Abstract
The past decade has seen a technological revolution in human genetics that has empowered population-level investigations into genetic associations with phenotypes. Although these discoveries rely on genetic variation across individuals, association studies have overwhelmingly been performed in populations of European descent. In this review, we describe limitations faced by single-population studies and provide an overview of strategies to improve global representation in existing data sets and future human genomics research via diversity-focused, multiethnic studies. We highlight the successes of individual studies and meta-analysis consortia that have provided unique knowledge. Additionally, we outline the approach taken by the Population Architecture Using Genomics and Epidemiology (PAGE) study to develop best practices for performing genetic epidemiology in multiethnic contexts. Finally, we discuss how limiting investigations to single populations impairs findings in the clinical domain for both rare-variant identification and genetic risk prediction.
Keywords: diversity, genomics, PAGE, multiethnic, fine mapping, transethnic
1. INTRODUCTION
1.1. Foundations of Large-Scale Genomic Studies
We are now entering the second decade of large-scale genome-to-phenome studies of complex human traits and diseases. Each new genetic finding can shed light on the underlying mechanisms of disease and ultimately inform targeted prevention and treatment strategies. Tremendous progress has been made in cataloging thousands of variants associated with numerous complex phenotypes through genome-wide association studies (GWASs). This work was enabled by large-scale investments in the early 2000s that became the foundation for subsequent discoveries. First, the International HapMap Project (61) created a common reference panel for mapping globally shared common genetic variation and revealed population-specific patterns of correlated variants, known as linkage disequilibrium (LD). These data were used to design the first set of genotyping platforms that assayed hundreds of thousands to millions of single-nucleotide polymorphisms (SNPs), thereby enabling cost-effective measurement of common genetic variation in thousands of participants. Strategies that leverage SNPs on genotyping arrays to infer unobserved variants at other positions by imputing LD structures in reference sequencing panels empowered an even broader exploration of variants with relatively cheap technology (21, 122).However, because allele frequencies and LD patterns differ across ancestries, the accuracy of imputation depends on how representative the genotyping markers and available reference sequencing panels are of the study population (21, 122).
Another logistical feat was powering GWAS discoveries through the assembly of massive consortia to bring together the hundreds of thousands of participants necessary for robust identification of genetic variants of complex disease. It is now abundantly clear that most common human diseases have highly complex genetic architectures, with hundreds or even thousands of genetic variants contributing to risk (17, 108, 126). The polygenicity of common complex traits means that most individual associated variants contribute a subtle effect and explain only a small proportion of the overall phenotype heritability. Consequently, achieving the necessary statistical power to detect the remaining variant–phenotype associations requires very large sample sizes (77). Given that GWASs mainly evaluate common variants, it is likely that much of the missing heritability is explained by a combination of rare variants [minor allele frequency (MAF) < 0.01], weak effects that require massive studies to uncover, and structural variants that are often poorly tagged by genotyping platforms. Thus, fully understanding the genetic architecture of complex diseases will require large-scale whole-genome sequencing studies to investigate all types of genetic variation in globally inclusive populations (detailed in Section 1.3).
1.2. European Bias in Large-Scale Genomic Research
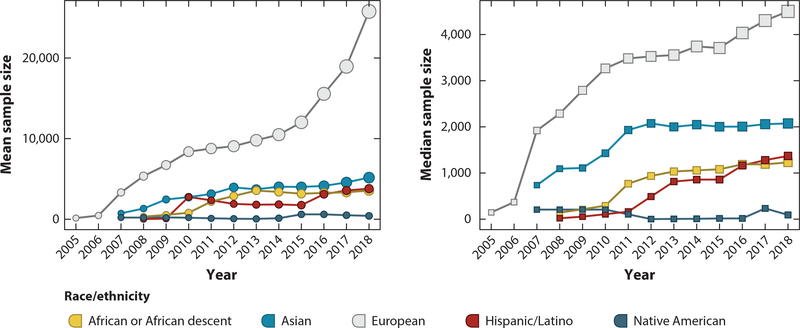
Despite impressive achievements in identifying genetic associations, the vast majority of results have been reported in populations of European ancestry, a limitation acknowledged by researchers a decade ago (19, 89). This Euro-centric bias has persisted, with more than 80% of GWAS participants being of European descent (94) and the largest studies performed in Europeans (Figure 1). Over the past decade, the mean and median sample sizes of published GWASs have skyrocketed for European-descent populations, driven mainly by cheaper technology and large-scale biobanking efforts such as the UK Biobank. However, the sample sizes of non-European descent studies have stagnated, resulting in limited statistical power for genomic discovery. There are myriad historical, technical, and logistical reasons for this lack of global diversity in genomic research. The first decade of GWASs focused predominantly on cohorts sampled in Europe or ongoing prospective US cohorts, and in 2001 federal guidelines were implemented that required the inclusion of women and minorities in clinical research, recognizing this gap in diversity. Consequently, many of today’s ongoing cohorts comprise predominantly European-descent participants, and even the participants of the more recent Genotype-Tissue Expression (GTEx) project are 85.2% of European origin (49). Several factors contributing to this bias have been detailed in other reviews and are briefly summarized here. Past mistreatment of marginalized communities has resulted in the documented mistrust of biomedical research and difficulty in recruiting more diverse populations (2). Other reports have cited logistical issues, such as the difficulty of recruiting underrepresented communities (118). Importantly, there is also a persistent lack of diversity among biomedical researchers leading, designing, and informing genomic studies (26, 84). Other recent reviews provide more complete perspectives on various strategies for increasing resources, improving the research culture, and modifying infrastructure to incorporate diversity into research (16, 56, 57, 99). Here, we focus on exemplars that highlight the scientific and clinical importance of diversifying genomic research.
Figure 1.
Cumulative mean and median sample sizes by racial/ethnic group across all traits in published genome-wide association studies from 2005 to 2018, as curated by the National Human Genome Research Institute–European Bioinformatics Institute GWAS Catalog. While the mean and median sample sizes of European-descent studies have grown over the past 13 years, all other groups have remained relatively stagnant. This is especially true for mega-scale biobanking efforts, such as the UK Biobank, which significantly raises the mean sample size. This limits statistical power for discovery and contributes to the resulting information bias in the published literature.
1.3. Global Perspectives on Human Variation
Differences in disease burden across ancestrally diverse populations are a major cause of health disparities. In the United States, African Americans experience the highest prevalence of hypertension and cardiovascular conditions and suffer the highest mortality rates for cardiovascular disease and renal failure (22, 25, 102). Mexican Americans in particular, but also African Americans, have a greater risk of developing liver disease than non-Hispanic European Americans (64, 101, 104). Non-Hispanic European American women have the highest incidence of breast cancer, but African American women are more likely to die from the disease, as their breast tumors are typically more aggressive and less responsive to treatments (90, 116). While lifestyle, cultural norms, health-care access, and socioeconomic status are undeniably important contributors to the disproportionate disease burden across racial/ethnic groups, many of the health disparities persist even after accounting for differences in social and environmental risk factors. This suggests that population-specific disease susceptibility also has innate biological, and thus genetic, causes that interact in a complex way with environmental factors.
Although most genetic studies have focused on common genetic variants, population genomics theory and empirical evidence from large, diverse sequencing efforts indicate that the vast majority of human genetic variation is rare and is expected to be population specific (48). This was affirmed by both the US National Heart, Lung, and Blood Institute’s Exome Sequencing Project study of European American and African American exomes (42) and the 1000 Genomes Project (1). As such, addressing European bias in genetic research is a major imperative, as we are currently unable to fully discover the genetic underpinnings of disease and the proportion that contributes to disparity.
The examples above highlight the importance of characterizing genetic variation among individuals of diverse ancestral backgrounds to gain a better understanding of differential susceptibility to disease, or variability in therapeutic response. While the focus of this review has thus far been on perspectives within the United States, it is important to acknowledge that global genetic variation is not well captured by populations in America. In fact, there is more genetic diversity across the more than 2,000 ethnolinguistic groups in Africa than anywhere else in the world because of population demographic history (e.g., population bottlenecks, short- and long-range migrations, and admixture) and dramatic variations in climate, diet, and exposures to infectious disease (20). However, much of what is currently known about genetic diversity and its contribution to disease comes from only a few ethnolinguistic groups (mostly from western Africa), severely limiting our understanding.
Resistance to malaria is a well-established example of strong selective pressure that has influenced genetic diversity in African populations. This disease is a major cause of mortality in sub-Saharan Africa, resulting in more than 1 million deaths (primarily children) each year (73). Genetic adaptations resulting in malaria resistance have become established in endemic regions, frequently accompanied by consequences in the homozygous state. For instance, the HbS mutation in the β-globin gene, which causes sickle cell disease in homozygous individuals, also confers protection against malarial infection in heterozygous carriers (50, 73). Similarly, variations in the G6PD gene are found in high frequency in African as well as Mediterranean and Asiatic populations, with patterns of variation consistent with recent positive selection. Even though deleterious mutations in G6PD cause diseases such as chronic hemolysis, high levels of frequency for such mutations are believed to be maintained in certain populations in response to selective pressure caused by malaria (85). This hypothesis is consistent with observations of high correlations between low-activity G6PD alleles and a decreased prevalence of malaria (100, 103, 114).
Importantly, for the benefits of precision medicine to be realized on a global scale, genetic epidemiological and pharmacogenetic studies must be more inclusive. Many examples demonstrating this necessity have already been identified. For example, human immunodeficiency virus (HIV) infection remains a major global health burden; nearly 37 million people are living with this disease, 53% of whom are in eastern and southern Africa. Even with the remarkable advancements in combination antiretroviral therapy, nearly a million people die every year from acquired immune deficiency syndrome (AIDS)–related illnesses worldwide 117). Although central to first-line antiretroviral therapy, efavirenz is associated with a high frequency of side effects and adverse drug reactions, including dizziness, insomnia, rash, hepatotoxicity, lipodystrophy, and several neuropsychiatric symptoms (including suicidal thoughts). Most of the variability in drug response is due to genetic variation in the metabolizing enzyme CYP2B6 (29). The c.516G>T and c.983T>C variants are predictive of reduced enzyme activity and remain the most prominent predictors of plasma concentrations (113). The CYP2B6*6 allele is more frequent and relevant in African populations, where it was reported at frequencies of 32.8% and 46.9% in African Americans and a Ghanaian population, respectively. This same allele was found to be present in 25.6% of European populations and in 15.9–18.0% of Asians (69). Importantly, many genetic variants have also been identified in African populations that confer resistance to this devastating disease, including those in killer immunoglobin-like receptors (KIRs) (62), interferon regulatory factor 1 (IRF-1) (8),and tripartite motif-containing protein 5α (TRIM5α) (74). Such findings can provide important insights for the development of new drugs.
Some successful initiatives have promoted genomic research in diverse global populations, including the 1000 Genomes Project (1), the Human Heredity and Health in Africa (H3Africa; http://h3africa.org) initiative (52), and the Mexico National Institute of Genomic Medicine (INMEGEN) (86). This list is by no means exhaustive, but it does demonstrate the importance of discovery of genetic variation across human populations to enable advances in precision medicine.
The 1000 Genomes Project was developed to provide a comprehensive description of common human genetic variation by applying whole-genome sequencing to a diverse set of individuals from multiple populations (1). Distinguished scientists from institutes at countries around the world, including China, Italy, Japan, Kenya, Nigeria, Peru, the United Kingdom, and the United States, reconstructed the genomes of 2,504 individuals from 26 populations using a combination of low-coverage whole-genome sequencing, deep exome sequencing, and dense microarray genotyping. They described a variable distribution of genetic variation across the global sample, with data freely available to the public and research community on various platforms, including through the project website and through Amazon Web Services, a cloud-computing system hosted by online retailer Amazon.com.
The INMEGEN project studied genomic variation within Mexico from more than 1,000 individuals representing 20 indigenous and 11 mestizo populations and described striking genetic variability (86). Indeed, several populations within Mexico displayed more differentiation than was observed between Europeans and East Asian reference populations.
The H3 Africa initiative was created to address the lack of large-scale genomic studies in Africa (52). Such work will contribute to large-scale pharmacogenomic studies in Africa that can provide a deeper understanding of variation in drug response. Although precision medicine may be most effective and beneficial to regions with high genetic diversity, such as Africa, many additional challenges exist in resource-poor environments. Implementation of pharmacogenomic practices is therefore unlikely to result in a sustainable health program in Africa without substantial new efforts. The African Pharmacogenomics Consortium aims to coordinate and to be the main driver in establishing pharmacogenomics guidelines in Africa.
2. MULTIETHNIC POPULATIONS MUST BE CONSIDERED AT EVERY STAGE OF A STUDY, FROM RECRUITMENT TO DATA MANAGEMENT AND ANALYSES
2.1. Increasing Diversity in Study Populations
For reasons outlined above, an intentional paradigm shift in genomic research is necessary to capture and leverage all aspects of diversity in the study of common and rare diseases. So far, studies aimed at being more inclusive have done so via two common approaches: (a) creating a new cohort or case–control study with recruitment in more diverse communities or geographic locations, and (b) performing a transethnic meta-analysis that gathers results from multiethnic participants in existing studies. We provide examples of these approaches and discuss study design specifics for researchers interested in conducting multiethnic studies and using algorithms that address greater genetic diversity.
Long-standing studies such as the Women’s Health Initiative (124) and the Multiethnic Cohort Study (72) have recruited hundreds of thousands of participants. Recruiting such large study populations in cosmopolitan settings in racially/ethnically diverse regions of the United States helps to ensure that many diverse groups are represented. Several smaller multiethnic studies have employed targeted recruitment strategies to increase representation of non-European groups, including the Multi-Ethnic Study of Atherosclerosis (15), the Atherosclerosis Risk in Communities Study (3), and the Consortium on Asthma Among African-Ancestry Populations in the Americas (82). Additionally, studies such as the Jackson Heart Study and the Hispanic Community Health Study/Study of Latinos have recruited participants from specific understudied ethnic groups (105, 110). In each context, and in each community, it is important to develop trust and address concerns to ensure that historically disadvantaged communities do not suffer incidental harm and that work is performed collaboratively with the community and all stakeholders.
In parallel, there has been interest in moving beyond single-study discoveries to combining numerous smaller studies in a meta-analysis framework, which combines summary statistics to gain power. The first major multiethnic meta-analysis, with comparable sample sizes across multiple ethnic groups, was performed by the EVE Consortium in a genetic susceptibility study of asthma that involved 10 studies from three ethnic groups in the Americas (European Americans, African Americans, and Latin Americans) (115). This study found both improved power at shared loci (e.g., 17q21, with strong shared signals across groups) and new population-specific associations (e.g., PYHIN1 in populations of African descent). There are numerous similar ongoing efforts with type 2 diabetes within the Diabetes Genetics Replication and Meta-Analysis Consortium (87), various psychiatric traits, and many other disease domains.
A new opportunity has been presented by the recent growth of biobanks linked with electronic health records. Covering the breadth of the patient base in multiple health systems provides a way to recruit relatively quickly and increase communication with communities that may be less likely to volunteer for a traditional cohort study. Similarly, as recruitment can be less labor intensive in this approach than in traditional epidemiological contexts, it is possible to develop sizable studies relatively quickly, as can be seen in the large repositories of national-level initiatives (such as the UK Biobank and Million Veteran Program) as well as the large and diverse patient bases at cosmopolitan academic medical centers [such as the Icahn School of Medicine at Mount Sinai (BioMe); the University of California, Los Angeles; the University of Colorado; and Vanderbilt University (BioVU)], which have genotyped tens to hundreds of thousands of patients. The UK Biobank includes more than 500,000 UK participants, and more than 35,000 individuals are of nonwhite British descent, providing a reasonably large sample size for transethnic analyses. Despite access to a diverse study population, UK Biobank studies to date have continued to focus primarily on individuals with mostly European ancestry, so there remain many opportunities for discovery.
2.2. Genotyping Ascertainment
Combining results across ethnic groups can be challenging given genetic, environmental, sociocultural, and study-based heterogeneity. Therefore, the field must develop and test new tools specifically designed for these contexts. The Population Architecture Using Genomics and Epidemiology (PAGE; http://pagestudy.org) study was formed with the goal of developing best practices for transethnic studies. This study has been continuously funded by the National Institutes of Health since 2008 to study genomic variation in order to advance our understanding of the population architecture of genetic traits and disease in the presence of ancestral diversity. The first phase of the study, which ran from 2008 to 2013 and was funded by the National Human Genome Research Institute and the National Institute of Mental Health, examined putative causal genetic variants across approximately 100,000 African Americans, Asian Americans, Native Americans, Hispanics/Latinos, and Native Hawaiians from four centers representing nine large United States–based cohorts. Two genotyping approaches were employed: targeted genotyping of selected SNPs identified in GWASs of common disease, and a large-scale array-based effort using the Metabochip to facilitate transethnic fine mapping of several diseases of public health importance. The Metabochip array is a custom genotyping array designed for replication and fine mapping of cardiometabolic traits (121).Early PAGE work demonstrated that, while most risk loci identified from GWASs and populations of primarily European ancestry are shared across one or more ethnic groups, the underlying causal variants and their effects vary across populations (21).
The second phase of PAGE, from 2013 to 2019, was funded by the National Human Genome Research Institute and the National Institute on Minority Health and Health Disparities and focused on how ancestry-specific differences in allele frequencies and LD could explain differences in risks of common traits and conditions. However, in 2013 there was only limited availability of genotyping arrays that could comprehensively capture variation across multiple genetic ancestries simultaneously. The majority of arrays for diverse populations were developed for a single group at a time, not a multiethnic sample. To address this issue, PAGE investigators partnered with the Consortium on Asthma Among African-Ancestry Populations in the Americas, Illumina, and other academic centers to develop the Multi-Ethnic Genotyping Array (MEGA). This platform utilized data from phase 3 of the 1000 Genomes Project with equal representation of non-European ancestries and was designed to have comparable imputation accuracy across all major populations, regardless of a given population’s level of admixture (14, 63, 122). To augment the capture of low-frequency variants, enhanced exome content was selected from available non-European exome sequencing studies (specifically, studies of Hispanic/Latino and African-American populations). MEGA is now commercially available for the wider research community (https://www.pagestudy.org/mega).
The current generation of genotyping arrays encompasses both population-specific and multiethnic products. To address a growing interest in biobank study design, especially with a diverse catchment area, an assortment of arrays have been developed to capture variation across multiple populations at once, including Illumina’s MEGA, Genome Screening Array, and upcoming Global Diversity Array as well as Affymetrix’s Precision Medicine Research Array. For homogeneous study designs, population-specific arrays have also been developed over the past several years to cover both continental and country-level populations, such as Illumina’s H3Africa Consortium Array (60) and Infinium OmniZhongHua arrays, as well as Affymetrix’s Biobank Genotyping and Axiom World arrays. It should be noted that, despite this progress, the development of these arrays is dependent upon available reference panels, and therefore many regions of the world are underrepresented. For example, the African variation present on MEGA is largely representative of western Africa and therefore does not offer equal coverage in eastern African populations.
2.3. Imputation
Imputation of genotyped samples to improve resolution and capture variation is now standard practice for many genomic studies, facilitated by the availability of large-scale, publicly available reference data and the development of faster, improved imputation methods (33). However, the accuracy of imputation is highly dependent on the selection and availability of reference data representative of the study population of interest (119). After the International HapMap Project, the 1000 Genomes Project aimed to provide a comprehensive description of common human genetic variation by applying whole-genome sequencing to 2,504 individuals from 26 populations. These efforts revealed that, in order to study rarer genetic variants, international efforts would be necessary to aggregate and harmonize whole-exome and whole-genome sequencing data. Indeed, these efforts have already begun with the Haplotype Reference Consortium (HRC) and Genome Aggregation Database (gnomAD) projects. Following the 1000 Genomes Project, the HRC created a large reference imputation panel by combining sequences from multiple cohorts with the aim of improving genotype imputation in other GWAS cohorts. In comparison with past panels, this resource improves imputation accuracy—particularly for low-frequency variants—and has led to several new discoveries.
The Michigan Imputation Server and Sanger Imputation Server have further advanced the feasibility of using imputation to increase coverage, providing user-friendly computational servers for imputation using a wide range of reference panels. These servers now include panels from International HapMap Project phase 2 (N = 60), 1000 Genomes Project phase 1 (N = 1,092) and phase 3 (N = 2,504), the Consortium on Asthma Among African-Ancestry Populations in the Americas (N = 883), and the HRC (N = 32,470) (61, 83). Despite these achievements, the reference panels remain biased toward European populations, with the majority of the largest panel (HRC) being of European descent. Therefore, studies of populations of non-European descent must rely on much smaller reference panels, which hinders the imputation of low-frequency and rare variation. The development of large-scale multiethnic panels is vital to addressing this source of ascertainment bias. The African Genome Variation Project (50) demonstrates the need for representative reference panels to not only capture population-specific variation but also reflect the shorter LD blocks found in African genomes compared with non-African groups. Future releases of sequence data as reference panels from such multicenter efforts as the Trans-Omics for Precision Medicine (TOPMed) program of the National Heart, Lung, and Blood Institute; H3Africa; Genome Asia 100K (http://www.genomeasia100k.com); the Singapore Sequencing Malay Project (http://phg.nus.edu.sg/StatGen/public_html/SSMP/SSMP_index.html); and the Genome Sequencing Program of the National Human Genome Research Institute will help address this issue by increasing multiethnic representation.
2.4. Association Methods
In studies of associations between genetic variants and a trait of interest, nonhomogeneous populations are a classic source of concern for false-positive associations. More specifically, because allelic variation differs across ancestral population groups, even slight differences in ancestral composition between cases and controls can result in a false-positive association between a variant and disease. Identification of this source of confounding, known as population stratification, resulted in a focus on populations that were presumed to be more ancestrally homogeneous, and statistical methods were modeled under this assumption (97). However, methods have since been developed to explicitly account for population substructure, allowing for the pooling of multiethnic samples. The most commonly used method is principal component analysis, as estimated in Plink or EIGENSTRAT (96). Including all of the samples in the analysis enables the resulting eigenvectors to estimate broad population structure as orthogonal linear variables, which can then be included in regression models to address possible confounding. More recent methods, as detailed below, directly estimate the genetic relationship between samples, allowing the model to assess phenotypic differences between genotypes beyond what may be expected from correlated genomes.
The PAGE studies were characterized by varying levels of known and cryptic relatedness and used distinct strategies of participant recruitment. Specifically, the Women’s Health Initiative, the Multiethnic Cohort Study, and BioMe used population- or clinic-based recruitment, and the Hispanic Community Health Study/Study of Latinos used a household sampling study design. These differences may have led to heterogeneity in covariate and phenotype associations with variants. PAGE evaluated potential heterogeneity in association analyses using the Genetic Estimation and Inference in Structured Samples (GENESIS) package (30–32), which uses a linear mixed model and accounts for the correlation among genetically similar samples through a kinship matrix that estimates the known and cryptic relatedness in the presence of population structure, admixture, and population-associated heterogeneity. This approach was compared with SUGEN (76), which uses a modified version of generalized estimating equations, creates extended families by connecting the households who share first-degree relatives, and allows for heterogeneity in both phenotypes and covariates across racial/ethnic and study groups. Both methods were able to appropriately account for population stratification and relatedness while ensuring adequate statistical power for the detection of novel association.
Beyond the standard SNP-level associations, looking at haplotype associations can be informative, including examination of identity-by-descent or local ancestry segments. Instead of examining a cross section across two chromosomes on a base-pair level, these methods examine segments of shared haplotypes along the chromosome to identify tracts with a common origin that result in a shared phenotype. Identity-by-descent methods are best applied in founder populations with a more recent common ancestor, such as the characterization of Steel syndrome in Puerto Rico (12). Admixture mapping relies on recent ancestry from two or more distinct populations, such as African American or Hispanic/Latino populations (18, 54). Methods such as RFMix (79) and Local Ancestry in Admixed Populations Using Linkage Disequilibrium (LAMP-LD) (9) estimate local ancestry, assigning haplotypes along the genome to their ancestral populations. Once local ancestry is estimated, admixture mapping tests the enrichment of a particular ancestral haplotype in cases versus controls (107).A cases-only study approach can also be used in which enrichment is tested against the overall genome-wide average for that ancestry. These methods have been successful at identifying regions associated with asthma (46), blood pressure (109), end-stage renal disease (65), and obesity (27), among others. Admixture and identity-by-descent mapping offer an alternative to a traditional GWAS framework, explicitly leveraging the unique haplotypes of admixed or founder populations.
2.5. Fine Mapping After a Genome-Wide Association Study
Fine mapping generally refers to a suite of tools to narrow associated regions and identify potential causal variants using summary statistics from GWAS or sequencing data. While many complex-trait loci replicate consistently across European-ancestry populations, tag SNPs from genotyping arrays are not expected to be causal, and loci containing multiple independent signals can be difficult to distinguish within homogeneous study populations (59, 120). Ancestrally diverse study populations are much more conducive to narrowing association signals and identifying multiple independent associations within genomic regions, primarily because geographic isolation over the course of human evolution has led to different LD structures by continental ancestry (6, 7, 55). Populations with ancestral admixture exhibit widely differing stretches of ancestry-specific LD that, evaluated in combination, can be extremely beneficial for limiting the number of potential functional LD proxies tagged by a GWAS index variant.
Tools have been developed to incorporate combined GWAS summary statistics, LD, and functional annotations to identify the most likely causal variants within a genetic locus. Methods developed to specifically leverage the benefits of transethnic study populations have demonstrated success compared with meta-analysis and fine-mapping attempts in European-ancestry study populations. For example, Fast Probabilistic Annotation Integrator (fastPAINTOR) incorporates bioinformatics and epigenomic data with GWAS summary statistics from multiple traits to select the most likely causal variant(s) within an association signal (68).Simulation studies demonstrated an improvement in credible-set reduction with the incorporation of functional annotations such as DNA accessibility. However, as discussed below, these resources are sorely lacking in data for non-European-ancestry populations, meaning their true benefits cannot currently be assessed in diverse study populations.
2.6. Examples from PAGE
Type 2 diabetes is a highly polygenic disorder for which transethnic association studies have been particularly meaningful. Early efforts by PAGE investigators found that causal variants at well-established risk loci identified in Europeans were likely shared across populations (53). A 2014 transethnic meta-analysis of four GWASs of ancestry-specific type 2 diabetes identified multiple associations unreported in European-only analyses, thereby highlighting opportunities for new discovery in diverse groups (34). Fine mapping of glycemic traits in PAGE African American and Hispanic/Latino participants utilized the Metabochip to narrow known associations and further demonstrated the importance of transethnic analyses via identification of an African-ancestry-specific independent association at the G6PC2 locus (13).These efforts in type 2 diabetes demonstrate that the most successful application of these resources will require not only multiethnic study populations but also bioinformatics resources built on globally representative reference populations.
In a recent flagship paper, Wojcik et al. (123) implemented single-variant genome-wide association testing for 26 clinical and behavioral phenotypes in SUGEN using phenotype-specific models. This work found 27 novel loci at the genome-wide significant threshold (Pcond < 5 × 10−8) after conditioning on all previously identified variants, due partly to both the diversity of the study population and the enrichment for population-specific variants on the MEGA array. As an example, the newly discovered CREB3L2/7q33 locus, associated with total cholesterol levels (rs73729087; P= 1.52 × 10−8, N= 33, 185, MAF = 0.05), may not have been discovered in European-ancestry populations given its rare frequency in those groups (MAF = 0.005). This variant is more common in PAGE racial/ethnic groups, including African Americans (P = 1.77×10−6, N = 10, 137, MAF= 0.11) and Hispanics/Latinos (P= 2.58 × 10−3 mapped independent signals (secondary variants) within known loci, further enriching our understanding of the genetic architecture of traits. To test for secondary signals, the study screened for statistical associations that remained genome-wide significant (Pcond < 5 × 10−8) after adjusting for all known tag SNPs (the adjusted model), identifying 38 new associations located within 1 Mb of a previously known variant. These results indicate that even in regions of known significance, novel ancestry-specific signals can be discovered in diverse, multiethnic study populations.
3. DOWNSTREAM ANALYSES RELY ON BIASED RESOURCES
3.1. Availability of Multiomic Resources
Despite continued success in cataloging variants associated with complex phenotypes, translation of GWAS findings into new biological insights has been complicated, in part because the most significantly associated variant is typically not the variant with the biological effect, but instead is in high LD with the causal variant. Additionally, most associations identify a large number of correlated variants in noncoding regions of the genome (91). As such, it is hypothesized that most GWAS associations affect gene regulation and are fundamentally more difficult to interpret because gene expression is tissue specific and modulated by other contextual factors (98). Targeted allele-specific assays are necessary, but these approaches are expensive and labor intensive. Functional follow-up to identify causal variants, relevant genes, and the underlying biological mechanism can be aided by bioinformatic investigation using other sequencing-based omics data sets in reference samples.
To aid in the characterization of candidate functional variants, integrative multiomic resources are now emerging, accompanied by the development of new analytical methods (10, 91, 98). For instance, GTEx, an ongoing effort to build a resource of tissue-specific gene expression and regulation, has collected 1,641 postmortem samples covering 54 body sites from 175 individuals (49). Approaches that leverage reference data to develop transcriptome imputation models of genetically regulated expression have been developed to utilize bioinformatics data, including a variety of machine-learning approaches to estimate combined-variant effects on gene expression. These trained models are then used to impute genetically regulated gene expression in GWAS data sets with genotypes and phenotypes (43, 51, 112). Because transcriptome measurement with RNA sequencing remains prohibitively expensive for GWAS-scale sample sizes, these innovative approaches have opened the door for better characterization of GWAS associations and have even led to novel discoveries. However, while GTEx is the most comprehensive transcriptome data set to date, the tissue donors were predominantly white (85.2%), and evidence suggests that this is likely to significantly hinder the performance of models developed in other ethnic groups (66). Diverse resources such as TOPMed may begin to address this issue, but models will be restricted to transcriptome measurements in blood, leaving the majority of publicly available expression data not globally representative.
3.2. Clinical Databases and Frequencies
In clinical-annotation pipelines, allele frequency estimates offer the most powerful filter available, by removing or down-prioritizing variants with a nonnegligible population frequency. The value of these data has been improved significantly by efforts driven by the scientific community to make these allele frequency resources publicly available, from the 1000 Genomes Project (N = 2,535) to the Exome Sequencing Project (N = 6,503) and now to the larger Bravo (N = 62,784), Exome Aggregation Consortium (N = 60,706), and gnomAD (N = 123,136) data sets. As clinical sequencing becomes routine, medical professionals face a new challenge in that patients with non-European ancestry have a significantly longer list of candidate variants for a suspected genetic disorder. Determining the pathogenicity of a rare variant is compounded by clinical laboratories labeling putatively deleterious nonsynonymous calls as variants of unknown significance, which occurs at higher rates for individuals of non-European descent (24). Accordingly, a review of genetic variants reported as pathogenic and causal for hypertrophic cardiomyopathy showed that these variants were overrepresented in African Americans, and further examination determined that many of them were benign (78). Such misclassification could be avoided with the inclusion of even a small number of individuals of African descent. Therefore, it is imperative that geneticists sequence and investigate a much broader ensemble of populations that are representative of the rich diversity of patients both in the United States and globally. If we do not, a biased picture will emerge of which variants are important, widening existing disparities and diminishing the benefits of genomic medicine for underserved populations.
To address misclassification, MEGA includes a curation of clinically relevant variants from numerous publicly available databases, such as ClinVar (http://www.ncbi.nlm.nih.gov/clinvar), Online Mendelian Inheritance in Man (OMIM; https://omim.org), and PharmGKB (14). Gignoux and colleagues (45, 88) estimated the allele frequencies of these variants in 11 region-level and 99 population-level groups, and these data are now available as a public resource through the University of Chicago’s Geography of Genetic Variants browser (80; https://popgen.uchicago.edu/ggv).
Variable allele frequencies across populations can strongly influence the discovery of clinically relevant variants. One example is the association of population-specific APOL1 variants with chronic kidney disease. Specifically, these variants are present only in populations of African ancestry, members of which are also twice as likely as European Americans to develop end-stage renal disease (41, 44). Discovery in an African American cohort was enabled by higher allele frequencies in that population, thus yielding adequate statistical power to detect the association. Risk variants for chronic kidney disease in APOL1 likely rose in frequency as an adaptation against trypanosomiasis (sleeping disease) in sub-Saharan Africa (70). These variants were later found to associate with higher rates and faster progression of kidney disease in other groups with African ancestry, including Hispanics/Latinos (41, 44, 70, 88, 93). However, Hispanic/Latino populations exhibit highly diverse genetic ancestry, and therefore APOL1 associations replicated in some groups (e.g., Dominicans, who have a substantial proportion of African ancestry) but not others (e.g., Mexicans, who do not). Thus, even ancestry-specific findings can have broad implications for populations who are outside of the discovery group but have shared ancestry (88). These results highlight the importance of moving beyond self-identified racial/ethnic categorizations and the need to model fine-scale genetic ancestry to carry out the next generation of complex- and Mendelian-disease studies (11).
3.3. Genetic Risk Scores
Another frequent translation use for GWASs of complex traits is the development of risk scores that can be utilized in both clinical prognosis and treatment plans. Genetic variation is incorporated into traditional risk score calculations or used to calculate a genetic risk score (GRS) that does not incorporate environmental or demographic risk factors, often with potential clinical benefits (67). Below, we briefly explore two issues at the forefront of GRS research: which variants to include, and the portability of a GRS from the discovery population (nearly always of European ancestry) to nationally or globally representative populations.
Developing a successful GRS depends on the proportion of variance for a particular phenotype that is explained by identified genetic variants. In research, it has become accepted practice to incorporate all measured variants (regardless of correlation structure) to calculate the proportion of variance explained, as this technique tends to improve prediction accuracy in complex traits (67, 126). Across many disease domains, this technique is becoming an accepted downstream application of GWASs; with large studies, the effect sizes become precise enough for accurate assessment of risk. Given sufficiently large study samples, European-ancestry GWAS tag SNPs can be used for GRS construction for global populations. However, this does not guarantee equally good performance across populations. Within a relevant association signal, a European tag SNP will differentially capture other SNPs (any of which could be the true causal variant) when compared with other ancestral populations. Biased SNP selection can lead to a model that provides different risk estimates based on ancestry, which can in turn exacerbate health inequities by impairing risk prediction for populations already underserved in the American health-care system.
However, the problem is far more pernicious than just hampered prediction accuracy. Martin et al. (81) recently demonstrated that—simply due to the effects of genetic drift on allele frequencies and LD patterns across populations—a GRS ascertained using standard methods in one population can yield unpredictable biases in the distributions of scores in other groups, and the distributions can therefore fluctuate dramatically across traits. In a research context, such biases can be accounted for by standard normalization or ranking of GRS scores by population when recruitment has been performed in a genetically clustered fashion. However, in a medical context, there is no guarantee that the ancestry of the patient will perfectly match the study used for GRS generation, and bias can easily result in either misclassification or a GRS that benefits only specific individuals. The only acceptable method for developing a clinically applicable GRS is to ensure that scores can be calculated accurately for everyone, meaning that the genomic data used must be globally representative, and any genetically informed personalized medicine approach that fails to take this issue into account is at risk of major misinterpretation of the underlying data.
4. CLOSING REMARKS
As demonstrated by the work of the PAGE study and other investigators, the inclusion of ancestrally diverse study populations in all aspects of genomic research and methods development is not only a scientific imperative but also essential for the equitable application of results (95). With support from the National Human Genome Research Institute and the National Institute on Minority Health and Health Disparities, PAGE has focused specifically on addressing the well-documented underrepresentation of US minority populations in genomic research by fostering productive collaboration with existing cohorts (56). The studies have attempted to address some of the noted historical biases throughout the research pipeline, including measurement and analysis of population-level genetic data. To address historical bias in genotyping platforms toward European variation, PAGE investigators and collaborators (the Consortium on Asthma Among African-Ancestry Populations in the Americas, Illumina, and other academic centers) designed a new array with comparable efficiencies in detecting genetic variation across all major continental populations, a tool that is now available to the scientific community (https://www.pagestudy.org/mega).The application of this platform to ancestrally diverse PAGE study participants has aided in the discovery of ancestry-specific disease-associated variation and improved understanding of the underlying biology of known genomic regions associated with risk. To date, PAGE has more than 80 published papers, many of which describe novel discoveries and fine mapping, generalization, and replication of previous findings for complex traits—for example, for lipids (35, 127), type 2 diabetes (53), adiposity (36, 38, 47), kidney function (40), coronary heart disease (125), blood pressure (39), electrocardiogram traits (4, 5), several cancers (28, 75, 92, 106), glucose and insulin levels (13, 37), inflammation (58, 71), and menopause/menarche (23, 111). PAGE has demonstrated the importance of multiethnic genomic studies in conjunction with careful consideration of recruitment, genotyping, and statistical methods development, leading to the discovery and refinement of disease-related loci and a better understanding of these complex traits in diverse populations.
Study population inclusivity will also help to ensure that, when included in clinical practice, the instruments developed using genetic findings will be more informative for the entire population, both within the United States and globally. Since the majority of human genome variation is rare and population specific, and an appreciable fraction of this rare variation is likely to have functional consequences, a consensus has emerged that properly powered multifactorial disease studies will require genetic analysis of individual-level genome-wide data from hundreds of thousands to millions of individuals across diverse ethnic groups. Consideration is needed of multiethnic groups throughout the research process, from recruitment to translation of findings. The current use of genetics to inform prevention and therapeutic strategies without these considerations will likely further exacerbate health disparities. At this pivotal time in medical history, PAGE advocates for increased representation of underrepresented populations and the continued development of tools to maximize the accurate measurement of global genetic variation.
ACKNOWLEDGMENTS
S.A.B and U.P. were supported by National Institutes of Health grant U01HG007376.
K.E.N. was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant 1R01DK101855. C.J.H. was supported by National Heart, Lung, and Blood Institute training grant T32 HL007055. C.R.G. was supported by National Heart, Lung, and Blood Institute grant R01HL104608 and National Human Genome Research Institute grant U01HG009080. T.C.M. and G.L.W. were supported by National Institutes of Health grants U01HG007419 and R56HG010297. E.E.K. was supported by National Institutes of Health grant U01HG007417. I.C. was supported by National Institutes of Health grant U01 HG007397 and National Cancer Institute grant U01 CA164973.
The PAGE program is funded by the National Human Genome Research Institute with cofunding from the National Institute on Minority Health and Health Disparities. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The PAGE Consortium thanks the staff and participants of all PAGE studies for their important contributions. We thank Rasheeda Williams and Margaret Ginoza for providing assistance with program coordination. The complete list of PAGE members can be found at http://www.pagestudy.org.
Footnotes
DISCLOSURE STATEMENT
C.R.G. owns stock in 23andMe, Inc. The authors are not aware of any other affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.1000 Genomes Proj. Consort. 2015. A global reference for human genetic variation. Nature 526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adashi EY, Walters LB, Menikoff JA. 2018. The Belmont Report at 40: reckoning with time. Am. J. Public Health 108:1345–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Investig ARIC. 1989. The Atherosclerosis Risk in Communities (ARIC) study: design and objectives.Am. J. Epidemiol 129:687–702 [PubMed] [Google Scholar]
- 4.Avery CL, Sethupathy P, Buyske S, He Q, Lin D-Y, et al. 2012. Fine-mapping and initial characterization of QT interval loci in African Americans. PLOS Genet. 8:e1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery CL, Wassel CL, Richard MA, Highland HM, Bien S, et al. 2017. Fine mapping of QT interval regions in global populations refines previously identified QT interval loci and identifies signals unique to African and Hispanic descent populations. Heart Rhythm 14:572–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baharian S, Barakatt M, Gignoux CR, Shringarpure S, Errington J, et al. 2016. The great migration and African-American genomic diversity. PLOS Genet. 12:e1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai H, Guo X, Narisu N, Lan T, Wu Q, et al. 2018. Whole-genome sequencing of 175 Mongolians uncovers population-specific genetic architecture and gene flow throughout North and East Asia. Nat. Genet 50:1696–704 [DOI] [PubMed] [Google Scholar]
- 8.Ball TB, Ji H, Kimani J, McLaren P, Marlin C, et al. 2007. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS 21:1091–101 [DOI] [PubMed] [Google Scholar]
- 9.Baran Y, Pasaniuc B, Sankararaman S, Torgerson DG, Gignoux C, et al. 2012. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28:1359–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, et al. 2014. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 24:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belbin GM, Nieves-Colón MA, Kenny EE, Moreno-Estrada A, Gignoux CR. 2018. Genetic diversity in populations across Latin America: implications for population and medical genetic studies. Curr. Opin. Genet. Dev 53:98–104 [DOI] [PubMed] [Google Scholar]
- 12.Belbin GM, Odgis J, Sorokin EP, Yee M-C, Kohli S, et al. 2017. Genetic identification of a common collagen disease in Puerto Ricans via identity-by-descent mapping in a health system. eLife 6:e25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bien SA, Pankow JS, Haessler J, Lu YN, Pankratz N, et al. 2017. Transethnic insight into the genetics of glycaemic traits: fine-mapping results from the Population Architecture Using Genomics and Epidemiology (PAGE) consortium. Diabetologia 60:2384–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bien SA, Wojcik GL, Zubair N, Gignoux CR, Martin AR, et al. 2016. Strategies for enriching variant coverage in candidate disease loci on a multiethnic genotyping array. PLOS ONE 11:e0167758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, et al. 2002. Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol 156:871–81 [DOI] [PubMed] [Google Scholar]
- 16.Bonham VL, Green ED, Pérez-Stable EJ. 2018. Examining how race, ethnicity, and ancestry data are used in biomedical research. JAMA 320:1533–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. 2015. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet 96:37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustamante CD, Burchard EG, De La Vega FM. 2011. Genomics for the world. Nature 475:163–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell MC, Tishkoff SA. 2008. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genom. Hum. Genet 9:403–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet 74:106–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carnethon MR, Pu J, Howard G, Albert MA, Anderson CAM, et al. 2017. Cardiovascular health in African Americans: a scientific statement from the American Heart Association. Circulation 136:e393–423 [DOI] [PubMed] [Google Scholar]
- 23.Carty CL, Spencer KL, Setiawan VW, Fernandez-Rhodes L, Malinowski J, et al. 2013. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture Using Genomics and Epidemiology (PAGE) study. Hum. Reprod 28:1695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caswell-Jin JL, Gupta T, Hall E, Petrovchich IM, Mills MA, et al. 2018. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet. Med 20:234–39 [DOI] [PubMed] [Google Scholar]
- 25.Cent. Dis. Control Prev. 2017. National diabetes statistics report, 2017. Rep., Cent. Dis. Control Prev, Atlanta: https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf [Google Scholar]
- 26.Check Hayden E 2015. Racial bias continues to haunt NIH grants. Nature 527:286–87 [DOI] [PubMed] [Google Scholar]
- 27.Cheng C-Y, Kao WHL, Patterson N, Tandon A, Haiman CA, et al. 2009. Admixture mapping of 15, 280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLOS Genet. 5:e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng I, Caberto CP, Lum-Jones A, Seifried A, Wilkens LR, et al. 2011. Type 2 diabetes risk variants and colorectal cancer risk: the multiethnic cohort and PAGE studies. Gut 60:1703–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Čolić A, Alessandrini M, Pepper MS. 2015. Pharmacogenetics of CYP2B6, CYP2A6 and UGT2B7 in HIV treatment in African populations: focus on efavirenz and nevirapine. Drug Metab. Rev 47:111–23 [DOI] [PubMed] [Google Scholar]
- 30.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, et al. 2016. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet 98:165–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conomos MP, Miller MB, Thornton TA. 2015. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol 39:276–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conomos MP, Reiner AP, Weir BS, Thornton TA. 2016. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet 98:127–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deelen P, Menelaou A, van Leeuwen EM, Kanterakis A, van Dijk F, et al. 2014. Improved imputation quality of low-frequency and rare variants in European samples using the “Genome of the Netherlands.” Eur. J. Hum. Genet 22:1321–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diabetes Genet. Replication Meta-Anal. (DIAGRAM) Consort., Asian Genet. Epidemiol. Netw. Type2 Diabetes (AGEN-T2D) Consort., South Asian Type 2 Diabetes (SAT2D) Consort., Mex. Am. Type 2 Diabetes (MAT2D) Consort., Type 2 Diabetes Genet. Explor. Next-Gener. Seq. Multi-Ethnic Samples (T2D-GENES) Consort., et al. 2014. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet 46:234–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumitrescu L, Carty CL, Taylor K, Schumacher FR, Hindorff LA, et al. 2011. Genetic determinants of lipid traits in diverse populations from the Population Architecture Using Genomics and Epidemiology (PAGE) study. PLOS Genet. 7:e1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernández-Rhodes L, Gong J, Haessler J, Franceschini N, Graff M, et al. 2017. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture Using Genomics and Epidemiology (PAGE) study reveals evidence for multiple signals at established loci. Hum. Genet 136:771–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fesinmeyer MD, Meigs JB, North KE, Schumacher FR, Bůžková P, et al. 2013. Genetic variants associated with fasting glucose and insulin concentrations in an ethnically diverse population: results from the Population Architecture Using Genomics and Epidemiology (PAGE) study. BMC Med. Genet 14:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fesinmeyer MD, North KE, Ritchie MD, Lim U, Franceschini N, et al. 2013. Genetic risk factors for BMI and obesity in an ethnically diverse population: results from the Population Architecture Using Genomics and Epidemiology (PAGE) study. Obesity 21:835–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franceschini N, Carty CL, Lu Y, Tao R, Sung YJ, et al. 2016. Variant discovery and fine mapping of genetic loci associated with blood pressure traits in Hispanics and African Americans. PLOS ONE 11:e0164132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franceschini N, Shara NM, Wang H, Voruganti VS, Laston S, et al. 2012. The association of genetic variants of type 2 diabetes with kidney function. Kidney Int. 82:220–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freedman BI, Limou S, Ma L, Kopp JB. 2018. APOL1-associated nephropathy: a key contributor to racial disparities in CKD. Am. J. Kidney Dis 72:S8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, et al. 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493:216–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, et al. 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet 47:1091–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, et al. 2010. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329:841–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gignoux C, Sorokin E, Wojcik G, Belbin G, Bien S, et al. 2018. The global landscape of pharmacogenomic variation. Paper presented at the Annual Meeting of the American Society of Human Genetics, San Diego, CA, Oct. 16–20 [Google Scholar]
- 46.Gignoux CR, Torgerson DG, Pino-Yanes M, Uricchio LH, Galanter J, et al. 2019. An admixture mapping meta-analysis implicates genetic variation at 18q21 with asthma susceptibility in Latinos. J. Allergy Clin. Immunol 143:957–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong J, Schumacher F, Lim U, Hindorff LA, Haessler J, et al. 2013. Fine mapping and identification of BMI loci in African Americans. Am. J. Hum. Genet 93:661–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, et al. 2011. Demographic history and rare allele sharing among human populations. PNAS 108:11983–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Consort GTEx. 2013. The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45:580–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, et al. 2015. The African genome variation project shapes medical genetics in Africa. Nature 517:327–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gusev A, Ko A, Shi H, Bhatia G, Chung W, et al. 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48:245–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H3Africa Consort. 2014. Enabling the genomic revolution in Africa. Science 344:1346–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haiman CA, Fesinmeyer MD, Spencer KL, Buzková P, Voruganti VS, et al. 2012. Consistent directions of effect for established type 2 diabetes risk variants across populations: the Population Architecture Using Genomics and Epidemiology (PAGE) consortium. Diabetes 61:1642–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, et al. 2014A genetic atlas of human admixture history. Science 343:747–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. PNAS 113:E440–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hindorff LA, Bonham VL, Brody LC, Ginoza MEC, Hutter CM, et al. 2018. Prioritizing diversity in human genomics research. Nat. Rev. Genet 19:175–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hindorff LA, Bonham VL, Ohno-Machado L. 2018. Enhancing diversity to reduce health information disparities and build an evidence base for genomic medicine. Pers. Med 15:403–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hodonsky CJ, Jain D, Schick UM, Morrison JV, Brown L, et al. 2017. Genome-wide association study of red blood cell traits in Hispanics/Latinos: the Hispanic Community Health Study/Study of Latinos. PLOS Genet. 13:e1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E. 2014. Identifying causal variants at loci with multiple signals of association. Genetics 198:497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Illumina. 2019. Human consortia. Illumina. https://www.illumina.com/science/consortia/human-consortia.html
- 61.Int. HapMap Consort. 2003. The International HapMap Project. Nature 426:789–96 [DOI] [PubMed] [Google Scholar]
- 62.Jennes W, Verheyden S, Demanet C, Adjé-Touré CA, Vuylsteke B, et al. 2006. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol 177:6588–92 [DOI] [PubMed] [Google Scholar]
- 63.Johnston HR, Hu Y-J, Gao J, O’Connor TD, Abecasis GR, et al. 2017. Identifying tagging SNPs for African specific genetic variation from the African diaspora genome. Sci. Rep 7:46398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kallwitz ER, Daviglus ML, Allison MA, Emory KT, Zhao L, et al. 2015. Prevalence of suspected nonalcoholic fatty liver disease in Hispanic/Latino individuals differs by heritage. Clin. Gastroenterol. Hepatol 13:569–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kao WHL, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, et al. 2008. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet 40:1185–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keys KL, Mak ACY, White MJ, Eckalbar W, Dahl AJ, et al. 2019. On the cross-population portability of gene expression prediction models. bioRxiv 552042 10.1101/552042 [DOI] [Google Scholar]
- 67.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, et al. 2018. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50:1219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kichaev G, Roytman M, Johnson R, Eskin E, Lindström S, et al. 2017Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics 33:248–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein K, Lang T, Saussele T, Barbosa-Sicard E, Schunck W-H, et al. 2005. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet. Genom 15:861–73 [DOI] [PubMed] [Google Scholar]
- 70.Ko W-Y, Rajan P, Gomez F, Scheinfeldt L, An P, et al. 2013. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am. J. Hum. Genet 93:54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kocarnik JM, Richard M, Graff M, Haessler J, Bien S, et al. 2018. Discovery, fine-mapping, and conditional analyses of genetic variants associated with c-reactive protein in multiethnic populations using the Metabochip in the Population Architecture Using Genomics and Epidemiology (PAGE) study. Hum. Mol. Genet 27:2940–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, et al. 2000. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol 151:346–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwiatkowski DP. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet 77:171–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lama J, Planelles V. 2007. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology 4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim U, Wilkens LR, Monroe KR, Caberto C, Tiirikainen M, et al. 2012. Susceptibility variants for obesity and colorectal cancer risk: the multiethnic cohort and PAGE studies. Int. J. Cancer 131:E1038–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin D-Y, Tao R, Kalsbeek WD, Zeng D, Gonzalez F, et al. 2014. Genetic association analysis under complex survey sampling: the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet 95:675–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. 2009. Finding the missing heritability of complex diseases. Nature 461:747–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, et al. 2016. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375:655–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maples BK, Gravel S, Kenny EE, Bustamante CD. 2013. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet 93:278–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marcus JH, Novembre J. 2017. Visualizing the geography of genetic variants. Bioinformatics 33:594–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, et al. 2017. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100:635–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, et al. 2016. A continuum of admixture in the Western Hemisphere revealed by the African diaspora genome. Nat. Commun 7:12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, et al. 2016. A reference panel of 64, 976 haplotypes for genotype imputation. Nat. Genet 48:1279–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mills MC, Rahal C. 2019. A scientometric review of genome-wide association studies. Commun. Biol 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miwa S, Fujii H. 1996. Molecular basis of erythroenzymopathies associated with hereditary haemolytic anemia: tabulation of mutant enzymes. Am. J. Hematol 51:122–32 [DOI] [PubMed] [Google Scholar]
- 86.Moreno-Estrada A, Gignoux CR, Fernández-López JC, Zakharia F, Sikora M, et al. 2014. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 344:1280–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, et al. 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet 44:981–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nadkarni GN, Gignoux CR, Sorokin EP, Daya M, Rahman R, et al. 2018. Worldwide frequencies of APOL1 renal risk variants. N. Engl. J. Med 379:2571–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Need AC, Goldstein DB. 2009. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 25:489–94 [DOI] [PubMed] [Google Scholar]
- 90.Newman LA. 2015. Disparities in breast cancer and African ancestry: a global perspective. Breast J. 21:133–39 [DOI] [PubMed] [Google Scholar]
- 91.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLOS Genet. 6:e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park SL, Fesinmeyer MD, Timofeeva M, Caberto CP, Kocarnik JM, et al. 2014. Pleiotropic associations of risk variants identified for other cancers with lung cancer risk: the PAGE and TRICL consortia. J. Natl. Cancer Inst 106:dju061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parsa A, Kao WHL, Xie D, Astor BC, Li M, et al. 2013. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med 369:2183–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Popejoy AB, Fullerton SM. 2016. Genomics is failing on diversity. Nature 538:161–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popejoy AB, Ritter DI, Crooks K, Currey E, Fullerton SM, et al. 2018. The clinical imperative for inclusivity: race, ethnicity, and ancestry (REA) in genomics. Hum. Mutat 39:1713–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38:904–9 [DOI] [PubMed] [Google Scholar]
- 97.Price AL, Zaitlen NA, Reich D, Patterson N. 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet 11:459–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roadmap Epigenom. Consort., Kundaje A, Meuleman W, Ernst J, Bilenky M, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518:317–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rotimi CN, Bentley AR, Doumatey AP, Chen G, Shriner D, Adeyemo A. 2017. The genomic landscape of African populations in health and disease. Hum. Mol. Genet 26:R225–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruwende C, Khoo SC, Snow RW, Yates SN, Kwiatkowski D, et al. 1995. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature 376:246–49 [DOI] [PubMed] [Google Scholar]
- 101.Saab S, Manne V, Nieto J, Schwimmer JB, Chalasani NP. 2016. Non-alcoholic fatty liver disease in Latinos. Clin. Gastroenterol. Hepatol 14:5–12 [DOI] [PubMed] [Google Scholar]
- 102.Saran R, Robinson B, Abbott KC, Agodoa LYC, Albertus P, et al. 2017. US Renal Data System 2016 Annual Data Report: epidemiology of kidney disease in the United States. Am. J. Kidney Dis 69(Suppl. 1):A7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saunders MA, Hammer MF, Nachman MW. 2002. Nucleotide variability at g6pd and the signature of malarial selection in humans. Genetics 162:1849–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schneider ALC, Lazo M, Selvin E, Clark JM. 2014. Racial differences in nonalcoholic fatty liver disease in the U.S. population. Obesity 22:292–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sempos CT, Bild DE, Manolio TA. 1999. Overview of the Jackson Heart Study: a study of cardiovascular diseases in African American men and women. Am. J. Med. Sci 317:142–46 [DOI] [PubMed] [Google Scholar]
- 106.Setiawan VW, Haessler J, Schumacher F, Cote ML, Deelman E, et al. 2012. HNF1B and endometrial cancer risk: results from the PAGE study. PLOS ONE 7:e30390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shriner D 2017. Overview of admixture mapping. Curr. Protoc. Hum. Genet 94:1.23.1–8 [DOI] [PubMed] [Google Scholar]
- 108.Simons YB, Bullaughey K, Hudson RR, Sella G. 2018. A population genetic interpretation of GWAS findings for human quantitative traits. PLOS Biol. 16:e2002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sofer T, Baier LJ, Browning SR, Thornton TA, Talavera GA, et al. 2017. Admixture mapping in the Hispanic Community Health Study/Study of Latinos reveals regions of genetic associations with blood pressure traits. PLOS ONE 12:e0188400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, et al. 2010. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol 20:629–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spencer KL, Malinowski J, Carty CL, Franceschini N, Fernández-Rhodes L, et al. 2013. Genetic variation and reproductive timing: African American women from the Population Architecture Using Genomics and Epidemiology (PAGE) study. PLOS ONE 8:e55258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su Y-R, Di C, Bien SA, Huang L, Dong X, et al. 2018. A mixed-effects model for powerful association tests in integrative functional genomics: an application to a large-scale genome-wide association study of colorectal cancer. Am. J. Hum. Genet 102:904–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Swart M, Evans J, Skelton M, Castel S, Wiesner L, et al. 2015. An expanded analysis of pharmacogenetics determinants of efavirenz response that includes 3′-UTR single nucleotide polymorphisms among black South African HIV/AIDS patients. Front. Genet 6:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, et al. 2001. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293:455–62 [DOI] [PubMed] [Google Scholar]
- 115.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, et al. 2011. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat. Genet 43:887–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. 2015. Global cancer statistics, 2012. CA Cancer J. Clin 65:87–108 [DOI] [PubMed] [Google Scholar]
- 117.UNAIDS. 2018. Global HIV & AIDS statistics – 2018 fact sheet. Fact Sheet, UNAIDS, Geneva. http://www.unaids.org/en/resources/fact-sheet
- 118.UyBico SJ, Pavel S, Gross CP. 2007. Recruiting vulnerable populations into research: a systematic review of recruitment interventions. J. Gen. Intern. Med 22:852–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vergara C, Parker MM, Franco L, Cho MH, Valencia-Duarte AV, et al. 2018. Genotype imputation performance of three reference panels using African ancestry individuals. Hum. Genet 137:281–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Visscher PM, Brown MA, McCarthy MI, Yang J. 2012. Five years of GWAS discovery. Am. J. Hum. Genet 90:7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, et al. 2012. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLOS Genet. 8:e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wojcik GL, Fuchsberger C, Taliun D, Welch R, Martin AR, et al. 2018. Imputation-aware tag SNP selection to improve power for large-scale, multi-ethnic association studies. G3 8:3255–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, et al. 2019. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Women’s Health Init. Study Group. 1998. Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials 19:61–109 [DOI] [PubMed] [Google Scholar]
- 125.Zhang L, Buzkova P, Wassel CL, Roman MJ, North KE, et al. 2013. Lack of associations of ten candidate coronary heart disease risk genetic variants and subclinical atherosclerosis in four US populations: the Population Architecture Using Genomics and Epidemiology (PAGE) study. Atherosclerosis 228:390–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Qi G, Park J-H, Chatterjee N. 2018. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet 50:1318–26 [DOI] [PubMed] [Google Scholar]
- 127.Zubair N, Graff M, Luis Ambite J, Bush WS, Kichaev G, et al. 2016. Fine-mapping of lipid regions in global populations discovers ethnic-specific signals and refines previously identified lipid loci. Hum. Mol. Genet 25:5500–12 [DOI] [PMC free article] [PubMed] [Google Scholar]