Abstract
Background
Cervical cancer (CC) is a common cancer with a poor prognosis due to the chemoresistance of CC cells to cisplatin. This study aimed to investigate the biological significance of lncRNA prostate cancer-associated transcript 6 (PCAT6) in the carcinogenesis of CC.
Materials and Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out to measure the abundance of PCAT6, miR-543 and zinc finger E-box binding protein 1 (ZEB1) in CC tissues and cells. The combination between miR-543 and lncRNA PCAT6 or ZEB1 was predicted by Starbase and was verified by dual-luciferase reporter assay, RNA-pull down assay and RNA immunoprecipitation (RIP) assay. Cell proliferation and chemoresistance to cisplatin were detected by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cell apoptosis and metastasis were determined by flow cytometry, Western blot and transwell migration and invasion assays.
Results
The abundance of ZEB1 protein was measured by Western blot assay. Murine xenograft model was established to confirm the function of lncRNA PCAT6 in vivo. The abundance of lncRNA PCAT6 was enhanced in CC tissues and cells compared with that in corresponding normal tissues and normal cervical epithelial cells Ect1/E6E7. MiR-543 was a target of PCAT6 and was negatively regulated by PCAT6. PCAT6 accelerated the proliferation, metastasis and the chemoresistance of CC cells to cisplatin while suppressed the apoptosis of CC cells. The overexpression of PCAT6 reversed the inhibitory effects of miR-543 accumulation on the proliferation, metastasis and chemoresistance of CC cells to cisplatin and the promoting impact on the apoptosis of CC cells. ZEB1 was a direct target of miR-543, and it functioned as the downstream gene of PCAT6/miR-543 to exert its oncogenic role in CC. PCAT6 promoted the growth of murine xenograft tumor through miR-543/ZEB1 axis in vivo.
Conclusion
LncRNA PCAT6 facilitated the proliferation, metastasis and chemoresistance of CC cells to cisplatin while impeded the apoptosis of CC cells via PCAT6/miR-543/ZEB1 axis. PCAT6/miR-543/ZEB1 axis might be a promising target for CC therapy.
Keywords: cervical cancer, lncRNA PCAT6, miR-543, ZEB1, chemoresistance
Introduction
There were 569,847 new cases of cervical cancer (CC) globally in 2018, accounting for 3.2% of all cancers.1 Human papillomavirus (HPV) infection is the leading cause of CC.2 Currently, surgery, chemotherapy and radiotherapy are the primary therapeutic methods for patients with CC.3 Therefore, it is pivotal for us to understand the molecular mechanism behind CC occurrence and progression.
Long noncoding RNAs (lncRNAs) are a class of non-coding RNAs (ncRNAs). They are implicated in many physiological and pathological processes.4–6 LncRNAs have been reported to bind to microRNAs (miRNAs) to exert their functions.7–9 LncRNA prostate cancer-associated transcript 6 (PCAT6) was identified as a regulator in prostate cancer.10 PCAT6 was abnormally regulated in multiple cancers, including colon cancer, gastric cancer and cervical cancer.11–13 Nevertheless, the biological role of PCAT6 in CC and the modulatory mechanism remain poorly understood.
MiRNAs are another class of ncRNAs, and they regulate the gene expression through binding to the 3ʹ untranslated region (3ʹ UTR) of their corresponding message RNAs (mRNAs).14–16 MiRNAs were implicated in cell proliferation, metastasis and apoptosis.17,18 MiR-543 have been reported to be abnormally modulated in a variety of cancers.19 For instance, Bing et al claimed that miR-543 was down-regulated in endometrial cancer, and it suppressed the oncogenicity of endometrial cancer through negatively regulating the abundance of FAK and TWIST1.20 Song et al claimed that miR-543 suppressed the metastasis of ovarian cancer cells by targeting MMP7.21 Liu et al reported that the abundance of miR-543 was reduced in CC tissues and cells, and miR-543 suppressed the growth and metastasis and promoted the apoptosis of CC cells by TRPM7.22 However, the underlying mechanism by which miR-543 regulating occurrence and progression of CC remains poorly understood.
Zinc finger E-box binding protein 1 (ZEB1) and ZEB2 are transcriptional regulators that promote epithelial-to-mesenchymal transition (EMT), and they play a crucial role in physiological and pathological processes.23–25 Larsen et al demonstrated that ZEB1 accelerated EMT in lung cancer.26 Preca et al claimed that ZEB1/HAS2 axis accelerated EMT of breast cancer cells.27 Herein, we concentrated on the biological role of ZEB1 in CC.
In this study, we first determined the expression of lncRNA PCAT6 in CC tissues and cells. Knockdown experiments were conducted to assess the role of PCAT6 on the growth, apoptosis, metastasis and drug resistance of CC cells. Besides, we explored the downstream genes of PCAT6 to further illustrate the mechanism by which PCAT6 facilitating the progression of CC.
Materials and Methods
Tissue Specimens
Forty-four CC tissues and paired control tissues were from patients in Liyang branch of jiangsu provincial people’s Hospital. The usage of human material was approved by the Ethics Committee of Liyang branch of jiangsu provincial people’s Hospital. Written informed consents were provided by all patients prior to surgical resection.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The reverse transcription was conducted using M-MLV reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) and All-in-One TM miRNA First-strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was regarded as the control for the quantification of PCAT6, miR-543 and ZEB1 using the 2−ΔΔCt method.28 The primers were presented as follows: PCAT6 (Forward, 5ʹ-CAGGAACCCCCTCCTTACTC-3ʹ; Reverse, 5ʹ-CTAGGGATGTGTCCGAAGGA-3ʹ), miR-543 (Forward, 5ʹ-AAACATTCGCG-3ʹ; Reverse, 5ʹ-AAGAAGTGCAC-3ʹ), ZEB1 (Forward, 5ʹ-TTCACAATTACTCACCTGTC-3ʹ; Reverse, 5ʹ-CCACACTCAGTGCATTTGAA-3ʹ), U6 (Forward, 5ʹ-CCTGCGCAAGGATGAC-3ʹ; Reverse, 5ʹ-GTGCAGGGTCCGAGGT-3ʹ), GAPDH (Forward, 5ʹ-CTGGGCTACACTGAGCACC-3ʹ; Reverse, 5ʹ-AAGTGG TCGTTGAGGGCAATG-3ʹ).
Cell Culture
CC cell lines SiHa, HeLa, ME180 and C-33A and normal cervical epithelial cell line Ect1/E6E7 were obtained from BeNa Culture Collection (Beijing, China). Cells were cultivated in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 10% penicillin (100 units/mL)/streptomycin (100 μg/mL) at 37°C in a 5% CO2 incubator with saturated humidity.
Cell Transfection
CC cells were transfected with the following small RNAs or plasmids using Lipofectamine 2000 (Invitrogen). Three small interfering RNA against PCAT6 (si-PCAT6#1, si-PCAT6#2, si-PCAT6#3), siRNA negative control (si-NC), empty vector (Vector), PCAT6 overexpression plasmid (PCAT6), short hairpin RNA negative control (sh-NC), shRNA against PCAT6 (sh-PCAT6) and ZEB1 overexpression plasmid (ZEB1) were obtained from Genepharma (Shanghai, China). MiRNA negative control (miR-NC), miR-543, anti-miR-NC and anti-miR-543 were purchased from Ribobio (Guangzhou, China).
Dual-Luciferase Reporter Assay
PCAT6-WT and PCAT6-MUT plasmids were constructed through cloning the seed sequences in PCAT6 (wild-type or mutant type) to the pmirGLO vector (Promega, Madison, WI, USA). SiHa and HeLa cells were co-transfected with PCAT6-WT or PCAT6-MUT and miR-543 or miR-NC. After incubating for 48 h, the CC cells were collected and the luciferase activity was examined by dual-luciferase assay system (Promega).
ZEB1 3ʹ UTR-WT or ZEB1 3ʹ UTR-MUT was generated through inserting the 3ʹ UTR of ZEB1 (wild-type or mutant type) to pmirGLO vector. The confirmation of the target relationship between ZEB1 and miR-543 was conducted following the similar approach.
RNA-Pull Down Assay
Bio-miR-543 and Bio-NC were generated through conjugating biotin with miR-543 or NC, and were transfected into CC cells. RNA-pull down assay was conducted as described previously.29 QRT-PCR was carried out to detect the abundance of PCAT6 and ZEB1 in the bound fractions.
RNA Immunoprecipitation (RIP)
The EZMagna RIP kit (Millipore, Billerica, MA, USA) was used to evaluate the target relationship between miR-543 and PCAT6. SiHa and HeLa cells were lysed. The supernatant was mixed with protein A/G beads conjugated with antibodies that recognized Argonaute-2 (Ago2) or Immunoglobulin G (IgG). The immunoprecipitated RNAs were used for identification by qRT-PCR assay.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
10 µL MTT (Invitrogen; 5 mg/mL) was added to the culture medium after transfection for 24 hrs, 48 hrs and 72 hrs, and cells were incubated for an additional 4 hrs. After discarding the cell supernatant, dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO, USA) was added to the wells to dissolve the formazan. The optical density at 570 nm was measured using a plate reader.
For detection of the chemoresistance of CC cells to cisplatin, CC cells were treated with cisplatin at a concentration of 12.5, 25, 50, 100 or 200 μM for 48 hrs, and the MTT assay was carried out as described above. The IC50 (inhibitory concentration 50%) value was defined when cell viability was 50%.
Cell Apoptosis Analysis
Annexin V-FITC Apoptosis Detection Kit (Invitrogen) was used to determine the apoptosis of CC cells. CC cells were harvested and re-suspended in 500µL Annexin V-fluorescein isothiocyanate (FITC) binding buffer. 5 µL Annexin V-FITC and 5 µL propidine iodide (PI; Solarbio, Beijing, China) were mixed with the CC cells in the dark for 10 min. The apoptotic cells (FITC+, PI±) were recognized through using flow cytometry system (BD Biosciences, San Jose, CA, USA).
Western Blot Assay
SiHa and HeLa cells were lysed and quantified using RIPA lysis solution (Beyotime, Shanghai, China) and the bicinchoninic acid (BCA) assay kit (Pierce, Rockford, IL, USA). 30 µg protein samples were loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). After being blocked with 5% non-fat dry milk for 1 h at room temperature, the membranes were incubated with antibodies against B cell leukemia/lymphoma 2 (Bcl-2; ab185002, Abcam, Cambridge, MA, USA), cleaved-caspase 3 (ab2302, Abcam), ZEB1 (ab228986, Abcam) and GAPDH (ab181602, Abcam) at 4°C overnight. The PVDF membranes were then incubated with secondary antibody combined with horseradish peroxidase (HRP) (ab205718, Abcam). The protein signal was detected with enhanced chemiluminescence (ECL) system (Pierce).
Transwell Migration and Invasion Assays
For migration assay, CC cells suspended in serum-free medium were seeded into the upper chambers, the culture medium supplemented with 10% FBS was added to the lower chambers. The CC cells in the lower surface of the upper chamber were stained and counted under an inverted optical microscope. The invasion assay was conducted using the upper chambers pre-coated with 1 mg/mL of Matrigel (BD Biosciences).
Murine Xenograft Assay
Animal experiments were performed with the permission of the Animal Research Committee of Liyang Branch of Jiangsu Provincial People’s Hospital and also followed with the guidelines of the NIH. A total of 12 mice were randomly divided into 2 groups. SiHa cells stably transfected with sh-PCAT6 or sh-NC were suspended in PBS buffer. 200µL cell suspension was subcutaneously injected into the flank of mice. The dimension of tumors was detected every 4 d, and the weight of tumors was recorded after injection for 28 d.
Statistical Analysis
All data were indicated as mean ± standard deviation (SD). The comparison significances between two groups were assessed by Student’s t-test, and the comparison significances among multiple groups were evaluated by one-way analysis of variance (ANOVA) followed by Turkey’s test. The correlation between the expression of miR-543, PCAT6 and ZEB1 was analyzed through Spearman correlation coefficient. P value less than 0.05 was regarded as statistically significant.
Results
The Abundance of lncRNA PCAT6 and miR-543 Is Aberrantly Regulated in CC Tissues and Cells
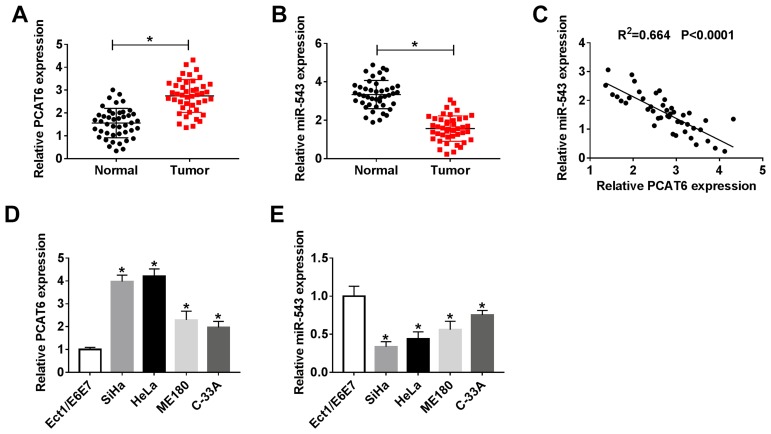
To investigate the potential role of lncRNA PCAT6 and miR-543 in CC, we first examined the expression of PCAT6 and miR-543 in CC tissues and corresponding normal tissues. The abundance of PCAT6 was elevated in CC tissues compared with that in corresponding normal tissues, while the level of miR-543 was lower in CC tissues than that in adjacent normal tissues (Figure 1A and B). The expression of miR-543 was negatively correlated with the level of PCAT6 in CC tissues (Figure 1C). Meanwhile, we measured the abundance of PCAT6 and miR-543 in CC cells. As expected, the level of PCAT6 was elevated in CC cells compared with that in normal cervical epithelial cells Ect1/E6E7, while the abundance of miR-543 was down-regulated in CC cells compared with that in Ect1/E6E7 cells (Figure 1D and E).
Figure 1.
The abundance of lncRNA PCAT6 and miR-543 is aberrantly regulated in CC tissues and cells. (A and B) The expression of lncRNA PCAT6 and miR-543 was determined in CC tissues (n=44) and adjacent normal tissues (n=44) by qRT-PCR. (C) Spearman correlation analysis was carried out to evaluate the correlation between the level of miR-543 and the abundance of lncRNA PCAT6 in CC tissues. (D and E) The expression of lncRNA PCAT6 and miR-543 was examined in CC cells and normal cervical epithelial cells Ect1/E6E7 by qRT-PCR. *P<0.05.
LncRNA PCAT6 Could Sponge miR-543
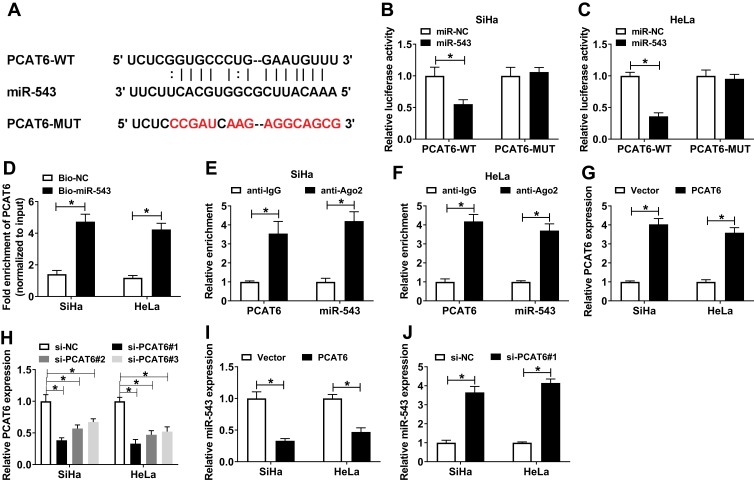
We wondered whether lncRNA PCAT6 could sponge and negatively regulate the expression of miR-543 in CC cells. Starbase bioinformatic software predicted that miR-543 was a target of PCAT6. The putative binding sites between PCAT6 and miR-543 are showed in Figure 2A. To confirm the combination between PCAT6 and miR-543, we constructed luciferase reporter vector with the sequence of PCAT6 wild-type or mutant binding sites, named as PCAT6-WT or PCAT6-MUT, and co-transfected with miR-543 or miR-NC into SiHa and HeLa cells. The results revealed that the luciferase activity was significantly decreased with the overexpression of miR-543 in PCAT6-WT group compared with that in PCAT6-MUT group, indicating that PCAT6 could bind to miR-543 in SiHa and HeLa cells (Figure 2B and C). RNA-pull down experiment further revealed that PCAT6 could bind to miR-543 in CC cells (Figure 2D). Besides, RIP experiment suggested that lncRNA PCAT6 could bind to the RISC complex, likely via the relationship with miR-543 (Figure 2E and F). We assessed the overexpression and knockdown efficiency of PCAT6 overexpression plasmid (PCAT6) and PCAT6 small interfering RNA (si-PCAT6#1, si-PCAT6#2 and si-PCAT6#3) in SiHa and HeLa cells. QRT-PCR results showed that the enrichment of PCAT6 was notably increased in CC cells transfected with PCAT6, and the interference of PCAT6 with PCAT6 small interfering RNA prominently decreased the expression of PCAT6 in CC cells (Figure 2G and H). We chose si-PCAT6#1 and si-PCAT6#2 for the following experiments because of their higher knockdown efficiency in CC cells. As mentioned in Figure 2I and J, the expression of miR-543 was down-regulated in CC cells transfected with PCAT6, and it was enhanced by the transfection of si-PCAT6#1. These findings demonstrated that miR-543 was a direct target of PCAT6 and was negatively modulated by PCAT6.
Figure 2.
LncRNA PCAT6 could sponge miR-543. (A) The binding sites between miR-543 and lncRNA PCAT6 were predicted by Starbase online software. (B and C) Luciferase activity was determined in SiHa and HeLa cells co-transfected with PCAT6-WT or PCAT6-MUT and miR-NC or miR-543. (D) RNA-pull down experiment was conducted to verify the combination between miR-543 and lncRNA PCAT6 in CC cells. (E and F) The combination between miR-543 and lncRNA PCAT6 was confirmed by RIP experiment. (G and H) The overexpression or knockdown efficiency of PCAT6 overexpression plasmid or small interfering RNA against PCAT6 (si-PCAT6#1, si-PCAT6#2, si-PCAT6#3) was assessed in SiHa and HeLa cells by qRT-PCR. (I) The expression of miR-543 was examined in SiHa and HeLa cells transfected with Vector or PCAT6 by qRT-PCR. (J) QRT-PCR was performed to detect the abundance of miR-543 in the two CC cells transfected with si-NC or si-PCAT6#1. *P<0.05.
LncRNA PCAT6 Plays an Oncogenic Role in the Progression of CC
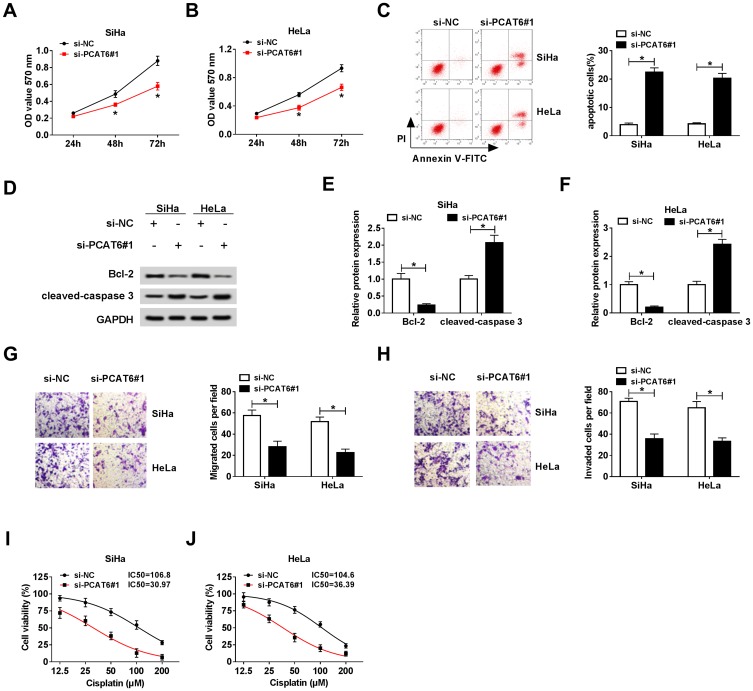
To illustrate the biological role of PCAT6 in CC cells, MTT assay, flow cytometry, Western blot and transwell migration and invasion assays were conducted to measure the proliferation, apoptosis, metastasis and chemoresistance of CC cells transfected with si-NC, si-PCAT6#1 or si-PCAT6#2. As showed in Figure 3A and B, we found that PCAT6 depletion conspicuously suppressed the proliferation of CC cells. Flow cytometry indicated that the apoptosis of CC cells was promoted with the intervention of PCAT6 in CC cells (Figure 3C). Western blot demonstrated that the level of anti-apoptotic protein Bcl-2 was reduced, but the enrichment of cleaved-caspase 3 was elevated in SiHa and HeLa cells transfected with si-PCAT6#1 and si-PCAT6#2 (Figure 3D–F). In addition, the interference of PCAT6 inhibited the migration and invasion of CC cells (Figure 3G and H). MTT assay showed that the IC50 (inhibitory concentration 50%) value was reduced by the transfection of si-PCAT6#1 and si-PCAT6#2 in CC cells, indicating that PCAT6 depletion decreased the chemoresistance of CC cells to cisplatin (Figure 3I and J). These data revealed that PCAT6 played an oncogenic role to promote the proliferation, metastasis and chemoresistance of CC cells to cisplatin and inhibit the apoptosis of CC cells.
Figure 3.
LncRNA PCAT6 plays an oncogenic role in the progression of CC. (A and B) Cell proliferation was examined in SiHa and HeLa cells transfected with si-NC or si-PCAT6#1 by MTT assay. (C) Flow cytometry was performed to assess the apoptosis of CC cells transfected with si-NC or si-PCAT6#1. (D–F) The abundance of apoptosis-related proteins (Bcl-2 and cleaved-caspase 3) in CC cells transfected with si-NC or si-PCAT6#1 was measured by Western blot assay and was quantified by gray analysis. (G and H) Cell migration and invasion were measured in CC cells transfected with si-NC or si-PCAT6#1 by transwell migration and invasion assays. (I and J) Si-NC or si-PCAT6#1 transfected SiHa and HeLa cells were treated with cisplatin at a concentration of 12.5, 25, 50, 100 or 200 μM, and MTT assay was performed to detect the chemoresistance of the above CC cells. *P<0.05.
The Overexpression of PCAT6 Alleviates the Inhibitory Effects of miR-543 Accumulation on the Proliferation, Metastasis and Chemoresistance and the Promoting Effect on the Apoptosis of CC Cells
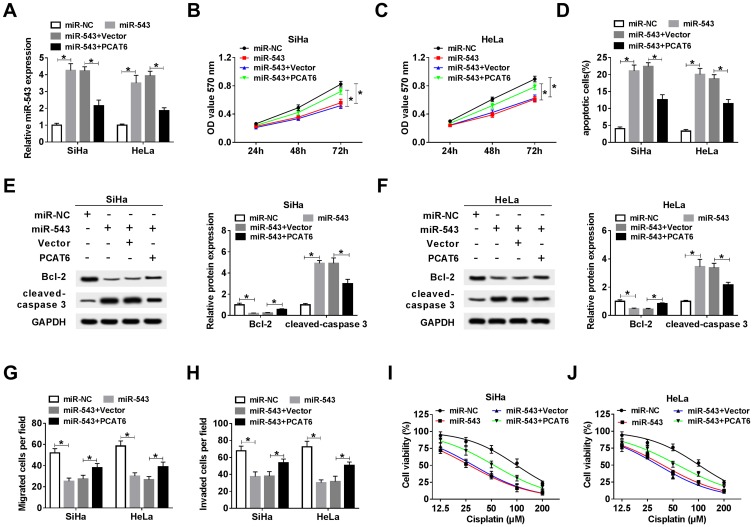
As mentioned above, miR-543 was a direct target of PCAT6 in CC cells. We wondered whether miR-543 was involved in PCAT6-mediated progression of CC. SiHa and HeLa cells were transfected with miR-NC, miR-543, miR-543 + Vector or miR-543 + PCAT6. We first measured the abundance of miR-543 in the above CC cells. The abundance of miR-543 was elevated with the overexpression of miR-543, and it was decreased with the addition of PCAT6 in CC cells (Figure 4A). As indicated in Figure 4B and C, the accumulation of miR-543 inhibited the proliferation of CC cells, and the overexpression of PCAT6 counteracted the inhibitory impact of miR-543 accumulation on the proliferation of CC cells. In addition, the transfection of PCAT6 overexpression plasmid reversed the promoting effect of miR-543 accumulation on the apoptosis of CC cells (Figure 4D). Meanwhile, Western blot assay revealed the same results, the abundance of cleaved-caspase 3 was enhanced with the addition of miR-543, and it was down-regulated by the co-transfection of miR-543 and PCAT6 in CC cells (Figure 4E and F). The expression of anti-apoptotic protein Bcl-2 showed an inverse trend to cleaved-caspase 3 in CC cells, indicating that PCAT6 suppressed the apoptosis of CC cells through sponging miR-543. Transwell migration and invasion assays suggested that the overexpression of PCAT6 alleviated the inhibitory impact of miR-543 accumulation on the metastasis of CC cells (Figure 4G and H). We assessed the chemoresistance of CC cells to cisplatin by MTT assay. MiR-543 overexpression reduced the chemoresistance of CC cells to cisplatin, and the addition of PCAT6 attenuated the effect of miR-543 accumulation on the chemoresistance of CC cells to cisplatin (Figure 4I and J). Collectively, lncRNA PCAT6 promoted the development of CC through sponging miR-543.
Figure 4.
The overexpression of PCAT6 alleviates the inhibitory effects of miR-543 accumulation on the proliferation, metastasis and chemoresistance and the promoting effect on the apoptosis of CC cells. CC cells transfected with miR-NC, miR-543, miR-543 + Vector or miR-543 + PCAT6 were used for the following experiments. (A) The abundance of miR-543 was measured in CC cells by qRT-PCR. (B and C) MTT assay was carried out to detect the proliferation of CC cells. (D) Flow cytometry was performed to determine the apoptosis of CC cells. (E and F) Western blot assay was conducted to measure the abundance of Bcl-2 and cleaved-caspase 3 in CC cells. (G and H) Transwell migration and invasion assays were conducted in CC cells, and the number of migration and invasion cells was counted. (I and J) Cell viability was detected in the above CC cells treated with cisplatin at a concentration of 12.5, 25, 50, 100 or 200 μM. *P<0.05.
ZEB1 Is a Direct Target of miR-543
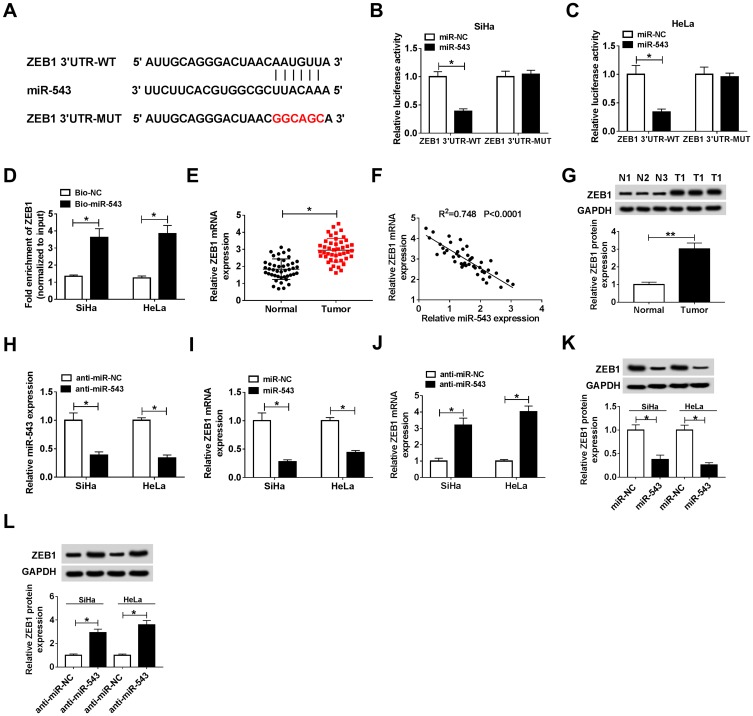
How does miR-543 affect the proliferation, apoptosis, metastasis and drug resistance of CC cells? Bioinformatic analysis predicted that ZEB1 could bind to miR-543 (Figure 5A). The combination between ZEB1 and miR-543 was then verified by dual-luciferase reporter assay and RNA-pull down assay in SiHa and HeLa cells. The luciferase activity was dramatically decreased with the co-transfection of ZEB1 3ʹ UTR-WT and miR-543, indicating that ZEB1 was a direct target of miR-543 in CC cells (Figure 5B and C). RNA-pull down assay revealed that ZEB1 could bind to miR-543 in SiHa and HeLa cells (Figure 5D).
Figure 5.
ZEB1 is a direct target of miR-543. (A) ZEB1 was predicted to be a target of miR-543 based on the information of Starbase. (B and C) Dual-luciferase reporter assay was conducted to confirm the combination between miR-543 and ZEB1 in SiHa and HeLa cells. (D) RNA-pull down experiment was conducted to verify the combination between miR-543 and ZEB1 in SiHa and HeLa cells. (E) The expression of ZEB1 was examined in CC tissues (n=44) and normal tissues (n=44) by qRT-PCR. (F) The correlation between the level of ZEB1 and the abundance of miR-543 was assessed in CC tissues. (G) Western blot assay was performed to measure the abundance of ZEB1 in CC tissues and adjacent normal tissues. (H) The knockdown efficiency of miR-543 inhibitor (anti-miR-543) was evaluated in CC cells by qRT-PCR. (I and K) The abundance of ZEB1 mRNA and protein was examined in CC cells transfected with miR-NC or miR-543 by qRT-PCR and Western blot. (J and L) The abundance of ZEB1 mRNA and protein was examined in CC cells transfected with anti-miR-NC or anti-miR-543 by qRT-PCR and Western blot assay. *P<0.05, **P<0.01.
We analyzed the mRNA and protein expression of ZEB1 in CC tissues by qRT-PCR and Western blot, and we found the mRNA and protein expression of ZEB1 was higher in CC tissues than that in corresponding normal tissues (Figure 5E and G). In addition, a conspicuous inverse correlation was found between the expression of miR-543 and the level of ZEB1 in CC tissues (Figure 5F). To illustrate the modulatory relationship between miR-543 and ZEB1 in CC cells, we first evaluated the knockdown efficiency of anti-miR-543 in CC cells. As mentioned in Figure 5H, the level of miR-543 was prominently decreased by the transfection of anti-miR-543 in CC cells. The abundance of ZEB1 mRNA and protein was reduced with the overexpression of miR-543 in SiHa and HeLa cells, and miR-543 depletion up-regulated the mRNA and protein level of ZEB1 in the two CC cells by qRT-PCR and Western blot assays (Figure 5I–L). The above results indicated that ZEB1 was a direct target of miR-543 in CC cells, and the expression of ZEB1 was negatively modulated by miR-543.
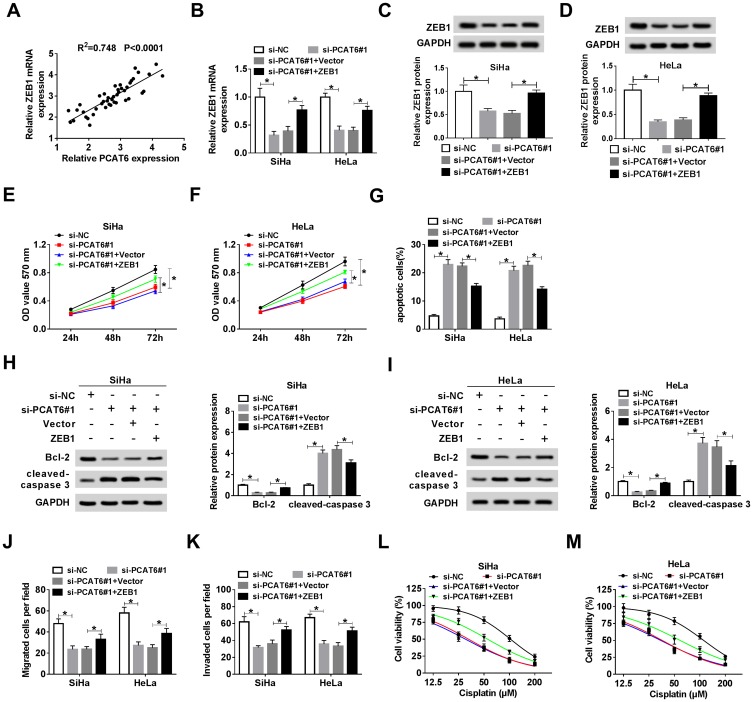
The Accumulation of ZEB1 Attenuates the Suppressive Impacts of PCAT6 Interference on the Proliferation, Metastasis and Chemoresistance and the Promoting Effect on the Apoptosis of CC Cells
As mentioned above, the expression of ZEB1 and PCAT6 was elevated in CC tissues compared with that in adjacent normal tissues. As expected, there was a positive correlation between the expression of ZEB1 mRNA and the enrichment of PCAT6 in CC tissues (Figure 6A). To investigate whether ZEB1 was involved in PCAT6-mediated proliferation, metastasis, apoptosis and drug resistance of CC cells, we transfected si-NC, si-PCAT6#1, si-PCAT6#1 + Vector or si-PCAT6#1 + ZEB1 into CC cells. The abundance of ZEB1 mRNA and protein was down-regulated with the depletion of PCAT6, and it was recovered with the addition of ZEB1 overexpression plasmid in CC cells (Figure 6B–D). As showed in Figure 6E and F, the overexpression of ZEB1 alleviated the inhibitory effect of PCAT6 depletion on the proliferation of CC cells. Meanwhile, flow cytometry and Western blot assay showed that the accumulation of ZEB1 attenuated the promoting impact of PCAT6 intervention on the apoptosis of CC cells (Figure 6G–I). Transwell migration and invasion assays revealed that the transfection of ZEB1 overexpression plasmid alleviated the suppressive effect of PCAT6 depletion on the metastasis of CC cells (Figure 6J and K). The IC50 value was down-regulated by the transfection of si-PCAT6#1, and it was partly recovered with the co-transfection of si-PCAT6#1 and ZEB1 in CC cells (Figure 6L and M). Taken together, PCAT6 promoted the progression of CC at least partly through miR-543/ZEB1 axis.
Figure 6.
The accumulation of ZEB1 attenuates the suppressive impacts of PCAT6 interference on the proliferation, metastasis and chemoresistance and the promoting effect on the apoptosis of CC cells. (A) The liner relationship between the expression of ZEB1 and the level of PCAT6 was analyzed in CC tissues. CC cells transfected with si-NC, si-PCAT6#1, si-PCAT6#1 + Vector or si-PCAT6#1 + ZEB1 were used for the following experiments. (B–D) The abundance of ZEB1 mRNA and protein was measured in CC cells by qRT-PCR and Western blot. (E and F) MTT assay was conducted to measure the proliferation of CC cells. (G) Cell apoptosis was evaluated in the two CC cells by flow cytometry. (H and I) The abundance of apoptosis-related proteins (Bcl-2 and cleaved-caspase 3) in CC cells was determined by Western blot assay. (J and K) The migration and invasion of CC cells were examined by transwell migration and invasion assays. (L and M) MTT assay was carried out to detect the viability of the above CC cells treated with cisplatin at a concentration of 12.5, 25, 50, 100 or 200 μM. *P<0.05.
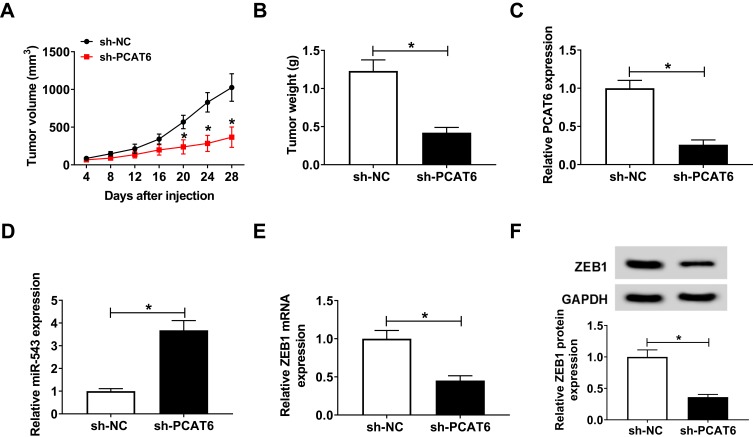
PCAT6 Depletion Inhibits CC Tumor Growth in vivo
We established murine xenograft model using SiHa cells stably transfected with sh-NC or sh-PCAT6 to confirm the effect of PCAT6/miR-543/ZEB1 axis in vivo. The transfection of sh-PCAT6 also inhibited the proliferation, metastasis, chemoresistance while promoted the apoptosis of SiHa cells (Figure S1), which was in agreement with the effects of si-PCAT6 transfection. Tumor volume was measured every 4 days, and the tumor was weighted after four-week inoculation. As indicated in Figure 7A and B, tumors from the sh-PCAT6 group were lower than that in sh-NC group. The expression of PCAT6, miR-543 and ZEB1 was detected in excisional tumor tissues by qRT-PCR or Western blot. The level of PCAT6 was down-regulated with the transfection of sh-PCAT6 (Figure 7C). The abundance of miR-543 was dramatically enhanced in sh-PCAT6 group compared with that in sh-NC group (Figure 7D). The abundance of ZEB1 mRNA and protein was reduced in sh-PCAT6 group compared with that in sh-NC group (Figure 7E and F). Therefore, lncRNA PCAT6 promoted the growth of murine xenograft tumor partly through the miR-543/ZEB1 axis in vivo.
Figure 7.
PCAT6 depletion inhibits CC tumor growth in vivo. SiHa cells stably transfected with sh-NC or sh-PCAT6 were subcutaneously injected into the murine (n=6). (A) Tumor volume was recorded every 4 d. (B) Murine xenograft tumor was weighed following four-week inoculation. (C) The expression of PCAT6 was measured in resected tumor tissues by qRT-PCR. (D) The level of miR-543 was determined in resected tumor tissues by qRT-PCR. (E and F) The abundance of ZEB1 mRNA and protein was examined in resected tumor tissues by qRT-PCR and Western blot. *P<0.05.
Discussion
Although the treatment methods of CC have been improved, the 5-year survival rate of CC patients remains dismal. Therefore, uncovering the pathogenesis is crucial for CC therapy. Our study first presented the vital role of PCAT6/miR-543/ZEB1 axis in CC. LncRNA PCAT6 facilitated the proliferation, metastasis and chemoresistance of CC cells to cisplatin while impeded the apoptosis of CC cells.
LncRNA PCAT6 has been reported to be up-regulated in multiple cancers. Lv et al demonstrated that the abundance of lncRNA PCAT6 was aberrantly enhanced in CC, and PCAT6 accelerated the proliferation and metastasis and inhibited apoptosis of CC cells.13 Shi et al claimed that PCAT6 was up-regulated in non-small-cell lung cancer, and PCAT6 served as an oncogene to promote the growth and metastasis of non-small-cell lung cancer cells.30 Wu et al reported that PCAT6 was abnormally up-regulated in colorectal cancer, and lncRNA PCAT6 elevated the chemoresistance of colorectal cancer cells to 5-fluorouracil (5-FU) via miR-204/HMGA2/PI3K axis.31 We concentrated on the role of PCAT6 in CC, and we wondered whether PCAT6 served as a competing endogenous RNA (ceRNA) to exert its function in CC. Consistent with the above findings, lncRNA PCAT6 was up-regulated in CC tissues and cells compared with that in adjacent normal tissues and normal cervical epithelial cells Ect1/E6E7. Loss-of-function experiments revealed that PCAT6 promoted the proliferation, metastasis and drug resistance while inhibited the apoptosis of CC cells.
MiR-543 was predicted as a target of lncRNA PCAT6, and the combination between miR-543 and PCAT6 was verified by dual-luciferase reporter assay. MiR-543 was negatively regulated by lncRNA PCAT6 in CC cells. The accumulation of lncRNA PCAT6 alleviated the inhibitory effects of miR-543 overexpression on the proliferation, metastasis and drug resistance of CC cells and the promoting impact on the apoptosis of CC cells.
To illustrate the precise mechanism by which miR-543 regulating the progression of CC, we aimed to find the downstream component of miR-543 in CC cells. ZEB1 is a crucial transcriptional regulator in EMT,32,33 and it could enhance the abundance of Vimentin and decline the enrichment of E-cadherin. The high expression of ZEB1 is correlated with the poor outcomes in cancers, including proliferation, metastasis and chemoresistance.34,35 Herein, ZEB1 was identified as a direct target of miR-543 in CC cells. To elucidate the modulatory relationship between miR-543 and ZEB1, SiHa and HeLa cells were transfected with miR-NC, miR-543, anti-miR-NC or anti-miR-543. The expression of ZEB1 was inversely modulated by miR-543 in CC cells. Subsequently, we found that the overexpression of ZEB1 could attenuate the suppressive effects of PCAT6 depletion on the proliferation, metastasis and chemoresistance and the promoting impact on the apoptosis of CC cells. In addition, lncRNA PCAT6 promoted the growth of CC tumor via miR-543/ZEB1 in vivo.
Taken together, we demonstrated that the overexpression of PCAT6 down-regulated the expression of miR-543 in CC cells, thereby enhanced the level of ZEB1, ultimately elevated the chemoresistance of CC cells to cisplatin, promoted the proliferation and metastasis while inhibited the apoptosis of CC cells. The PCAT6/miR-543/ZEB1 axis might be an underlying therapeutic target for the treatment of CC.
Disclosure
The authors report no financial conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Chelimo C, Wouldes TA, Cameron LD, et al. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J Infect. 2013;66(3):207–217. doi: 10.1016/j.jinf.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 3.Divine LM, Huh WK. Tertiary prevention of cervical cancer. Clin Obstet Gynecol. 2014;57(2):316–324. doi: 10.1097/GRF.0000000000000020 [DOI] [PubMed] [Google Scholar]
- 4.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eades G, Zhang YS, Li QL, et al. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5(2):134–141. doi: 10.5306/wjco.v5.i2.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34(2):613–620. doi: 10.1007/s13277-013-0658-6 [DOI] [PubMed] [Google Scholar]
- 7.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Long B, Zhou LY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596 [DOI] [PubMed] [Google Scholar]
- 10.Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W, Su G, Huang X, et al. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle. 2019;18(1):69–83. doi: 10.1080/15384101.2018.1558872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Sun JY, Jin YF, et al. PCAT6 participates in the development of gastric cancer through endogenously competition with microRNA-30. Eur Rev Med Pharmacol Sci. 2018;22(16):5206–5213. doi: 10.26355/eurrev_201808_15718 [DOI] [PubMed] [Google Scholar]
- 13.Lv XJ, Tang Q, Tu YQ, et al. Long noncoding RNA PCAT6 regulates cell growth and metastasis via Wnt/beta-catenin pathway and is a prognosis marker in cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23(5):1947–1956. doi: 10.26355/eurrev_201903_17233 [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 16.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 17.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036 [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 19.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287(51):42695–42707. doi: 10.1074/jbc.M112.387761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bing L, Hong C, Li-Xin S, et al. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecol Obstet. 2014;290(3):533–541. doi: 10.1007/s00404-014-3219-3 [DOI] [PubMed] [Google Scholar]
- 21.Song N, Liu H, Ma X, et al. Placental growth factor promotes metastases of ovarian cancer through MiR-543-regulated MMP7. Cell Physiol Biochem. 2015;37(3):1104–1112. doi: 10.1159/000430235 [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Gan L, Zhang J. miR-543 inhibits cervical cancer growth and metastasis by targeting TRPM7. Chem Biol Interact. 2019;302:83–92. doi: 10.1016/j.cbi.2019.01.036 [DOI] [PubMed] [Google Scholar]
- 23.Eger A, Aigner K, Sonderegger S, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24(14):2375–2385. doi: 10.1038/sj.onc.1208429 [DOI] [PubMed] [Google Scholar]
- 24.Vandewalle C, Comijn J, De Craene B, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33(20):6566–6578. doi: 10.1093/nar/gki965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14(4):481–487. doi: 10.1080/15384101.2015.1006048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen JE, Nathan V, Osborne JK, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126(9):3219–3235. doi: 10.1172/JCI76725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preca BT, Bajdak K, Mock K, et al. A novel ZEB1/HAS2 positive feedback loop promotes EMT in breast cancer. Oncotarget. 2017;8(7):11530–11543. doi: 10.18632/oncotarget.14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 29.Lal A, Thomas MP, Altschuler G, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7(11):e1002363. doi: 10.1371/journal.pgen.1002363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:(177–187. doi: 10.1016/j.ebiom.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H, Zou Q, He H, et al. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8(5):2484–2495. doi: 10.1002/cam4.2019.8.issue-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Tillo E, Siles L, De Barrios O, et al. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am J Cancer Res. 2011;1(7):897–912. [PMC free article] [PubMed] [Google Scholar]
- 33.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003;22(10):2443–2452. doi: 10.1093/emboj/cdg225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazarova D, Bordonaro M. ZEB1 mediates drug resistance and EMT in p300-Deficient CRC. J Cancer. 2017;8(8):1453–1459. doi: 10.7150/jca.18762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Y, Zheng X, Zhou J, et al. ZEB1 promotes the progression and metastasis of cervical squamous cell carcinoma via the promotion of epithelial-mesenchymal transition. Int J Clin Exp Pathol. 2015;8(9):11258–11267. [PMC free article] [PubMed] [Google Scholar]