Abstract
Background
Inflammatory bowel disease (IBD) is a lifelong digestive disease characterized by periods of severe inflammation and remission. To our knowledge, this is the first study showing a variable effect on ileitis severity from human gut microbiota isolated from IBD donors in remission and that of healthy controls in a mouse model of IBD.
Methods
We conducted a series of single-donor intensive and nonintensive fecal microbiota transplantation (FMT) experiments using feces from IBD patients in remission and healthy non-IBD controls (N = 9 donors) in a mouse model of Crohn’s disease (CD)-like ileitis that develops ileitis in germ-free (GF) conditions (SAMP1/YitFC; N = 96 mice).
Results
Engraftment studies demonstrated that the microbiome of IBD in remission could have variable effects on the ileum of CD-prone mice (pro-inflammatory, nonmodulatory, or anti-inflammatory), depending on the human donor. Fecal microbiota transplantation achieved a 95% ± 0.03 genus-level engraftment of human gut taxa in mice, as confirmed at the operational taxonomic unit level. In most donors, microbiome colonization abundance patterns remained consistent over 60 days. Microbiome-based metabolic predictions of GF mice with Crohn’s or ileitic-mouse donor microbiota indicate that chronic amino/fatty acid (valine, leucine, isoleucine, histidine; linoleic; P < 1e-15) alterations (and not bacterial virulence markers; P > 0.37) precede severe ileitis in mice, supporting their potential use as predictors/biomarkers in human CD.
Conclusion
The gut microbiome of IBD remission patients is not necessarily innocuous. Characterizing the inflammatory potential of each microbiota in IBD patients using mice may help identify the patients’ best anti-inflammatory fecal sample for future use as an anti-inflammatory microbial autograft during disease flare-ups.
Keywords: fecal microbiota transplantation, autologous, remission, Crohn’s disease, amino acids, biomarkers
By conducting a series of fecal microbiota transplantation experiments, we show a variable effect on ileitis severity from human gut microbiota isolated from IBD donors in remission and that of healthy controls in a mouse model of IBD.
INTRODUCTION
Inflammatory bowel disease (IBD) is a lifelong chronic disorder of the gastrointestinal tract that causes periods of severe inflammation alternating with periods of remission. Several studies have indicated that environmental factors including abnormal gut microbiota composition (“dysbiosis”) seem to trigger disease and periods of inflammation. Disruptions in the gut microbiota or their metabolic functions are proposed as critical pathogenic elements in various immune disorders such as IBD1–4 that could foster pro-inflammatory imbalances and cause relapse-remission in IBD. However, many aspects of the functional role of the gut microbiota in IBD, especially during remission, remain unknown.
Crohn’s disease (CD) and ulcerative colitis (UC) are the prototypical forms of IBD in humans. Although CD and UC differ in presentation, location, and level of transmural gut wall inflammation, both are characterized by alternating periods of active (flare-up) and inactive (remission) states. Because IBD individuals often feel healthy during remission, it is possible to hypothesize that the gut microbiota is innocuous to the intestinal mucosa/wall during periods of IBD inactivity (remission). The validation/use of IBD spontaneous mouse models is needed to functionally characterize the gut microbiota in IBD patients. Such models could help quantify the causal association between dysbiosis and IBD in remission and determine if feces from healthy donors or IBD remission patients could be used for heterologous (person-person) and autologous (self) transplantation as therapy during flare-ups. A recent meta-analysis indicated that fecal microbiota transplantations (FMT) induce remission in 33%–52% of IBD patients.5 Despite such benefit, it is unclear why FMT is not always successful.6, 7 It is possible that the microbiota from healthy non-IBD donors, or IBD remission patients, is not necessarily innocuous to the gut. Testing such a hypothesis in humans is not possible; therefore, the use of animal models is desirable.
Previous FMT studies have investigated the transfer of human gut microbiota (pooled or unpooled donor samples) using germ-free (GF) mice without disease (eg, C57BL/6J, Swiss Webster) or used immunodeficient and knock-out genetic mouse lines.8–12 Despite the valuable knowledge gained from acute and chemically induced inflammation models, there is a lack of mouse models of spontaneous IBD, in particular models of CD ileitis (inflammation of distal small intestine). As a model, SAMP1/YitFc mice spontaneously develop CD-like “cobblestone” ileitis with a penetrance of 100%,13 with the advantage that bacterial colonization is not a prerequisite for ileitis induction (ie, the disease still manifests in GF SAMP mice). Because gut microbes are known to modulate the severity of disease in specific pathogen-free (SPF) mice,14–16 characterizing the inflammatory potential of human gut microbiotas isolated in the absence of inflammation using ileitis-prone mice may help identify “super donors” or a patient’s best anti-inflammatory fecal sample for future use as autologous FMT therapy.
Herein, we propose to use the spontaneous SAMP mouse model, prone to developing CD-like ileitis, to determine whether the gut microbiota from 9 human donors (6 with IBD in remission and 3 healthy non-IBD) could modulate the progression of ileitis in GF SAMP mice. The 2 main objectives of our study were (1) to determine the effect of gut microbiota isolated from IBD patients in remission and healthy non-IBD controls on the ileitis severity in transplanted GF SAMP mice and (2) to characterize GF SAMP mice as a model to reproducibly harbor human gut microbiota. We used a long-term repeat dose (intensive weekly) inoculation protocol as recommended in humans17, 18 to ensure the proper assessment of the functional effects of the microbiota on the mouse ileitis phenotype and the correspondence with the clinical diagnosis of human donors. Lastly, we tested study reproducibility using less-intense (single-dose) FMT. Herein, we describe the validation steps for our FMT strategy, the colonization efficiency, and the discovery that some patients in remission and some healthy non-IBD individuals harbor pro-inflammatory microbiotas. Metabolic predictions derived from microbiome data also indicated there is a strong association between amino acid metabolic alterations in the gut and CD-like ileitis in FMT SAMP mice, which would support recently reported connections between plasma amino acid concentrations and human IBD.19, 20 These findings emphasize that the role of the gut microbiota in IBD pathogenesis is complex and requires investigation of the effect of an individual human gut microbiome on the regulation of IBD as a future model for donor selection.
MATERIALS AND METHODS
Animals and Preventing External and Bedding Microbial Confounders
This experiment tested groups (5 to 6 mice/group) of age-matched and sex-matched 7-week-old GF SAMP mice.21 All mice were individually caged using our GF-grade nested isolation (NesTiso) caging system22 and maintained on nonedible Aspen bedding (N = 96 mice; 54 were transplanted with human feces; the remaining served as controls) (see study design and caging system in Figs. 1A and 1B). The SAMP mice are a substrain of AKR/J initially developed in Japan (Yakult Central Institute for Microbial Research, Tokyo, Japan). These mice have a genetic, immunological disposition to spontaneously develop intestinal disease.23, 24 Establishment of chronic intestinal inflammation occurs within a well-defined time course with 100% penetrance.13 Mice were kept on a 12 hour light and 12 hour dark cycle in species-appropriate temperature and humidity-controlled rooms and fed autoclaved GF-grade 40 to 50 kGy irradiated pellet food (PMI Nutrition Int’l., LLC. Labdiet Charles River; Vac-Pac Rodent 6/5 irradiated, 5% kcal% fat) and water (double-autoclaved, non-acidic) was given ad libidum. Protocols on animal handling, housing, and transplant of human microbiota into GF mice were approved by the IACUC and the Institutional Review Board at CWRU, following the National Research Council Guide for the Care and Use of Laboratory Animals. Measures to control for bedding-dependent microbial bias/overgrowth were implemented in all experiments, as previously described.22 See details in the online supplementary material.
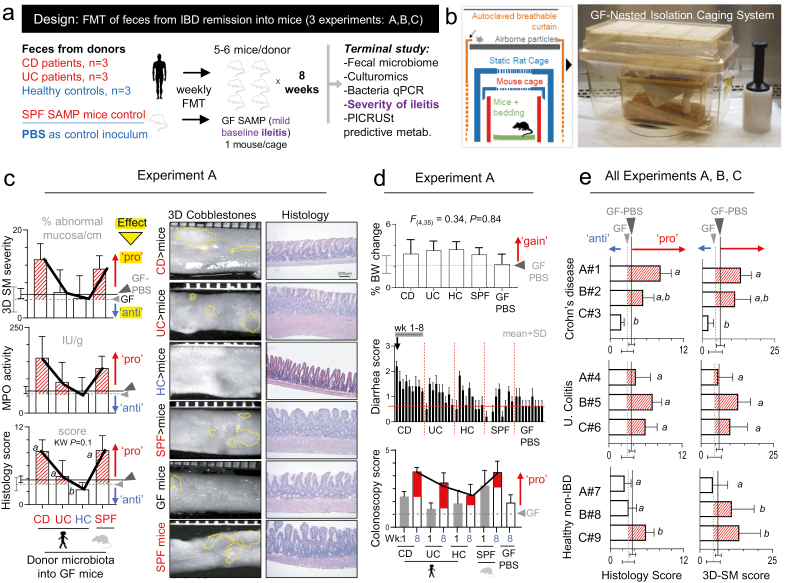
FIGURE 1.
Effect of human gut microbiota transplantation on chronic ileitis in GF SAMP mice depends on donor and not disease status. A, Study design for Exp. A, B, C using weekly FMT (N = 9 donors; n = 6 mice/ human donor). B, Nested isolation (NesTiso) caging system used in this study.22 C, Effect of donor microbiota on the 60-day severity of Crohn’s disease–like ileitis in GF transplanted SAMP mice and GF-PBS control mice. Validated methods were used to assess severity of cobblestone ileitis (dotted yellow lines) in mice, with corresponding representative photomicrographs. As reference, all plots have data from nontransplanted GF SAMP and GF SAMP PBS control mice (GF; grey dashed lines. GF PBS; solid black line) to illustrate the effect of donor microbiota on ileitis. “Pro,” pro-inflammatory effect (arrows and red shaded area above GF-reference line); “anti,” anti-inflammatory (area below line). SD around GF and GF PBS reference dashed, and solid lines represent a range for nonmodifying microbiotas. Thick black line connecting error bars from each column illustrates reproducible patterns across diagnostic tests. D, Effect of donor microbiota on body weight (cumulative change over 60 days), colonoscopy inflammatory scores for weeks 1 and 8 (day 60), and weekly diarrhea scores. E, Effect of gut microbiota from 9 donors (eg, A#1) on ileitis severity (histology scores; pro-inflammatory, nonmodifier [neutral] or anti-inflammatory) after 60 days shown for GF SAMP mice transplanted with human feces. Y-axis; mean ± SD of histological scores in mice for each donor across disease status. A#1 indicates experimental group (ie, A; Exp. A) and arbitrary donor code number (ie, #1). The same superscript letter indicates nonsignificant differences; different letters represent pairwise P < 0.05
Fecal Microbiota Transplantation and Intestinal Inflammation
The GF SAMP mice develop milder ileitis (herein referred to as “mild baseline ileitis”) compared with their SPF counterparts.21 It is also known that SPF SAMP mice have a pro-inflammatory gut microbiota that triggers mouse ileitis.14–16 Herein, we used GF SAMP mice to assess the effect of human microbiota on ileitis severity after transplantation using gut microbiota (feces) isolated from IBD patients in remission (CD and UC) or healthy non-IBD controls (HC). As an additional control, an SPF recipient group received a composite (pool) of feces from our SPF SAMP mouse colony. In all experiments, a systematic approach was used to randomize mice. All IBD donors were in complete remission (assessed by clinical, endoscopic radiologic, and/or biochemical parameters) and were not taking biologics or corticosteroids. No donors reported taking supplements/vitamins, antibiotics, or probiotics. Each mouse received weekly oral gavages (0.20 mL/10 g of body weight; 108–9 CFU/mouse) for 60 days. The GF SAMP (littermates) who were administered a “sham” gavage (phosphate-buffered saline [PBS], 7% dimethyl sulfoxide [DMSO] mixture) were used as negative controls (GF-PBS recipient group). Experiments were repeated 3 times, each time with a different set of donors (9 human donors; Exp. A, B, C; n = 3 donors per experiment). We also performed a single-dose transplantation experiment (Exp. D) in GF mice using a random original CD donor (CD “donor B#2”). Bodyweight and diarrhea score were quantified weekly. Ileitis severity in the FMT mice was assessed at the end of experiments using histological and 3D stereomicroscopic (SM) assessment of intestinal inflammation in terminal ilea, and studied myeloperoxidase (MPO) activity adjusted according to the percentage of abnormal stereomicroscopic mucosa using our validated weighted methodology system as previously described.21, 25
Gut Microbiome 16S rRNA Gene Sequencing Analysis
Illumina MiSeq paired-end rRNA gene sequencing (V3-V4 region) was used to determine the bacterial composition of fecal genomic DNA extracted from mouse and donor specimens (N = 403 samples). Processing of frozen fecal specimens, DNA extraction, library preparation, sequencing, quality control, and primary bioinformatics analysis were performed by the Genomics Core, CWRU, and the Beijing Genomics Institute in Shenzen, China. The selection of operational taxonomic units (OTUs) was performed using the QIIME1 open reference picking and USEARCH (v7.0.1090) to perform clustering at 97% similarity. Operational taxonomic unit representative sequences were taxonomically classified using Ribosomal Database Project (RDP) Classifier v.2.2 trained on the Greengenes database, using 1 confidence value as cutoff. Based on OTU abundance information, the relative abundance of each OTU in each sample was calculated, and the principal component analysis (PCA) of OTU was done with the relative abundance value. The software used in this step was package “ade4” of software R(v3.1.1). Downstream analyses of OTU tables used principles based on QIIME1.26 See details in online supplementary materials.
Metabolic Pathway Analysis
After unassigned OTUs were removed and normalized by 16S rRNA copy number, the number of each KEGG gene was counted and abundance estimated at each KEGG hierarchy level using PICRUSt (Phylogenic Investigation of Communities by Reconstruction of Unobserved States); 27 figures were generated using STAMP (v2.1.3).28
Independent Verification of Microbiota Colonization by Fecal Quantitative Polymerase Chain Reaction and Culturomics
Enumeration of fecal bacterial families by real-time polymerase chain reaction (qPCR) was quantified for verification purposes21 using β-actin and universal bacterial primers as internal controls. For culturomics of complex communities and single-colony Sanger sequencing, we implemented our “parallel lanes plating” method.16
Statistical Methods
Blinding was enforced using noninformative codes in all experimental and analytical stages; codes were revealed after analysis. Data fulfilling assumptions for parametric statistics were tested using univariate summary statistics. Alternative nonparametric tests were used for data with unfulfilled normality distributional assumptions. Data were expressed as SEMs, and 95% confidence intervals (CIs) were reported when appropriate. An alpha level of 0.05 was considered significant. Statistical analyses and graphics were performed using STATA, R (R-Project, Vienna, Austria), and Graph Pad Prism (La Jolla, CA, USA) software. Statistical analysis of OTU normalized 0.00017 + log2 transformed data tables was conducted using STATA v13.0 and R software v. 3.4.2 packages. Differences in fecal bacterial composition for all experiments were assessed by V3-V4 16S rRNA gene sequences using the Illumina MiSeq platform to generate 25,000 ± 178 reads per sample; average length 252 bp passed quality control filter.
RESULTS
Human Microbiota Can Modulate Intestinal Inflammation But Requires Proper Donor Selection
To determine whether the microbiota from IBD in remission was innocuous to intestinal inflammation, we conducted 60-day experiments using young, ileitis-prone GF SAMP mice (mild baseline ileitis) transplanted with human feces (IBD in remission; CD, UC, or healthy control; HC, see Supplementary Table 1 for donor characteristics) or SPF SAMP mouse feces (Figs. 1A and 1B). We used histological and 3D SM assessment and quantified MPO activity in the ileum to infer ileitis severity21, 25 (Fig. 1C). Collectively, our data show that transplantation of human gut microbiota into GF SAMP mice is well tolerated as it did not induce acute mortality and served to statistically determine the effect of gut microbiota in SAMP ileitis severity. To ensure reproducibility and prevent bias, all histological assessments were conducted in a blinded fashion (Supplementary Fig. 1). Although colonoscopy comprehensively supported that FMT did not induce spontaneous clinical colitis (Fig. 1D), histological analysis of the ileum demonstrated that some human donors induce significant ileitis in SAMP mice. Specifically, some human donors had a pro-inflammatory ileitis effect on groups of 6 mice, and others had an anti-inflammatory (suppressive) or non-modifying (neutral) effect. Of importance, the promotion of ileitis in mice occurred independently of whether the donor was an IBD patient in remission or a healthy control (Fig. 1E), indicating that the gut microbiome cannot be assumed to be innocuous (not pro-inflammatory) because it is obtained from a seemingly “healthy” donor and that the microbiome of IBD in remission can be pro-inflammatory. Histology also confirmed that the GF control mice, which exhibited mild ileitis at day 60, had worsened ileal inflammation following transplantation with feces from SAMP donor mice that were euthanized for comparative purposes at the same age (GF control receiving PBS, 3.1 ± 1.1; GF transplanted with SPF SAMP feces, 7.7 ± 4.2; SPF donors, 9.5 ± 1.9, K-W, P < 0.05).
Our studies demonstrate that the microbiome of IBD patients in remission and that of healthy donors have variable effects on CD-like inflammation, confirming that the gut microbiome of IBD patients is not necessarily innocuous to the gut wall during periods of remission. Findings demonstrate that the SAMP mice transplanted with human feces is an appropriate tool to study FMT-associated pro-inflammatory phenotypes relevant to IBD.
Microbiome Analysis Confirms Quantitative Parallel Between Human Donor and Recipient Mice
To gain insight into similarities and differences in microbial communities between donor and the adequacy of engraftment into mice after FMT over time,29 we performed 16S rRNA gene amplicon sequencing of fecal samples isolated from matching donors and recipient GF mice (Supplementary Fig. 2). Illumina sequencing of high-quality fecal genomic DNA with analysis of the normalized relative abundance bacterial taxa data across mice and donor groups showed that the mouse samples clustered together under distinct principal component analysis (PCA) clouds depending on donor (Figs. 2A and 2B). These findings illustrate that (1) each donor had a distinct microbiota pattern as expected and (2) mice resembled the donor microbiome composition. The PCAs for all samples over time, in the context of each donor, are presented in Supplementary Figure 3.
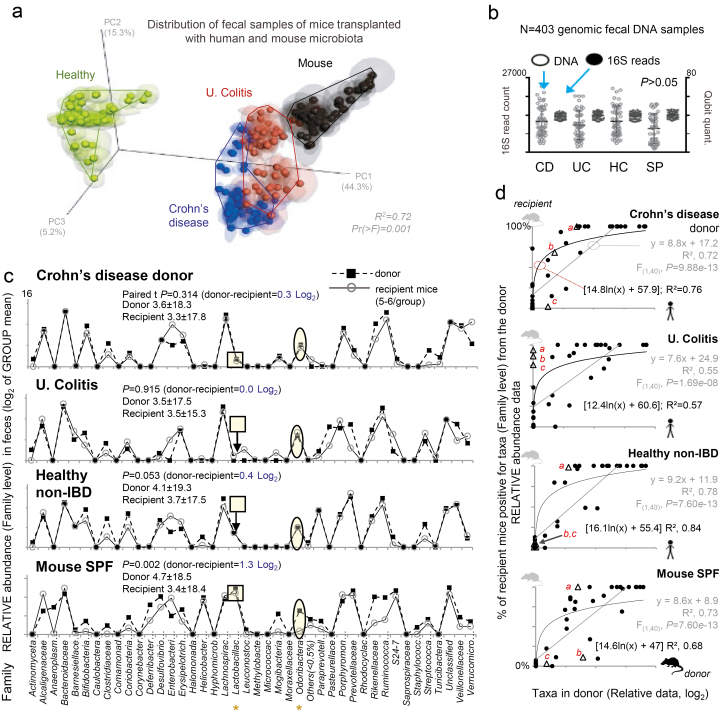
FIGURE 2.
Microbiome sequencing data illustrate quantitative parallel between donor and recipient. Normalized relative 16S rRNA gene sequencing data (log2, family-level) from Exp. A (42 bacterial families accounts for 95% of total read counts). “Others,” taxa with abundance <0.5% in all samples. A, Principal coordinates analysis (PCoA) based on 16S data from recipient mice. Phylogeny-based weighted UniFrac; jackknifed analysis on multiple rarefactions at single depth of 90. Notice SPF donor-recipient mice clusters near CD and UC mice, as expected.29 B, Distribution of fecal genomic DNA and 16S microbiome data for donor and replicated murine recipient fecal samples for Exp. A over 60 days. Scatterplots illustrate the high quality of 16S microbiome and fecal genomic DNA data (16S sampling depth 25,000 ± 178) and suggest that the total load of bacteria was comparable between mice (see qPCR validation Fig. 5). C, Line plots comparing donor inocula to that of the mean abundance for 9 fecal samples for each recipient mouse. Paired t test statistics showed no or minor differences between donor and recipient mice (CD, UC, HC, SP; 0.3, 0, 0.4, and 1.3 Log2). Notice the parallel between human and mouse microbiota and subtle abundance differences (shaded squares and circles). D, Linear and exponential correlation (agreement) between human taxa and percentage of mice colonized (less data dispersion, higher R2 if exponential). Compared with Enterobacteriaceae (trianglesa; strong colonizer, high in all plots), some low abundant donor taxa (b, cMogibacteriaceae and Leuconostraceae) can colonize most mice, but pattern depends on donor. See parallel line plots for other donors and recipient mice in Supplementary Fig. 4
To delineate specific abundance differences and determine patterns of taxa recovery not inferable from PCA, we used line-based abundance plots22 and Kappa agreement statistics. These analyses illustrated a mirrored/parallel pattern of donor bacterial abundance and recovery in the mouse feces, with similar quantitative taxa signatures recovered across donor groups (3.98 ± 18.41), demonstrating (1) the optimal colonization of mice by human microbiotas and (2) the reproducibility of the resemblance of the human microbiota line-plot profiles in the feces of mice over time (Fig. 2C, see Supplementary Fig. 4).
Lastly, the use of “probability of recovery in mice” statistics indicated that the abundance of any given taxon in the human donor feces is strongly correlated with the percentage of mice and mouse fecal samples that contain any given bacterial taxon after the FMT. In other words, bacteria taxa present with high abundance in the feces of humans colonized a larger number of mice compared with bacterial taxa present at low abundance in the human feces (significant positive correlation of 78%, F(1,40) = P < 0.9e-13). Collectively, correlation analyses (linear and logarithmic R2 values) illustrate the potential for optimal recovery of the human microbiome in SAMP mice following the intensive FMT (Fig. 2D).
Binary Time-series Microbiome Analyses and Colonization Patterns
Binary analysis (presence/absence) of normalized relative abundance recovery profiles of murine taxa revealed that the engraftment of human microbial taxa (family, genus, and species) into GF SAMP mice could be up to 100% efficient, that is, that all the taxa in the inoculum are detectable in the transplanted mice over 60 days (see heat maps in Fig. 3 and Supplementary Figs. 5–8). Computed recovery rates showed that across mice groups, the potential for human gut taxa colonization in GF mice was 95% ± 0.03 at the genus level and up to 100% at the species level (Supplementary Table 2). Using an unbiased approach to determine OTU recovery and excluding mouse samples with ≤5 reads, mouse OTU absolute read count data support a conservative recovery rate of ~90% when a read threshold of >20 in the inoculum is considered (Fig. 4A).
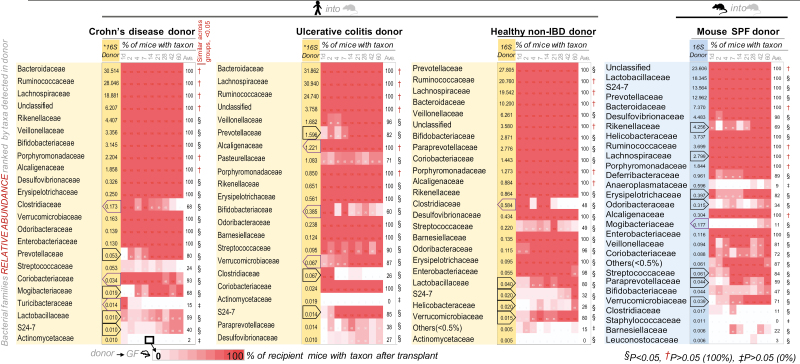
FIGURE 3.
Binary microbiome sequencing analyses illustrate various colonization patterns of human gut microbiota in SAMP FMT mice. Heat map depicts normalized relative abundance 16S rRNA sequencing data for taxa (family-level) present in donor inoculum (16S Donor) and the percentage of mice with taxon colonized over time for Exp. A (9 fecal sample time points; days 1 through 60). “Ave.,” donor group average for the percentage of mice with taxon for all mice across the 9 time points. Notice changes in percentage of mice with taxon over time for various taxa; increase or decrease are highlighted by red and purple polygons in 16S Donor column (eg, reduction of Clostridiaceae in all groups indicated by left pointing purple polygon). Statistical significance indicated by symbols under “Similar across groups, <0.05” column; number of mice that carry the taxon detected in the inoculum is significantly different to the number of FMT mice that does not carry the taxon (absence vs presence). P > 0.05 indicates taxa recovery was equal across groups; †if present in 6 of 6 mice in all groups, ‡if absent in 6 of 6 mice in all groups. §P < 0.05, if at least 1 of the groups was different (2 x 4 χ 2, P < 0.0001; eg, Deferribacteraceae/Mucispirillum schaedleri in group that received SPF mouse fecal inoculum). §Collectively, the taxa for which the engraftment in mice was dependent on the donor sample (eg, Prevotellaceae). Note that Eubacteriaceae was detected by 16S microbiome Illumina analysis in CD and UC donors at the species-level (Eubacterium dolichum spp., see Supplementary Figs. 5–8)
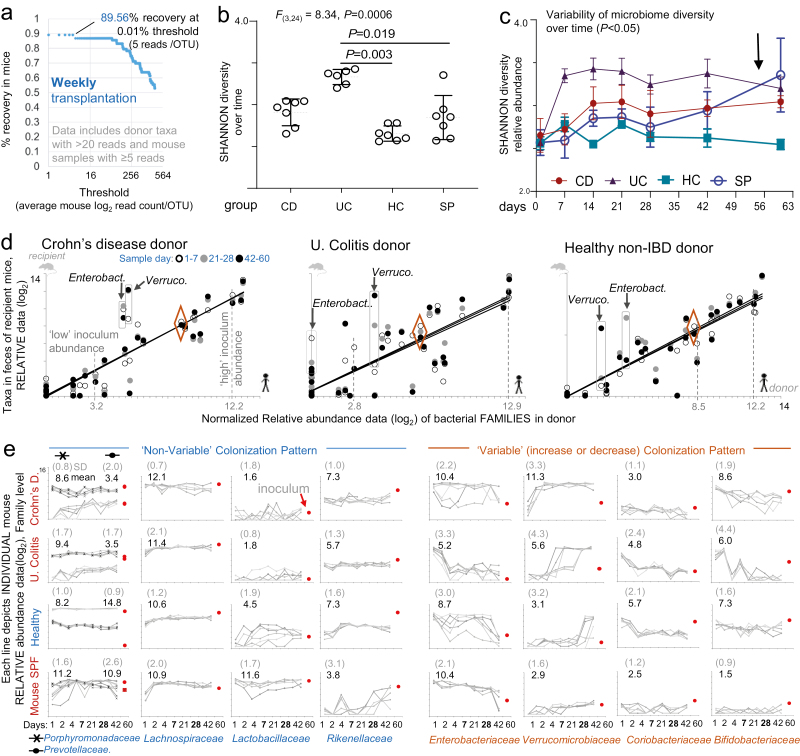
FIGURE 4.
Percentage of recovery, variability of microbiome diversity, and colonization patterns over time. Weekly transplantation. Normalized relative abundance data (Log2, family-level, Exp. A). A, Percentage of taxa recovery estimates in mice (y-axis) compared with OTU counts (complete unassigned taxa dataset) in donor with >20 reads over various thresholds (x-axis; average log2 of read count/OTU for all transplanted mice). B, Cumulative dot plots for Shannon diversity (richness) for recipient mouse (mean, SD for each mouse; 7 time points/mouse over 60-day length of study). Shannon SD is highly reproducible (constrained within 1 unit of range) in all groups. ANOVA; pairwise Turkeys statistics. C, Shannon diversity line plots illustrate variability over time and differences across groups (ANOVA/Tukey statistics; days 1 to 28; P < 0.05). D, Correlation line plots for average 16S data for the indicated time periods illustrate parallel positive correlation between donor and FMT mice (eg, transplantation of high or medium/low abundance donor taxa; Lachnospiracheae and Coriobacteriaceae, dashed red lines). Rectangles indicate differences in abundance (not present/absence), which influence Shannon estimations. Diamonds indicate Porphyromonadaceae as example of a taxa with nonvariable colonizing pattern across donors. See Supplementary Figure 9 for logarithmic slope and R2 values. E, Individual mouse abundance line plots over time for representative taxa across FMT groups for Exp. A. “Mean,” mean of taxa abundance in mice over time; “SD,” average of SD for all time points. Notice the dynamic variability of microbiome engraftment and the unique patterns for each donor. See colonization patterns for other donors in Supplementary Figure 10.
Shannon diversity illustrated that transplanted mice within groups are very similar to each other over time, further demonstrating data reproducibility at the mouse level (P > 0.05). Of clinical relevance, microbiome diversity differences occurred in our intensive FMT SAMP model primarily between groups (eg, CD vs UC), which is as expected due to donor differences and not differences due to chance or sequencing errors (ie, narrow dispersion in dot plots, Figs. 4B and 4C). Because Shannon indexes do not indicate the sources of data variability, we conducted variability analysis using correlation statistics.
Spearman and Pearson statistics, linear univariate equations (y = B0 + B1X1), and R2 dispersion statistics and line-abundance plots (donor vs recipient mouse) identified the leading causes of quantitative variance (ie, which and how bacterial taxa varied dynamically) both between and within the recipient mouse groups over time (Fig. 4D). Our analyses revealed remarkable reproducibility in patterns oftaxon variability across transplanted mice, for example, Verrucomicrobiaceae (eg, Akkermansia muciniphila) and Enterobacteriaceae (Fig. 4E, see detailed significant differ- 5.85 ences in Supplementary Figs. 9, 10). We show that the majority of bacterial colonization potential in SAMP mice has a reproducible pattern but that temporal patterns depend on the donor. Our findings indicate that the colonization of an entire microbiota community has a self-arrangement feature that allows the successful colonization of taxa at higher, lower, or transiently higher abundance. Analyzing the read abundance, quantile breakpoints data, and count-of-mice-colonized-by-each taxa (frequency) statistics over time, we illustrate that some taxa have reproducible and predictable patterns of colonization in mice, but these vary depending on the donor microbiome (Supplementary Fig. 11).
Bacterial Enumeration and qPCR Confirm Microbiome Colonization Dynamics
To quantitatively validate our 16S rRNA microbiome findings, we processed a new set of fecal sample aliquots from mice and donors for bacterial enumeration using culture methods and by using real-time quantitative PCR (qPCR) as 2 independent diagnostic strategies to verify colonization dynamics.
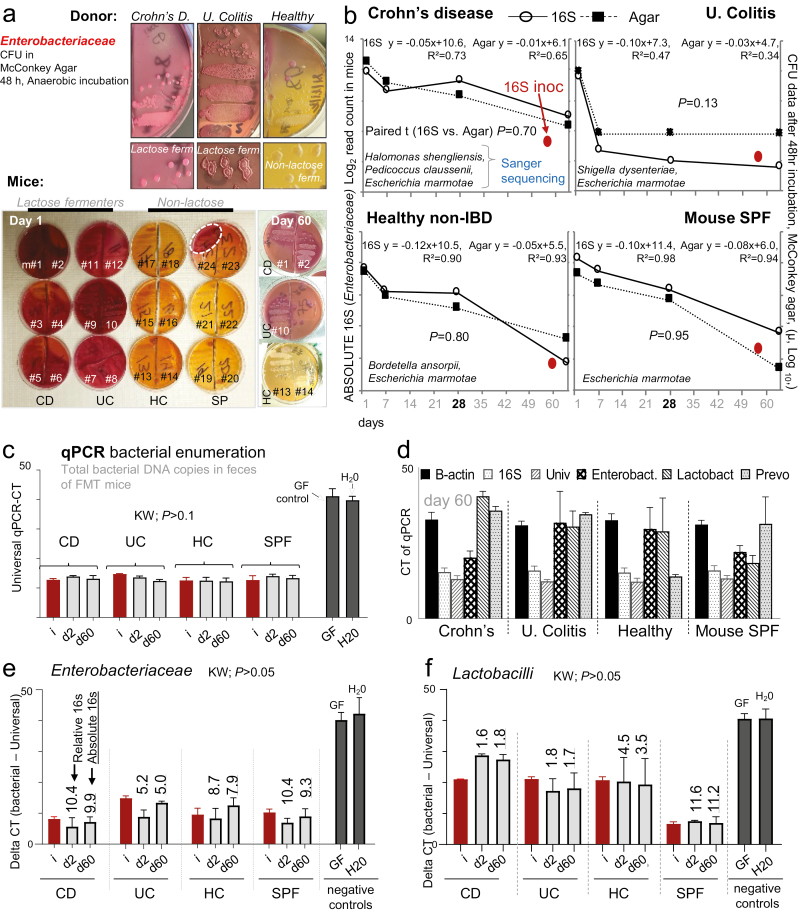
Because several species of Enterobacteriaceae, particularly when present at low abundance, cannot always be detected or differentiated with conventional sequencing primers, we used a rapid community quantitative culture (“parallel lanes plating”)16 method as a gold standard for cultivable species to verify findings derived from microbiome sequencing. Previous analysis has supported the use of coliforms as a surrogate for community variability in SAMP mice.16 Therefore, we focused on Enterobacteriaceae as a surrogate indicator of microbiome composition and dynamic abundance profiles over time (eg, crescendo-decrescendo in UC mice, Fig. 4E). By using single-colony Sanger sequencing (250bp 16S rRNA gene V4 region) and McConkey agar (medium that allows qualitative enumeration of lactose fermentation) focusing on lactobacilli and Streptococcus spp. counts as comparators using selective agar, we confirmed that cultivable Enterobacteriaceae reductions of 1000-fold over time (2 to 3 Log10 units) follow the relative abundance normalized patterns observed for the microbiome sequencing data (25,000 reads total/sample). Coliform culture profiling based on fermentation of lactose was highly reproducible within mice transplanted with the same microbiota, which persisted until day 60 (ie, all 6 mice transplanted exhibited almost identical coliforms patterns; Figs. 5A and 5B).
FIGURE 5.
Bacterial enumeration, Sanger sequencing, and qPCR confirm colonization dynamics in SAMP mice. Community quantitative culture using parallel lanes plating of fecal Enterobacteriaceae on McConkey agar over time, Exp. A. A, Colony appearance of eight 10-fold fecal serial dilutions/sample (2 samples/plate). Note similarity/differences (agar color/colony types) between donor and mice for days 1 and 60 across groups. McConkey plates illustrate lactose fermenting pattern that is conserved within mice depending on donor; lactose-fermenters (turn agar dark pink, CD and UC), nonlactose fermenters (turn agar yellow, SPF and HC). B, Single-colony Sanger sequencing guided enumeration of coliforms and line plots illustrate that relative changes of microbial abundance for Enterobacteriaceae microbiome sequencing data (solid line) parallels the CFU McConkey data (dashed line) over the length of the study. Solid red circle, Enterobacteriaceae abundance in donor inoculum. C, Quantification of DNA copies in the feces of donor in column “i” and transplanted mice for days 2 and 60 (y-axis). Y-axis, raw CT-values after 50 log2 qPCR amplification cycles. Notice, minimal variation across groups vs negative controls. D, Quantification qPCR CT-values (DNA copy abundance) for Enterobacteriaceae, Lactobacilli, and Prevotella in the context of 2 generic/universal bacterial primers (16S, Univ.) and the host gut cell DNA copies using β-actin in FMT mice for day 60. Notice the comparative abundance of β-actin across groups (E and F). Quantification of delta qPCR CT-values (DNA copy abundance) for Enterobacteriaceae and Lactobacilli (data from panel 5D) after subtraction of (ΔCT) universal primer CT values. Kruskal-Wallis; KW, P > 0.05; no differences between mice and donor. Supplementary Figure 12 shows additional qPCR analysis and Prevotella. To contextualize qPCR findings, numbers above bars indicate mean read-abundance values derived from Illumina microbiome data
To further confirm the microbiome findings, we used generic and taxa specific bacterial primers21 for Enterobacteriaceae, Lactobacilli, and Prevotella to perform qPCR enumeration of bacterial DNA copy numbers in donor and recipient mouse feces for days 0 (ie, before FMT), 2, and 60 after FMT (Supplementary Table 3). Two 16S universal primers to identify most bacteria showed that there was no difference in total bacterial load counts between the mouse groups and the donors (Fig. 5C and 5D). Controlling for host DNA using β-actin as a reference, we verified the microbiome crescendo-decrescendo colonization abundance for Enterobacteriaceae over time, which also reflected the culture enumeration data (Fig. 5E and 5F, Supplementary Fig. 12).
In conclusion, 3 independent diagnostics (microbiome sequencing, quantitative culture paired to Sanger sequencing, and qPCR) confirmed that GF SAMP mice successfully allow the colonization of human gut microbiomes, suggesting that intensive FMT in SAMP is suitable as a model to characterize the functionality of the human microbiota.
Single-dose Human Gut Microbiota FMT in SAMP Mice Verifies Intensive FMT Outcomes
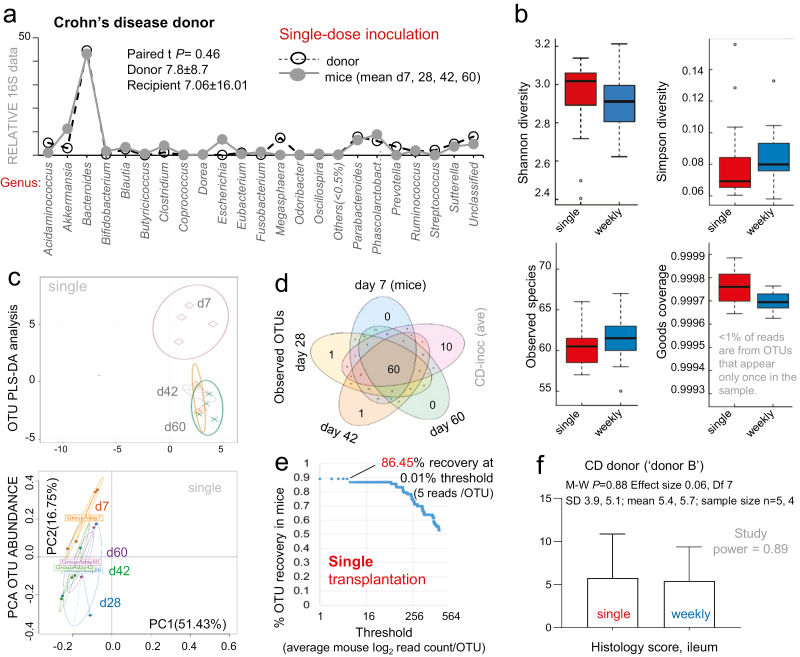
Lastly, we sought to determine if a single-dose FMT protocol could resemble the microbiome and inflammatory profiles described with weekly FMT. To address this, we repeated the same 60-day study design using a single-dose inoculation, using intensive (weekly) FMT as a comparator. We used the remaining collection of fecal sample aliquots from a randomly selected CD donor (CD donor “B#2,” Fig. 1E). Alpha and beta diversity, PCA statistics, correlation analysis, percentage of OTU recovery, and ileal histology confirmed that both dosing protocols generate equivalent gut microbiome engraftment and ileitis profiles in SAMP mice (Fig. 6A–D; Supplementary Fig. 13). Engraftment recovery analysis also indicated that the percentage of taxa recovery (genus level 95.6%) and ileitis severity did not change in mice using a single-dose FMT (Fig. 6E and 6F). Collectively, this demonstrates that taxa recovery in GF SAMP mice is not dependent on the intensiveness of the FMT, thus improving the viability of SAMP as a humanized FMT model for human donor screening.
FIGURE 6.
Effect of single-dose FMT on bacterial colonization, percentage of recovery and ileitis in GF mice. Single-dose transplantation using CD donor (donor B#2). Normalized relative abundance 16S rRNA data (genus and OTU level) for days 7, 28, 42, and 60. A, Line plots of mean sequencing data (genus-level, log2) comparing donor inocula to mean abundance for each recipient mouse. B, Boxplots for alpha diversity indices for recipient mice groups. Good’s coverage (1 – [F1 / N]; where F1 is N equals the number of reads in the sample) shows that <1% of reads are from OTUs that appear only once in the sample. C, PCA-based OTU abundance plots illustrate the maximum separation between OTU abundance in donor inocula and each transplanted mouse group over time. Each dot represents one sample. To delineate differences in microbiome composition illustrated by PCA-based analyses, we conducted further variability analysis using correlation statistics (Supplementary Fig. 13). D, Venn diagrams show the number of shared OTUS for single-dose inoculated mice to that of donor inoculum (mean of taxa replicates analyzed in 6 technical replicates interpreted in series). E, Percentage of OTU-level taxa recovery in single-dose transplanted mice. Notice that recovery is comparable to weekly transplanted mice, shown in Figure 4A. F, Histological scores for single vs weekly transplantation on CD-like ileitis severity in GF SAMP mice at day 60.
Amino/Fatty Acid Metabolic Predictors and Not Bacterial Virulence Markers Precede CD-Like Ileitis
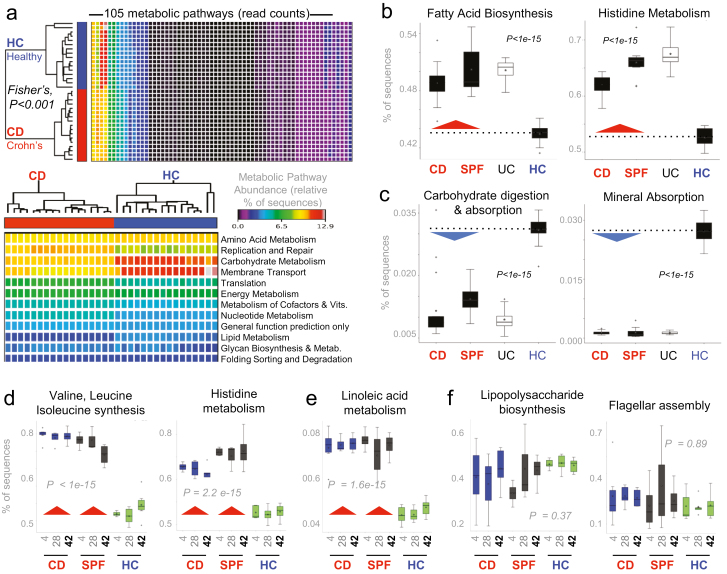
To increase our understanding of the pro-inflammatory role of potential metabolic abnormalities of the IBD gut microbiota in remission, we used the predictive bioinformatic tool PICRUSt to infer the metabolic pathways present in the feces of transplanted mice. Of statistical relevance, analysis of mice that exhibited marked ileitis after FMT with CD and SPF microbiota showed significantly different microbial metabolic functions that could be associated with promotion of inflammation in the ileum (compared with HC mice that exhibited less ileitis; see Fig. 1E). Pathways enriched in ileitic mice were especially evident for branched-chain amino acids (BCAA: valine, leucine, isoleucine) and linoleic acid (Fig. 7A–C, Supplementary Fig. 14). Notable differences were also evident for fatty acid biosynthesis, histidine metabolism, carbohydrate digestion and absorption, and mineral absorption pathways in the bacterial microbiome at various levels; metabolic pathways were inferred to be chronically dysfunctional in CD and SPF mice compared with HC recipient mice before the development of chronic ileitis. Such significant differences in bacterial gene sequence abundances were reproducibly present as early as day 4 after FMT and throughout the study on days 28 and 42 (P < 1e-15, study power >0.8, Fig. 7D and 7E). The identification of a metabolically abnormal gut microbiota soon after FMT, that remains stable over time, indicates that metabolic dysfunctions could be primarily driven by the transplanted microbiota and precede ileitis in SAMP mice. Of interest, the amount of bacterial virulence factor pathways that are known to worsen IBD, namely lipopolysaccharide and flagellar assembly, were not enriched in the feces of ileitic (CD and SPF) mice compared with noninflamed (HC) SAMP mice (Fig. 7F, P > 0.37). Collectively, findings indicate that gut microbiota metabolic functions could play a primary biological role promoting chronic intestinal inflammation in IBD.
FIGURE 7.
Metabolic microbiome predictive analysis reveals chronically abnormal pathways in FMT SAMP mice. Statistical analyses of metagenomic profiles (STAMP)28 of relative abundance values for various KEGG metabolic pathways encoded in mice fecal samples (Exp. A). A, Heatmap illustrates that metabolic predictions are reproducible across samples and that the CD and HC group cluster together with apparent differences for some pathways. B, Boxplots of sequence abundance (ANOVA with multiple tests correction by Benjamin-Hochberg FDR) for representative KEGG pathways deemed relevant for driving severity of inflammation in experimental CD. Boxplots show that some paths are enriched while others are deficient in the microbiota, irrespective of whether it originates from a human with CD or a mouse with severe CD-ileitis. C–E, Boxplots of sequence abundance illustrates the consistent, inverse differences in microbiota functionality for CD donor mice (severely inflamed mice; CD donor A#1, see Fig. 1C) and SPF donor mice to mice that received feces from the healthy non-IBD donor (HC donor A#7; see Fig. 1C) over time (days 4, 28, and 42). Plots illustrate that BCAAs (valine, leucine, isoleucine), histidine, and linoleic acid remain chronically synthesized and degraded (Supplementary Fig. 14) through the study period, but that there is no difference in virulence markers (LPS, flagellin; panel E) between groups (post hoc study power calculations achieved >0.80).45 Chronically abnormal metabolic pathways in the gut microbiota could be chronic drivers or biomarkers that precede CD-like ileitis.
DISCUSSION
This study represents a collective effort to determine (1) whether the inbred GF SAMP mouse line could be a useful model for the engraftment and colonization of the human gut microbiota from IBD in remission and (2) whether the gut microbiota from IBD in remission could modulate the severity of CD-like ileitis in transplanted mice. After analyzing 403 fecal microbiome samples, our studies in GF SAMP mice indicated that the detection of inflammatory microbiotas in humans is independent on the IBD status of the person and cannot be easily or intuitively predicted. These findings may explain why FMT from a healthy donor into a recipient IBD patient is not always effective,6, 7 as compared with other diseases such as Clostridium difficile infection where FMT cures the majority of patients.30–36 In our study, the attenuation of ileitis in mice transplanted with gut microbiota from 2 specific donors (donors C#3 and A#7) supports the concept that some individuals function as “super anti-inflammatory donors” as analogous to other disease scenarios.37, 38 Our results also highlight that pro-inflammatory microbiota from a healthy control donor can be transferred by FMT and therefore calls for the need to functionally characterize the pro-inflammatory potential of human donors using mice to lessen the potential harm and improve potential benefit in IBD.
To our knowledge, the SAMP mouse is the only mouse line that presents with a CD-like cobblestone ileitis that follows a spontaneous, chronic inflammatory process that occurs independently of microbial colonization.13 In our FMT experiments, a ~90% OTU level engraftment colonization of human gut taxa in GF SAMP mice was achieved, as confirmed by 3 independent diagnostics (microbiome sequencing, quantitative culturomics, and qPCR). Microbiome analysis also supports a protocol of single-dose or intensive (weekly) FMT in SAMP to facilitate the characterization of the pro-inflammatory potential of human gut microbiota. Collectively, the SAMP FMT model using GF mice herein described represents a functional tool for the identification of IBD remission and healthy non-IBD donor fecal samples that could have anti-inflammatory potential. In the IBD susceptible host, our findings hold important implications with respect to maintaining or achieving remission status and for clinical studies in the selection of anti-inflammatory FMT donor specimens. The potential for an IBD patient to serve as a self donor is clinically relevant because infectious pathogen transmission39, 40 from FMT donor to recipient remains the most significant concern in heterologous FMT.41, 42
Our predictive PICRUSt metagenome data provide preliminary insight that the gut microbiota can harbor bacteria that have enriched metabolic pathways toward the biosynthesis or altered metabolism of amino acids and fatty acids. Our findings with respect to amino and fatty acids (valine, leucine, isoleucine, histidine; linoleic) are intriguingly aligned with recent reports in human samples in which plasma amino acid concentrations methionine and histidine and the BCAAs valine, leucine, and isoleucine correlate significantly with disease activity19, 20, 43 and linoleic metabolism.19, 44 It is of interest to note that the pro-inflammatory effect from the samples studied was not associated with the pathways related to known pro-inflammatory virulence factors from bacteria such as lipopolysaccharide and expression of flagellin. Acting as an internal control, the latter highlights that metabolic pathways could indeed be associated with the prediction or manifestation of inflammation in humans. Collectively, our metabolic based analyses suggest that alterations in metabolic pathways could be intrinsically present in the microbiota, can be detected early (>day 4 after FMT), persist over time, and precede the development of microbiota-driven severe chronic CD-like ileitis. It is not known whether chronic early alterations in metabolic pathways could promote inflammation in humans.
Although this study was conducted with 9 human donors, our study supports as a proof of principle that the SAMP FMT based model described can be used to characterize human donor gut microbiota and identify predictive metabolic or virulence mechanisms that could control or promote CD-like inflammation independently of IBD status. Our study also suggests that a single-donor experimental approach is needed for the individualized analysis of human fecal microbiota. Future studies on metabolomics and metagenomics and prospective studies following humans over time are desired to elucidate the mechanisms by which early metabolic microbial alterations could promote IBD.
In conclusion, our engraftment studies in GF SAMP mice indicate that the gut microbiome of IBD remission patients is not necessarily innocuous and that healthy non-IBD donor microbiota can have pro-inflammatory activity in an IBD-susceptible host. Transplantation of human gut microbiota into SAMP ileitis-prone GF mice represents a useful tool to investigate the functionality of the human gut microbiome in chronic diseases in which “dysbiosis” has been suggested as an important pathogenic factor, such as IBD. Time series functional analysis to identify anti-inflammatory microbiotas of IBD patients during remission could be tremendously valuable as it would abolish the risk of person-to-person transmission of pathogens, which is a major concern in heterologous FMT.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank John D. Ward, Alexandra Warner, Heather Wang, Alicia DePlatchett, Jonathan Craven, and Rachael Murphy for their technical and administrative support, Dr. Gurkan Bebek for bioinformatics consultation, and Dr. Wei Xin for the histological scoring of ileitis severity. AB would especially like to thank and acknowledge Mr. and Mrs. Raffner for their unwavering support.
Glossary
Abbreviations:
- CD
Crohn’s disease
- Exp.
Experiment
- FMT
fecal microbiota transplantation
- GF
germ-free
- HC
healthy control non-IBD healthy individual (fecal microbiota transplantation group)
- IBD
inflammatory bowel disease
- MPO
myeloperoxidase
- NesTiso
nested isolation caging system
- OUT
operational taxonomic unit
- SAMP
SAMP1/YitFC
- SM
stereomicroscopy
- SPF
specific pathogen-free
- UC
ulcerative colitis
Author Contribution: AB, AR, and FC conceptualized and supervised the study. AB and AR wrote the final manuscript with input from FC. AR, AG, NA, PM, LB, AO, AL, LM, JE, HE, ML, DF, and CB provided technical/experimental/intellectual support. AB and AR performed statistical analysis and interpretation of data. AO performed all histological analysis. FC, AR, and AB acquired funding. All authors edited and approved the final version of the paper.
Supported by: NIH grant DK055812, DK091222, and DK097948 (to FC), T32DK083251 and F32DK117585 (to AB), and P01DK091222 Germ-free and Gut Microbiome Core and R21DK118373 (to ARP). Authors acknowledge the support of the Mouse Models, the Histology Imaging, and Tissue Biorepository Cores of the NIH P30 Silvio O. Conte Cleveland Digestive Diseases Research Core Center.
Data availability: The microbiome data that supported the findings of this study have been deposited in figshare and can be accessed via the following links: https://figshare.com/s/0aebf552dc5e74cc9e4d, https://figshare.com/s/b893a5222fa6f15230fb, https://figshare.com/s/831fa0ff5238817c7ecb, and https://figshare.com/s/ce079cb9769c5aff16a0. Additional detailed protocols are available upon request or freely available as Supplementary Materials.
REFERENCES
- 1. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. [DOI] [PubMed] [Google Scholar]
- 2. Basson A, Trotter A, Rodriguez-Palacios A, et al. Mucosal Interactions between genetics, diet, and microbiome in inflammatory bowel disease. Front Immunol. 2016;7:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wright EK, Kamm MA, Teo SM, et al. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm Bowel Dis. 2015;21:1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buttó LF, Haller D. Functional relevance of microbiome signatures: the correlation era requires tools for consolidation. J Allergy Clin Immunol. 2017;139:1092–1098. [DOI] [PubMed] [Google Scholar]
- 5. Paramsothy S, Paramsothy R, Rubin DT, et al. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2017;11:1180–1199. [DOI] [PubMed] [Google Scholar]
- 6. Vaughn BP, Vatanen T, Allegretti JR, et al. Increased intestinal microbial diversity following fecal microbiota transplant for active Crohn’s disease. Inflamm Bowel Dis. 2016;22:2182–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui B, Feng Q, Wang H, et al. Fecal microbiota transplantation through mid-gut for refractory Crohn’s disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol. 2015;30:51–58. [DOI] [PubMed] [Google Scholar]
- 8. Britton GJ, Contijoch EJ, Mogno I, et al. Microbiotas from humans with inflammatory bowel disease alter the balance of Gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity. 2019;50:212–224.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagao-Kitamoto H, Shreiner AB, Gillilland MG 3rd, et al. Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell Mol Gastroenterol Hepatol. 2016;2:468–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shepherd ES, DeLoache WC, Pruss KM, et al. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557:434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brooks PT, Brakel KA, Bell JA, et al. Transplanted human fecal microbiota enhanced Guillain Barré syndrome autoantibody responses after Campylobacter jejuni infection in C57BL/6 mice. Microbiome. 2017;5:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pizarro TT, Pastorelli L, Bamias G, et al. SAMP1/YitFc mouse strain: a spontaneous model of Crohn’s disease-like ileitis. Inflamm Bowel Dis. 2011;17:2566–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bamias G, Okazawa A, Rivera-Nieves J, et al. Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J Immunol. 2007;178:1809–1818. [DOI] [PubMed] [Google Scholar]
- 15. Menghini P, Di Martino L, Lopetuso LR, et al. A novel model of colitis-associated cancer in SAMP1/YitFc mice with Crohn’s disease-like ileitis. PLoS One. 2017;12:e0174121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodriguez-Palacios A, Harding A, Menghini P, et al. The artificial sweetener splenda promotes gut proteobacteria, dysbiosis, and myeloperoxidase reactivity in Crohn’s disease-like ileitis. Inflamm Bowel Dis. 2018;24:1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. [DOI] [PubMed] [Google Scholar]
- 18. Costello SP, Hughes PA, Waters O, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiba T, Suzuki K, Matsumoto T. Plasma-free amino acid profiles in crohn’s disease: relationship with the Crohn disease activity index. Clin Med Insights-Ga. 2018;11: 1– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakano M, Tominaga K, Hoshino A, et al. Therapeutic efficacy of an elemental diet for patients with Crohn’s disease and its association with amino acid metabolism. Saudi J Gastroenterol. 2017;23:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodriguez-Palacios A, Kodani T, Kaydo L, et al. Stereomicroscopic 3D-pattern profiling of murine and human intestinal inflammation reveals unique structural phenotypes. Nat Commun. 2015;6:7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez-Palacios A, Aladyshkina N, Ezeji JC, et al. ‘Cyclical Bias’ in microbiome research revealed by a portable germ-free housing system using nested isolation. Sci Rep. 2018;8:3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blakeley-Ruiz JA, Erickson AR, Cantarel BL, et al. Metaproteomics reveals persistent and phylum-redundant metabolic functional stability in adult human gut microbiomes of Crohn’s remission patients despite temporal variations in microbial taxa, genomes, and proteomes. Microbiome. 2019;7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodriguez-Palacios A, Aladyshkina N, Cominelli F. Stereomicroscopy and 3D-target myeloperoxidase intestinal phenotyping following a fecal flora homogenization protocol. Protocol Exchange. 2015. ISSN 2043-0116. doi:10.1038/protex.2015.065. http://www.nature.com/protocolexchange/protocols/3863 [Google Scholar]
- 26. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parks DH, Tyson GW, Hugenholtz P, et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. 2018;9:2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. [DOI] [PubMed] [Google Scholar]
- 31. Hamilton MJ, Weingarden AR, Sadowsky MJ, et al. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:761–767. [DOI] [PubMed] [Google Scholar]
- 32. Brandt LJ, Aroniadis OC, Mellow M, et al. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:1079–1087. [DOI] [PubMed] [Google Scholar]
- 33. Aroniadis OC, Brandt LJ, Greenberg A, et al. Long-term follow-up study of fecal microbiota transplantation for severe and/or complicated Clostridium difficile infection: a multicenter experience. J Clin Gastroenterol. 2016;50:398–402. [DOI] [PubMed] [Google Scholar]
- 34. Hocquart M, Lagier JC, Cassir N, et al. Early fecal microbiota transplantation improves survival in severe Clostridium difficile infections. Clin Infect Dis. 2018;66:645–650. [DOI] [PubMed] [Google Scholar]
- 35. Juul FE, Garborg K, Bretthauer M, et al. Fecal microbiota transplantation for primary Clostridium difficile infection. N Engl J Med. 2018;378:2535–2536. [DOI] [PubMed] [Google Scholar]
- 36. Debast SB, Bauer MP, Kuijper EJ; European Society of Clinical Microbiology and Infectious Diseases European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20(Suppl 2):1–26. [DOI] [PubMed] [Google Scholar]
- 37. Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149(1):102–109.e106. [DOI] [PubMed] [Google Scholar]
- 38. Wilson BC, Vatanen T, Cutfield WS, et al. The super-donor phenomenon in fecal microbiota transplantation. Front Cell Infect Microbiol. 2019;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McSeveney M. FDA In Brief: FDA warns about potential risk of serious infections caused by multi-drug resistant organisms related to the investigational use of Fecal Microbiota for Transplantation. Vol. 2019 U.S. Food & Drug Administration; https://www.fda.gov/news-events/fda-brief/fda-brief-fda-warns-about-potential-risk-serious-infections-caused-multi-drug-resistant-organisms (14 June 2019, date last accessed). [Google Scholar]
- 40. Hohmann EL. Case 25-2014: a man with ulcerative colitis and bloody diarrhea. N Engl J Med. 2014;371:1848–1849. [DOI] [PubMed] [Google Scholar]
- 41. Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther. 2012;36:503–516. [DOI] [PubMed] [Google Scholar]
- 42. Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2014;8:1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang X, Deeke SA, Ning Z, et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat Commun. 2018;9:2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ueda Y, Kawakami Y, Kunii D, et al. Elevated concentrations of linoleic acid in erythrocyte membrane phospholipids in patients with inflammatory bowel disease. Nutr Res. 2008;28:239–244. [DOI] [PubMed] [Google Scholar]
- 45. Faul F, Erdfelder E, Lang AG, et al. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.