleavage of cell wall polysaccharides.97 Therefore, the decrease in TBL44 gene expression induced by OSRG at this stage can reduce the level of the acetylation of wall polymers, making them more sensitive to hydrolytic enzymes, which will favor the passage of the primordium.
4.4. Toward the possible mechanisms of oligosaccharin effects
The analysis of transcriptome changes caused by the presence of OSRG in the IAA-containing medium used for buckwheat hypocotyl cultivation suggested the major pathways that lead to an increased number of adventitious roots developed in explants. Although these data are not sufficient to definitively establish the complete mechanisms and chain of events involved, they suggest some highlights regarding the molecular events associated with oligosaccharin action (Figure 7). We concentrate on components potentially associated with OSRG action and do not consider the expression of “classical” auxin-induced genes.
Figure 7.
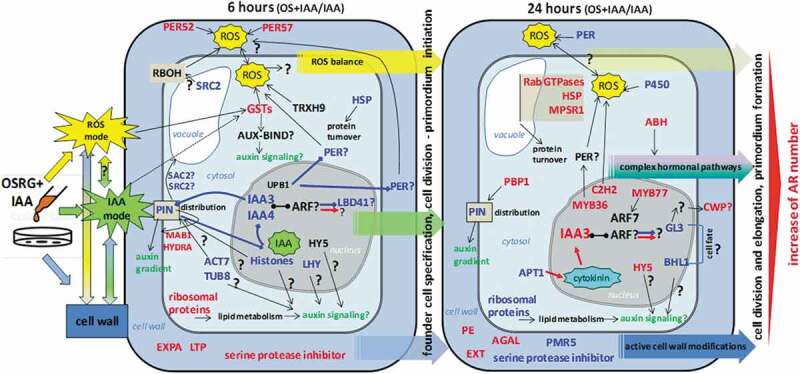
Simplified model summarizing the effect of OSRG on gene expression in the course of IAA-induced adventitious root formation in buckwheat hypocotyls. Differentially expressed genes in IAA+OSRG compared to IAA samples are presented after 6 and 24 of treatment; upregulated genes (FC>2) are indicated in red and downregulated genes (FC<0.5), in blue; if a gene did not show DE but its product may play an important role in the examined processes or interactions with other genes (proteins) according to the literature, it is indicated in black. To designate the association of gene products with processes or other gene products, black arrows are used. If a positive or negative association is clear (based on the literature), it is designated by a red or blue arrows, respectively. Most of the designated genes and their products are discussed in the manuscript. OSRG cannot pass through the plasma membrane as such, but it can affect targets associated with ROS and IAA regulatory modes. The ROS mode is more active at the early stage of the response (6 h). The ROS mode comprises cell wall peroxidases, RBOH, and others. Both of these modes could be associated with the changes in cell wall components. One of the types of targets that could be involved in the intracellular modulation of both ROS and auxin action is the glutathione transferases (GST). OSRG is suggested to affect the PIN distribution and, as a result, the auxin signaling pathway (IAA mode). The important regulators whose activity can be modulated by OSRG include IAA3, IAA4, UPB1, and LBD41 at the early stage and IAA3, MYB77, HY5, C2H2, GL3, and BHL1 at the later stage. The presence of OSRG increases the number of IAA-induced adventitious roots (ARs).
Apparently, oligosaccharin either stimulates an increase in the number of competent cells that give rise to adventitious roots or contributes to the maintenance of the auxin-initiated root primordia, leading to their full development since not all primordia initiated after auxin treatment develop into emergent roots.98 Herewith, oligosaccharin molecules are too large to be able to penetrate inside the cells; for example, labeled nonasaccharide is not taken up by cultured plant cells.99 OSRG rather acts on some targets located in the cell wall or on the outer plasma membrane side. Thus, OSRG does not affect gene transcription directly but provides conditions that change the regulatory modes involved in adventitious root development. There are two major regulatory modes involved in root branching that can be subjected to OSRG action: the ROS and auxin modes, which can also interact with each other. Notably, the ROS mode is manifested more strongly at the earlier stage, during founder cell specification and primordium initiation. One of the most prominent types of direct or indirect targets that could be involved in the intracellular modulation of both ROS and auxin action is the glutathione transferases, which seem to be quite important in plant development, but their exact influence on root initiation and development remains unclear. Both of these modes could contribute to the observed regulation of cell wall components, which also have an impact on cell proliferation and elongation.
No indication of a specific receptor or corresponding signaling cascade for OSRG is provided by our data. Rather, oligosaccharin can be suggested to interact with other effector receptors located at the plasma membrane or with the membrane itself. For example, the acidic oligosaccharins (oligogalacturonids) are responsible for the phosphorylation of the subclass of remorins, which are plant-specific proteins located in lipid rafts of the plasma membrane.100 The interactions of OSRG with membrane components may be increased by ROS or by members of the ROS regulatory mode. Changes in the lipid constituents of the membrane can be anticipated from the upregulation of the gene encoding HYDRA1, involved in the metabolism of sterols, in the presence of OSRG. Sterols are important components of the membrane that regulate its permeability and lateral mobility and influence the distribution of the PIN1 and PIN2 auxin efflux carriers.47 The detected changes in the expression of genes encoding the enzymes involved in lipid modifications of proteins can also contribute to such effects. Upregulation of the expression of ribosomal proteins at the early stage could influence lipid metabolism and link auxin-regulated developmental patterning with endomembrane trafficking.28 Fine-tuning of PIN recycling in the presence of OSRG is also suggested by changes in the expression of the genes encoding several proteins involved in vesicle trafficking and in the internalization of PIN proteins into vacuole.
Active chromatin reorganization and remodeling of genetic material are induced by OSRG, but again, the mediator is unknown. The behavior of the IAA3 and IAA4 genes was also noteworthy, which differed from the behavior observed under treatment with IAA alone. This fact may indicate some changes in auxin signaling and its influence on primordium initiation under OSRG action. Among the transcription factors that were upregulated under OSRG treatment, the most intriguing factors were HY5 and MYB77, both of which were activated at a late stage and could be involved in the regulation of the “late response” determining the emergence of adventitious roots on the surface of explants.
The results obtained in the current study contribute to the understanding of the mechanism of adventitious root formation in plants and demonstrate that OSRG is involved in the signaling pathway for the regulation of rhizogenesis, providing a number of promising scientific “primordia” for further investigation.
Funding Statement
This work was partially supported by the Russian Foundation for Basic Research (grant 17-04-01539, isolation and characterization of OSRG, transcriptome analysis) and by Russian Science Foundation (grant 18-76-10018, expression data analysis). The part of work (data interpretation) was performed by Larskaya I., Gorshkov O., Mokshina N., Trofimova O., Mikshina P., Gogoleva N, and Gorshkova T. at financial support from the government assignment for FRC Kazan Scientific Center of RAS. The study was carried out on the equipment of the CSF-SAC FRC KSC RAS.
Author Contributions
TG conceived, planned the project and supervised the research, wrote manuscript. IL prepared plant material, isolated, purified oligosaccharin fraction, drafted the manuscript. PM performed biochemical analysis of carbohydrate fraction, drafted the manuscript. OT prepared and treated buckwheat explants. NM isolated mRNA, prepared cDNA libraries, and drafted the manuscript. OG performed bioinformatics work, drafted the manuscript. NG performed Illumina HiSeq experiments. AK performed RNA-SEQ data analysis. Discussed the data: TG, IL, OG, NM, PM. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Atkinson JA, Rasmussen A, Traini R, Voß U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ.. Branching out in roots: uncovering form, function, and regulation. Plant Physiol. 2014;166:1–17. doi: 10.1104/pp.114.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steffens B, Rasmussen A.. The physiology of adventitious roots. Plant Physiol. 2016;170:603–617. doi: 10.1104/pp.15.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3331. [DOI] [PubMed] [Google Scholar]
- 4.Banda J, Bellande K, von Wangenheim D, Goh T, Guyomarch S, Laplaze L, Bennett MJ. Lateral root formation in Arabidopsis: a well-ordered LRexit. Trends Plant Sci. 2019;24:826–839. doi: 10.1016/j.tplants.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Napsucialy-Mendivil S, Dubrovsky JG. Genetic and phenotypic analysis of lateral root development in Arabidopsis thaliana. In: Ristova D, Barbez E, editors. Root development. Methods in molecular biology. New York (USA): Humana Press; 2018. p. 47–75. doi: 10.1007/978-1-4939-7747-5_4. [DOI] [PubMed] [Google Scholar]
- 6.Guan L, Murphy AS, Peer WA, Gan L, Li Y, Cheng Z-M. Physiological and molecular regulation of adventitious root formation. Crit Rev Plant Sci. 2015;34:506–521. doi: 10.1080/07352689.2015.1090831. [DOI] [Google Scholar]
- 7.De Klerk GJ, Keppel M, TerBrugge J, Meekes H. Timing of the phases in adventitious root formation in apple microcuttings. J Exp Bot. 1995;46:965–972. doi: 10.1093/jxb/46.8.965. [DOI] [Google Scholar]
- 8.Drew RA, McComb JA, Considine JA. Rhizogenesis and root growth ofCarica papaya L. invitro in relation to auxin sensitive phases and use of riboflavin. Plant Cell Tissue Organ Cult. 1993;33:1–7. doi: 10.1007/BF01997591. [DOI] [Google Scholar]
- 9.Druege U, Hilo A, Pérez-Pérez JM, Klopotek Y, Acosta M, Shahinnia F, Zerche S, Franken P, Hajirezaei MR. Molecular and physiological control of adventitious rooting in cuttings: phytohormone action meets resource allocation. Ann Bot. 2019;123:929–949. doi: 10.1093/aob/mcy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagnussat GC, Lanteri ML, Lombardo MC, Lamattina L. Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 2004;135:279–286. doi: 10.1104/pp.103.038554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagnussat GC, Lanteri ML, Lamattina L. Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 2003;132:1241–1248. doi: 10.1104/pp.103.022228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu L, Yu J, Liao W, Yu J, Zhang M, Dawuda MM. Calcium and calmodulin are involved in nitric oxide-induced adventitious rooting of cucumber under simulated osmotic stress. Front Plant Sci. 2017;8:1684. doi: 10.3389/fpls.2017.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li SW, Leng Y, Shi RF. Transcriptomic profiling provides molecular insights into hydrogen peroxide-induced adventitious rooting in mung bean seedlings. BMC Genomics. 2017;18:188. doi: 10.1186/s12864-017-3576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellincampi D, Salvi G, De Lorenzo G, Cervone F, Marfà V, Eberhard S, Darvill A, Albersheim P. Oligogalacturonides inhibit the formation of roots on tobacco explants. Plant J. 1993;4:207–213. doi: 10.1046/j.1365-313X.1993.04010207.x. [DOI] [Google Scholar]
- 15.Zabotina OA, Zabotin AI. Biologically active oligosaccharide functions in plant cell. In: Gordon NS, editor. Oligosaccharides: sources, properties and applications. Paris (France): Nova Press:; 2011. p. 209–243. [Google Scholar]
- 16.Kollárová K, Zelko I, Henselová M, Capek P, Liškova D. Growth and anatomical parameters of adventitious roots formed on mung bean hypocotyls are correlated with galactoglucomannan oligosaccharides structure. Scientific World J. 2012;2012:797815. doi: 10.1100/2012/797815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larskaya IA, Gorshkova TA. Plant oligosaccharides – outsiders among elicitors? Biochemistry (Moscow). 2015;80:881–900. doi: 10.1134/S0006297915070081. [DOI] [PubMed] [Google Scholar]
- 18.Eberhard S, Doubrava N, Marfa B, Mohnen V, Southwick D, Darvill A, Albersheim P. Pectic cell wall fragments regulate tobacco thin-cell-layer explant morphogenesis. Plant Cell. 1989;1:747–755. doi: 10.1105/tpc.1.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zabotina OA, Gurjanov OP, Ibragimova NN, Ayupova DA, Lozovaya VV. Rhizogenesis in buckwheat thin-cell-layer explants: effect of plant oligosaccharides. Plant Sci. 1998;135:195–201. doi: 10.1016/S0168-9452(98)00022-3. [DOI] [Google Scholar]
- 20.Ishii K, Kinoshita I, Shigenaga H. The effects of oligosaccharides on tissue culture of white birch and black pine. Trans Jpn For Soc. 1992;103:485–486. [Google Scholar]
- 21.Savatin DV, Ferrari S, Sicilia F, De Lorenzo G. Oligogalacturonide auxin antagonism does not require posttranscriptional gene silencing or stabilization of auxin response repressors in Arabidopsis. Plant Physiol. 2011;157:1163–1174. doi: 10.1104/pp.111.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki KI, Matsubara Y. Purification of pectate oligosaccharides showing root-growth-promoting activity in lettuce using ultrafiltration and nanofiltration membrane. J Biosci Bioeng. 2000;89:495–497. doi: 10.1016/S1389-1723(00)89104-5. [DOI] [PubMed] [Google Scholar]
- 23.Miranda JH, Williams RW, Kerven G. Galacturonic acid-induced changes in strawberry plant development in vitro. In Vitro Cell Dev Biol Plant. 2007;43:639–643. doi: 10.1007/s11627-007-9052-7. [DOI] [Google Scholar]
- 24.Bellincampi D, Cardarelli M, Zaghi D, Serino G, Salvi G, Gatz C, Cervone F, Altamura MM, Costantino P, De Lorenzo G. Oligogalacturonides prevent rhizogenesis in rolB-transformed tobacco explants by inhibiting auxin-induced expression of the rolB gene. Plant Cell. 1996;8:477–487. doi: 10.1105/tpc.8.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauro ML, De Lorenzo G, Costantino P, Bellincampi D. Oligogalacturonides inhibit the induction of late but not of early auxin-responsive genes in tobacco. Planta. 2002;215:494–501. doi: 10.1007/s00425-002-0772-y. [DOI] [PubMed] [Google Scholar]
- 26.Larskaya IA, Barisheva TS, Zabotin AI, Gorshkova TA. Character of oligosaccharin OS-RG participation in the IAA-induced formation of adventitious roots. Russ J Plant Physiol. 2015;62:171–178. doi: 10.1134/S1021443715020120. [DOI] [Google Scholar]
- 27.Bellincampi D, Dipierro N, Salvi G, Cervone F, De Lorenzo G. Extracellular H2O2 induced by oligogalacturonides is not involved in the inhibition of the auxin-regulated rolB gene expression in tobacco leaf explants. Plant Physiol. 2000;122:1379–1385. doi: 10.1104/pp.122.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SW, Shi RF, Leng Y. De novo characterization of the mung bean transcriptome and transcriptomic analysis of adventitious rooting in seedlings using RNA-Seq. PLoS One. 2015;10:e0132969. doi: 10.1371/journal.pone.0132969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villacorta-Martín C, Sánchez-García AB, Villanova J, Cano A, van de Rhee M, de Haan J, Acosta M, Passarinho P, Pérez-Pérez JM. Gene expression profiling during adventitious root formation in carnation stem cuttings. BMC Genomics. 2015;16:789. doi: 10.1186/s12864-015-2003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Druege U, Franken P, Lischewski S, Ahkami AH, Zerche S, Hause B, Hajirezaei MR. Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in petunia cuttings. Front Plant Sci. 2014;5:494. doi: 10.3389/fpls.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quan J, Meng S, Guo E, Zhang S, Zhao Z, Yang X. De novo sequencing and comparative transcriptome analysis of adventitious root development induced by exogenous indole-3-butyric acid in cuttings of tetraploid black locust. BMC Genomics. 2017;18:179. doi: 10.1186/s12864-017-3554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei K, Wang LY, Wu LY, Zhang CC, Li HL, Tan LQ, Cao HL, Cheng H. Transcriptome analysis of indole-3-butyric acid-induced adventitious root formation in nodal cuttings of Camellia sinensis (L.). PLoS One. 2014;9:e107201. doi: 10.1371/journal.pone.0107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moscatiello R. Transcriptional analysis of calcium-dependent and calcium-independent signalling pathways induced by oligogalacturonides. J Exp Bot. 2006;57:2847–2865. doi: 10.1093/jxb/erl043. [DOI] [PubMed] [Google Scholar]
- 34.Davidsson P, Broberg M, Kariola T, Sipari N, Pirhonen M, Palva ET. Short oligogalacturonides induce pathogen resistance-associated gene expression in Arabidopsis thaliana. BMC Plant Biol. 2017;17:19. doi: 10.1186/s12870-016-0959-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lozovaya VV, Zabotina OA, Rumyantzeva NI, Malihov RG, Zihareva MV. Stimulation of root development of buckwheat thin cell-layer explants by pectic fragments from pea stem cell wall. Plant Cell Rep. 1993;12:530–533. doi: 10.1007/BF00236102. [DOI] [PubMed] [Google Scholar]
- 36.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 37.Logacheva MD, Kasianov AS, Vinogradov DV, Samigullin TH, Gelfand MS, Makeev VJ, Penin AA. De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genomics. 2011;12:30. doi: 10.1186/1471-2164-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA‐seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott RW, Moore WE, Effland MJ, Millett MA. Ultraviolet spectrophotometric determination of hexoses, pentoses, and uronic acids after their reactions with concentrated sulfuric acid. Anal Biochem. 1967;21:68–80. doi: 10.1016/0003-2697(67)90084-X. [DOI] [PubMed] [Google Scholar]
- 41.Ito A, Ito T. Absorption spectra of deoxyribose, ribosephosphate, ATP and DNA by direct transmission measurements in the vacuum-UV (150-190 nm) and far-UV (190-260 nm) regions using synchrotron radiation as a light source. Photochem Photobiol. 1986;44:355–358. doi: 10.1111/j.1751-1097.1986.tb04675.x. [DOI] [PubMed] [Google Scholar]
- 42.Li SW, Shi RF, Leng Y, Zhou Y. Transcriptomic analysis reveals the gene expression profile that specifically responds to IBA during adventitious rooting in mung bean seedlings. BMC Genomics. 2016;17:43. doi: 10.1186/s12864-016-2372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omelyanchuk NA, Wiebe DS, Novikova DD, Levitsky VG, Klimova N, Gorelova V, Weinholdt C, Vasiliev GV, Zemlyanskaya EV, Kolchanov NA, et al. Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci Rep. 2017;7:2489. doi: 10.1038/s41598-017-02476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batoko H, Zheng HQ, Hawes C, Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell. 2000;12:2201–2217. doi: 10.2307/3871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrowski M, Hetmann A, Jakubowska A. Indole-3-acetic acid UDP-glucosyltransferase from immature seeds of pea is involved in modification of glycoproteins. Phytochem. 2015;117:25–33. doi: 10.1016/j.phytochem.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 46.Ohbayashi I, Huang S, Fukaki H, Song X, Sun S, Morita MT, Tasaka M, Millar AH, Furutani M. Mitochondrial pyruvate dehydrogenase contributes to auxin-regulated organ development. Plant Physiol. 2019;180:896–909. doi: 10.1104/pp.18.01460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Short E, Leighton M, Imriz G, Liu D, Cope-Selby N, Hetherington F, Smertenko A, Hussey PJ, Topping JF, Lindsey K. Epidermal expression of a sterol biosynthesis gene regulates root growth by a non-cell-autonomous mechanism in Arabidopsis. Development. 2018;145:dev160572. doi: 10.1242/dev.160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nováková P, Hirsch S, Feraru E, Tejos R, van Wijk R, Viaene T, Heilmann M, Lerche J, De Rycke R, Feraru MI, et al. SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:2818–2823. doi: 10.1073/pnas.1324264111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oufattole M, Park JH, Poxleitner M, Jiang L, Rogers JC. Selective membrane protein internalization accompanies movement from the endoplasmic reticulum to the protein storage vacuole pathway in Arabidopsis. Plant Cell. 2005;17:3066–3080. doi: 10.1105/tpc.105.035212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vigneault F, Lachance D, Cloutier M, Pelletier G, Levasseur C, Séguin A. Members of the plant NIMA‐related kinases are involved in organ development and vascularization in poplar, Arabidopsis and rice. Plant J. 2007;51:575–588. doi: 10.1111/j.1365-313X.2007.03161.x. [DOI] [PubMed] [Google Scholar]
- 51.Zeidman R, Jackson CS, Magee AI. Protein acyl thioesterases (Review). Mol Membr Biol. 2009;26:32–41. doi: 10.1080/09687680802629329. [DOI] [PubMed] [Google Scholar]
- 52.Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lytle BL, Song J, de la Cruz NB, Peterson FC, Johnson KA, Bingman CA, Phillips GN, Volkman BF. Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds. Proteins. 2009;76:237–243. doi: 10.1002/prot.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chrost B, Kolukisaoglu U, Schulz B, Krupinska K. An alpha-galactosidase with an essential function during leaf development. Planta. 2007;225:311–320. doi: 10.1007/s00425-006-0350-9. [DOI] [PubMed] [Google Scholar]
- 55.Tehseen M, Cairns N, Sherson S, Cobbett CS. Metallochaperone-like genes in Arabidopsis thaliana. Metallomics. 2010;2:556–564. doi: 10.1039/C003484C. [DOI] [PubMed] [Google Scholar]
- 56.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol. 2016;26:640–646. doi: 10.1016/j.cub.2015.12.066. [DOI] [PubMed] [Google Scholar]
- 59.Bernhardt C. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- 60.Mittler R, Kim YS, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2002;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, preins with a TBL and DUF231 domain. Plant Cell. 2011;23:4041–4053. doi: 10.1105/tpc.111.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian Q, Reed J. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. [DOI] [PubMed] [Google Scholar]
- 63.Bellini C, Pacurar DI, Perrone I. Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol. 2014;65:639–666. doi: 10.1146/annurev-arplant-050213-035645. [DOI] [PubMed] [Google Scholar]
- 64.Verstraeten I, Schotte S, Geelen D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front Plant Sci. 2014;5:495. doi: 10.3389/fpls.2014.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA–ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signaling. Phil Trans R Soc B. 2012;367:1461–1468. doi: 10.1098/rstb.2011.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DelloIoio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 67.Chapman EJ, Estelle M. Cytokinin and auxin intersection in root meristems. Genome Biol. 2009;10:210. doi: 10.1186/gb-2009-10-2-210. [DOI] [Google Scholar]
- 68.Zhang X, Chen Y, Lin X, Hong X, Zhu Y, Li W, He W, An F, Guo H. Adenine phosphoribosyl transferase 1 is a key enzyme catalyzing cytokinin conversion from nucleobases to nucleotides in Arabidopsis. Mol Plant. 2013;6:1661–1672. doi: 10.1093/mp/sst071. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt RE, Ainley WM, Nagao RT, Conner TW, Key JL. Expression of the Arabidopsis AtAux2–11 auxin-responsive gene in transgenic plants. Plant Mol Biol. 1993;22:731–749. doi: 10.1007/BF00027361. [DOI] [PubMed] [Google Scholar]
- 70.Caldelari D, Sternberg H, Rodríguez-Concepción M, Gruissem W, Yalovsky S. Efficient prenylation by a plant geranylgeranyltransferase-I requires a functional CaaL box motif and a proximal polybasic domain. Plant Physiol. 2001;126:1416–1429. doi: 10.1104/pp.126.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson CD, Narasimha Chary S, Chernoff EA, Zeng Q, Running MP, Crowell DN. Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol. 2005;139:722–733. doi: 10.1104/pp.105.065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng C, Andressson E, Maslak A, Mock HP, Mattsson O, Mundy L. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004;167:1099–1107. doi: 10.1016/j.plantsci.2004.06.014. [DOI] [Google Scholar]
- 73.Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- 74.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- 76.Benjamins R, Galvan Ampudia CS, Hooykaas PJJ, Offringa R. PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 2003;132:1623–1630. doi: 10.1104/pp.103.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abas L, Benjamins R, Malenica N, Paciorek T, Wišniewska J, Moulinier–Anzola JC, Sieberer T, Friml J, Luschnig C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol. 2006;8:249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- 78.Torres-Martínez HH, Rodríguez-Alonso G, Shishkova S, Dubrovsky JG. Lateral root primordium morphogenesis in angiosperms. Front Plant Sci. 2019;10:206. doi: 10.3389/fpls.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manzano C, Pallero-Baena M, Casimiro I, De Rybel B, Orman-Ligeza B, Van Isterdael G, Beeckman T, Draye X, Casero P, Del Pozo JC. The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiol. 2014;165:1105–1119. doi: 10.1104/pp.114.238873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orman-Ligeza B, Parizot B, de Rycke R, Fernandez A, Himschoot E, Van Breusegem F, Bennett MJ, Périlleux C, Beeckman T, Draye X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development. 2016;143:3328–3339. doi: 10.1242/dev.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawarazaki T, Kimura S, Iizuka A, Hanamata S, Nibori H, Mishikawa M, Imai A, Abe M, Kaya H, Kuchutsu K. A low temperature-inducible protein AtSRC2 enhances the ROS-producing activity of NADPH oxidase AtRbohF. Biochim Biophys Acta Mol Cell Res. 2013;1833:2775–2780. doi: 10.1016/j.bbamcr.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 82.Dixon DP, Edwards R. Glutathione transferases. Arabidopsis Book. 2010;8:e0131. doi: 10.1199/tab.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bilang J, Macdonald H, King PJ, Sturm A. A soluble auxin-binding protein from Hyoscyamus muticus is a glutathione S-transferase. Plant Physiol. 1993;102:29–34. doi: 10.1104/pp.102.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- 85.Sylvestre-Gonon E, Law SR, Schwartz M, Robe K, Keech O, Didierjean C, Dubos C, Rouhier N, Hecker A. Functional, structural and biochemical features of plant serinyl-glutathione transferases. Front Plant Sci. 2019;10:608. doi: 10.3389/fpls.2019.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 87.Singh R, Singh S, Parihar P, Mishra RK, Tripathi DK, Singh VP, Chauhan DK, Prasad SM. Reactive oxygen species (ROS): beneficial companions of plants’ developmental processes. Front Plant Sci. 2016;7:1299. doi: 10.3389/fpls.2016.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim JH, Cho SK, Oh TR, Ryu MY, Yang SW, Kim WT. MPSR1 is a cytoplasmic PQC E3 ligase for eliminating emergent misfolded proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2017;114:E10009–E10017. doi: 10.1073/pnas.1713574114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ibáñez S, Ruiz-Cano H, Fernández MÁ, Sanchez-Garcia AB, Villanova J, Micol JL, Perez-Perez JM. A network-guided genetic approach to identify novel regulators of adventitious root formation in Arabidopsis thaliana. Front Plant Sci. 2019;10:461. doi: 10.3389/fpls.2019.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Genetics. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cho J, Paszkowski J. Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife. 2017;6:e30038. doi: 10.7554/eLife.30038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell. 2013;25:3329–3346. doi: 10.1105/tpc.113.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lucas M, Kenobi K, von Wangenheim D, Voβ U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl.Acad.Sci USA. 2013;110:5229–5234. doi: 10.1073/pnas.1210807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolf S, Mouille G, Pelloux J. Homogalacturonan methyl-esterification and plant development. Mol Plant. 2009;2:851–860. doi: 10.1093/mp/ssp066. [DOI] [PubMed] [Google Scholar]
- 96.Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006;47:788–792. doi: 10.1093/pcp/pcj043. [DOI] [PubMed] [Google Scholar]
- 97.Biely P, Mackenzie C, Puls J, Schneider H. Cooperativity of esterases and xylanases in the enzymatic degradation of acetylxylan. Biotechnology. 1986;4:731–733. doi: 10.1038/nbt0886-731. [DOI] [Google Scholar]
- 98.Mitsuhashi-Kato M, Shibaoka H, Shimokoriyama M. The nature of the dual effect of auxin on root formation in Azukia cuttings. Plant Cell Physiol. 1978;19:1535–1542. doi: 10.1093/oxfordjournals.pcp.a075738. [DOI] [Google Scholar]
- 99.Smith RC, Fry SC. Endotransglycosylation of xyloglucans in plant cell suspension cultures. Biochem J. 1991;279:529–535. doi: 10.1042/bj2790529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marín M, Ott T. Phosphorylation of intrinsically disordered regions in remorin proteins. Front Plant Sci. 2012;3:86–91. doi: 10.3389/fpls.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


