Abstract
Cardiovascular disease is an enormous socioeconomic burden worldwide and remains a leading cause of mortality and disability despite significant efforts to improve treatments and personalize healthcare. Heart failure is the main manifestation of cardiovascular disease and has reached epidemic proportions. Heart failure follows a loss of cardiac homeostasis, which relies on a tight regulation of gene expression. This regulation is under the control of multiple types of RNA molecules, some encoding proteins (the so-called messenger RNAs) and others lacking protein-coding potential, named noncoding RNAs. In this review article, we aim to revisit the notion of regulatory RNA, which has been thus far mainly confined to noncoding RNA. Regulatory RNA, which we propose to abbreviate as regRNA, can include both protein-coding RNAs and noncoding RNAs, as long as they contribute, directly or indirectly, to the regulation of gene expression. We will address the regulation and functional role of messenger RNAs, microRNAs, long noncoding RNAs, and circular RNAs (ie, regRNAs) in heart failure. We will debate the utility of regRNAs to diagnose, prognosticate, and treat heart failure, and we will provide directions for future work.
Keywords: biomarkers, epigenetics, heart failure, RNA, transcriptome analysis
Introduction: the Ever-Growing Burden of Heart Failure
Heart failure is an ever-growing problem for which new therapeutic interventions are still required. This is despite remarkable advances made in our understanding of heart failure and in our management of the condition. So what is the problem with heart failure? There are two major issues: (1) vast numbers of patients have the condition; and (2) despite treatment advances, the prognosis remains poor. The estimated prevalence of heart failure in industrialized societies is 1% to 2% of all adults, rising to >10% of those >70 years of age. In Europe, the lifetime risk for an individual 55 years of age is estimated at 33% for men and 28% for women,1 and in the United States the lifetime risk is 20% for those 40 years of age or more.2 In the United States, more than 650 000 incident cases are identified annually. Although this figure has remained relatively stable, improvements in management and subsequent survival have led to the increasing prevalence of heart failure in the US population3 and in most other industrialized countries;4 developing countries face an epidemic yet to materialize fully.
In this article, we describe the place of RNAs in aspects of heart failure and their potential as novel clinical and therapeutic biomarkers. We introduce the concept of regulatory RNAs (regRNAs) and gather the current knowledge of the role of RNA molecules with regulatory functions (ie, regRNAs) in several pathophysiological pathways associated with heart failure, namely cardiac hypertrophy, inflammation, regeneration, and angiogenesis. We emphasize the translational potential of regRNAs and propose directions for future work. Because of the large amount of available data in the field and space limitations for this article, we unfortunately could not cite several interesting works.
The RNA Family: Regulatory RNAs Revisited
For many years, the scientific community has mostly dedicated attention to a mere ≈2% of the human genome corresponding to protein-coding exons of messenger RNAs (mRNAs). Ribosomal RNAs, transfer RNAs, small nuclear and nucleolar RNAs, although known for many years, displayed mainly housekeeping functions dedicated to mRNA maturation and protein synthesis. From the beginning of this century, however, high-throughput RNA-sequencing analysis of the genomic functional elements has clearly shown that more than 70% of the human genome is transcribed. It is now clear that the genome portions previously indicated as “junk DNA” are transcribed, giving origin to a large and heterogeneous family of RNA molecules with a variety of functions. Indeed, RNAs have surfaced as a promising tool in the field of diagnostics, therapeutics, and personalized medicine. Figure 1 displays the categories of RNAs known so far and constituting the “RNA family.” Several categories possess regulatory roles and can be grouped into a subfamily that we propose to name regRNAs.
Figure 1.
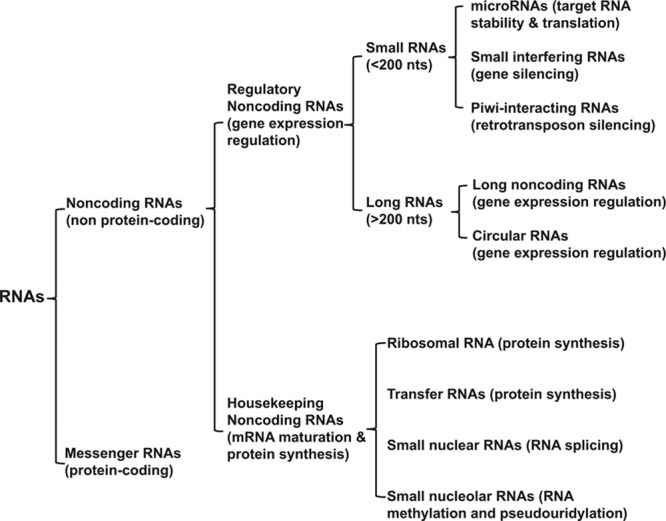
The RNA family and regulatory RNAs. Nts indicates nucleotides.
Regulatory RNAs are composed mostly of noncoding RNAs (ncRNAs). MicroRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) have gained increased interest in the cardiovascular community for their ability to modulate cellular responses. These should be intended as operative categories, given that the distinction between coding and noncoding RNAs is at times blurred, with several cases of RNA species displaying dual roles encoding microproteins (less than 100 amino acids). Accordingly, the genomewide analysis of heart translatomes, characterized using ribosome profiling, identified hundreds of microproteins, expressed from both lncRNAs and circRNAs.5 Some of these microproteins are expressed from functionally characterized lncRNAs, suggesting their contribution to biological functions previously assigned to the lncRNA. On the other hand, protein-coding sequences display evidence of other embedded functions, as indicated by constraints on synonymous codon usage.
Regulatory RNAs can be part of complex structures or interact with RNA-binding proteins, DNA, and other RNA types, regulating their activity. They display distinctive temporal and spatial patterns of expression, indicating a precise regulation of their expression. They can be subjected to methylation, the most important being N6-methyladenosine. Furthermore, given that certain regRNAs demonstrate increased stability in the blood and that dysregulated numbers represent different disease states, they may function as important disease biomarkers. Noncoding RNAs are classified for practical reasons according to their size, less or more than 200 nucleotides long, into short- and long-ncRNAs, respectively (Figure 1). The main mechanisms of action of ncRNAs are summarized in Figure 2.
Figure 2.
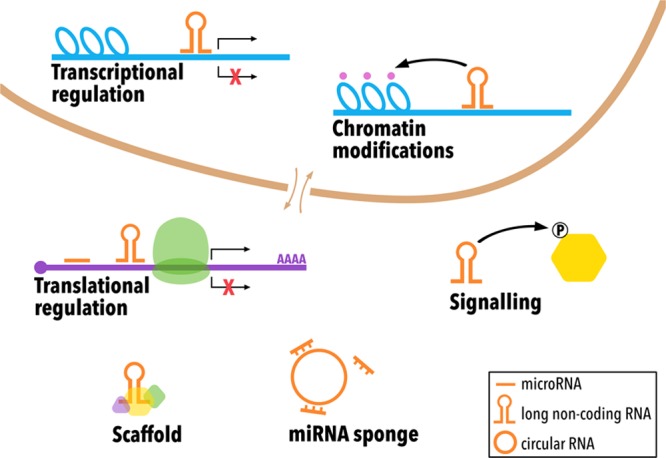
Main mechanisms of action of ncRNAs. Noncoding RNAs (microRNAs, long non-coding RNAs, and circular RNAs), represented in orange, act in different ways to regulate gene expression, such as chromatinmodification, transcriptional and translational activation and repression, protein signalling, scaffold, and miRNA sponge. miRNA indicates microRNA.
Short Noncoding RNAs
Short ncRNAs include different RNA categories, such as small interfering RNAs, piwi-interacting RNAs, and miRNAs, that are the most widely studied.
In most circumstances, miRNAs are transcribed as long precursors in which miRNA-containing hairpins are cleaved by the sequential actions of DROSHA-DGCR8 (Drosha Ribonuclease III; DiGeorge Syndrome Critical Region Gene 8) microprocessor in the nucleus and of DICER-TRBP (Dicer 1, Ribonuclease III; transactivation response element RNA-binding protein) complex in the cytoplasm. The product is a ≈22-nt mature miRNA that binds to Argonaute proteins to form the RNA-induced silencing complex, which mediates target-gene silencing by mRNA degradation or translation inhibition. MiRNA binding sites have been identified in 3’ and 5’ untranslated regions, introns as well as exonic regions of protein-coding mRNAs and lncRNAs. Moreover, miRNAs targeting of the 5′- untranslated region can even induce gene expression, and a role in both negative and positive gene transcriptional regulation by intranuclear miRNAs is also emerging.6
Regardless of their mechanism of action, each miRNA can regulate hundreds of mRNAs, often targeting whole pathways, and each transcript can be targeted by more than 1 miRNA, in a combinatorial way. In addition, miRNA families have identical seed-sequences, likely sharing most targets. The result is a complex network, often displaying a high degree of redundancy, that may be necessary to stabilize central pathways.
Long Noncoding RNAs
The classification of lncRNAs is not standardized, and their nomenclature is often related to experimental features, such as tissue specificity, mechanism of action, or molecular function. One possible classification is according to their genomic location related to protein-coding genes. For example, natural antisense lncRNAs are expressed from the opposite DNA-strand of mRNAs, whereas long intergenic noncoding RNA (lincRNAs) are localized in intergenic regions.
Although there are a few examples of lncRNAs displaying a conserved primary sequence, conservation is generally weak. However, different computational methods indicate that the secondary structure of lncRNAs is often conserved, suggesting that structural constraints may be more relevant than the nucleotide sequence in maintaining function.
Indeed, while many lncRNAs act in trans, leaving the site of transcription and executing a variety of cellular functions throughout the cell, others work in cis, affecting the expression and/or chromatin state of nearby genes. Among cis-acting lncRNAs, some regulate the expression of neighboring genes through recruitment of regulatory factors to the originating loci, thereby modulating their function. A prototype of this class of lncRNAs is represented by XIST (X-inactive specific transcript). For other lncRNAs such as Airn or Blustr, the process of transcription or splicing of the lncRNA and not the transcript itself elicits the regulation of gene expression. For yet other lncRNAs such as lincRNA-p21, the functional elements are only the DNA sequences within the promoter, regardless of the encoded RNA or its generation.
Despite these specificities, mechanisms of action and functions of an ever-increasing number of lncRNAs have started to be elucidated, notably in the cardiovascular field.7 One of the best-studied features of lncRNA is their role in the epigenetic regulation of gene expression. Some nuclear lncRNAs, such as XIST, work as scaffolds for chromatin modification enzymes, modulating positively or negatively, reversibly or irreversibly, the expression of individual genes and whole chromosomal regions. LncRNAs can bring together transcriptional repressors and activators, modulating the rates of RNA Polymerase II initiation and elongation, interacting with a variety of complexes, such as Polycomb Repressive Complex 1 or 2. Some nuclear lncRNAs have been implicated in maintaining nuclear bodies, such as nuclear speckles, built on the architectural lncRNA, NEAT1.8 Other lncRNAs regulate gene expression posttranscriptionally, modulating mRNA maturation and stability through different mechanisms. For example, some antisense lncRNAs can base-pair with sense-mRNAs, thereby altering splice-site recognition and spliceosome recruitment.9
Other lncRNAs, such as MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), do not hybridize with the target mRNA but can affect mRNA splicing through cooperation with the splicing machinery.10 LncRNAs can induce both stabilization and destabilization of target mRNA transcripts. Some lncRNAs act as a molecular scaffold, such as H19, which favors mRNA degradation mediated by the protein KSRP of myogenin.11 Others bind their complementary sense transcripts and mask miRNA binding sites, such as BACE1 antisense that protects BACE1 from miR-485-5p inhibition.12 Several lncRNAs act as competing endogenous RNAs (ceRNAs), sponging miRNAs and possibly other regulatory factors. As an example, linc-MD1 is encoded by the genomic locus containing the miR-206 and miR-133b and works as ceRNA for miR-133 and miR-135.13 LncRNAs can also affect gene expression at the posttranslational level, inducing ubiquitin-mediated proteolysis. For example, HOTAIR, in addition to its epigenetic function, can bind to E3 ubiquitin-ligases bearing RNA-binding domains, along with their respective ubiquitination substrates, facilitating their ubiquitination and degradation.14
A particular class of lncRNAs is represented by circRNAs, that are the product of back-splicing events and that can accumulate at relatively high levels, at least in part, because of their resistance to exonucleases.15 One prominent function of circRNAs is to act as ceRNA (miRNA sponges), but they can also modulate RNA transcription, splicing, turnover, translation, and may even have protein coding potential.15 Regardless of the specific ceRNA nature, miRNA/ceRNA stoichiometry is essential for effective miRNA sequestration. Indeed, although virtually all ncRNAs display at least 1 putative miRNA binding-site, only a few are expressed at levels sufficiently high to operate as ceRNAs.
Overall, regRNAs constitute a diverse class of RNA molecules that significantly affect gene expression and are of paramount importance for the maintenance of cardiac homeostasis. Multiple regRNAs are dysregulated in the failing heart, and many of them functionally contribute to the development and progression of heart failure.
Epitranscriptome
RNAs are subjected to diverse types of chemical modifications, which affect their fate and impact cellular responses, a field called epitranscriptomics. Although chemical modifications on mRNA are known for decades, studies show that ncRNAs also undergo different modifications (Figure 3). However, investigations on regRNA modifications and their effects are restricted because of technical limitations. It has been suggested that the epitranscriptome in regRNAs affects their regulatory activity, stability, protection from degradation, and recruitment of proteins to specific sites of the regRNA.16 The epitranscriptome provides another layer of complexity to the transcriptome.
Figure 3.
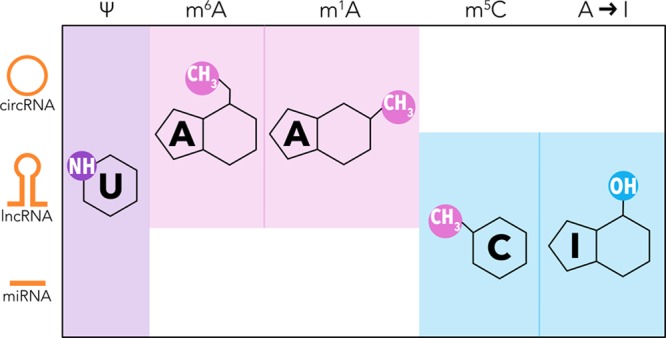
Most frequent posttranscriptional modifications in regRNAs. Simplified chemical structure of modified RNA nitrogenous bases: purple, pseudouridine [y], reported in the three ncRNAs; pink, N6-methyladenosine [m6A] and N1-methyladenosine [m1A], observed in circRNA and lncRNA; blue, 5-methylcytosine [m5C] and adenosine to inosine editing [A to I], occuring in lncRNA and miRNA. circRNA indicates circular RNA; lncRNA, long non-coding RNA; miRNA, microRNA; and regRNA, regulatory RNA.
Regulatory RNAs In the Circulation
Although RNA molecules are usually considered unstable, particularly in the circulation because of the abundance of exonucleases, different types of RNA can be found intact. Both coding and ncRNAs are protected from degradation through different means, such as packed in lipid vesicles (eg, exosomes and microvesicles), bound to high-density lipoproteins or proteins, especially Argonaute 1 and 2 (which is the case of miRNAs), and by their circular structure. CircRNAs are significantly more stable than linear RNAs because of their lack of free ends, poly(A) tails, and 5′ caps. The stability of RNAs in the circulation reinforces their intercellular communication and regulatory roles. Likewise, their presence in extracellular vesicles further supports such a biological function. There is increasing evidence that cells package and export different molecules, including regRNAs, in extracellular vesicles. These are taken up by recipient cells, where their cargo can induce expressional and functional changes.
The spectrum of regRNAs in the circulation is highly dynamic, according to the health status of an individual. Numerous regRNAs have been described to have distinct profiles associated with specific pathophysiological states. There are many reports of miRNAs, lncRNAs, and circRNAs for which altered levels correlate with different cardiovascular diseases such as heart failure. Circulating regRNA profiles have also been reported to change in response to environmental and lifestyle factors, such as exercise, diet, and circadian rhythm.17
RegRNAs, particularly miRNAs, have characteristics that make them prominent clinical biomarkers: stability, ease of access, the possibility to be measured in a routine lab, and specificity to pathophysiological states. Indeed, different miRNAs or profiles of miRNAs have been described as potential biomarkers for virtually all cardiovascular diseases, as well as lncRNAs and circRNAs, although less extensively. Circulating regRNAs could serve not only as prognostic and diagnostic biomarkers for disease but also for treatment selection and monitoring, and as therapeutic targets.18
Regulatory RNAs In Cardiac Hypertrophy
MiRNAs
Cardiac hypertrophy is a hallmark of heart failure. Numerous miRNA-mediated mechanisms underlying cardiac hypertrophy have been identified (Table 1).
Table 1.
Regulatory RNAs and RNA Binding Proteins Associated With Cardiac Hypertrophy
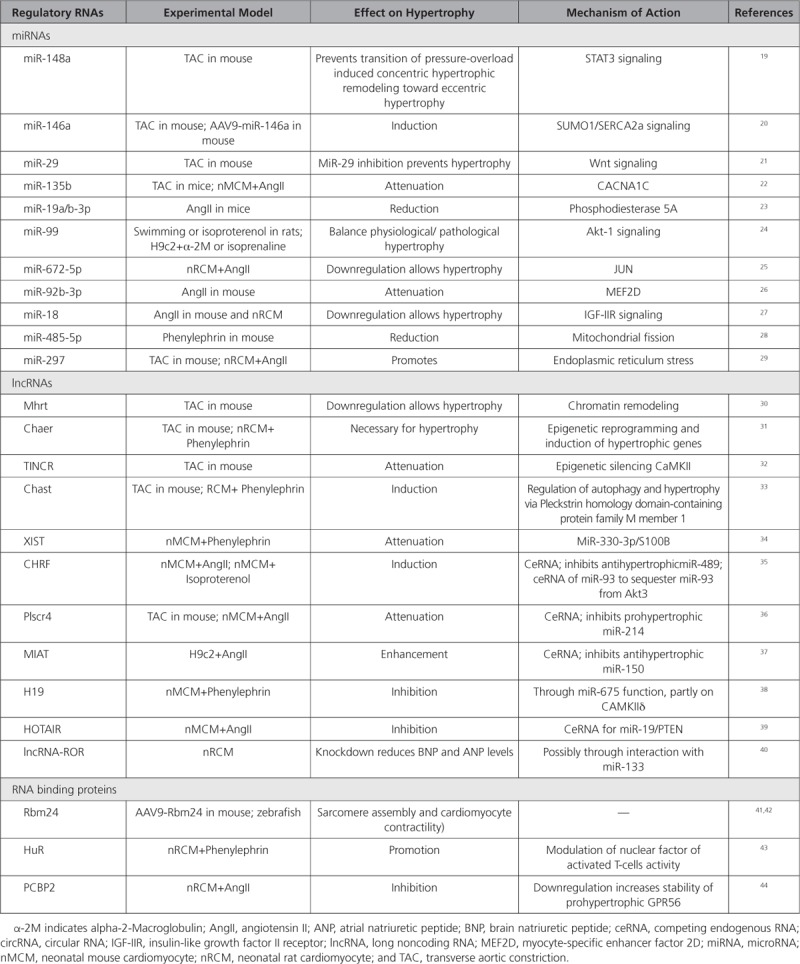
MiRNA-148a was shown to control the transition from concentric hypertrophy toward dilation upon pressure overload in mice.19 Adeno-associated viral delivery of miR-148a protected the mouse heart from pressure overload-induced systolic dysfunction, whereas antagomir-mediated silencing of miR-148a caused cardiac dilation and failure. MiR-146a induced cardiac dysfunction during maladaptive hypertrophy through the suppression of cardiac SUMO1 expression.20 Overexpression of miR-146a using AAV9 (adeno associated virus 9) vectors reduced SUMO1 expression, SERCA2a SUMOylation, and cardiac contractility both in vitro and in vivo. It is interesting that transdifferentiation of fibroblasts triggers miR-146a upregulation and secretion through extracellular vesicles. This transfer was identified as the causative mechanism of miR-146a upregulation in failing cardiomyocytes. MiR-29 promotes pathologic hypertrophy of cardiac myocytes and overall cardiac dysfunction.21 In a mouse model of cardiac pressure overload, both global genetic deletion of miR-29 and antimiR-29 infusion prevented cardiac hypertrophy and fibrosis and improved cardiac function. Targeted deletion of miR-29 in cardiomyocytes in vivo also prevented cardiac hypertrophy and fibrosis, implying that the role of miR-29 in cardiomyocytes dominates over that in nonmyocyte cell types. The effects of miR-29 on cardiomyocyte hypertrophy were at least partly mediated by several targets involved in Wnt signaling.
Overexpression of miR-135b may attenuate pathological cardiac hypertrophy by targeting CACNA1C.22 MiR-19a/b-3p was found to be antihypertrophic in a model of angiotensin II-induced hypertrophy, an effect involving the phosphodiesterase PDE5A, a leading factor contributing to cGMP signaling and cardiac hypertrophy.23 The miR-99 family fine-tunes Egr-1/Akt1 signaling and thereby balance cardiomyocyte physiological and pathological hypertrophic responses.24 MiR-672-5p, which is downregulated in hypertrophic cardiomyocytes, reduces cardiomyocyte hypertrophy in vitro by inhibiting JUN expression.25 Furthermore, cardiac hypertrophy was shown to be attenuated in angiotensin II-infused mice that received tail vein injection of miR-92b-3p mimic.26 Myocyte-specific enhancer factor 2D was identified as a potential target gene mediating this effect. Cardiac-specific overexpression of miR-18 via AAV2 protected against cardiac dilatation during hypertension-induced heart failure.27 Here, the p53-miR-18-HSF2-IGF-IIR axis was claimed to be involved. In a mouse model of cardiac hypertrophy induced by phenylephrine, the administration of miR-485-5p agomir significantly decreased phenylephrine-induced SUMO E3 ligase, mitochondrial anchored protein ligase, and hypertrophic markers atrial natriuretic peptide and β-myosin heavy chain, and protected against cardiac dysfunction.28 MiR-297 promotes cardiomyocyte hypertrophy by inhibiting the expression of Sigma-1 receptor, a ligand-regulated endoplasmic reticulum chaperone involved in cardiac hypertrophy, and activation of endoplasmic reticulum stress signaling.29
MicroRNA-Associated Feed-Forward Loops Associated With Cardiomyocyte Hypertrophy
MicroRNA-associated feed-forward loops (miR-FFL) are 3-gene modules. Mining published expression data led to the identification of 22 miR-FFL motifs dysregulated in cardiac hypertrophy.45 These modules included 17 miRNAs, including miR-20a and -b, miR-21, let-7b, miR-16, miR-9, miR-106a, miR-15, miR-93, miR-125b, miR-34b, miR-1, miR-30a, miR-130b, miR-92b, miR-335, and miR-425. Most of these miRNAs are known to be associated with cardiomyocyte hypertrophy and cardiac dysfunction.
Long Noncoding RNAs
Long Noncoding RNAs Functioning Through (Epi)Genetic Mechanism
Whereas more than a decade of research into the roles of miRNAs in cardiomyocyte hypertrophy has resulted in a vast amount of data, the implications of lncRNAs in cardiomyocyte hypertrophy are still limited (Table 1).
The first study implicating a lncRNA in cardiac hypertrophy identified a new antisense transcript of myosin heavy chain 7 formed by alternative splicing, called MHRT (Myosin Heavy Chain Associated RNA Transcript).30 MHRT is highly expressed in the mouse and human adult heart and functions through the modification of Brg1 (brahma-related gene 1)-mediated chromatin remodeling. The MHRT-Brg1 feedback circuit appears crucial for cardiac function and is conserved in humans. More recently, it was suggested that MHRT acts together with myocardin in a regulatory feedback mechanism in the regulation of cardiac hypertrophy.46 MHRT was found to affect the acetylation of myocardin by HDAC5 and proposed to inhibit thereby cardiac hypertrophy induced by myocardin. Moreover, myocardin also directly activated MHRT transcription through binding to the CarG box.
Another identified lncRNA with a function in epigenetic reprogramming of hypertrophic cardiomyocytes is Chaer.31 Chaer modulates the chromatin remodeling function of polycomb repressor complex 2 through direct interaction with its catalytic subunit, and in this way affects histone methylation and the expression of cardiac hypertrophic genes. TINCR(Terminal differentiation-induced ncRNA) has been linked to the epigenetic silencing of CaMKII via the direct targeting of the methyltransferase enhancer of zeste 2 that associates with the CaMKIIδ promotor.32 Enforced expression of TINCR attenuated cardiac hypertrophy in pressure-overloaded mice. Both Chaer and TINCR are annotated in human, but whether the epigenetic mechanisms described in mice are conserved in humans remains to be demonstrated.
The lncRNA Chast(cardiac hypertrophy-associated transcript), on the other hand, functions through a cis-regulatory action directly on the gene located on the opposite strand: pleckstrin homology domain-containing protein family M member 1, an autophagy regulator. Through a yet unknown mechanism, Chast induced cardiomyocyte hypertrophy in vitro and in vivo, whereas silencing of Chast attenuated cardiac remodeling after pressure overload.33
An important subclass of lncRNAs with epigenetic functions is that of the enhancer lncRNAs, derived from the enhancer regions of the genome that among other processes regulate gene expression reprogramming during cardiomyocyte hypertrophy.47
Long Noncoding Rnas With Proposed Scaffold-Based Mechanisms
Searching for competitive endogenous RNA interactions between lncRNAs and miRNAs indirectly affecting mRNAs has become an exciting area of research attesting the complexity of the cross-talks between different regRNA species.
The lncRNA XIST was upregulated in hypertrophic murine hearts and phenylephrine-treated cardiomyocytes.34 Inhibition of XIST induced cardiomyocyte hypertrophy while overexpression of XIST attenuated cardiomyocyte hypertrophy induced by phenylephrine in vitro. These effects may involve a sponging effect of XIST for miR-330-3p, leading to enhanced S100B expression and cardiomyocyte hypertrophy.
The lncRNA CHRF(cardiac hypertrophy related factor) is also upregulated in cardiomyocyte hypertrophy where it scavenges miR-489, thereby indirectly regulating the expression levels of the myeloid differentiation primary response gene 88.35 The lncRNA Plscr4 may down-regulate cardiac hypertrophy through the miR-214-Mfn2 axis.36 Also, the MIAT(myocardial infarction associated transcript) was found to contribute to cardiomyocyte hypertrophy in vitro through a miR-150-5p/P300 axis.37 The lncRNA H19 was shown to inhibit cardiomyocyte hypertrophy in vitro, and these effects were completely dependent on its encoded miR-675 and may partly involve the downstream target CAMKIIδ.38 LncRNA HOTAIR may function as a miR-19-sponge to modulate PTEN levels.39 Human large intergenic noncoding RNA ROR (lncRNA-ROR) was shown to reduce ANP and BNP protein levels in cardiomyocytes in vitro and may do so via its interaction with miR-133.40
RNA Binding Proteins
Dedicated RNA binding proteins (RBPs) are pivotal to the functioning of regRNAs. It is important to note that knowledge emerges on an intricate RBP-dependent interplay between miRNA-based gene silencing of mRNAs and alternative splicing of pre-mRNAs, a crucial posttranscriptional mechanism implicated in the cellular stress response. For example, the RBP family Rbfox regulates both tissue-specific pre-mRNA splicing and miRNA processing, and both Rbfox1 and -2 were recently linked to heart failure, the latter through regulation of miR-34a.48 Moreover, the recognition of the importance of tightly controlled alternative pre-mRNA splicing in cardiac homeostasis is emerging. RNA binding proteins play a major role in RNA splicing. The first identified RBP with a role in alternative splicing during heart failure was alternative splicing factor 1 (ASF/SF2), regulating the remodeling-induced splicing switch of CaMKIIδ.49 The RNA binding protein RBM24 is a pivotal cardiac splice factor, which governs sarcomerogenesis in the heart by controlling the expression of alternative protein isoforms.41 Although a role in hypertrophy is unclear, AAV9-mediated cardiomyocyte Rbm24 overexpression in mice was recently shown to slightly increase heart weights, but this may have been a result of increased fibrosis in these mice.42 RBM38, a homolog of RBM24, has been implicated in RNA splicing, RNA stability, and RNA translation. Yet, Rbm38 deficiency did not affect cardiac hypertrophy in vivo and was found to be dispensable during pressure overload-induced cardiac remodeling in mice.50
The RNA binding protein HuR (human antigen R) interacts with specific AU-rich domains in target mRNAs and is highly expressed in many cell types, including cardiomyocytes. Upon hypertrophic stimulation, HuR undergoes cytoplasmic translocation, indicative of its activation, in cultured cardiac myocytes.43 HuR activation was necessary for Gq-mediated hypertrophic growth of neonatal rat ventricular myocytes and may function through the activation of the transcription factor nuclear factor of activated T cells.
The RNA-binding protein PCBP2 (Poly(C)-binding protein 2) is downregulated in failing human hearts and mouse hypertrophied hearts.44 PCBP2 knockdown promoted angiotensin II-induced hypertrophy of neonatal cardiomyocytes and H9C2 cells, whereas PCBP2 overexpression promoted opposite effects. Furthermore, PCBP2 inhibited prohypertrophic GPR56 expression by promoting its mRNA degeneration in cardiomyocytes.
Regulatory RNAs In Cardiac Inflammation
Cardiac and systemic inflammation is associated with chronic heart failure as well as being a contributing factor to metabolic syndrome, type 2 diabetes, atherosclerosis, aortic aneurysm, and aging. Several regRNAs are modulated and participate in the regulation of cardiac inflammation (Table 2).
Table 2.
Regulatory RNAs Associated With Cardiac Inflammation
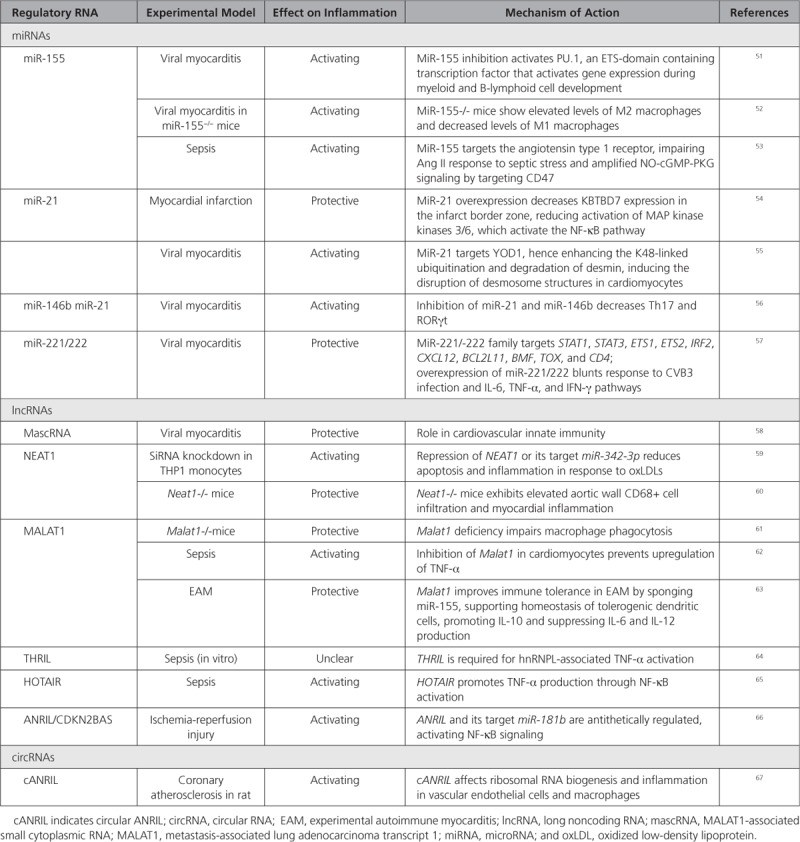
MiRNAs
Translational miRNA expression studies sustain causal links between miRNA dysregulation and cardiac inflammation and have identified potential miRNA therapeutic targets. In human myocarditis samples, a microarray screen identified 107 miRNAs differentially expressed as compared with control hearts, of which 21 were upregulated and 37 were down-regulated.51 MiR-155, miR-146b, and miR-21 were upregulated, and inhibition of miR-155,52 miR-21, and miR-146b,56 reduced myocardial inflammation and necrosis in coxsackievirus-B3 or autoimmune myocarditis in mice. Likewise, miR-155 inhibits PU.1 and SOCS1 in the heart and per se derepresses the proinflammatory cytokines, facilitating activation of T-cells and monocytes.68 In addition, miR-155 amplifies nitric oxide/cGMP signaling in the heart and increases cardiac inflammation upon a septic shock in mice and human hearts.53 Combined inhibition of miR-21 and miR-146a reduced cardiac dysfunction after myocardial infarction in a model of viral myocarditis and decreased the expression levels of T-helper cell 17 and RAR-related orphan receptor gamma RORγt in mice.56
MicroRNA-21 is particularly increased in the border and infarct zones during the early inflammatory phase of remodeling following myocardial infarction in mice and prevents excessive inflammation and cardiac dysfunction.54 Indeed, miR-21 knockout mice have elevated baseline levels of inflammatory cytokines, CD11b+ monocytes and macrophages in the heart and, after myocardial infarction, they exhibit higher mortality rates, worse cardiac dysfunction, and increased infarct and scar areas as compared with wild-type mice.54
Systemic inhibition of the miR-221/-222 cluster in mice infected by coxsackievirus-B3 increased the cardiac viral load and aggravated cardiac inflammation and injury.57 Regulation of inflammation by miR-221/-222 involves the target genes ETS1/2, IRF2, BCL2L11, TOX, BMF, and CXCL12. It is interesting to note that the miR-221/-222 cluster regulates both inflammation and cardiac virulence.
Long Noncoding RNAs
Besides miRNAs, lncRNAs are also implicated in immune modulation in the heart, and some regulatory networks involving miRNAs and lncRNAs have been linked to cardiac inflammation.
A translational study exploring the involvement of lncRNAs in antiviral capacity in patients with coxsackievirus-B3 cardiomyopathy conferred beneficial immunoregulatory functions to the lncRNA MALAT1 and its enzymatic processing product MALAT1-associated small cytoplasmic RNA.58 MALAT1-associated small cytoplasmic RNA overexpression in cardiomyocytes induced antiviral immunity genes and rendered cells resistant to virus infection. MALAT1-associated small cytoplasmic RNA ablation in monocytes significantly increased the expression of proinflammatory proteins, such as FASLG, FAS, TNF-α, and IL6. In autoimmune myocarditis in mice, MALAT1 induces tolerogenic dendritic cells and regulatory T cells, via miR-155/dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin/IL10 axis.63 MALAT1 also inhibits immune activation and atherosclerosis development in ApoE-deficient mice.61
The lncRNA HOTAIR is intergenic and antisense to the HOXC gene. Inhibiting HOTAIR improved cardiac function during lipopolysaccharide-induced sepsis in mice, presumably via inhibition of TNF-α and NFκB/p65 phosphorylation.65 The lncRNA ANRIL/INK4/CDKN2B-AS1A is encoded by a risk haplotype at the locus 9p21.3 identified in genome-wide association studies as conveying risk for coronary artery disease and myocardial infarction as well as other inflammatory conditions. ANRIL has a circRNA counterpart (cANRIL) thought to modulate proinflammatory gene expression in vascular endothelial cells (ECs), inciting coronary atherosclerosis.69
Other lncRNAs, which may benefit from further investigation in cardiac inflammatory disease, include CARL (cardiac and apoptosis-related lncRNA), and THRIL (TNFα and hnRNPL related immunoregulatory lincRNA). The first is involved in macrophage activation and NF-kB signaling and inhibits mitochondrial fission and apoptosis in cardiomyocytes.70 The second regulates expression of key immune response genes, including TNF-α.64
Circular RNAs have also been associated with inflammation. Expression levels of the circRNA MICRA in inflammatory blood cells of patients after acute myocardial infarction are associated with the development of heart failure.71 cANRIL modulates the maturation of ribosomal RNAs and confers protection from atherogenesis, providing evidence that circularization of lncRNAs (ANRIL in this particular case), can affect RNA processing and disease development.67
Regulatory RNAs In Cardiac Regeneration
Regeneration pathways are highly preserved between species. However, because of early postnatal cell cycle exit of cardiomyocytes and failure to activate traditional regeneration programs upon injury, the adult mammalian heart exhibits limited regenerative capacities. Common mechanisms of successful cardiac regeneration seen in lower vertebrates and neonatal mice include controlled inflammation and matrix remodeling centering around cardiomyocyte dedifferentiation and proliferation. In contrast, new cardiomyocyte formation resulting from endogenous differentiation of progenitor cells after injury has lately been put into question. Therefore, strategies to induce cardiomyocyte proliferation and to facilitate cardiomyogenic differentiation of stem/progenitor cells for therapeutic application or to directly reprogram nonmyocytes into cardiomyocytes are currently under intense investigation. All these processes require sophisticated gene regulation involving regRNAs (Table 3).
Table 3.
Regulatory RNAs Associated With Cardiac Regeneration
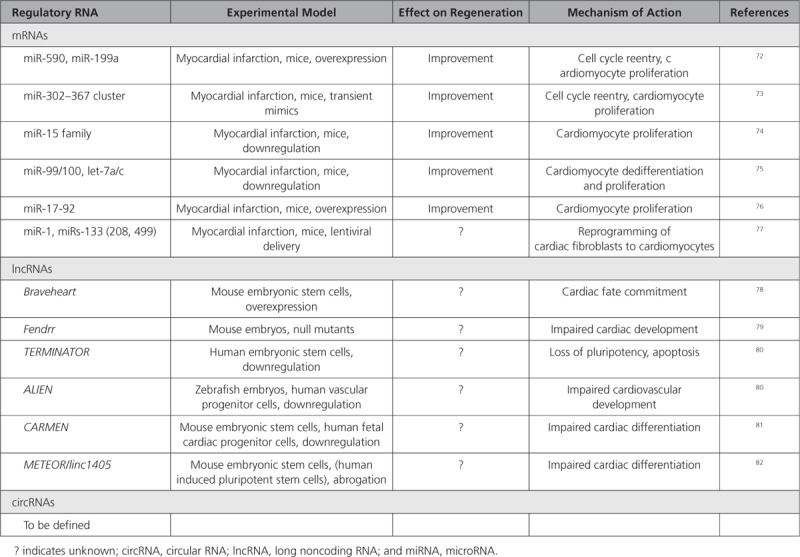
MiRNAs
Screening for miRNAs that regulate postnatal cardiomyocyte proliferation revealed that miR-590 and miR-199a induce cell cycle reentry and proliferation of adult cardiomyocytes, thereby improving outcome after myocardial infarction.72 Similarly, miR-302–367 cluster promotes cardiomyocyte proliferation, and its transient activation was associated with reduced scarring and improved function after myocardial infarction in mice.73 In contrast, the miR-15 family induces postnatal cell cycle exit, and its inhibition from the early postnatal stage onwards allows for retained regeneration of the adult mouse heart.74 Dedifferentiation and proliferation of cardiomyocytes with consecutive regeneration after myocardial infarction could also be induced in adult mice upon miR-99/100 and let-7a/c inhibition.75 These data support the therapeutic targeting of miRNAs to activate preserved regeneration programs in the adult mammalian heart.
RegRNAs play essential roles during cardiac development, which could open new avenues for cell therapy and reprogramming. The muscle-specific myomiR families miR-1 and miR-133 are highly preserved between species and their tight regulation is required for balancing proliferation and differentiation during cardiogenesis, supporting a role for miRNAs in gene dosage during development. Both miR-1 and miR-133 have been used for reprogramming of cardiac fibroblasts to cardiomyocytes.77 As another example of miRNAs exerting multiple functions, the miR-17/92 cluster induces cardiomyocyte proliferation in the adult mouse.76
Long Noncoding RNAs
Of interest to cardiac regeneration are also lncRNAs. Among the first identified was Braveheart, which regulates embryonic cardiovascular progenitor cell differentiation in the mouse via epigenetic modulation of cardiac transcription factor expression.78 Similar functions were shown for Fendrr, which is indispensable for normal cardiogenesis in mice.79 LncRNAs are less preserved than miRNAs, and whereas a human homolog for Braveheart is missing, TERMINATOR, ALIEN,80 CARMEN,81 and METEOR/linc140582 have been implicated in human stem and progenitor cell differentiation. LncRNAs also regulate cardiomyocyte proliferation. Cardiomyocyte dedifferentiation and cell cycle re-entry occur in adult mouse and human cardiomyocytes as part of a myocardial stress response guided by lncRNAs.83
Although circRNAs are dynamically regulated during human cardiac development, their identities, regulation, and roles have still to be revealed.
Recent initiatives that synthesize publicly available gene expression data in cardiac regeneration provide valuable resources for further research in the field (eg, https://regendbase.org).
Regulatory RNAs In the Vascular System
Changes in both the coronary and peripheral vasculature are key to the development of heart failure. Coronary vessel remodeling contributes to impaired myocardial contractility, while peripheral vascular remodeling results in decreased compliance, increased resistance and decreased capacitance in the arterial circulation, thus increasing the afterload and exacerbating the heart failure. Dysfunction of ECs and consequent alterations in vascular smooth muscle cells are central to this altered vascular structure and function.
Prevention or reversal of this remodeling may help to restore homeostasis and protect against heart failure, as would harnessing the mechanisms of angiogenesis to promote neovascularization. The role of regRNAs in these phenotypic changes in vascular cells is increasingly recognized (Table 4).
Table 4.
Regulatory RNAs in the Vascular System
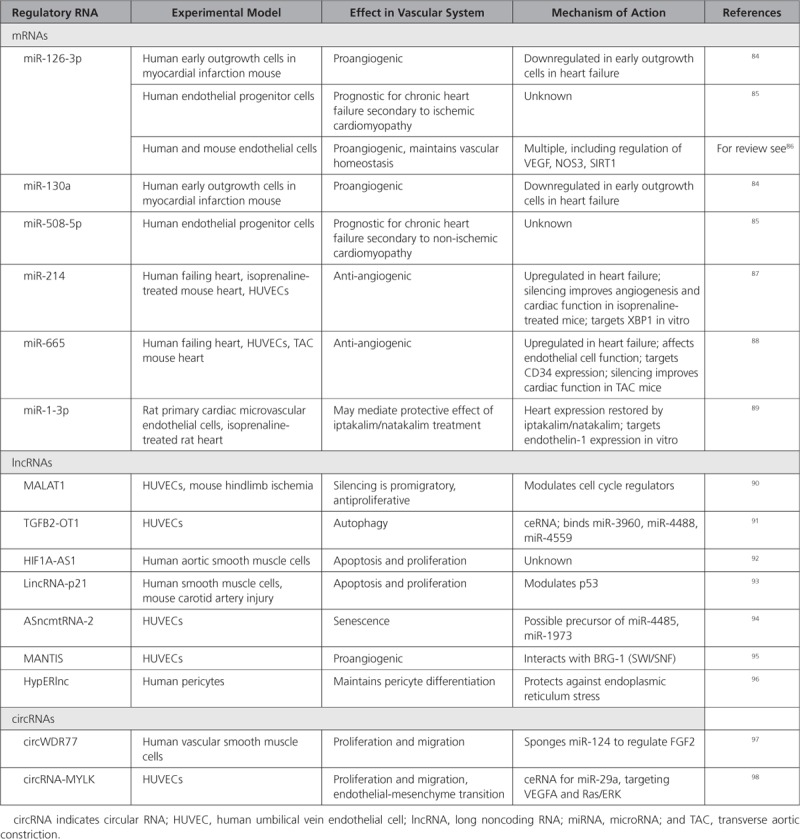
Messenger RNAs
A global change in the localization of mRNA molecules in ECs has been shown to regulate blood vessel sprouting and EC migration during angiogenesis, and mRNA transcript allocation controls tissue morphogenesis.99
MiRNAs
Angiogenic early outgrowth cells from chronic heart failure patients show reduced miR-126 and miR-130a, which impairs their ability to improve cardiac function,84 while levels of miR-126 and miR-508-5p in endothelial progenitor cells are prognostic for chronic heart failure in ischemic and nonischemic cardiomyopathy patients respectively.85 Mir-21487 and miR-66588 are both increased in failing human hearts and in the circulation. Inhibition of each of these miRNAs improved cardiac function and angiogenesis in mouse models of hypertrophy.87,88 Recently, it was postulated that the KATP channel openers iptakalim and natakalim may exert protective effects on the endothelium via a miR-1-3p/ET-1 pathway, although the evidence at this stage is mostly in vitro.89
More broadly, numerous miRNAs (eg, miR-126, miR-17-92a cluster, miR-143/145, miR-210, miR-24, miR-15a/16, and miR-221/-222) have been shown to have important roles in angiogenesis and are thus potential therapeutic targets and biomarkers of heart failure.
Long Noncoding RNAs
Long noncoding RNAs have also come into view as regulators of numerous pathways involved in the pathophysiology of the vascular system associated with heart failure. In vitro silencing of MALAT1 switched ECs from a proliferative to a promigratory phenotype, and its inhibition in vivo reduced vascular growth.90 HIF 1 alpha-antisense RNA 192 and lincRNA-p2193 are implicated in vascular smooth muscle cell regulation while TGFB2-OT191 or mitochondrial ASncmtRNA-294 play roles in inflammation or senescence of vascular ECs, respectively. MANTIS controls transcription factor expression in ECs via interaction with BRG-1, part of the SWI/SNF complex, to promote angiogenic functions.95 HypERlnc is expressed in pericytes and is downregulated in hearts of patients with heart failure.96 Silencing of HypERlnc in pericytes in vitro resulted in dedifferentiation, attributed to increased endoplasmic reticulum stress.
Circular RNAs are expressed and regulated in the vascular system, although their role in heart failure development is still poorly known. CircWDR77 targets FGF-2 to modulate vascular smooth muscle cell proliferation and migration by sponging miR-124.97 In another study, circRNA-MYLK acted as a ceRNA for miR-29a, contributing to endothelial-mesenchymal transition by activating VEGFA/VEGFR2 pathway.98 It is also worth mentioning that endothelial lncRNAs and circRNAs are regulated by hypoxia and may contribute to regulating fibrosis.
Translational Relevance of Regulatory RNAs
The discovery of novel regRNAs implicated in heart failure has progressed extremely rapidly over the past few years, yet has not come to an end. The question is how to translate research findings into biologically and clinically relevant information.
Given their relative cell-type specific expression and tight temporal and spatial regulation, regRNAs make attractive therapeutic targets and biomarkers for heart failure. The potential clinical applications of regRNAs are diverse, particularly as tools for personalized medicine. The identification of regRNA profiles in heart failure may lead to new preventive measures, diagnostics, and therapies. It may also have an added value to existing tools and biomarkers, giving molecular snapshots of specific moments throughout the evolution of a disease, and, thus, providing additional information to guide clinical decisions. Understanding the interactions between the different types of RNA and how they affect gene expression will help fill critical gaps in our knowledge, enabling such translational applications.
Although there is compelling evidence for the potential of regRNAs in clinical applications, our mechanistic insights into the molecular pathophysiology underlying heart failure have not yet emerged into new therapeutic or diagnostic options based on regRNAs. This is partly because of technical and nontechnical hindrances. For example, comparing and reproducing results from different studies is challenging. Translational transcriptomics studies in the cardiovascular field, as well as other fields, must rely on a consensus of best practices and experimental standards for all steps of the process, such as study design, sample type and size, methods of normalization and analysis, specificity of the detection and targeting of regRNAs.100 Extensive progresses in these technical aspects are paramount for the use of RNA-based diagnostic and therapeutic approaches.
Conclusion and Future Directions
It has become increasingly evident that RNAs possess a plethora of regulatory roles, some of which remain to be unraveled. A better knowledge of the role of regRNAs may ultimately foster the development of novel RNA-based therapeutic approaches (Figure 4).
Figure 4.

Future directions. circRNA indicates circular RNA; lncRNA, long non-coding RNA; miRNA, microRNA; and regRNA, regulatory RNA.
While the role of miRNAs in heart failure development and progression has been extensively studied, the knowledge on lncRNAs is far scarcer. Although deep RNA-sequencing experiments revealed an extensive regulation of lncRNAs in the failing heart, only a few lncRNAs have been functionally involved in heart failure processes. Even less well known, circRNAs also appear to be regulated during heart failure, while their role in the heart has still to be fully uncovered.
A few reports highlight the importance of RNA binding proteins that are necessary to maintain cardiac homeostasis through tight control of pre-mRNA splicing. Dysregulation of these RNA binding proteins has been observed in the failing heart, and further research is needed to systematically characterize their relevance in heart failure and evaluate their therapeutic and diagnostic potential. Since these proteins act at a very early stage of gene expression (pre-mRNA splicing), it is plausible that the study of their regulation may provide early indicators of subtle changes in gene regulatory networks that contribute to heart failure development.
Secondary analyzes of publicly-available high throughput datasets (eg, RNA-sequencing) with constantly improved bioinformatics tools have the potential to discover interesting novel targets through data sharing and collaborative work. Any new target requires extensive characterization and validation using independent methods before being considered for further development as a drug or a biomarker.
Now that also more direct mRNA targets of miRNAs become validated, pathway analysis possibilities based on systems biology concepts emerge. As such, the identification of miR-FFLs represents a new era in disease research and may help shed light on the diversity of miRNA-mediated pathways identified so far.
The development of bioinformatics has greatly contributed to a better knowledge of the role of regRNAs in heart failure. Intriguingly, most of the lncRNA studies described scaffold-based mechanisms (“sponge effect”), in which they prevent miRNAs from exerting their roles on mRNAs. It is unclear whether this tripartite mechanism is indeed widely effective in cardiac cells. Deciphering the functions of lncRNAs is not trivial. The identification of putative miRNA binding sites is relatively easy using current in silico tools, as it is based on sequence complementarity. Identifying interactions of lncRNAs with proteins or DNA, on the other hand, requires more complicated and time-consuming biochemical assays, especially when performed in an unbiased manner. Thus, more efforts are required to deepen our understanding of the role of regRNA networks in heart failure.
Sources of Funding
This publication is based upon work from the EU-CardioRNA COST ACTION CA17129, supported by COST (European Cooperation in Science and Technology). The open access fee of this publication was supported by EU-CardioRNA COST ACTION CA17129. Y.D. is funded by the Ministry of Higher Education and Research, the Society for Research on Cardiovascular Disease, and the National Research Fund of Luxembourg (grant C17/BM/11613033). C.P.C.G. is funded by the Eurostars project MIPROG (grant EUROSTARS E! 9686). G.M.K. received funding from the Swiss National Science Foundation (310030_156953). S.H. received funding from the European Union Commission’s Seventh Framework programme under grant agreement N° 305507 (HOMAGE), N° 602904 (FIBROTARGETS) and N° 261409 (MEDIA) and N° 278249 (EU MASCARA); the Marie-Curie Industry Academy Pathways and Partnerships (CARDIOMIR) N°285991, FP7-Health-2013-Innovations-1 N° 602156 (HECATOS); the Netherlands Organization for Scientific Research (NWO) Vidi 91796338; the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation, CVON2016-Early HFPEF, 2015-10, and CVON She-PREDICTS, grant 2017–21, CVON-Arena-PRIME. B.S. is supported by grants from the Netherlands Heart Foundation (Dekker 2014T105 and CVON-SHE-PREDICTS-HF), Netherlands Organisation for Scientific Research (NWO vidi 917.14.363) and Health Foundation Limburg. F.M. is supported by Ministero della Salute (Ricerca Corrente, 5X1000). C.E. received funding from a British Heart Foundation programme grant “microRNAs from cardiac surgery to basic science–and back?” (RG/15/5/31446) and a British Heart Foundation Chair in Cardiovascular Science (CH/15/1/31199). E.L.R. received funding from CVON RECONNECT Talent programme grant, a Netherlands CardioVascular Research Initiative supported by the Hartstichting (Dutch Heart Foundation).
Disclosures
None.
Footnotes
References
- 1.Bleumink GS, Knetsch AM, Sturkenboom MC, Straus SM, Hofman A, Deckers JW, Witteman JC, Stricker BH. Quantifying the heart failure epidemic: prevalence, incidence rate, lifetime risk and prognosis of heart failure. The Rotterdam Study. Eur Heart J. 2004;25:1614–1619. doi: 10.1016/j.ehj.2004.06.038. doi: 10.1016/j.ehj.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 2.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr, Drazner MH, Fonarow GC, Geraci SA, Horwich T, Januzzi JL, et al. American College of Cardiology Foundation; American Heart Association Task Force on Practice Guidelines. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62:e147–e239. doi: 10.1016/j.jacc.2013.05.019. doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Jr, Colvin MM, Drazner MH, Filippatos GS, Fonarow GC, Givertz MM, et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. Circulation. 2017;136:e137–e161. doi: 10.1161/CIR.0000000000000509. doi: 10.1161/CIR.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 4.Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ, Falk V, González-Juanatey JR, Harjola VP, Jankowska EA, et al. Task Force Members; Document Reviewers. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2016;18:891–975. doi: 10.1002/ejhf.592. doi: 10.1002/ejhf.592. [DOI] [PubMed] [Google Scholar]
- 5.van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, et al. The translational landscape of the human heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiedler J, Baker AH, Dimmeler S, Heymans S, Mayr M, Thum T. Noncoding RNAs in vascular disease—from basic science to clinical applications: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res. 2018;114:1281–1286. doi: 10.1093/cvr/cvy121. doi: 10.1093/cvr/cvy121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakagawa S, Yamazaki T, Hirose T. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol. 2018;8:180150. doi: 10.1098/rsob.180150. doi: 10.1098/rsob.180150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, Puppo M, Briata P, Gherzi R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A. 2014;111:E5023–E5028. doi: 10.1073/pnas.1415098111. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. Scaffold function of long noncoding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, Dimmeler S, Squire I Cardiolinc network. Circular RNAs in heart failure. Eur J Heart Fail. 2017;19:701–709. doi: 10.1002/ejhf.801. doi: 10.1002/ejhf.801. [DOI] [PubMed] [Google Scholar]
- 16.Gatsiou A, Stellos K. Dawn of Epitranscriptomic Medicine. Circ Genom Precis Med. 2018;11:e001927. doi: 10.1161/CIRCGEN.118.001927. doi: 10.1161/CIRCGEN.118.001927. [DOI] [PubMed] [Google Scholar]
- 17.Gomes CPC, de Gonzalo-Calvo D, Toro R, Fernandes T, Theisen D, Wang DZ, Devaux Y Cardiolinc™ network. Noncoding RNAs and exercise: pathophysiological role and clinical application in the cardiovascular system. Clin Sci (Lond) 2018;132:925–942. doi: 10.1042/CS20171463. doi: 10.1042/CS20171463. [DOI] [PubMed] [Google Scholar]
- 18.de Gonzalo-Calvo D, Vea A, Bär C, Fiedler J, Couch LS, Brotons C, Llorente-Cortes V, Thum T. Circulating noncoding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur Heart J. 2019;40:1643–1650. doi: 10.1093/eurheartj/ehy234. doi: 10.1093/eurheartj/ehy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raso A, Dirkx E, Philippen LE, Fernandez-Celis A, De Majo F, Sampaio-Pinto V, Sansonetti M, Juni R, El Azzouzi H, Calore M, et al. Therapeutic delivery of miR-148a suppresses ventricular dilation in heart failure. Mol Ther. 2019;27:584–599. doi: 10.1016/j.ymthe.2018.11.011. doi: 10.1016/j.ymthe.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh JG, Watanabe S, Lee A, Gorski PA, Lee P, Jeong D, Liang L, Liang Y, Baccarini A, Sahoo S, et al. miR-146a suppresses SUMO1 expression and induces cardiac dysfunction in maladaptive hypertrophy. Circ Res. 2018;123:673–685. doi: 10.1161/CIRCRESAHA.118.312751. doi: 10.1161/CIRCRESAHA.118.312751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sassi Y, Avramopoulos P, Ramanujam D, Grüter L, Werfel S, Giosele S, Brunner AD, Esfandyari D, Papadopoulou AS, De Strooper B, et al. Cardiac myocyte miR-29 promotes pathological remodeling of the heart by activating Wnt signaling. Nat Commun. 2017;8:1614. doi: 10.1038/s41467-017-01737-4. doi: 10.1038/s41467-017-01737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu Q, Li A, Chen X, Qin Y, Sun X, Li Y, Yue E, Wang C, Ding X, Yan Y, et al. Overexpression of miR-135b attenuates pathological cardiac hypertrophy by targeting CACNA1C. Int J Cardiol. 2018;269:235–241. doi: 10.1016/j.ijcard.2018.07.016. doi: 10.1016/j.ijcard.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Liu K, Hao Q, Wei J, Li GH, Wu Y, Zhao YF. MicroRNA-19a/b-3p protect the heart from hypertension-induced pathological cardiac hypertrophy through PDE5A. J Hypertens. 2018;36:1847–1857. doi: 10.1097/HJH.0000000000001769. doi: 10.1097/HJH.0000000000001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramasamy S, Velmurugan G, Rekha B, Anusha S, Shanmugha Rajan K, Shanmugarajan S, Ramprasath T, Gopal P, Tomar D, Karthik KV, et al. Egr-1 mediated cardiac miR-99 family expression diverges physiological hypertrophy from pathological hypertrophy. Exp Cell Res. 2018;365:46–56. doi: 10.1016/j.yexcr.2018.02.016. doi: 10.1016/j.yexcr.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Wu F. A new miRNA regulator, miR-672, reduces cardiac hypertrophy by inhibiting JUN expression. Gene. 2018;648:21–30. doi: 10.1016/j.gene.2018.01.047. doi: 10.1016/j.gene.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 26.Hu ZQ, Luo JF, Yu XJ, Zhu JN, Huang L, Yang J, Fu YH, Li T, Xue YM, Feng YQ, et al. Targeting myocyte-specific enhancer factor 2D contributes to the suppression of cardiac hypertrophic growth by miR-92b-3p in mice. Oncotarget. 2017;8:92079–92089. doi: 10.18632/oncotarget.20759. doi: 10.18632/oncotarget.20759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CY, Pai PY, Kuo CH, Ho TJ, Lin JY, Lin DY, Tsai FJ, Padma VV, Kuo WW, Huang CY. p53-mediated miR-18 repression activates HSF2 for IGF-IIR-dependent myocyte hypertrophy in hypertension-induced heart failure. Cell Death Dis. 2017;8:e2990. doi: 10.1038/cddis.2017.320. doi: 10.1038/cddis.2017.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Ponnusamy M, Liu C, Tian J, Dong Y, Gao J, Wang C, Zhang Y, Zhang L, Wang K, et al. MiR-485-5p modulates mitochondrial fission through targeting mitochondrial anchored protein ligase in cardiac hypertrophy. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2871–2881. doi: 10.1016/j.bbadis.2017.07.034. doi: 10.1016/j.bbadis.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Bao Q, Zhao M, Chen L, Wang Y, Wu S, Wu W, Liu X. MicroRNA-297 promotes cardiomyocyte hypertrophy via targeting sigma-1 receptor. Life Sci. 2017;175:1–10. doi: 10.1016/j.lfs.2017.03.006. doi: 10.1016/j.lfs.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao M, Chen G, Lv F, Liu Y, Tian H, Tao R, Jiang R, Zhang W, Zhuo C. LncRNA TINCR attenuates cardiac hypertrophy by epigenetically silencing CaMKII. Oncotarget. 2017;8:47565–47573. doi: 10.18632/oncotarget.17735. doi: 10.18632/oncotarget.17735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Liu X, Chen L, Chen W, Zhang Y, Chen J, Wu X, Zhao Y, Wu X, Sun G. The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem Biophys Res Commun. 2018;505:807–815. doi: 10.1016/j.bbrc.2018.09.135. doi: 10.1016/j.bbrc.2018.09.135. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 36.Lv L, Li T, Li X, Xu C, Liu Q, Jiang H, Li Y, Liu Y, Yan H, Huang Q, et al. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol Ther Nucleic Acids. 2018;10:387–397. doi: 10.1016/j.omtn.2017.12.018. doi: 10.1016/j.omtn.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Liu Y, Guo X, Sun G, Ma Q, Dai Y, Zhu G, Sun Y. Long noncoding RNA myocardial infarction–associated transcript is associated with the microRNA–150–5p/P300 pathway in cardiac hypertrophy. Int J Mol Med. 2018;42:1265–1272. doi: 10.3892/ijmm.2018.3700. doi: 10.3892/ijmm.2018.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, An X, Li Z, Song Y, Li L, Zuo S, Liu N, Yang G, Wang H, Cheng X, et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 39.Lai Y, He S, Ma L, Lin H, Ren B, Ma J, Zhu X, Zhuang S. HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy. Mol Cell Biochem. 2017;432:179–187. doi: 10.1007/s11010-017-3008-y. doi: 10.1007/s11010-017-3008-y. [DOI] [PubMed] [Google Scholar]
- 40.Jiang F, Zhou X, Huang J. Long noncoding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS One. 2016;11:e0152767. doi: 10.1371/journal.pone.0152767. doi: 10.1371/journal.pone.0152767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon KL, Tan KT, Wei YY, Ng CP, Colman A, Korzh V, Xu XQ. RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc Res. 2012;94:418–427. doi: 10.1093/cvr/cvs095. doi: 10.1093/cvr/cvs095. [DOI] [PubMed] [Google Scholar]
- 42.van den Hoogenhof MMG, van der Made I, de Groot NE, Damanafshan A, van Amersfoorth SCM, Zentilin L, Giacca M, Pinto YM, Creemers EE. AAV9-mediated Rbm24 overexpression induces fibrosis in the mouse heart. Sci Rep. 2018;8:11696. doi: 10.1038/s41598-018-29552-x. doi: 10.1038/s41598-018-29552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slone S, Anthony SR, Wu X, Benoit JB, Aube J, Xu L, Tranter M. Activation of HuR downstream of p38 MAPK promotes cardiomyocyte hypertrophy. Cell Signal. 2016;28:1735–1741. doi: 10.1016/j.cellsig.2016.08.005. doi: 10.1016/j.cellsig.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Si Y, Ma N, Mei J. The RNA-binding protein PCBP2 inhibits Ang II-induced hypertrophy of cardiomyocytes though promoting GPR56 mRNA degeneration. Biochem Biophys Res Commun. 2015;464:679–684. doi: 10.1016/j.bbrc.2015.06.139. doi: 10.1016/j.bbrc.2015.06.139. [DOI] [PubMed] [Google Scholar]
- 45.Qu W, Shi S, Sun L, Zhang F, Zhang S, Mu S, Zhao Y, Liu B, Cao X. Construction of a microRNA–associated feed–forward loop network that identifies regulators of cardiac hypertrophy and acute myocardial infarction. Int J Mol Med. 2018;42:2062–2070. doi: 10.3892/ijmm.2018.3790. doi: 10.3892/ijmm.2018.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y, Xu Y, Liang C, Xing W, Zhang T. The mechanism of myocardial hypertrophy regulated by the interaction between mhrt and myocardin. Cell Signal. 2018;43:11–20. doi: 10.1016/j.cellsig.2017.11.007. doi: 10.1016/j.cellsig.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Viganò V, Stirparo GG, Latronico MV, Hasenfuss G, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110:20164–20169. doi: 10.1073/pnas.1315155110. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, Gao C, Wei C, Xue Y, Shao C, Hao Y, Gou LT, Zhou Y, Zhang J, Ren S, et al. RBFox2-miR-34a-Jph2 axis contributes to cardiac decompensation during heart failure. Proc Natl Acad Sci U S A. 2019;116:6172–6180. doi: 10.1073/pnas.1822176116. doi: 10.1073/pnas.1822176116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 50.van den Hoogenhof MMG, van der Made I, Beqqali A, de Groot NE, Damanafshan A, van Oort RJ, Pinto YM, Creemers EE. The RNA-binding protein Rbm38 is dispensable during pressure overload-induced cardiac remodeling in mice. PLoS One. 2017;12:e0184093. doi: 10.1371/journal.pone.0184093. doi: 10.1371/journal.pone.0184093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corsten MF, Papageorgiou A, Verhesen W, Carai P, Lindow M, Obad S, Summer G, Coort SL, Hazebroek M, van Leeuwen R, et al. MicroRNA profiling identifies microRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis. Circ Res. 2012;111:415–425. doi: 10.1161/CIRCRESAHA.112.267443. doi: 10.1161/CIRCRESAHA.112.267443. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Zhang M, Li X, Tang Z, Wang X, Zhong M, Suo Q, Zhang Y, Lv K. Silencing MicroRNA-155 attenuates cardiac injury and dysfunction in viral myocarditis via promotion of M2 phenotype polarization of macrophages. Sci Rep. 2016;6:22613. doi: 10.1038/srep22613. doi: 10.1038/srep22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasques-Nóvoa F, Laundos TL, Cerqueira RJ, Quina-Rodrigues C, Soares-Dos-Reis R, Baganha F, Ribeiro S, Mendonça L, Gonçalves F, Reguenga C, et al. MicroRNA-155 amplifies nitric oxide/cGMP signaling and impairs vascular angiotensin ii reactivity in septic shock. Crit Care Med. 2018;46:e945–e954. doi: 10.1097/CCM.0000000000003296. doi: 10.1097/CCM.0000000000003296. [DOI] [PubMed] [Google Scholar]
- 54.Yang L, Wang B, Zhou Q, Wang Y, Liu X, Liu Z, Zhan Z. MicroRNA-21 prevents excessive inflammation and cardiac dysfunction after myocardial infarction through targeting KBTBD7. Cell Death Dis. 2018;9:769. doi: 10.1038/s41419-018-0805-5. doi: 10.1038/s41419-018-0805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye X, Zhang HM, Qiu Y, Hanson PJ, Hemida MG, Wei W, Hoodless PA, Chu F, Yang D. Coxsackievirus-induced miR-21 disrupts cardiomyocyte interactions via the downregulation of intercalated disk components. PLoS Pathog. 2014;10:e1004070. doi: 10.1371/journal.ppat.1004070. doi: 10.1371/journal.ppat.1004070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu YL, Wu W, Xue Y, Gao M, Yan Y, Kong Q, Pang Y, Yang F. MicroRNA-21 and -146b are involved in the pathogenesis of murine viral myocarditis by regulating TH-17 differentiation. Arch Virol. 2013;158:1953–1963. doi: 10.1007/s00705-013-1695-6. doi: 10.1007/s00705-013-1695-6. [DOI] [PubMed] [Google Scholar]
- 57.Corsten MF, Heggermont W, Papageorgiou AP, Deckx S, Tijsma A, Verhesen W, van Leeuwen R, Carai P, Thibaut HJ, Custers K, et al. The microRNA-221/-222 cluster balances the antiviral and inflammatory response in viral myocarditis. Eur Heart J. 2015;36:2909–2919. doi: 10.1093/eurheartj/ehv321. doi: 10.1093/eurheartj/ehv321. [DOI] [PubMed] [Google Scholar]
- 58.Gast M, Schroen B, Voigt A, Haas J, Kuehl U, Lassner D, Skurk C, Escher F, Wang X, Kratzer A, et al. Long noncoding RNA MALAT1-derived mascRNA is involved in cardiovascular innate immunity. J Mol Cell Biol. 2016;8:178–181. doi: 10.1093/jmcb/mjw003. doi: 10.1093/jmcb/mjw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Xia JW, Ke ZP, Zhang BH. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J Cell Physiol. 2019;234:5319–5326. doi: 10.1002/jcp.27340. doi: 10.1002/jcp.27340. [DOI] [PubMed] [Google Scholar]
- 60.Gast M, Rauch BH, Haghikia A, Nakagawa S, Haas J, Stroux A, Schmidt D, Schumann P, Weiss S, Jensen L, et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovasc Res. 2019;115:1886–1906. doi: 10.1093/cvr/cvz085. doi: 10.1093/cvr/cvz085. [DOI] [PubMed] [Google Scholar]
- 61.Gast M, Rauch BH, Nakagawa S, Haghikia A, Jasina A, Haas J, Nath N, Jensen L, Stroux A, Böhm A, et al. Immune system-mediated atherosclerosis caused by deficiency of long noncoding RNA MALAT1 in ApoE-/-mice. Cardiovasc Res. 2019;115:302–314. doi: 10.1093/cvr/cvy202. doi: 10.1093/cvr/cvy202. [DOI] [PubMed] [Google Scholar]
- 62.Zhuang YT, Xu DY, Wang GY, Sun JL, Huang Y, Wang SZ. IL-6 induced lncRNA MALAT1 enhances TNF-α expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur Rev Med Pharmacol Sci. 2017;21:302–309. [PubMed] [Google Scholar]
- 63.Wu J, Zhang H, Zheng Y, Jin X, Liu M, Li S, Zhao Q, Liu X, Wang Y, Shi M, et al. The long noncoding RNA MALAT1 induces tolerogenic dendritic cells and regulatory T cells via miR155/dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin/IL10 axis. Front Immunol. 2018;9:1847. doi: 10.3389/fimmu.2018.01847. doi: 10.3389/fimmu.2018.01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H, Liu J, Li W, Liu G, Li Z. LncRNA-HOTAIR promotes TNF-α production in cardiomyocytes of LPS-induced sepsis mice by activating NF-κB pathway. Biochem Biophys Res Commun. 2016;471:240–246. doi: 10.1016/j.bbrc.2016.01.117. doi: 10.1016/j.bbrc.2016.01.117. [DOI] [PubMed] [Google Scholar]
- 66.Guo F, Tang C, Li Y, Liu Y, Lv P, Wang W, Mu Y. The interplay of lncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-κB signalling pathway. J Cell Mol Med. 2018;22:5062–5075. doi: 10.1111/jcmm.13790. doi: 10.1111/jcmm.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. Circular noncoding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heymans S, Corsten MF, Verhesen W, Carai P, van Leeuwen RE, Custers K, Peters T, Hazebroek M, Stöger L, Wijnands E, et al. Macrophage microRNA-155 promotes cardiac hypertrophy and failure. Circulation. 2013;128:1420–1432. doi: 10.1161/CIRCULATIONAHA.112.001357. doi: 10.1161/CIRCULATIONAHA.112.001357. [DOI] [PubMed] [Google Scholar]
- 69.Song CL, Wang JP, Xue X, Liu N, Zhang XH, Zhao Z, Liu JG, Zhang CP, Piao ZH, Liu Y, et al. Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell Physiol Biochem. 2017;42:1202–1212. doi: 10.1159/000478918. doi: 10.1159/000478918. [DOI] [PubMed] [Google Scholar]
- 70.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 71.Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, Burkhardt R, Thiery J, Wagner DR, Devaux Y. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 72.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 73.Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, Snitow M, Morley M, Li D, Petrenko N, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 2015;7:279ra238. doi: 10.1126/scitranslmed.3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110:187–192. doi: 10.1073/pnas.1208863110. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, et al. miR-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131:1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. 2015;89(Pt A):98–112. doi: 10.1016/j.yjmcc.2015.09.016. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 82.Alexanian M, Maric D, Jenkinson SP, Mina M, Friedman CE, Ting CC, Micheletti R, Plaisance I, Nemir M, Maison D, et al. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat Commun. 2017;8:1806. doi: 10.1038/s41467-017-01804-w. doi: 10.1038/s41467-017-01804-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, Luu TDA, Ackers-Johnson M, Foo RS. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8:225. doi: 10.1038/s41467-017-00319-8. doi: 10.1038/s41467-017-00319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jakob P, Doerries C, Briand S, Mocharla P, Kränkel N, Besler C, Mueller M, Manes C, Templin C, Baltes C, et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: role for impaired in vivo neovascularization and cardiac repair capacity. Circulation. 2012;126:2962–2975. doi: 10.1161/CIRCULATIONAHA.112.093906. doi: 10.1161/CIRCULATIONAHA.112.093906. [DOI] [PubMed] [Google Scholar]
- 85.Qiang L, Hong L, Ningfu W, Huaihong C, Jing W. Expression of miR-126 and miR-508-5p in endothelial progenitor cells is associated with the prognosis of chronic heart failure patients. Int J Cardiol. 2013;168:2082–2088. doi: 10.1016/j.ijcard.2013.01.160. doi: 10.1016/j.ijcard.2013.01.160. [DOI] [PubMed] [Google Scholar]
- 86.Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J Mol Cell Cardiol. 2016;94:107–121. doi: 10.1016/j.yjmcc.2016.03.015. doi: 10.1016/j.yjmcc.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 87.Duan Q, Yang L, Gong W, Chaugai S, Wang F, Chen C, Wang P, Zou MH, Wang DW. MicroRNA-214 is upregulated in heart failure patients and suppresses XBP1-mediated endothelial cells angiogenesis. J Cell Physiol. 2015;230:1964–1973. doi: 10.1002/jcp.24942. doi: 10.1002/jcp.24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fan J, Li H, Nie X, Yin Z, Zhao Y, Zhang X, Yuan S, Li Y, Chen C, Wang DW. MiR-665 aggravates heart failure via suppressing CD34-mediated coronary microvessel angiogenesis. Aging (Albany NY) 2018;10:2459–2479. doi: 10.18632/aging.101562. doi: 10.18632/aging.101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang S, Guo X, Long CL, Li C, Zhang YF, Wang J, Wang H. SUR2B/Kir6.1 channel openers correct endothelial dysfunction in chronic heart failure via the miR-1-3p/ET-1 pathway. Biomed Pharmacother. 2019;110:431–439. doi: 10.1016/j.biopha.2018.11.135. doi: 10.1016/j.biopha.2018.11.135. [DOI] [PubMed] [Google Scholar]
- 90.Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 91.Huang S, Lu W, Ge D, Meng N, Li Y, Su L, Zhang S, Zhang Y, Zhao B, Miao J. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–2183. doi: 10.1080/15548627.2015.1106663. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S, Zhang X, Yuan Y, Tan M, Zhang L, Xue X, Yan Y, Han L, Xu Z. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long noncoding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surg. 2015;47:439–446. doi: 10.1093/ejcts/ezu215. doi: 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- 93.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bianchessi V, Badi I, Bertolotti M, Nigro P, D’Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A, Lauri A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in endothelial cells. J Mol Cell Cardiol. 2015;81:62–70. doi: 10.1016/j.yjmcc.2015.01.012. doi: 10.1016/j.yjmcc.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 95.Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Günther S, et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017;136:65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bischoff FC, Werner A, John D, Boeckel JN, Melissari MT, Grote P, Glaser SF, Demolli S, Uchida S, Michalik KM, et al. Identification and functional characterization of hypoxia-induced endoplasmic reticulum stress regulating lncRNA (HypERlnc) in pericytes. Circ Res. 2017;121:368–375. doi: 10.1161/CIRCRESAHA.116.310531. doi: 10.1161/CIRCRESAHA.116.310531. [DOI] [PubMed] [Google Scholar]
- 97.Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017;494:126–132. doi: 10.1016/j.bbrc.2017.10.068. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 98.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 99.Costa G, Tarannum N, Herbert SP. mRNA Localisation in endothelial cells regulates blood vessel sprouting. bioRxiv. 2018 doi: 10.1101/374850. [Google Scholar]
- 100.Gomes CPC, Agg B, Andova A, Arslan S, Baker A, Bartekova M, Beis D, Betsou F, Wettinger SB, Bugarski B, et al. Catalyzing Transcriptomics Research in Cardiovascular Disease: The CardioRNA COST Action CA17129. Noncoding RNA. 2019;5:E31. doi: 10.3390/ncrna5020031. doi: 10.3390/ncrna5020031. [DOI] [PMC free article] [PubMed] [Google Scholar]


