Abstract
Personality traits are the relatively enduring patterns of thoughts, feelings, and behaviors that reflect the tendency to respond in certain ways under certain circumstances. Twin and family studies have demonstrated that personality traits are moderately heritable, and can predict various lifetime outcomes, including psychopathology. The Research Domain Criteria (RDoC) characterizes psychiatric diseases as extremes of normal tendencies, including specific personality traits. This implies that heritable variation in personality traits, such as neuroticism, would share a common genetic basis with psychiatric diseases, such as major depressive disorder (MDD). Despite considerable efforts over the past several decades, the genetic variants that influence personality are only beginning to be identified. We review these recent and increasingly rapid developments, which focus on the assessment of personality via several commonly used personality questionnaires in healthy human subjects. Study designs covered include twin, linkage, candidate gene association studies, genome-wide association studies (GWAS) and polygenic analyses. Findings from genetic studies of personality have furthered our understanding about the genetic etiology of personality, which, like neuropsychiatric diseases themselves, is highly polygenic. Polygenic analyses have demonstrated genetic correlations between personality and psychopathology, confirming that genetic studies of personality can help to elucidate the etiology of several neuropsychiatric diseases.
Keywords: EPQ, gene, GWAS, NEO, personality, TPQ, neuroticism, extraversion, agreeableness, conscientiousness, openness
Genetic studies of human personality
Personality traits are the relatively enduring patterns of thoughts, feelings, and behaviors that reflect the tendency to respond in certain ways under certain circumstances (Roberts 2009 p 140). Strong phenotypic correlations have been reported over the years between personality traits and a wide array of psychopathological conditions (Khan et al. 2005); however, it is unknown whether these correlations have a genetic or environmental basis.
Personality traits can be closely aligned with domains within the Research Domain Criteria (RDoC), which views psychiatric disorders as extremes of normal tendencies, and is intended to foster a biological analysis of behavior. While numerous genetic studies have examined psychiatric diseases, relatively less work has been done on the genetic basis of RDoC traits such as personality. Twin studies have demonstrated that personality traits, as measured by self-report questionnaires (Cervone and Pervin 2009), are moderately heritable (Kandler et al. 2017; Bratko et al. 2017) and have a relatively stable trajectory over time after early adulthood (Kupper et al. 2011). However, the exact genetic basis of personality is still poorly understood.
We review findings from twin and family studies of heritability, followed by linkage studies, candidate gene association studies, and GWAS. We summarize the most robust loci associated with personality. We only included genetic studies of personality if they used validated, standardized, self-report measures of personality traits (Box 1) in healthy adults of any ethnic origin. We conclude by discussing polygenic methods, which provide mounting evidence that the association between personality traits and psychopathology has a genetic basis. Future directions for research are also suggested.
BOX 1. SELF-REPORTED QUESTIONNAIRES TO CAPTURE THE MULTIFACETED TRAITS OF PERSONALITY.
The Eysenck Personality Questionnaire (EPQ; Eysenck and Eysenck 1975) defines three traits of personality: psychoticism (characterized by aggressiveness and interpersonal hostility), extraversion (manifested in outgoing, energetic behavior), neuroticism (typified by emotional stability).
The Tridimensional Personality Questionnaire (TPQ; Cloninger, 1986; Cloninger, Przybeck, & Svrakic, 1991) consists of three traits of personality (or temperaments) that are based on the biochemical bases of temperament: novelty seeking (or the intense exploration towards novel stimulation or potentially rewarding cues, and active avoidance of punishment), harm avoidance (or the tendency to respond intensely to aversive cues, and to learn to avoid punishment and novelty), reward dependence (or the rapid response to rewarding cues and/or relief of punishment). Later, the Temperament and Character Inventory (TCI; Cloninger et al. 1993; Cloninger 1994) included a fourth temperament dimension, persistence (or perseverance in spite of fatigue or frustration), and three character traits - self-directedness (the ability to modify behavior in order to achieve personal goals), cooperativeness (the tendency to exhibit agreeable relations with others) and self-transcendence (associated with experiencing spiritual aspects of the self).
The Five Factor model (Costa and McCrae 1992) is based on biological mechanisms shaping five higher-order traits (referred to as ‘the Big Five’ personality traits): neuroticism (proneness to experience negative affect), extraversion (motivation to engage with others), openness to experience (inventive or curious behavior), agreeableness (friendliness and compassion toward others), and conscientiousness (attentive and organized behavior). The NEO-PI questionnaire (NEO; and its derivatives, such as the NEO-PI-R and NEO-FFI) is the most commonly used questionnaire for genetic studies.
Developments in the field, including the availability of large datasets such as UK Biobank, and bioinformatics techniques such as gene pathway analysis, have furthered our understanding of the genetic etiology of personality. Ultimately, genetic studies of personality may enhance our understanding of neuropsychiatric diseases and thus foster novel treatment approaches (So et al. 2017).
Twin & family studies: heritability estimates
Twin studies of personality have shown that identical twins are more concordant than non-identical twins, yielding narrow-sense heritability estimates in the range of 40% (Johnson et al. 2008; van den Berg et al. 2014; Polderman et al. 2015; Vukasović and Bratko 2015), suggesting that a moderate proportion for the variance of personality traits can be attributed to additive genetic effects. Family and adoption studies, which estimate the resemblance between parent/biological child, or relatives separated by adoption, have yielded slightly lower estimates (~30%; Bouchard and Loehlin 2001; Rijsdijk and Sham 2002).
Heritability does not appear to vary by sex (estimated by comparing the resemblance between opposite-sex versus same-sex dizygotic twins; Vink et al. 2012). For example, a meta-analysis of data from over 29,000 twin pairs (van den Berg et al. 2014) showed that the heritability of neuroticism scores was at 48%, and that the same proportion of variance in neuroticism can be attributed to genetic factors in both in men and women.
Environmental differences can increase or decrease the importance of genetic factors, and may also reflect gene-environment interactions (Flint and Willis-Owen 2010; Kendler and Myers 2010). Twin data demonstrate that genetic influences contribute to personality stability and are relatively constant with age. Personality stability is defined as the degree to which the relative differences between individuals are preserved over time, which is typically assessed with a test-retest correlation. In contrast, the environmental influence on personality increases with age (Briley and Tucker-Drob 2014).
All questionnaires show roughly similar levels of heritability, albeit with variable estimates from study to study (see Table 1). Moreover, heritability estimates across personality traits do not reflect significantly different levels of genetic influence. For example, a recent meta-analysis reported heritability estimates for the Big Five (NEO) traits, ranging from 31% to 41% (Vukasović and Bratko 2015; also see Jang et al. 1996). The moderate variability in heritability estimates is to be expected since heritability is not only a property of a trait but is also influenced by differences among samples, methodology, including strong theoretical assumptions in twin studies (e.g. assumption of equal environment, exclusion of gene-environment interactions; Boomsma et al. 2002; Evans and Martin 2002; Rijsdijk and Sham 2002).
Table 1.
Overview of narrow-sense heritability estimates for the most commonly used personality questionnaires. These studies used a combination of twin and family based designs.
| Questionnaire | Heritability Estimate | Reference |
|---|---|---|
| EPQ | 35–57% | (Keller, et al., 2005; Zietsch, Verweij, Bailey, Wright, & Martin, 2010) |
| TPQ/TCI | 30–60% | (Gillespie, Cloninger, Heath, & Martin, 2003; Gillespie, Johnstone, Boyce, Heath, & Martin, 2001; Heath, Cloninger, & Martin, 1994; Heiman, Stallings, Hofer, & Hewitt, 2003; Keller, et al., 2005) |
| NEO-PI | 17–65% | (Riemann et al. 1997; Jang et al. 1998; Loehlin et al. 1998; Pilia et al. 2006; Vernon et al. 2008; Distel et al. 2009) |
Candidate Genes Association Studies
20 years ago, it was commonly assumed that the genetic architecture of personality might be relatively simple, or at least simpler than psychiatric diseases, with a few key genes explaining much of the observed heritable variance (e.g., Cloninger, Adolfsson, & Svrakic, 1996). This partially explains the enthusiasm for candidate gene association studies that continue to be performed even today, despite mounting evidence that personality is likely to be at least as polygenic as psychiatric diseases (Hart et al. 2013; Chabris et al. 2013). The candidate gene literature has been dominated by a small number of genes that were largely selected based on their involvement in key neurotransmitter systems (e.g. dopamine, DA; serotonin, 5-HT) that mediate the effects of many psychoactive drugs. These studies have produced inconsistent and inconclusive findings (Munafò and Flint 2011). In an attempt to summarize this complex literature, we have focused on the major findings obtained from meta-analyses, which are summarized in Table 2. Rather than exhaustively surveying the candidate gene literature, we have focused on the most heavily studied genes/polymorphisms, since these allow the most informative meta-analyses. These studies show that associations between personality traits and the proportion of variance explained by a specific variant is unlikely to be ≥ 3% (Visscher et al. 2017). As described below, GWAS studies have clearly indicated that true effect sizes are much smaller.
Table 2.
Meta-analyses of the most commonly studied polymorphisms in relation to personality. Only meta-analyses with more than ten studies are included in the table.
| Candidate gene | Form | Comparison | Reference | Number of studies (total sample size) | Trait; Questionnaire (effect size, P) |
|---|---|---|---|---|---|
| DRD4 | VNTR | +7R | (Schinka et al. 2002) | 14 (N = 2,720) | Novelty Seeking; TPQ, TCI,KSP (d = 0.00, P > 0.05) |
| L/S | 12 (N = 1,719) | Novelty Seeking; TPQ, TCI, NEO, KSP (d = 0.21, P > 0.05) | |||
| S/L | (Munafò et al. 2003) | 17 (N = NA) | Approach-related; NEO, TPQ, EPQ, KSP, TCI (d = .07, P > .05) | ||
| 10 (N = NA) | Avoidance-related; NEO, TPQ, EPQ, KSP, TCI (d = −.08, P > .05) | ||||
| +7R | (Munafò et al. 2008) | 36 (N = NA) | Approach-related*; TCI, TPQ, NEO, KSP (d = .04, P = .14) | ||
| C-521T | CC/CT-TT | 11 (N = NA) | Approach-related*; TCI, TPQ, NEO, EPI, EPQ (d = .25, P < .001) | ||
| 5HTT (SERT or SLC6A4) | 5HTTLPR | ll-sl/ss | (Munafò et al. 2003) | 22 (N = NA) | Avoidance-related* (d = −.11, P = .04) |
| 17 (N = NA) | Approach-related* (d = −.11; P > 0.05) | ||||
| sl-ss/ll | (Sen et al. 2004) | 26 (N = 5,629) | Neuroticism, Harm Avoidance; NEO, TPQ/TCI (β = 1.68, P = 0.087) | ||
| sl-ss/ll | (Schinka et al. 2004) | 26 (N = 7,657) | Neuroticism, Harm Avoidance; NEO, TPQ, EPQ, KSP, TCI, EPI (d = 0.10, P > 0.05) | ||
| sl-ss/ll | (Munafò et al. 2009) | 55** (N = 3,872) | Harm Avoidance; TPQ, TCI (d = 0.02, P = 0.37) | ||
| Neuroticism; EPQ (d = 0.01, P = 0.71) | |||||
| Neuroticism; NEO (d = 0.18, P < 0.001) | |||||
| ll/sl-ss | (Minelli et al. 2011) | 44 (N = 15,476) | Neuroticism, Harm Avoidance; NEO, TCI (d = −.04, P =.03) |
Abbreviations: VNTR = variable number tandem repeat or presence or absence of a 7-repeat (‘long’) allele in exon III; +7-R = presence or absence of a 7-repeat (‘long’) allele; S/L = short/long; d = cohen’s d effect size; NA = not listed; KSP = Karolinska Scales of Personality; C-521T = DRD4 promoter region; 16PF = 16 Personality Factor Scale; SSS = sensation Seeking Scale; BIS/BAS = Behavioral Inhibition/Activation Scale;
= calculated from BIS/BAS scale scores, that measures sensitivity to reward and punishment (reward responsiveness, drive, fun seeking); 5HTTLPR = 5HTTL polymorphism;
= 55 is the total number of studies in the full meta-analysis; the number of studies for each individual trait may be lower.
Over 60 studies have investigated whether variation in genes involved with DA transmission were associated with personality. Early publications reported associations between SNPs in the D4 dopamine receptor (DRD4) gene and extraversion or novelty seeking (e.g. Benjamin et al. 1996; Ebstein 2006; Roussos et al. 2009), but those effects were not replicated in two meta-analyses (Munafò et al. 2003, 2008). Some argued that the genetic effects may be sex-dependent, as polymorphisms in the tyrosine hydroxylase (TH) gene, the rate limiting enzyme in the biosynthesis of DA, were associated with novelty seeking (TCI) in healthy males but not females (Sadahiro et al. 2010). On the other hand, others hypothesized that early environmental exposure and polymorphisms in the DRD4 gene might either strengthen or dilute the observed associations with novelty seeking (for a review see: Ebstein 2006); but these hypotheses have yet to be rigorously tested in human populations.
Similarly, early publications reported that variation in the serotonin transporter gene 5HTT (SERT or SLC6A4), which is responsible for re-uptake of 5-HT from synapses, was associated with neuroticism (Schinka et al. 2004; Sen et al. 2004) and harm avoidance (TPQ), which is related to neuroticism (Lesch et al. 1996). However, meta-analyses of over 40 studies involving 5-HTTLPR variants show that this genotype is not consistently associated with neuroticism (EPQ) or harm avoidance (TPQ) (Munafò et al. 2009; Minelli et al. 2011). The difficulty of accurately genotyping of this locus may contribute to these inconsistent results (Yonan et al. 2006).
In hindsight it is easy to identify reasons why candidate gene association studies have produced unsatisfactory results (Hart et al. 2013; Chabris et al. 2013; Johnson et al. 2017); most notably, they were assumed that the loci under study would have larger effect sizes than have been observed in more recent GWAS, and were thus underpowered. Additional reasons for the lack of replication include confounding due to population stratification and selective reporting of positive results (Colhoun et al. 2003). Furthermore, the assumption that genes related to key neurotransmitter systems were critical for personality turned out to be overoptimistic (Munafò et al., 2003; Van Gestel & Van Broeckhoven, 2003b). Moreover, candidate gene association studies generally focused on coding regions rather than regulatory/non-coding regions; however most GWAS hits implicate non-coding regions (Boyle et al. 2017). Views about the polygenicity of personality and many other traits continue to evolve, with a recent paper suggesting that even the most important ‘core genes’ for a trait can only explain a minor fraction of the heritable variance (Boyle et al. 2017).
Genome wide scans
Linkage studies
Genome-wide linkage studies, which take advantage of close familial relationships to identify chromosomal regions, have also been used to study the genetics of personality. Amin et al. (2011) reported significant linkage results for conscientiousness (NEO) at 20p13. A later meta-analysis (N = 6,149) of four genome-wide linkage scans by Amin et al. (2013) identified 11q24 for openness to experience (NEO). Other suggestive linkage signals have been identified in independent studies for several chromosomal regions and different personality traits (see Flint & Willis-Owen 2010, Flint & Munafo 2012, and Amin et al., 2012 for a thorough review of the topic). Nonetheless, only a few genomic regions (e.g., 12q) have been consistently reported in multiple studies (Fullerton et al. 2003; Nash et al. 2004; Kuo et al. 2007; Wray et al. 2008; Gillespie et al. 2008). The estimated effect sizes did not suggest variants with large effects, which is consistent with the results from candidate gene studies. Although linkage studies are effective at mapping traits with monogenic or simple genetic architecture, they are suboptimal for the study of complex traits like personality (Ott et al. 2011).
Genome-wide association studies
GWAS interrogate hundreds of thousands to millions of SNPs across the genome (Visscher et al. 2017). Unlike candidate gene studies, GWAS are a hypothesis generating approach that do not assume any prior knowledge about the underlying biology associated with the trait. Unlike linkage studies, GWAS utilize unrelated individuals, which has the potential to identify smaller regions that contain one or a few genes. By convention (Risch and Merikangas 1996), the threshold for statistical significance is based on a Bonferroni correction for one million comparisons, yielding a threshold of P < 5.0 × 10−8. This stringent threshold requires large sample sizes and favors type II over type I errors. Table 3 summarizes all previous GWAS of personality (N = 16) at the time of our writing. Figure 1 provides an overview of the chronology of GWAS for human personality traits.
Table 3.
Comparison of previous GWAS of personality traits.
| Reference | Questionnaire | Trait | Study Design | Discovery Sample | GW association P < 5 × 10−8 | Nearby Gene (region) | Replication Sample | Replicated |
|---|---|---|---|---|---|---|---|---|
| (van den Oord et al. 2008) | EPQ (short) | Neuroticism | GWAS | 1,227 EUR | N | - | 1880 EUR | N |
| (Calboli et al. 2010) | EPQ (short) | Neuroticism | GWAS | 2,235 EUR | N | - | NA | - |
| (Verweij et al. 2010) | TPQ | HA, NS, RD, P | GWAS | 5,117 EUR | N | - | NA | - |
| (Terracciano et al. 2010a) | NEO-PI-R | Neuroticism | GWAS | 3,972 EUR (Sardinia) | N | 5,105 EUR | N | |
| Extraversion | N | |||||||
| Openness | N | |||||||
| Agreeableness | N | |||||||
| Conscientiousness | N | |||||||
| TPQ | HA | N | ||||||
| (Terracciano et al. 2011) | NEO-PI-R | Excitement-seeking | Meta-analysis | 7,860 EUR | 1 SNP: rs7600563 (P= 2 × 10−8) | CTNNA2 | 608, 1,545, 3,043 EUR | N |
| (Luciano et al. 2012) | EPQ | Neuroticism | Meta-analysis | 6,268 EUR | N | - | 527–6032 EUR | N |
| Extraversion | N | - | ||||||
| (Service et al. 2012) | TPQ | HA, NS, RD, P | Meta-analysis | ~ 11,000 EUR | N | - | NA | - |
| (de Moor et al. 2012) | NEO | Openness | Meta-analysis | 17,375 EUR | 2 SNP: rs1477268 (P= 2.8 × 10−8), rs2032794 (P=3.1×10−8) | RASA1 | 3,294 EUR | N |
| Conscientiousness | 1 SNP: rs2576037(P= 4.9×10−8) | KATNAL2 | N | |||||
| (Bae et al. 2013) | NEO-FFI | Agreeableness | GWAS | 4,595 EUR | 3 SNPs: rs9650241 (P= 8.12×10−10), rs2701448 (P= 9.46×10−10), kgp6080058 (P= 1.57×10−9) | Chr 8 | 1,279 EUR | N |
| (Kim et al. 2013) | NEO-PI-R | Openness | GWAS | 4,000 EAS | 1 SNP: rs214618 (P= 1.67 × 10−8) | PTPRD | 2,090 EAS | N |
| Neuroticism | N | |||||||
| (Kim et al. 2015a) | NEO-PI-R | Agreeableness | Meta-analysis | 3,898 EAS | N | - | 1,021 EAS | N |
| (de Moor et al. 2015) | IRT | Neuroticism | Meta-analysis | 63,661 EUR | 1 SNP: rs35855737; P= 9.26 × 10−9 | MAGI1 | 9,786 EUR | N |
| (Smith et al. 2016) | EPQ (short) | Neuroticism | Meta-analysis | 91,370 EUR (UK Biobank) 6,659 EUR; 8687 EUR |
9 SNPs: rs490647 (P= 3.8 × 10−8), rs4653663 (P= 2.0 × 10−8), rs12637928 (P= 4.3 × 10−8), rs62353264 (P= 3.7 × 10−8), rs12682352 (P= 1.5 × 10−15), rs12378446 (P= 9.4 × 10−9), rs4977844 (P= 3.2 × 10−8), rs111433752 (P= 9.3 × 10−12), rs1187264 (P= 1.2 × 10−8) |
GRIK3, ENAH, SRP9, PVRL3, TMEM192, KLHL2, MSMO1 >10 genes chr 8, PTRD, ELAVL2, >10 genes chr 17 (e.g. CRHR1) , CELF4, respectively |
NA | - |
| (Okbay et al. 2016) | IRT, EPQ (short) | Neuroticism | Meta-analysis | 170,911 EUR (GPC, UK Biobank) | 11 SNPs: rs2572431 (P= 4.2 × 10−16), rs193236081 (P= 6.3 × 10−11), rs10960103 (P= 2.1 × 10−10), rs4938021 (P= 4.0 × 10−10), rs139237746 (P= 2.6 × 10−9), rs1557341 (P= 5.6 × 10−9), rs12938775 (P= 8.5 × 10−9), rs12961969 (P= 2.2 × 10−8), rs35688236 (P= 2.4 × 10−8), rs2150462 (P= 2.7 × 10−8), rs12903563 (P= 2.9 × 10−8) | - | Y* | Y |
| (Lo et al. 2016) | NEO-FFI (23andMe); NEO, IRT, EPQ | Extraversion | Meta-analysis | 122,886 EUR (23andMe and GPC) |
4 SNPs: rs57590327 (P= 1.26 × 10−9), rs2164273 (P= 1.61 × 10−9), rs6481128 (P= 5.44 × 10−10), rs1426371 (P= 9.54 × 10−15) | GBE1, MTMR9, PCDH15, WSCD2 | 39,500 EUR (23andMe) 7,100 (deCODE) |
Y |
| Neuroticism | 2 SNPs: rs6981523 (P= 3.17 × 10−24), rs9611519 (P= 9.16 × 10−9) | XKR6, L3MBTL2/CHADL | 39,500 EUR (23andMe) 7,100 (deCODE) 91,370 (UK Biobank) |
Y | ||||
| (van den Berg et al. 2016) | IRT | Extraversion | Meta-analysis | 63,030 EUR | N | - | 9,783 EUR | - |
| Luciano et al. (2017) | EPQ (short) | Neuroticism | GWAS | 329,821 EUR | 116 SNPs | Y | Y | |
| (Nagel et al. 2017) | EPQ (short), NEO, NEO-FFI | Neuroticism | Meta-analysis | 449,484 EUR (UK Biobank, 23andMe, GPC1) | 136 SNPs | NA | - |
“quasi-replication”;
see Okbay et al. (2016).
Figure 1.
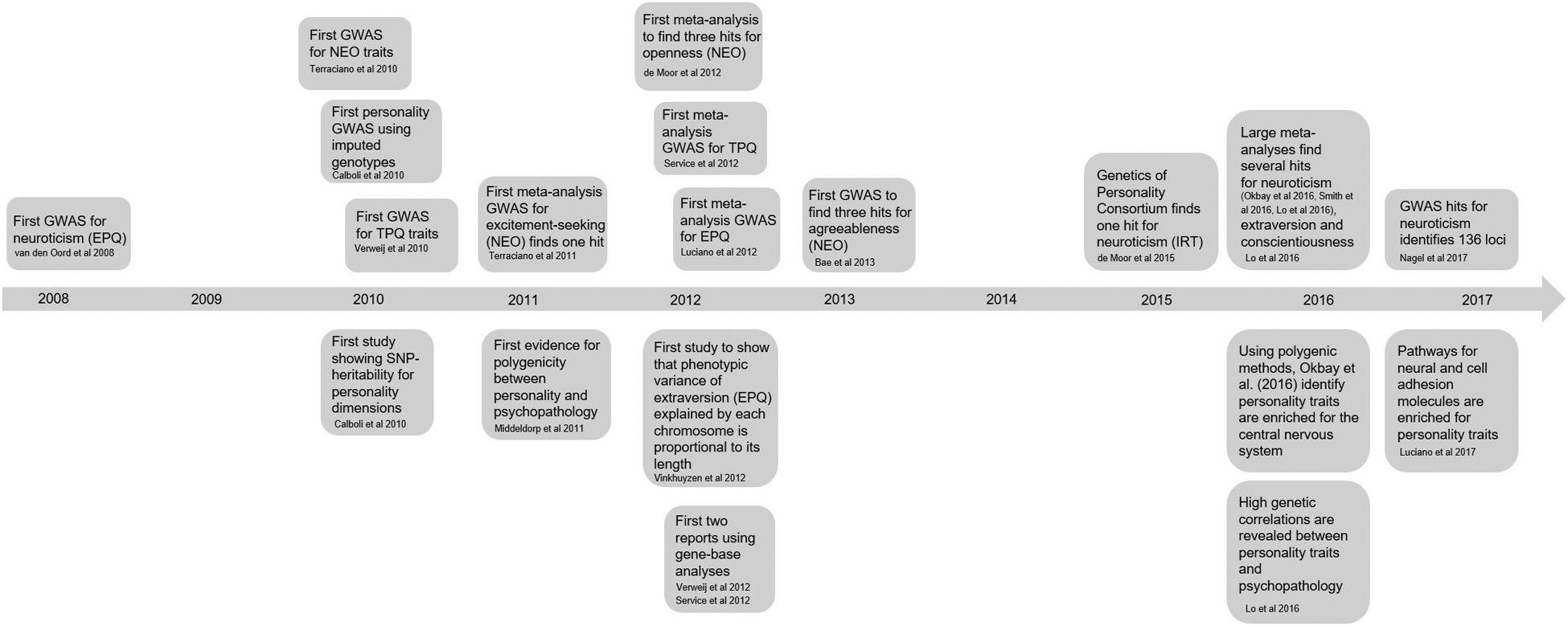
Timeline of major findings in personality genetics using GWAS (top panels) and polygenic (bottom panels) methods. Not all references in Table 3 are included in this figure.
The earliest GWAS for personality used samples in the range of 1,000–5,000 subjects, which proved to be underpowered (Shifman et al. 2008; van den Oord et al. 2008; Calboli et al. 2010; Verweij et al. 2010). Terracciano et al. (2010b) performed an unsuccessful GWAS for NEO-PI-R; in the absence of any significant results, the same group (Terracciano et al. 2011) conducted a meta-analysis for excitement-seeking (NEO-PI-R), which identified rs7600563 (P = 2.0 × 10−8). This SNP is located within an intron of the gene CTNNA2 (cadherin-associated protein alpha 2) and is within 200 kb of the gene LRRTM1; however, this association was not replicated in three independent samples.
Several other meta-analyses followed. Service et al. (2012) and Luciano et al. (2012) did not find any significant results for TPQ and EPQ, respectively. In contrast, de Moor et al. (2012) conducted a meta-analysis of NEO personality traits that showed significant associations for openness near the gene RASA1 (rs1477268, P = 2.8 × 10−8), and for conscientiousness in the gene KATNAL2 (rs2576037, P= 4.9×10−8). However, neither association could be replicated.
Bae et al. (2013) conducted a GWAS of NEO-FFI; although a significant association for agreeableness was detected, it was not replicated in an independent cohort. Kim et al. (2013) performed the first GWAS for personality traits (NEO-PI-R) in a non-European population of young women; however, none of the results were significant. The same group subsequently performed a meta-analysis of NEO-FFI data using four cohorts of Korean ancestry; again, none of the results were significant (Kim et al. 2015a).
More recent GWAS and meta-analyses of personality traits have used dramatically larger sample sizes. de Moor et al. (2015) and van den Berg et al. (2016) conducted meta-analyses of neuroticism and extroversion. Those studies sought to harmonize data obtained from different personality questionnaires by using ‘Item Response Theory’ (IRT; van den Berg et al. 2014). For neuroticism there was a significant association at rs35855737 (P = 9.26 × 10−9). This SNP is located in an intron of the MAGI1 gene, which is expressed in brain, and has been implicated in bipolar disorder, SCZ, and MDD (Etain et al. 2006; Karlsson et al. 2012; Ferentinos et al. 2014). However, the association with MAGI1 (rs35855737) failed to replicate in an independent cohort. van den Berg et al. (2016) did not identify any significant associations with extraversion.
Smith et al. (2016) performed a meta-analysis of neuroticism (EPQ) that identified nine significant loci (Table 3). Examples of the implicated loci include rs111433752, which is near the gene CRHR1, which regulates release of cortisol and is involved in anxiety-like behaviors in mice (Stetler and Miller 2011; Weber et al. 2016). Another example is rs490647, which is near the gene GRIK3 (glutamate receptor kainite 3), a gene that is highly expressed in post-mortem brains of complete suicides (Minelli et al. 2009).
Okbay et al. (2016) performed a large meta-analysis of GWAS data for neuroticism using UK Biobank and Genetics of Personality Consortium (GPC) cohorts. Eleven loci were identified (Table 3); two of these tagged inversion polymorphisms on Chromosomes 8 and 17.
Lo et al. (2016) conducted a meta-analysis of the Five Factor Model that included multiple cohorts using different personality instruments. They used data from the GPC, UK Biobank and two companies: 23andMe, Inc. and deCode Genetics. The authors identified six SNPs exceeded GWAS significance in the meta-analysis (Table 3). For example, extraversion showed an association at rs6481128, which is near the gene PCDH15, which encodes a member of the cadherin superfamily and is important for calcium-dependent cell–cell adhesion. Furthermore, rs9611519, on Chromosome 8, was associated with neuroticism; this locus is located in the L3MBTL2 gene. Additionally, rs2164273, which is an intronic variant of MTMR9, also on Chromosome 8, was associated with extraversion and inversely associated with neuroticism. This locus resides in the inversion region on Chromosome 8 found in the UK Biobank study (Obkay et al., 2016).
At the time of our writing, two studies using large cohorts from UKBiobank have identified multiple loci associated with neuroticism; however, these are not yet peer-reviewed and are therefore only briefly discussed. Luciano et al. (2017) performed a GWAS of neuroticism (EPQ) and reported 116 significant independent genetic loci distributed across the genome (N = 329,821). Two of the SNPs most strongly associated with neuroticism were rs6981523 and rs9611519, which were previously reported by Lo et al. (2016). Furthermore, rs2572431, which is on chromosome 8, was associated with neuroticism, replicating a finding by Okbay et al. (2016). Using UK Biobank participants and two additional cohorts (23andMe, GPC), Nagel et al. (2017) performed the largest meta-analysis of neuroticism ever conducted (N = 449,484), which identified 136 independent loci. The authors performed independent GWAS for different neuroticism subclusters (i.e. ‘depressed affect’ and ‘worry’), and identified that the two subclusters showed notable differences in genetic signal, establishing for the first time the genetic multidimensionality of neuroticism.
Summary of GWAS studies of Personality Traits
Personality traits are extremely polygenic and are influenced by many common alleles of small effect (e.g. Rietveld et al. 2014; also see Supplementary Tables 1 and 2). However, their effect sizes on molecular phenotypes can be large (Visscher et al. 2017). Neuroticism is currently the best-studied personality trait (Table 3); the Supplementary Table 1 shows the list of SNPs that replicated across studies. Figure 2 shows that the number of significant associations has increased as a function of sample size, which is consistent with a polygenic model.
Figure 2.
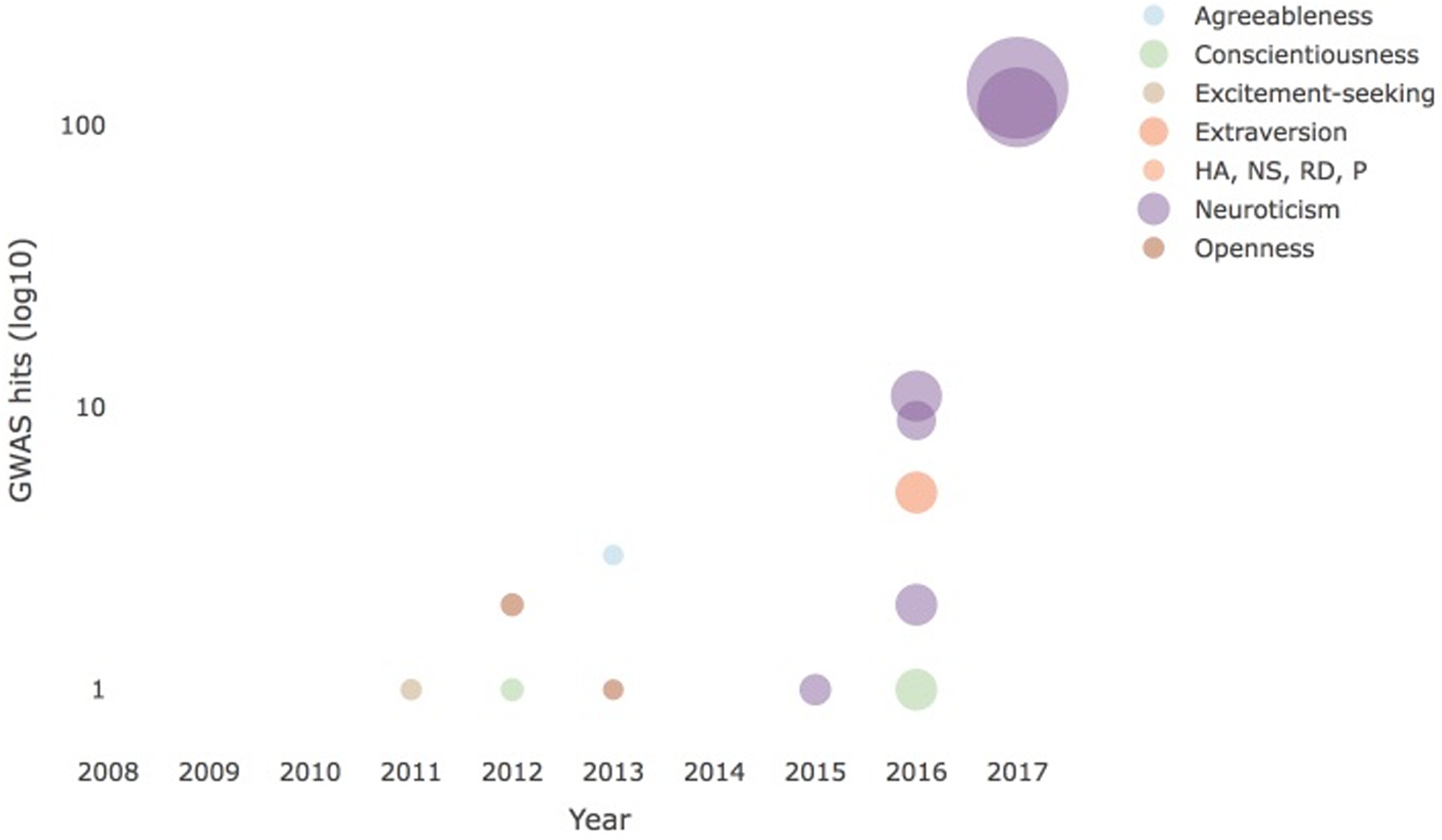
GWAS hits (log10 transformed) discovered as a function of sample size and personality trait.
Because of the need for ever-larger sample sizes, meta-analyses are widely used because they can integrate GWAS results obtained with similar but not identical measurement scales. However, meta-analyses potentially introduce their own problems such as differences in personality scales or properties of the cohorts, including sex, age and environmental factors (Plomin et al. 2016). For example, performing a meta-analysis of neuroticism that includes different scales would be problematic if the traits were not strongly genetically correlated. However, Okbay et al. (2016) reported a perfect genetic correlation (>100% ± 14%) between measures of neuroticism from the UK Biobank (Okbay et al. 2016) and the GPC study (de Moor et al. 2015). Furthermore, using IRT, different personality scales can be transformed into a common phenotype, which can then be subjected to GWAS. A newer approach does not focus on transforming phenotypes but instead jointly analyzes GWAS results from putatively similar phenotypes (Multi-Trait Analysis of GWAS, MTAG; Turley et al. 2017). MTAG was recently used to analyze GWAS results from neuroticism, depressive symptoms, and subjective well-being, and allowed identification of 66 new loci for neuroticism; the top SNP replicated in an independent sample (N= 8,197) and in previous studies (Supplementary Table 1).
Polygenic strategies for studying the genetics of personality
While the primary goal of GWAS for personality traits has been to identify specific loci, polygenic analyses can be performed using both significant and non-significant GWAS results. In this section, we will discuss these polygenic approaches (see Box 2).
BOX 2. MEASURING SNP HERITABILITY, GENE PATHWAYS, AND GENETIC OVERLAP.
SNP heritability (or ‘chip heritability’; hg2) is the proportion of phenotypic variance explained by additive contributions from large sets of genotyped and imputed SNPs, measured in a set of unrelated individuals. Using a Genomic-relatedness-matrix Restricted Maximum Likelihood (GREML) analysis, the Genetic Complex Trait Analysis software (GCTA; Yang et al. 2010) estimates the proportion of variation in a phenotype that is due to all the SNPs tested, and exploits the fact that genotypic similarity (i.e., “relatedness”, measured using genotyped SNPs from unrelated individuals) will be correlated with phenotypic similarity for heritable traits. Linkage Disequilibrium Regression Score (LDSC) is a more recent method that provides estimates of SNP heritability and takes linkage disequilibrium (LD) into account (Bulik-Sullivan et al. 2015b, a). An advantage of LDSC is that it only requires the results of the GWAS (the “summary statistics”) rather than the individual-level phenotype and genotype data that is required by GCTA.
Gene set and pathway analyses incorporate multiple genetic signals relevant to a specific gene that would not otherwise reach GWAS significance (Wray et al. 2014). This method requires a lower significance threshold (P < 2.8 × 10−6) because it only needs to correct for the number of genes being tested (approximately 18,000; de Leeuw et al. 2016).
Genomic profile risk scoring methods sum the effects of multiple SNPs to predict individual risk for a given trait (Wray et al. 2014; Rietveld et al. 2014). This approach can be used to assess the shared genetic basis of a target sample (e.g. personality) and a ‘discovery sample’ (e.g. psychiatric disorder). Because the discovery sample is independent from the target sample, any detected observation must be genetically driven since there are no shared environmental factors.
Genetic correlations estimate how much of the genetic influence on two traits/diseases is common to both. Unlike phenotypic correlations, which can be due to a combination of genetic and non-genetic factors, genetic correlations can only measure genetically driven correlations. GCTA uses bivariate GREML analysis to estimate the genetic variance of two traits and the genetic covariance between them that is captured by all SNPs tested (Lee et al. 2012). LDSC calculates pairwise genetic correlations for autosomal SNPs between two traits using the summary results of GWAS from two independent datasets (Bulik-Sullivan et al. 2015a).
SNP heritability
The earliest estimates of the heritability of personality traits came from twin studies (Table 1). More recently, the heritable variance attributable to measured SNPs (see Box 2) has been used to estimate the heritability of personality traits; this so-called “SNP heritability” ranges from 5–18% (Table 4). Whereas twin studies provide heritability estimates for both common and rare alleles, SNP heritability is based on available SNP information from unrelated individuals, which is typically limited to relatively common alleles (Yang et al. 2011a) excluding other factors such as dominance effects, gene-gene and gene-environment interactions; therefore, SNP heritability is expected to be lower than the narrow sense heritability obtained using twins (see Plomin and Dreary 2015 for an extended discussion).
Table 4.
SNP heritability estimates of personality traits.
| Reference | Questionnaire | Trait | Study Design | Discovery Sample | GWAS association P < 5×10−8 | SNP-heritability (%, SE) |
|---|---|---|---|---|---|---|
| (Vinkhuyzen et al. 2012) | EPQ | Extraversion | GCTA | ~ 12,000 EUR | N | 6% (3%) |
| Neuroticism | 12% (3%) | |||||
| (Verweij et al. 2012) | TPQ | HA | GCTA | ~ 8,000 EUR | NA | 6.6% (3.7%) |
| NS | 9.9% (3.6%) | |||||
| RD | 4.2% (3.6%) | |||||
| Persistence | 8.1% (3.7%) | |||||
| (Power and Pluess 2015) | NEO-PI-R | Neuroticism | GCTA | 5,011 EUR | NA | 15% (8%) |
| Openness | 21% (8%) | |||||
| (de Moor et al., 2015) | IRT | Neuroticism | GCTA | 63,661 EUR (NTR, QIMR cohorts) | 1 SNP | 14.7% (5.5%) |
| 15.7% (8.3%) | ||||||
| (van den Berg et al. 2016) | IRT | Extraversion | GCTA | 3,597 and 3,369 (NTR, QIMR cohorts) | N | 4.9% (0.8%) |
| (Okbay et al. 2016) | EPQ, NEO | Neuroticism | LDSC | 170,911 EUR | 11 SNPs | 9.1% (0.7%) |
| (Smith et al. 2016) | EPQ (short) | Neuroticism | GCTA | ~ 106,000 EUR (UK Biobank) | 9 SNPs | 15% (0.7%) |
| (Lo et al. 2016) | EPQ, NEO-FFI | Extraversion | LDSC | 59,176 EUR (23andMe) | 5 SNPs | 18.1% (1.0%) |
| Neuroticism | 2 SNPs | 11.9% (1.6%) | ||||
| Conscientiousness | 1 SNP | 9.6% (0.9%) | ||||
| Agreeableness | 8.5% (0.9%) | |||||
| Openness | 10.7% (0.9%) | |||||
| (Docherty et al. 2016) | EPQ | Neuroticism | GCTA | 9,633 EAS (MDD and control women) | NA | 10% (3%) |
| (Luciano et al. 2017) | EPQ (short) | Neuroticism | LDSC | 329,821 EUR (UK Biobank) | 116 SNPs | 10.8% (0.5%) |
| (Nagel et al. 2017) | EPQ (short), NEO, NEO-FFI | Neuroticism | LDSC | 449,484 EUR (UK Biobank, 23andMe, GPC1) | 136 SNPs | 10.0% (0.3%) |
Gene-set and pathway based analyses
Gene-set and pathway based analyses are intended to perform tests at the gene or pathway levels, rather than at the level of individual SNPs and are described in Box 2. The first two attempts to apply gene-base analyses to personality traits did not identify significant findings, likely due to insufficient power (Verweij et al. 2010; Service et al. 2012). Using GWAS results from larger datasets, Luciano et al. (2012) identified six genes for neuroticism (EPQ), five of which were in a region of high LD on Chromosome 15. One of those genes, SCAMP2 (P = 1.0 × 10−6), which is involved in norepinephrine transporter function and is a drug target of mood disorders (Matthies et al. 2009). That same year, de Moor et al. (2012) reported that the gene KATNAL2, which is involved in neurodevelopment (neuronal migration, axonal growth, dendritic pruning), was implicated in conscientiousness (P = 2.0 × 10−6). For extraversion, van den Berg et al. (2016) found one significant association for a long non-coding RNA site (P = 2.87 × 10−6). Using the largest cohort thus far (N = 329,000), Luciano et al. (2017) identified 249 genes significantly associated with neuroticism (P < 2.77 × 10−6). Intriguingly, three of these genes (XKR6 and L3MBTL2 and CHADL) are near SNPS that have been previously associated with neuroticism (Lo et al. 2016).
Gene-set analyses have repeatedly implicated genes involved in synapse and cell communication. For example, Kim et al. (2015b) identified an association between axon guidance gene sets and neuroticism (NEO); similarly, pathways comprising cell adhesion molecules were enriched for neuroticism. More recently, Luciano et al. (2017) implicated five pathways (P = 1.21 × 10−6) in processes for neuron spine and differentiation and cell adhesion in neuroticism (EPQ); Nagel et al. (2017) identified three specific pathways including neurogenesis (P = 4.4 × 10−9), behavioral response to cocaine (P = 1.84 × 10−7), and axon guidance (P = 5.26 × 10−8). Pathways that implicate synaptic communication were also implicated when using meta-analyzed data for depressive symptoms, neuroticism and subjective well-being (Turley et al. 2017); this relationship was strongest for depressive symptoms.
Genomic profile risk scoring
There are numerous studies showing correlations between personality traits and various forms of psychopathology. All of these correlations, which we term ‘phenotypic correlations’ are based on measurements of both traits in the same subjects and cannot easily separate the environmental versus the genetic basis of the correlations. For example, neuroticism is correlated with anxiety, depressive and substance use disorders (Kotov et al. 2010). Twin studies have extended these observations, and demonstrated that the overlap between personality traits and psychopathology has a genetic basis. For example, co-morbidity between neuroticism and both MDD and anxiety disorders is attributed to shared genetic vulnerability (Middeldorp et al. 2005; Kendler and Myers 2010).
Genomic profile risk scoring (PRS) is a method for investigating the polygenic nature of traits like personality and can also be used to explore the genetic basis of correlations between traits like personality and psychopathology (Box 2). Reassuringly, PRS for neuroticism significantly predicted neuroticism in independent cohorts (Luciano et al. 2012, 2017; de Moor et al. 2015; Nagel et al. 2017). Recently, polygenic scores based on joint GWAS for neuroticism, subjective well-being and depressive symptoms showed greater power at predicting neuroticism scores (Turley et al. 2017). Early evidence based on a meta-analysis of GWAS data (Middeldorp et al. 2011) revealed that both PRS for neuroticism, and PRS for extroversion, predicted BP status. Furthermore, PRS for neuroticism predicted MDD (Middeldorp et al. 2011; de Moor et al. 2015; Luciano et al. 2017), and depressive symptoms (Luciano et al. 2012). Similarly, PRS for neuroticism obtained using European subjects could predict MDD in Chinese women (Docherty et al. 2016); showing that PRS for neuroticism can even be useful when applied to populations with different ancestry. Lastly, PRS of neuroticism (calculated using summary data from the GPC) also predicted more general health risk outcomes, including high BMI, smoking, and coronary artery disease (Gale et al. 2016). In all these examples, the two traits being examined are not measured in the same individuals. Instead, the correlation is based on genetic similarity among SNPs between two independent cohorts. This approach removes confounding environmental influences and thus disentangles genetic and environmental influences.
Genetic correlations between personality traits, and overlap with psychopathology
Linkage Disequilibrium Regression Score (LDSC) is a method for performing genetic correlations (see Box 2 and Bulik-Sullivan et al. 2015a) that has been widely adopted, in part because it only requires the more readily available summary statistics produced by a GWAS. Smith et al. (2016) used LDSC to show a positive genetic correlation between neuroticism (EPQ) and MDD (64% ± 7%), and a much smaller genetic correlation with schizophrenia (SCZ; 22% ± 5%). Similarly, moderate and high genetic correlations were later identified between neuroticism (NEO, EPQ) and MDD, at 56%, 68% (± 7%) and 68% (Lo et al. 2016; Luciano et al. 2017; Nagel et al. 2017, respectively), consistent with genetic correlations found in twin research (Kendler, Neale, Kessler, Heath, & Eaves, 1993; Kendler, Gatz, Gardner, & Pedersen, 2006). Moreover, Okbay et al (2016) identified a negative genetic correlation between neuroticism and subjective well-being, at 75% (± 3%); and a positive genetic correlation between neuroticism and depressive symptoms, at 75% (± 3%), as has also been recently identified by Luciano et al. (2017; 82% ± 3%). The latest meta-analysis of neuroticism (Nagel et al. 2017) identified genetically distinguishable subclusters, namely ‘depressed affect’ and ‘worry’, both showing strong positive genetic correlations with major depressive (66–58%, respectively) and anxiety disorders (67–74%), and negative with subjective well-being (−55–52%).
Power and Pluess (2015) reported inter-correlation values between different personality traits (the International Personality Item Pool, IPIP, which is highly correlated with the NEO inventory, rs=0.70–0.82), identifying a strong genetic correlation between neuroticism and openness (82% ± 39%, P < 0.05). Using LDSC, Lo et al. (2016) identified positive inter-correlation values between different personality traits (NEO), which were generally no higher than 40% (Lo et al. 2016). Lo et al. (2016) subsequently conducted a principal component analysis to extract components of genetic variation, derived from the genetic correlation matrix between all personality traits and neuropsychiatric disorders. They found that all psychiatric disorders had positive correlations with the first principal component, while agreeableness and conscientiousness had negative correlations with the first principal component. The extraversion-introversion axis aligned with the second principal component. In general, they found that most personality traits (conscientiousness, agreeableness and extraversion) clustered together. Furthermore, some of the personality traits clustered with several neuropsychiatric disorders. For example, openness clustered with BP and SCZ. Intriguingly, ADHD clustered with personality traits more than psychiatric disorders, showing an especially high genetic correlation with extraversion (30%, SE not reported). This indicates that ADHD, or at least some ADHD subtypes, is genetically coincident with an extreme form of extraversion. Considering the heterogeneity of the ADHD disorder (e.g., subtypes and differences between child and adult forms), genetic studies of personality may particularly helpful for characterizing the etiology of different ADHD subtypes (Karalunas et al. 2014).
More recently, a positive genetic correlation between neuroticism and loneliness (40% ± 9%; Gao et al. 2017), and delay-discounting (18% ± 8%; Sanchez-Roige et al. 2017), have been reported. Correlations between genetic variants for personality traits and other traits may thus improve our understanding of the biological basis of psychiatric diseases and inform novel diagnostic approaches (Insel 2014).
Limitations and future directions
GWAS have furthered our understanding of the etiology of personality traits. The number of specific loci discovered will increase as ever-larger sample sizes are obtained. However, some attention should also be given to the limitations of these studies that cannot be addressed by sample size alone. First, the methods used to date are dominated by GWAS, which focus on additive effects, at the expense of non-additive genetic variance, such as dominance and epistasis. Additionally, by focusing on SNPs, genetic contributions from structural and rare variants are not captured. Moreover, longitudinal twin studies (Bleidorn et al. 2014) revealed a stable genetic foundation throughout adulthood, with more prominent environmental influences during early-middle adulthood. This observation suggests that both genetic and non-shared environmental factors influence personality traits (Plomin et al. 2016), but this has not been adequately tested. Third, even though polygenic methods have identified potential overlap between personality traits and psychopathology, additional methods are needed to identify the specific variants. For example, in a recent Phenome Wide Association Study (PheWAS), which tests the contribution of a genetic variant across multiple traits, the genes CADM2 and MSRA were associated with risk taking behavior and irritability, respectively, and aspects of the Big Five traits (Boutwell et al. 2017). In addition, a ‘proxy-phenotype’ method (Rietveld et al. 2014) has also been used to identify genetic overlap between personality and psychopathology. Okbay et al. (2016) identified a set of 170 candidate SNPs with suggestive evidence of association (P < 1 × 10−4) with subjective well-being, and tested these candidates for association with neuroticism. Using this method, the authors identified four SNPs showing overlap between neuroticism and subjective well-being.
GWAS associations do not unambiguously identify specific genes and thus do not directly lead to novel biological insights. Pathway analyses of GWAS will likely become more useful as pathway annotations continue to improve. The integration of data from GWAS and gene expression will present new opportunities and challenges in the discovery of the genes underlying implicated loci and pathways associated with personality (Fan et al. 2017). Using imputed transcripts from GWAS and comparing those with drug-induced changes in gene expression, So et al. (2017) recently demonstrated that the effectiveness of psychiatric medications was strongly associated with genomic information based on GWAS data for several psychiatric conditions. Aggregating genetic findings based on GWAS of personality traits may ultimately influence the development of novel medications for psychiatric conditions that are characterized by, for example, high levels of neuroticism.
Conclusion
Over the past decade, findings from GWAS and polygenic analyses have made major progress towards unraveling the genetic etiology of personality traits. And yet, the path from GWAS to biology and, ultimately, psychology is not straightforward. Functional validation of some of the identified potential candidate genes will prove difficult, as there are no direct animal analogues of personality traits. Nonetheless, continued inquiry into the genetic basis of personality has the potential to yield insight into the biology of psychiatric disease risk.
Supplementary Material
Funding
J.M.’s contributions were partially supported by the Peter Boris Chair in Addictions Research. C.-H. C. is supported by National Institute of Mental Health R01MH100351. S.S-R is supported by the Frontiers of Innovation Scholars Program (FISP), the Interdisciplinary Research Fellowship in NeuroAIDS (IRFN), and pilot award from P50DA037844.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Disclosure
The opinions and assertions expressed herein are those of the author(s) and do not necessarily reflect the official policy or position of the Uniformed Services University or the Department of Defense.
REFERENCES
- Amin N, Hottenga J-J, Hansell NK, et al. (2013) Refining genome-wide linkage intervals using a meta-analysis of genome-wide association studies identifies loci influencing personality dimensions. Eur J Hum Genet 21:876–882. doi: 10.1038/ejhg.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae HT, Sebastiani P, Sun JX, et al. (2013) Genome-Wide Association Study of Personality Traits in the Long Life Family Study. Front Genet. doi: 10.3389/fgene.2013.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin J, Li L, Patterson C, et al. (1996) Population and familial association between the D4 dopamine receptor gene and measures of Novelty Seeking. Nat Genet 12:81–84. doi: 10.1038/ng0196-81 [DOI] [PubMed] [Google Scholar]
- Bleidorn W, Kandler C, Caspi A (2014) The Behavioural Genetics of Personality Development in Adulthood-Classic, Contemporary, and Future Trends: Behavioural genetics of personality development. Eur J Personal 28:244–255. doi: 10.1002/per.1957 [DOI] [Google Scholar]
- Boomsma D, Busjahn A, & Peltonen L (2002) Classical twin studies and beyond. Nature Reviews Genetics 3:872–882. [DOI] [PubMed] [Google Scholar]
- Bouchard TJ, Loehlin JC (2001) Genes, evolution, and personality. Behav Genet 31:243–273. [DOI] [PubMed] [Google Scholar]
- Boutwell B, Hinds D, 23andMe Research Team, et al. (2017) Replication and characterization of CADM2 and MSRA genes on human behavior. Heliyon 3:e00349. doi: 10.1016/j.heliyon.2017.e00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, Li Y, Pritchard JK (2017) An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169:1177–1186. doi: 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratko D, Butković A, Vukasović Hlupić T (2017) Heritability of Personality. Psychol Top 26:1–24. [Google Scholar]
- Briley DA, Tucker-Drob EM (2014) Genetic and environmental continuity in personality development: A meta-analysis. Psychol Bull 140:1303–1331. doi: 10.1037/a0037091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, et al. (2015a) An atlas of genetic correlations across human diseases and traits. Nat Genet 47:1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Loh P-R, Finucane HK, et al. (2015b) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calboli FCF, Tozzi F, Galwey NW, et al. (2010) A Genome-Wide Association Study of Neuroticism in a Population-Based Sample. PLoS ONE 5:e11504. doi: 10.1371/journal.pone.0011504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervone D, Pervin L (2009) Personality: Theory and research, : John Wiley and Sons; Hoboken, NJ [Google Scholar]
- Chabris CF, Lee JJ, Benjamin DJ, et al. (2013) Why it is hard to find genes associated with social science traits: theoretical and empirical considerations. Am J Public Health 103 Suppl 1:S152–166. doi: 10.2105/AJPH.2013.301327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Svrakic DM, Przybeck TR (1993) A psychobiological model of temperament and character. Arch Gen Psychiatry 50:975–990. [DOI] [PubMed] [Google Scholar]
- Cloninger R (1994) The temperament and character inventory (TCI): a guide to its development and use Center for Psychobiology of Personality, Washington University, St. Louis, Mo. [Google Scholar]
- Colhoun HM, McKeigue PM, Smith GD (2003) Problems of reporting genetic associations with complex outcomes. The Lancet 361:865–872. doi: 10.1016/S0140-6736(03)12715-8 [DOI] [PubMed] [Google Scholar]
- Costa PT, McCrae RR (1992) The Five-Factor Model of Personality and Its Relevance to Personality Disorders. J Personal Disord 6:343–359. doi: 10.1521/pedi.1992.6.4.343 [DOI] [Google Scholar]
- de Leeuw CA, Neale BM, Heskes T, Posthuma D (2016) The statistical properties of gene-set analysis. Nat Rev Genet 17:353–364. doi: 10.1038/nrg.2016.29 [DOI] [PubMed] [Google Scholar]
- de Moor MHM, Costa PT, Terracciano A, et al. (2012) Meta-analysis of genome-wide association studies for personality. Mol Psychiatry 17:337–349. doi: 10.1038/mp.2010.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MHM, van den Berg SM, Verweij KJH, et al. (2015) Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA Psychiatry 72:642. doi: 10.1001/jamapsychiatry.2015.0554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty AR, Moscati A, Peterson R, et al. (2016) SNP-based heritability estimates of the personality dimensions and polygenic prediction of both neuroticism and major depression: findings from CONVERGE. Transl Psychiatry 6:e926. doi: 10.1038/tp.2016.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duberstein PR, Chapman BP, Tindle HA, et al. (2011) Personality and Risk for Alzheimer’s Disease in Adults 72 Years of Age and Older: A Six-Year Follow-Up. Psychol Aging 26:351–362. doi: 10.1037/a0021377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebstein RP (2006) The molecular genetic architecture of human personality: beyond self-report questionnaires. Mol Psychiatry 11:427–445. doi: 10.1038/sj.mp.4001814 [DOI] [PubMed] [Google Scholar]
- Etain B, Mathieu F, Rietschel M, et al. (2006) Genome-wide scan for genes involved in bipolar affective disorder in 70 European families ascertained through a bipolar type I early-onset proband: supportive evidence for linkage at 3p14. Mol Psychiatry 11:685–694. doi: 10.1038/sj.mp.4001815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eysenck H, Eysenck S (1975) Manual for the Eysenck Personality Questionnaire. Hodder Stoughton, London [Google Scholar]
- Evans DM, Gillespie NA, Martin NG (2002) Biometrical genetics. Biol Psychol 61:33–51. doi: 10.1016/S0301-0511(02)00051-0 [DOI] [PubMed] [Google Scholar]
- Fan Q, Wang W, Hao J, et al. (2017) Integrating genome-wide association study and expression quantitative trait loci data identifies multiple genes and gene set associated with neuroticism. Prog Neuropsychopharmacol Biol Psychiatry 78:149–152. doi: 10.1016/j.pnpbp.2017.05.017 [DOI] [PubMed] [Google Scholar]
- Ferentinos P, Rivera M, Ising M, et al. (2014) Investigating the genetic variation underlying episodicity in major depressive disorder: Suggestive evidence for a bipolar contribution. J Affect Disord 155:81–89. doi: 10.1016/j.jad.2013.10.027 [DOI] [PubMed] [Google Scholar]
- Flint J, Willis-Owen S (2010) The Genetics of Personality In: Vogel and Motulsky’s Human Genetics. Springer, Berlin, Heidelberg, pp 651–661 [Google Scholar]
- Fullerton J, Cubin M, Tiwari H, et al. (2003) Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am J Hum Genet 72:879–890. doi: 10.1086/374178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale CR, Hagenaars SP, Davies G, et al. (2016) Pleiotropy between neuroticism and physical and mental health: findings from 108 038 men and women in UK Biobank. Transl Psychiatry 6:e791. doi: 10.1038/tp.2016.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Davis LK, Hart AB, et al. (2017) Genome-Wide Association Study of Loneliness Demonstrates a Role for Common Variation. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 42:811–821. doi: 10.1038/npp.2016.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Zhu G, Evans DM, et al. (2008) A Genome-Wide Scan for Eysenckian Personality Dimensions in Adolescent Twin Sibships: Psychoticism, Extraversion, Neuroticism, and Lie. J Pers 76:1415–1446. doi: 10.1111/j.1467-6494.2008.00527.x [DOI] [PubMed] [Google Scholar]
- Hart AB, de Wit H, Palmer AA (2013) Candidate Gene Studies of a Promising Intermediate Phenotype: Failure to Replicate. Neuropsychopharmacology 38:802–816. doi: 10.1038/npp.2012.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR (2014) The NIMH Research Domain Criteria (RDoC) Project: precision medicine for psychiatry. Am J Psychiatry 171:395–397. doi: 10.1176/appi.ajp.2014.14020138 [DOI] [PubMed] [Google Scholar]
- Jang KL, Livesley WJ, Vernon PA (1996) Heritability of the big five personality dimensions and their facets: a twin study. J Pers 64:577–591. [DOI] [PubMed] [Google Scholar]
- Johnson AM, Vernon PA, Feiler AR (2008) Behavioral genetic studies of personality: An introduction and review of the results of 50+ years of research In: Boyle GJ, Matthews G, Saklofske DH (eds) The SAGE handbook of personality theory and assessment, Vol 1: Personality theories and models. Sage Publications, Inc, Thousand Oaks, CA, US, pp 145–173 [Google Scholar]
- Johnson EC, Border R, Melroy-Greif WE, et al. (2017) No Evidence That Schizophrenia Candidate Genes Are More Associated With Schizophrenia Than Noncandidate Genes. Biol Psychiatry. doi: 10.1016/j.biopsych.2017.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge TA, Rodell JB, Klinger RL, et al. (2013) Hierarchical representations of the five-factor model of personality in predicting job performance: integrating three organizing frameworks with two theoretical perspectives. J Appl Psychol 98:875–925. doi: 10.1037/a0033901 [DOI] [PubMed] [Google Scholar]
- Jylhä P, Mantere O, Melartin T, et al. (2010) Differences in neuroticism and extraversion between patients with bipolar I or II and general population subjects or major depressive disorder patients. J Affect Disord 125:42–52. doi: 10.1016/j.jad.2010.01.068 [DOI] [PubMed] [Google Scholar]
- Kandler C, Richter J, Zapko-Willmes A (2017) Genetic Basis of Traits In: Zeigler-Hill V, Shackelford TK (eds) Encyclopedia of Personality and Individual Differences. Springer International Publishing, Cham, pp 1–13 [Google Scholar]
- Kappe R, H van der Flier (2012) Predicting academic success in higher education: what’s more important than being smart? Eur J Psychol Educ 27:605–619. doi: 10.1007/s10212-011-0099-9 [DOI] [Google Scholar]
- Karalunas SL, Fair D, Musser ED, et al. (2014) Subtyping attention-deficit/hyperactivity disorder using temperament dimensions: toward biologically based nosologic criteria. JAMA Psychiatry 71:1015–1024. doi: 10.1001/jamapsychiatry.2014.763 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Karlsson R, Graae L, Lekman M, et al. (2012) MAGI1 Copy Number Variation in Bipolar Affective Disorder and Schizophrenia. Biol Psychiatry 71:922–930. doi: 10.1016/j.biopsych.2012.01.020 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Gatz M, Gardner CO, Pedersen NL (2006) Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry 63:1113–1120. doi: 10.1001/archpsyc.63.10.1113 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Myers J (2010) The genetic and environmental relationship between major depression and the five-factor model of personality. Psychol Med 40:801–806. doi: 10.1017/S0033291709991140 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, et al. (1993) A longitudinal twin study of personality and major depression in women. Arch Gen Psychiatry 50:853–862. [DOI] [PubMed] [Google Scholar]
- Khan AA, Jacobson KC, Gardner CO, et al. (2005) Personality and comorbidity of common psychiatric disorders. Br J Psychiatry J Ment Sci 186:190–196. doi: 10.1192/bjp.186.3.190 [DOI] [PubMed] [Google Scholar]
- Kim B-H, Kim H-N, Roh S-J, et al. (2015a) GWA meta-analysis of personality in Korean cohorts. J Hum Genet 60:455–460. doi: 10.1038/jhg.2015.52 [DOI] [PubMed] [Google Scholar]
- Kim H, Roh S-J, Sung YA, et al. (2013) Genome-wide association study of the five-factor model of personality in young Korean women. J Hum Genet 58:667–674. doi: 10.1038/jhg.2013.75 [DOI] [PubMed] [Google Scholar]
- Kim H-N, Kim B-H, Cho J, et al. (2015b) Pathway analysis of genome-wide association datasets of personality traits. Genes Brain Behav 14:345–356. doi: 10.1111/gbb.12212 [DOI] [PubMed] [Google Scholar]
- Kuo P-H, Neale MC, Riley BP, et al. (2007) A genome-wide linkage analysis for the personality trait neuroticism in the Irish affected sib-pair study of alcohol dependence. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 144B:463–468. doi: 10.1002/ajmg.b.30478 [DOI] [PubMed] [Google Scholar]
- Kupper N, Boomsma DI, de Geus EJC, et al. (2011) Nine-Year Stability of Type D Personality: Contributions of Genes and Environment: Psychosom Med 73:75–82. doi: 10.1097/PSY.0b013e3181fdce54 [DOI] [PubMed] [Google Scholar]
- Lahey BB (2009) Public Health Significance of Neuroticism. Am Psychol 64:241–256. doi: 10.1037/a0015309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, et al. (2012) Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinforma Oxf Engl 28:2540–2542. doi: 10.1093/bioinformatics/bts474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch K-P, Bengel D, Heils A, et al. (1996) Association of Anxiety-Related Traits with a Polymorphism in the Serotonin Transporter Gene Regulatory Region. Science 274:1527–1531. doi: 10.1126/science.274.5292.1527 [DOI] [PubMed] [Google Scholar]
- Lo M-T, Hinds DA, Tung JY, et al. (2016) Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet 49:152–156. doi: 10.1038/ng.3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Hagenaars SP, Davies G, et al. (2017) 116 independent genetic variants influence the neuroticism personality trait in over 329,000 UK Biobank individuals. bioRxiv 168906. doi: 10.1101/168906 [DOI] [Google Scholar]
- Luciano M, Huffman JE, Arias-Vásquez A, et al. (2012) Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 0:684–695. doi: 10.1002/ajmg.b.32072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies HJG, Han Q, Shields A, et al. (2009) Subcellular localization of the antidepressant-sensitive norepinephrine transporter. BMC Neurosci 10:65. doi: 10.1186/1471-2202-10-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp CM, Cath DC, Van Dyck R, Boomsma DI (2005) The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med 35:611–624. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, de Moor MHM, McGrath LM, et al. (2011) The genetic association between personality and major depression or bipolar disorder. A polygenic score analysis using genome-wide association data. Transl Psychiatry 1:e50. doi: 10.1038/tp.2011.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, Bonvicini C, Scassellati C, et al. (2011) The influence of psychiatric screening in healthy populations selection: a new study and meta-analysis of functional 5-HTTLPR and rs25531 polymorphisms and anxiety-related personality traits. BMC Psychiatry 11:50. doi: 10.1186/1471-244X-11-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, Scassellati C, Bonvicini C, et al. (2009) An association of GRIK3 Ser310Ala functional polymorphism with personality traits. Neuropsychobiology 59:28–33. doi: 10.1159/000202827 [DOI] [PubMed] [Google Scholar]
- Munafò, Flint J (2011) Dissecting the genetic architecture of human personality. Trends Cogn Sci 15:395–400. doi: 10.1016/j.tics.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Munafò MR, Clark TG, Moore LR, et al. (2003) Genetic Polymorphisms and Personality in Healthy Adults: A systematic review and meta-analysis. Mol Psychiatry 8:471–484. doi: 10.1038/sj.mp.4001326 [DOI] [PubMed] [Google Scholar]
- Munafò MR, Freimer NB, Ng W, et al. (2009) 5-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 150B:271–281. doi: 10.1002/ajmg.b.30808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò MR, Yalcin B, Willis-Owen SA, Flint J (2008) Association of the dopamine D4 receptor (DRD4) gene and approach-related personality traits: meta-analysis and new data. Biol Psychiatry 63:197–206. doi: 10.1016/j.biopsych.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Nagel M, Jansen PR, Stringer S, et al. (2017) GWAS Meta-Analysis of Neuroticism (N=449,484) Identifies Novel Genetic Loci and Pathways. bioRxiv 184820. doi: 10.1101/184820 [DOI] [PubMed] [Google Scholar]
- Nash M, Huezo-Diaz P, Williamson RJ, et al. (2004) Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet 13:2173–2182. doi: 10.1093/hmg/ddh239 [DOI] [PubMed] [Google Scholar]
- Okbay A, Baselmans BML, De Neve J-E, et al. (2016) Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 48:624–633. doi: 10.1038/ng.3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Kamatani Y, Lathrop M (2011) Family-based designs for genome-wide association studies. Nat Rev Genet 12:465–474. doi: 10.1038/nrg2989 [DOI] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, Knopik VS, Neiderhiser JM (2016) Top 10 Replicated Findings From Behavioral Genetics. Perspect Psychol Sci 11:3–23. doi: 10.1177/1745691615617439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, et al. (2015) Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47:702–709. doi: 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- Power RA, Pluess M (2015) Heritability estimates of the Big Five personality traits based on common genetic variants. Transl Psychiatry 5:e604. doi: 10.1038/tp.2015.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Conley D, Eriksson N, et al. (2014) Replicability and robustness of genome-wide-association studies for behavioral traits. Psychol Sci 25:1975–1986. doi: 10.1177/0956797614545132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Esko T, Davies G, et al. (2014) Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci 111:13790–13794. doi: 10.1073/pnas.1404623111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijsdijk FV, Sham PC (2002) Analytic approaches to twin data using structural equation models. Brief Bioinform 3:119–133. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The Future of Genetic Studies of Complex Human Diseases. Science. 273:1516–1517. [DOI] [PubMed] [Google Scholar]
- Roberts BW (2009) Back to the Future: Personality and Assessment and Personality Development. J Res Personal 43:137–145. doi: 10.1016/j.jrp.2008.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Bitsios P (2009) Cognitive and emotional processing in high novelty seeking associated with the L-DRD4 genotype. Neuropsychologia 47:1654–1659. doi: 10.1016/j.neuropsychologia.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Sadahiro R, Suzuki A, Shibuya N, et al. (2010) Association study between a functional polymorphism of tyrosine hydroxylase gene promoter and personality traits in healthy subjects. Behav Brain Res 208:209–212. doi: 10.1016/j.bbr.2009.11.035 [DOI] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, et al. (2017) Genetics of the Research Domain Criteria (RDoC): genome-wide association study of delay discounting. bioRxiv 146936. doi: 10.1101/146936 [DOI] [Google Scholar]
- Schinka JA, Busch RM, Robichaux-Keene N (2004) A meta-analysis of the association between the serotonin transporter gene polymorphism (5-HTTLPR) and trait anxiety. Mol Psychiatry 9:197–202. doi: 10.1038/sj.mp.4001405 [DOI] [PubMed] [Google Scholar]
- Schinka JA, Letsch EA, Crawford FC (2002) DRD4 and novelty seeking: results of meta-analyses. Am J Med Genet 114:643–648. doi: 10.1002/ajmg.10649 [DOI] [PubMed] [Google Scholar]
- Sen S, Burmeister M, Ghosh D (2004) Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet 127B:85–89. doi: 10.1002/ajmg.b.20158 [DOI] [PubMed] [Google Scholar]
- Service SK, Verweij KJH, Lahti J, et al. (2012) A genome-wide meta-analysis of association studies of Cloninger’s Temperament Scales. Transl Psychiatry 2:e116. doi: 10.1038/tp.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bhomra A, Smiley S, et al. (2008) A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry 13:302–312. doi: 10.1038/sj.mp.4002048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Escott-Price V, Davies G, et al. (2016) Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry 21:749–757. doi: 10.1038/mp.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So H-C, Chau CK-L, Chiu W-T, et al. (2017) Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci. doi: 10.1038/nn.4618 [DOI] [PubMed] [Google Scholar]
- Stetler C, Miller GE (2011) Depression and hypothalamic-pituitary-adrenal activation: a quantitative summary of four decades of research. Psychosom Med 73:114–126. doi: 10.1097/PSY.0b013e31820ad12b [DOI] [PubMed] [Google Scholar]
- Terracciano A, Esko T, Sutin AR, et al. (2011) Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking. Transl Psychiatry 1:e49. doi: 10.1038/tp.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Sanna S, Uda M, et al. (2010a) Genome-wide association scan for five major dimensions of personality. Mol Psychiatry 15:647–656. doi: 10.1038/mp.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, et al. (2010b) BDNF Val66Met is Associated with Introversion and Interacts with 5-HTTLPR to Influence Neuroticism. Neuropsychopharmacology 35:1083–1089. doi: 10.1038/npp.2009.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley P, Walters RK, Maghzian O, et al. (2017) MTAG: Multi-Trait Analysis of GWAS. bioRxiv 118810. doi: 10.1101/118810 [DOI] [Google Scholar]
- van den Berg SM, de Moor MHM, McGue M, et al. (2014) Harmonization of Neuroticism and Extraversion phenotypes across inventories and cohorts in the Genetics of Personality Consortium: an application of Item Response Theory. Behav Genet 44:295–313. doi: 10.1007/s10519-014-9654-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg SM, de Moor MHM, Verweij KJH, et al. (2016) Meta-analysis of Genome-Wide Association Studies for Extraversion: Findings from the Genetics of Personality Consortium. Behav Genet 46:170–182. doi: 10.1007/s10519-015-9735-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord EJCG, Kuo P-H, Hartmann AM, et al. (2008) Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatry 65:1062–1071. doi: 10.1001/archpsyc.65.9.1062 [DOI] [PubMed] [Google Scholar]
- Van Gestel S, Van Broeckhoven C (2003) Genetics of personality: are we making progress? Mol Psychiatry 8:840–852. doi: 10.1038/sj.mp.4001367 [DOI] [PubMed] [Google Scholar]
- Verweij KJH, Yang J, Lahti J, et al. (2012) Maintenance of genetic variation in human personality: testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evol Int J Org Evol 66:3238–3251. doi: 10.1111/j.1558-5646.2012.01679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Medland SE, et al. (2010) A genome-wide association study of Cloninger’s temperament scales: implications for the evolutionary genetics of personality. Biol Psychol 85:306–317. doi: 10.1016/j.biopsycho.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Bartels M, van Beijsterveldt TCEM, et al. (2012) Sex Differences in Genetic Architecture of Complex Phenotypes? PLOS ONE 7:e47371. doi: 10.1371/journal.pone.0047371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkhuyzen AAE, Pedersen NL, Yang J, et al. (2012) Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Transl Psychiatry 2:e102. doi: 10.1038/tp.2012.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, et al. (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 101:5–22. doi: 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukasović T, Bratko D (2015) Heritability of personality: A meta-analysis of behavior genetic studies. Psychol Bull 141:769–785. doi: 10.1037/bul0000017 [DOI] [PubMed] [Google Scholar]
- Weber H, Richter J, Straube B, et al. (2016) Allelic variation in CRHR1 predisposes to panic disorder: evidence for biased fear processing. Mol Psychiatry 21:813–822. doi: 10.1038/mp.2015.125 [DOI] [PubMed] [Google Scholar]
- Wills TA, Vaccaro D, McNamara G (1994) Novelty seeking, risk taking, and related constructs as predictors of adolescent substance use: An application of Cloninger’s theory. J Subst Abuse 6:1–20. doi: 10.1016/S0899-3289(94)90039-6 [DOI] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, et al. (2014) Research review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 55:1068–1087. doi: 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- Wray NR, Middeldorp CM, Birley AJ, et al. (2008) Genome-Wide Linkage Analysis of Multiple Measures of Neuroticism of 2 Large Cohorts From Australia and the Netherlands. Arch Gen Psychiatry 65:649. doi: 10.1001/archpsyc.65.6.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee H, Goddard ME, Visscher PM (2011a) GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet 88:76–82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, et al. (2011b) Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 43:519–525. doi: 10.1038/ng.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonan AL, Palmer AA, Gilliam TC (2006) Hardy-Weinberg disequilibrium identified genotyping error of the serotonin transporter (SLC6A4) promoter polymorphism. Psychiatr Genet 16:31–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


