Abstract
Background
Polymerase chain reaction (PCR) analysis using DNA from dried blood spot (DBS) samples on filter paper is a critical technique for spinal muscular atrophy (SMA) newborn screening. However, DNA extraction from DBS is time-consuming, and elimination of PCR inhibitors from DBS is almost impossible.
Methods
Exon 7 of the two homologous SMA-related genes, survival motor neuron (SMN) 1 and SMN2, of five SMA patients and five controls were amplified by PCR with a punched-out circle of the DBS paper. Two types of DNA preparation methods were tested; DNA-extraction (extracted DNA was added in a PCR tube) and non-DNA-extraction (a punched-out DBS circle was placed in a PCR tube). As for the DNA polymerases, two different enzymes were compared; TaKaRa Ex Taq™ and KOD FX Neo™. To test the diagnostic quality of PCR products, RFLP (Restriction fragment length polymorphism) analysis with DraI digestion was performed, differentiating SMN1 and SMN2.
Results
In PCR using extracted DNA, sufficient amplification was achieved with TaKaRa Ex Taq™ and KOD FX Neo™, and there was no significant difference in amplification efficiency between them. In direct PCR with a punched-out DBS circle, sufficient amplification was achieved when KOD FX Neo™ polymerase was used, while there was no amplification with TaKaRa Ex Taq™. RFLP analysis of the direct PCR products with KOD FX Neo™ clearly separated SMN1 and SMN2 sequences and proved the presence of both of SMN1 and SMN2 in controls, and only SMN2 in SMA patients, suggesting that the direct PCR products with KOD FX Neo™ were of sufficient diagnostic quality for SMA testing.
Conclusion
Direct PCR with DNA polymerases like KOD FX Neo™ has potential to be widely used in SMA newborn screening in the near future as it obviates the DNA extraction process from DBS and can precisely amplify the target sequences in spite of the presence of PCR inhibitors.
Keywords: spinal muscular atrophy, screening, dried blood spot, PCR, DNA extraction, DNA polymerase
INTRODUCTION
Spinal muscular atrophy (SMA) is an autosomal recessive disease exhibiting muscle atrophy and weakness of the whole body due to degeneration or loss of motor neurons in the anterior horn of the spinal cord. In 1995, SMA-related genes, survival motor neuron (SMN) 1 and SMN2 genes, were identified and successfully cloned7. SMN1 and SMN2 are highly homologous genes, but succeeding studies clarified that SMN1 is responsible for SMA13. According to our previous study9, the incidence rate of the infantile onset SMA is ~1/20,000 newborn babies, and its carrier rate is ~1/70 in Japan.
SMA cannot be definitely diagnosed by clinical symptoms alone and it should be confirmed by detection of SMN1 abnormalities1, 13. Advantages of such genetic test are minimal invasiveness and high accuracy for the disease diagnosis. Nevertheless, the availability of the genetic test has not been well recognized, resulting in delayed or missed diagnosis in some cases9. In addition, SMA was considered as a disease without treatment options, which might be related to the delayed diagnosis of SMA.
However, much progress has been made in therapeutic strategies for SMA. An antisense oligonucleotide drug for SMA, nusinersen, has recently been developed, and its clinical trials have shown promising clinical effects on the patients3,4. In 2016, nusinersen was approved for clinical use in the US, and in 2017, it was also approved in Japan. The clinical trial results also indicated that early start of treatment can produce more effective efficacy and thus delayed diagnosis would impair the potential of treating patients early.
Since early treatment is related to early diagnosis and recognition of the disease in patients, the feasibility of implementing newborn screening for SMA should be discussed9. Dried blood spot (DBS) on the filter paper is used for nationwide newborn screening to detect inborn errors of metabolism in Japan. Thus, inclusion of genetic test for SMA using DBS would enable us to carry out the nationwide newborn screening for this disorder on the currently available screening platform. Nationwide newborn screening for SMA can decrease the rate of delayed diagnosis of SMA in Japan.
For SMA newborn screening with genetic testing, polymerase chain reaction (PCR) using DBS is an essential technique. Previous studies, including ours, have shown successful DNA extraction from DBS paper5,8. However, the DNA extraction process was time consuming. In addition, DBS paper contains various PCR inhibitors by which some DNA polymerase does not work as expected12. Thus, we investigated the method of the optimal DNA preparation and DNA polymerase selection from DBS samples. In the current study, we compared the representative enzymes of two distinctive DNA polymerase groups. TaKa Ra Ex Taq™ is an advanced version of the most popular thermophilic DNA polymerase, Taq, which was isolated from Thermus aquaticus10. KOD FX Neo™ is also an advanced version of KOD DNA polymerase, which was isolated from Thermococcus kodacaraensis10. KOD has 3′→5′ exonuclease activity, so the fidelity of KOD polymerase is much better than Taq10.
MATERIALS AND METHODS
1. DNA preparation from DBS
The participants in this study were five SMA patients and five healthy controls (volunteers). The patients had been diagnosed by genetic test using freshly collected blood. Prior to this study, informed consent was obtained from all participants who provided their whole blood for the study. This study was approved by the Ethics Committee of Kobe University Graduate School of Medicine (No. 1089).
DBS samples were prepared as follows: EDTA-anticoagulated fresh blood was spotted on the filter paper (Toyo Roshi; Advantec MFS Inc. Tokyo, Japan) and allowed to air dry for more than 2 hours. DNA was prepared from DBS according to the 5 different methods of (A)–(E). (A), (B), (C) and (D) were grouped into the “DNA extraction” method, while (E) was grouped into the “non-DNA-extraction” method. The details of each method is as follows:
DNA extraction by a simple “boiling” method: a punched-out DBS circle with a 2mm-diameter (DBS disk) was placed in 50 μL of TE buffer and boiled at 95°C for 30 minutes. DNA was eluted in the solution. Two μL of the DNA solution was then added into PCR reaction mixture.
DNA extraction by a “rinsing and boiling” method: a DBS disk was rinsed with Milli-Q (MQ) water, and then placed in 50 μL of TE buffer and boiled at 95°C for 30 minutes. Two μL of the DNA solution was then added into PCR reaction mixture.
DNA extraction by a “protein fixation-1, rinsing and boiling” method: a DBS disk was placed in 100 μL of a mixed solution containing acetone, methanol, and water (7:7:2, volume ratio) (protein fixation)8. After protein fixation, the disk was rinsed with MQ water, and then placed in 50 μL of TE buffer and boiled at 95°C for 30 minutes. Two μL of the DNA solution was then added into PCR reaction mixture.
DNA extraction by a “protein fixation-2, rinsing and boiling” method: a DBS disk was placed in 100 μL of methanol (protein fixation). After protein fixation, the disk was rinsed with MQ water, and then placed in 50 μL of TE buffer and boiled at 95°C for 30 minutes. Two μL of the DNA solution was then added into PCR reaction mixture.
No DNA extraction process: A half of the DBS disk was directly placed into a PCR tube, that is, PCR was performed without DNA extraction.
2. PCR experiments with two different DNA polymerases
All PCR experiments were carried out using a Thermal Cycler GeneAtlas (Aztec Inc. Tokyo, Japan). DNA extracted from DNA solution of (A), (B), (C) and (D) (2μL each), and one half of the DBS disk (E) was used as template for PCR. Two μL of DNA solution of (A), (B), (C) and (D) contained ~100 ng DNA.
Two DNA polymerases, TaKaRa Ex Taq™ (Takara Bio Inc. Shiga, Japan) or KOD FX Neo™ (Toyobo Inc. Osaka, Japan) were used in this study. Each PCR experiment was performed with 50 μL of the reaction mixture according to the instruction manuals of the companies. PCR primer set of R111 (forward primer) 7 and X7-Dra (reverse primer) 11 was used in this study. The PCR conditions were as follows; 1). Initial denaturation: 94°C for 7 minutes; 2). PCR cycles: 94°C for 1 minute, 56°C for 1 minute and 72°C for 1 minute. The cycle numbers were 25, 30, 35, or 40 cycles; and 3). Final extension: 72°C for 7 minutes. The PCR products were electrophoresed on a 4% agarose gel and visualized with ethidium bromide.
3. PCR-RFLP analysis with DraI digestion
To test the diagnostic quality of the PCR products, PCR-RFLP (Restriction fragment length polymorphism) with DraI digestion was performed differentiating SMN1 and SMN2. DraI site was introduced into SMN2 product during the PCR11. The PCR product was digested by overnight incubation with DraI. More specifically, 8 μl of the PCR products was added to the enzyme solution (final volume, 20 μl) containing 1 × buffer M [final concentration] and 1 μl of DraI (15 U/μl) (Takara Biomedicals, Shiga), and the mixture was incubated overnight at 37°C. Subsequently, an aliquot of digested product was electrophoresed on a 4% agarose gel, and visualized with ethidium bromide.
After DraI digestion, the SMN2 product (187 bp) generated two fragments of 163 bp and 24 bp, while the SMN1 product (187 bp) did not undergo DraI digestion and retained the same product size as the non-digested fragment.
RESULTS
1. Comparison between DNA preparations and between DNA polymerases
The experiments for the comparison between DNA preparations and between DNA polymerases were done using a control sample.
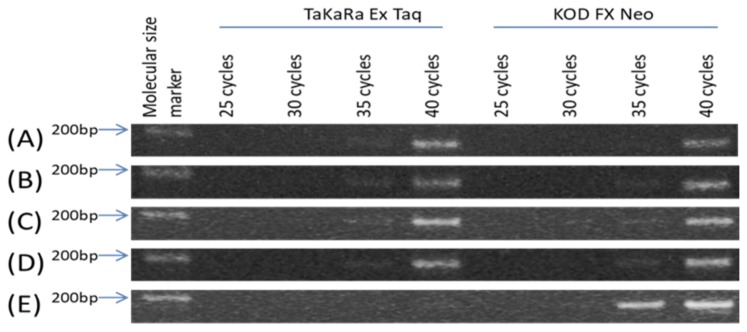
When TaKaRa Ex Taq™ polymerase was used, PCR with DNA preparation methods (A), (B), (C) and (D) generated high amount of PCR products at 40 cycles and the agarose gel electrophoresis showed clear bands of the PCR products. However, PCR with DNA preparation method (E) did not generate enough amount of PCR products at 40 cycles and the agarose gel electrophoresis showed no band of the PCR product (Fig. 1).
Fig. 1. Comparison between DNA preparations and between DNA polymerases.
DNA preparation methods were as follows: (A) DNA extraction by a simple “boiling” method, (B) DNA extraction by a “rinsing and boiling” method, (C) DNA extraction by a “protein fixation-1, rinsing and boiling” method, (D) DNA extraction by a “protein fixation-2, rinsing and boiling” method, and (E) No DNA extraction process. (A), (B), (C) and (D) were grouped into the “DNA extraction” method, while (E) was grouped into the “non-DNA-extraction” method. DNA polymerases tested in this study were TaKaRa Ex Taq™ and KOD FX Neo™.
When KOD FX Neo™ polymerase was used, PCR with DNA preparation methods (A) to (D) generated high amount of PCR products at 40 cycles. It should be noted that PCR with DNA preparation method (E) using KOD FX Neo™ also generated high amount of PCR products at 35 and 40 cycles.
The difference of PCR amplification efficiency with KOD FX Neo™ between (E) and others (A–D) may be due to the abundance of DNA. DNA in samples (E) was much more abundant than DNA in samples (A–D); one half of the DBS disk (E), which was used as template for PCR, was much more abundant in DNA than 2 μL of DNA solution of (A), (B), (C) and (D).
2. Test of the diagnostic quality for SMA
To test whether a combination of the method (E) and KOF FX Neo™ polymerase (i.e., direct PCR with KOF FX Neo™ polymerase) can produce sufficient and qualified PCR products for SMA diagnosis, we performed PCR-RFLP analysis with DraI digestion.
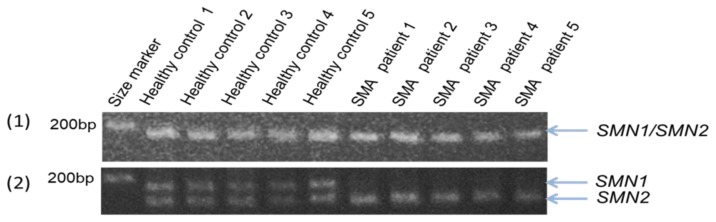
The agarose gel electrophoresis of the PCR products obtained at 40 cycles showed clear bands of SMN1 and/or SMN2 (Fig. 2, upper panel). The results of PCR-RFLP showed two bands of the PCR products from SMN1 and SMN2 in the five healthy volunteers, whereas only one band of the PCR products from SMN2 in five SMA patients (Fig. 2, lower panel). All SMA patients analyzed here had complete SMN1 deletion. These results were perfectly consistent with those of PCR-RFLP analysis using freshly collected blood samples (data not shown).
Fig. 2. RFLP using direct PCR products with KOD FX Neo™ polymerase.
(1) Before DraI digestion. The bands represent the mixture of PCR products from SMN1 and/or SMN2. (2) After DraI digestion. Upper bands represent PCR products from SMN1 and lower bands PCR products from SMN2. DraI digestion proved the presence of SMN1 and SMN2 in the PCR products of the healthy volunteers and the absence of SMN1 and the presence of SMN2 in the SMA patients. All SMA patients showed the absence of SMN1.
DISCUSSION
Here, we investigated whether PCR can be performed with or without DNA extraction process from DBS. When TaKaRa Ex Taq™ polymerase was used, DNA extraction process might be essential. Without DNA extraction process, PCR amplification was not successful even at 40 cycles. Whereas, when KOD FX Neo™ was used, PCR amplification was successful without DNA extraction process; PCR products at both 35 and 40 cycles were clearly detected on the gel electrophoresis (Fig. 1). These results indicate that KOD FX Neo™ is much less affected by PCR inhibitors presented in the DBS paper, and that we are able to perform direct PCR from the DBS paper without DNA extraction process.
We have known from our experience that KOD FX Neo™ polymerase is useful for long-range PCR targeting at amplification of more than 25Kb sized DNA such as SMN genes6. However, we used it in this study, not for long-range PCR (the PCR product sizes in this study was less than 200bp), but for direct PCR with a piece of DBS paper. According to the information from the company website (http://lifescience.toyobo.co.jp/), “the KOD FX Neo™ is able to amplify DNA from crude samples and can reduce influence of PCR inhibitors within soil, food, etc.” Our results clearly proved that KOD FX Neo™ polymerase is useful for the PCR amplification from DBS which can be considered as a kind of crude sample. Although the company emphasizes that KOD FX Neo™ overcomes PCR inhibitors in the crude samples, it may be empirical data, because we failed to get clear explanation about the mechanism of KOD FX Neo™ in the previous literatures.
If the diagnostic quality of direct PCR products is guaranteed, DNA extraction process from DBS can be omitted from the DBS-based SMA screening system. Bypassing the DNA extraction process can simplify the DBS-based SMA screening system and also reduce the required time for the screening. Thus, we performed RFLP analysis to test the diagnostic quality of the direct PCR products with KOD FX Neo™ polymerase. The results of RFLP analysis showed that SMN1 and SMN2 were present in all of five healthy volunteers, whereas only SMN2 in all of the five SMA patients. These findings were completely compatible with those of PCR-RFLP analysis with freshly collected blood samples, indicating that the diagnostic quality of direct PCR products was guaranteed.
In conclusion, direct PCR with DNA polymerases like KOD FX Neo™ can be widely used in newborn screening for SMA in the near future, because it can skip the process of DNA extraction from DBS and precisely amplify the target sequences in spite of the presence of PCR inhibitors. We previously established a DBS-based SMA screening with a modified competitive PCR (mCOP-PCR) technique2. In the previous protocol, a DNA extraction process was needed. However, the results of the current study may enable us to develop more sophisticated DBS-based newborn screening system for SMA, which can significantly reduce both process time and work load.
ACKNOWLEDGMENTS
This research was supported in part by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development, Grant No. 16ek0109086h0002 (title “Practical study for multicenter cooperative and investigator initiated clinical trial using valproic acid in childhood onset spinal muscular atrophy”).
Footnotes
DECLARATION OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
REFERENCES
- 1.Arnold WD, Kassar D, Kissel JT. Spinal muscμLar atrophy: Diagnosis and management in a new therapeutic era. Muscle Nerve. 2015;51:157–167. doi: 10.1002/mus.24497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ar Rochmah M, Harahap NIF, Niba ETE, et al. Genetic screening of spinal muscular atrophy using a real-time modified COP-PCR technique with dried blood-spot DNA. Brain Dev. 2017;39:774–782. doi: 10.1016/j.braindev.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Finkel RS, Chiriboga CA, Vajsar J, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 4.Finkel RS, Mercuri E, Darras BT, et al. ENDEAR Study Group. nusinersen versus sham control in infantile-onset spinal muscμLar atrophy. N Engl J Med. 2017;377:1723–32. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 5.Harahap NI, Harahap IS, Kaszynski RH, et al. Spinal muscular atrophy patient detection and carrier screening using dried blood spots on filter paper. Genet Test Mol Biomarkers. 2012;16:123–129. doi: 10.1089/gtmb.2011.0109. [DOI] [PubMed] [Google Scholar]
- 6.Kubo Y, Nishio H, Saito K. A new method for SMN1 and hybrid SMN gene analysis in spinal muscular atrophy using long-range PCR followed by sequencing. J Hum Genet. 2015;60:233–239. doi: 10.1038/jhg.2015.16. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre S, 1, Bürglen L, ReboμLlet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 8.Mikami A, Tajima T, Yamaguchi A, et al. Molecular diagnosis for steroid 21-hydroxylase deficiency by polymerase chain reaction with dried blood spots. Clin Pediatr Endocrinol. 1997;6:15–22. [Google Scholar]
- 9.Nishio H, Okamoto K, Saito T, Shinohara M. Spinal muscμLar atrophy: from molecular genetic diagnosis to neonatal screening BioClinica. 2018;33:70–73. [in Japanese] [Google Scholar]
- 10.Terpe K. Overview of thermostable DNA polymerases for classical PCR applications: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2013;97:10243–10254. doi: 10.1007/s00253-013-5290-2. [DOI] [PubMed] [Google Scholar]
- 11.van der Steege G, Grootscholten PM, van der Vlies P, et al. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995;345:985–986. [PubMed] [Google Scholar]
- 12.Wijaya YO, Niba ETE, Ar Rochmah M, et al. Nested PCR amplification secures DNA template quality and quantity in real-time mCOP-PCR screening for SMA. Kobe J Med Sci. 2019;65:E54–E58. [PMC free article] [PubMed] [Google Scholar]
- 13.Zerres K, Davies KE. Workshop report: ENMC International Workshop: Spinal Muscular Atrophies: recent progress and revised diagnostic criteria 17–19 April 1998, Soestduinen, The Netherlands. Neuromuscul Disord. 1999;9:272–278. doi: 10.1016/s0960-8966(99)00016-4. [DOI] [PubMed] [Google Scholar]