SUMMARY
Callosal projections are thought to play a critical role in coordinating neural activity between the cerebral hemispheres in placental mammals, but the rules that govern the arrangement of callosal synapses on the dendrites of their target neurons remain poorly understood. Here we describe a high-throughput method to map the functional organization of callosal connectivity by combining in vivo 3D random-access two-photon calcium imaging of the dendritic spines of single V1 neurons with optogenetic stimulation of the presynaptic neural population in the contralateral hemisphere. We find that callosal-recipient spines are more likely to cluster with non-callosal-recipient spines with similar orientation preference. These observations, based on optogenetic stimulation, were confirmed by direct anatomical visualization of callosal synaptic connections using post hoc expansion microscopy. Our results demonstrate, for the first time, that functional synaptic clustering in a short dendritic segment could play a role in integrating distinct neuronal circuits.
In Brief
Lee et al. examine the functional synaptic organization of callosal projections onto layer 2/3 pyramidal neurons in the primary visual cortices of mice. They find that callosal and intra-cortical inputs with similar orientation preference cluster together on short dendritic segments.
INTRODUCTION
The knowledge of how individual pyramidal neurons integrate the activity of the large number of synaptic inputs that converge on their dendritic field is critical for understanding the fundamental computations underlying cortical function. Previous studies of the visual cortex have provided evidence of a fine-scale dendritic organization for functional properties, including spatial clustering of inputs with similar preferences for orientation (Wilson et al., 2016) and for location in visual space (Scholl et al., 2017; lacaruso et al., 2017). Such a clustered arrangement may enable spatio-temporal coactivity to trigger dendritic non-linearities that shape neuronal output (Polsky et al., 2004; Losonczy et al., 2008; Branco et al., 2010). Missing from this picture has been the ability to specify the source of the inputs that contribute to these functionally defined dendritic clusters. Relatively few terminals from individual axons are thought to give rise to synaptic contacts on nearby dendritic spines, suggesting that functional clusters are likely to represent inputs from different axonal branches (Kasthuri et al., 2015; Lee et al., 2016). The location of the parent cell bodies remains unclear, as does the issue of whether these axons originate from the same or different neurons.
This same gap in our understanding has hampered our ability to elucidate how a specific set of inputs impacts the function of a given cortical area. For example, previous studies in rodents have shown that callosal inputs to the visual cortex play a crucial role in binocular perception (Restani et al., 2009; Cerri et al., 2010; Zhao et al., 2013). Callosal connections are known to be the source of a substantial fraction of the ipsilateral-eye responses in the binocular region of the visual cortex, where feedforward drive is dominated by inputs from the contralateral eye (Restani et al., 2009; Cerri et al., 2010). Although these studies clearly suggest an important role for callosal inputs in binocular vision, how callosal inputs are arranged within the dendritic field and how this organization is related to other functional inputs remain unknown.
Progress on both of these fronts requires overcoming the ultimate challenge of simultaneously documenting structure and function at the synaptic level. Heretofore, only functional connectomics—combining physiological imaging and large-scale electron microscopy (EM) — has been able to reveal the synaptic logic of local networks (Lee et al., 2016). However, it is difficult to extend this approach to synaptic inputs that arise from distant sources. To address this issue, we developed two new protocols to simultaneously map the functional properties and the source of input to a given dendritic spine labeled with the calcium indicator GCaMP6s. One approach relies on a high-throughput in vivo physiological mapping technique that combines 3D random-access two-photon scanning microscopy using acousto-optical deflection (AOD; Szalay et al., 2016) with channelrhodopsin-2-assisted synaptic mapping (CRASM) using red-shifted opsins (Petreanu et al., 2007; Rajasethupathy et al., 2015). The other approach relies on a correlative-imaging technique that combines conventional two-photon imaging and post hoc anatomical synapse identification with X10 expansion microscopy (ExM; Chen et al., 2013; Tillberg et al., 2016; Truckenbrodt et al., 2018).
Reliable presynaptic activation with CRASM allowed us to identify putative callosal spine inputs, and we found that these spines are strongly ipsilateral-eye biased, consistent with previous findings (Restani et al., 2009; Cerri et al., 2010; Zhao et al., 2013). Callosal-recipient spines tended to appear spatially close to non-callosal-recipient spines with a similar orientation preference, a clustered arrangement we confirmed with anatomical evidence of monosynaptic connectivity derived from our correlative imaging method. At a larger scale, we found that callosal inputs to a neuron were concentrated in certain dendritic branches; branch-wise biases significantly deviated from a uniform distribution and were reminiscent of the structured synaptic connectivity seen in hippocampal CA1 pyramidal neurons (Druckmann et al., 2014). Based on the results of these novel approaches, we suggest that both the coarse and fine spatial arrangement of synaptic inputs are relevant factors in the integration of different networks within a neuron’s dendritic field.
RESULTS
Mapping Tuning Properties and Source of Inputs to Dendritic Spines
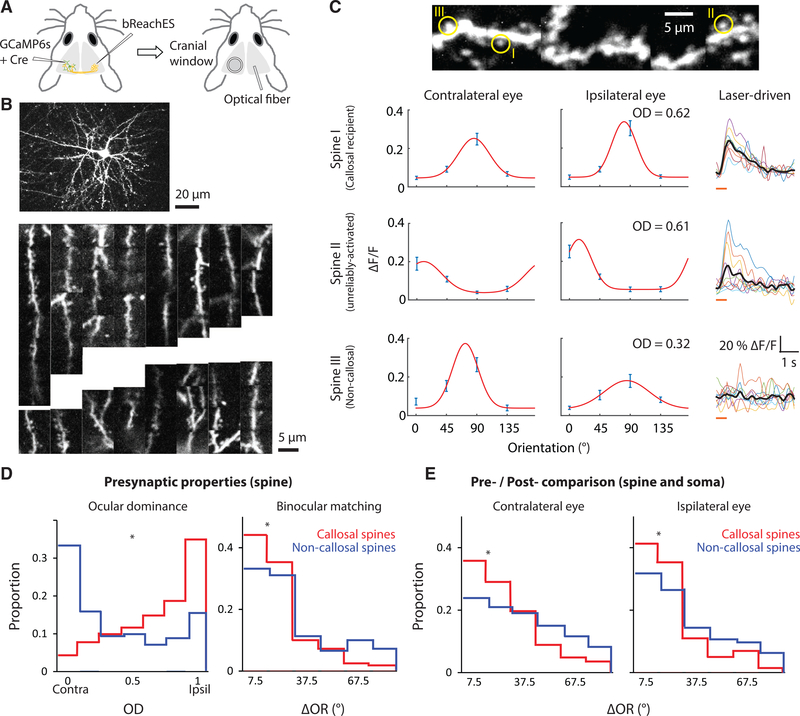
To map the functional properties and synaptic organization of callosal inputs within the dendritic field of a single layer-2/3 pyramidal neuron, we used in vivo AOD two-photon imaging of calcium responses in dendritic spines following sparse adeno-associated virus (AAV) expression of a genetically encoded calcium sensor (GCaMP6s) in the murine visual cortex (Chen et al., 2013; Wilson et al., 2016). To enable the use of an optogenetic method for synaptic mapping, we expressed red-shifted channelrhodopsin (bReaChES) in the contralateral hemisphere (Figures 1A and S1A–S1C). To make sure that we reliably and strongly activated callosal boutons in the contralateral hemisphere, we co-expressed GCaMP6s with bReaChES in a separate set of animals and imaged the calcium activity of boutons during optogenetic stimulation to determine the optimal parameters (Figures S1D–S1G; for details, see Methods). We then combined CRASM with AOD by applying optogenetic stimulation to the hemisphere with the bReaChES injection and two-photon imaging of the hemisphere with the GCaMP6s injection to visualize stimulation-related events in the postsynaptic spines (Figure 1B). Spine-specific signals induced by visual or optogenetic stimulation were isolated by applying a subtraction method (Figures S1H–S1L) similar to that of previous publications (Chen et al., 2013; Wilson et al., 2016; lacaruso et al., 2017). Optogenetic stimulation-related activity in dendritic spines was defined by a surge of calcium signaling, measuring at least 2 SD above baseline, within 1.5 s immediately following laser stimulation. Unexpectedly, nearly 40% percent of the spines in our sample were found to be activated by optogenetic stimulation of the contralateral hemisphere. Within this population, there was a wide range of reliability in single-trial responses to laser stimulation (Figure S1N), suggesting that this distribution included activation via mono- and multi-synaptic pathways. Given the uncertainty in the relation between reliability and connectivity, we asked whether the visual-response properties of dendritic spines could provide insight. ln order to characterize the visual-response properties of callosal- and non-callosal-recipient spines, we measured the response of individual spines to grating stimuli presented independently to the ipsilateral and contralateral eye (Figure 1C). Consistent with previous studies of callosal inputs (Restani et al., 2009; Cerri et al., 2010; Zhao et al., 2013), those spines that exhibited 100% activation reliability exhibited a strong bias to the ipsilateral eye, a striking difference from the contralateral bias found in dendritic spines that were not significantly activated (Figure S1M). This strong ipsilateral bias was maintained when the sample of activated spines was extended to include those spines with reliability greater than 80%, a threshold value that we then used to define spines as callosal recipient (16%). Spines that were not significantly activated by laser stimulation were classified as non-callosal-recipient spines (61%) and the remaining population of less-reliably activated spines were defined as unreliably activated spines (23%; Figure S1N). We found no significant difference in orientation tuning between callosal- and non-callosal-recipient spines (P = 0.461, Wilcoxon rank sum test), but we did note that the degree of matching of the orientation preference of the monocular responses in individual spines was significantly greater for callosal-recipient spines (Figure 1D). We also found that the orientation preference of both the contralateral and ipsilateral responses of callosal-recipient spines was better matched to the somatic orientation preference than the responses of non-callosal-recipient spines (Figure 1E).
Figure 1. Combining CRASM with AOD Imaging Enables High-Throughput Imaging of Dendritic Spines, Defining Their Visual Response Properties and Source of Synaptic Input.
(A) Injections of viruses expressing GCaMP6s and bReaChES were made in opposite hemispheres. Optogenetic laser stimulation was applied only to the bReaChES injection site.(B) Top: an example neuron labeled with GCaMP6s, found in binocular V1 and imaged with AOD microscopy. Bottom: dendritic segments widely distributed in space were simultaneously monitored during one experimental session. (C) Left: orientation tuning curves measured through each eye. Ocular dominance (OD) was derived from comparing the peaks of orientation tuning curves. Data are represented as mean ± SEM. Right: laser-driven activity of three example dendritic spines. Single trial laser-driven activities are indicated in color and average response in black. Orange line: duration of light stimulus. (D) A comparison of the response properties of callosal-recipient (n = 343 spines, 129 dendrites, 12 mice) and non-callosal-recipient spines (n = 1162 spines, 152 dendrites, 12 mice). These populations of spines differ in the distribution of ocular dominance values and in the degree to which individual spines exhibit matching orientation preference *p < 0.001, Wilcoxon rank sum test.(E) Pre- and post-synaptic difference in orientation preference between callosal (n = 343 spines, 129 dendrites, 12 mice) and non-callosal spines (n = 1162 spines, 152 dendrites, 12 mice). *p < 0.001, Wilcoxon rank sum test.
Functional Clustering of Callosal and Non-callosal Spines
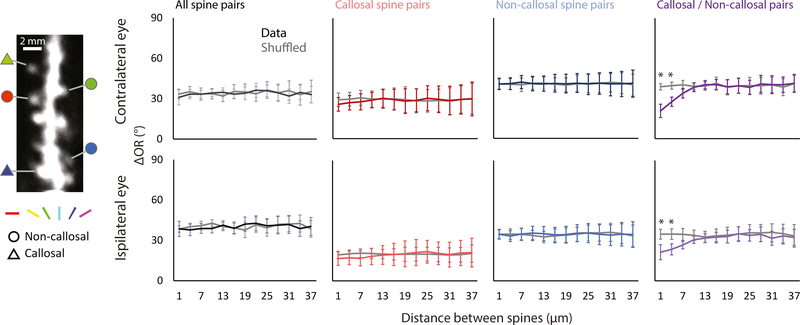
Having demonstrated that this approach allowed us to identify spines whose optogenetic activation and functional properties were consistent with callosal inputs, we wondered whether callosal inputs might exhibit fine-scale functional clustering of orientation preference within the dendritic field. As a first step, we computed a pairwise difference of orientation preferences versus distance along the dendritic shaft for all orientation selective spines and we confirmed the conclusions of previous studies in mice (Jia et al., 2010; Chen et al., 2013; lacaruso et al., 2017); we found no evidence of functional clustering. We also tested for functional clustering within the class of callosal-recipient spines and found no departure from a random distribution. A similar result was found for the non-callosal-recipient population (Figure 2). This left us with one additional possibility to test: could the callosal-recipient spines be clustered with non-callosal-recipient spines with a similar orientation preference? Surprisingly, this analysis across groups revealed a significant non-random spatial distribution at distances less than 5 μm that was not observed within either group. These findings were independent of the method used to evaluate significance from random shuffle, shuffling either within or across dendritic branches (Figure S2; see Methods). These results emphasize that the fine-scale functional organization of inputs within the dendritic field could play a significant role in the integration of inputs from different sources.
Figure 2. Orientation Preference-Based Functional Clustering of Callosal- and Non-callosal-Recipient Spine Pairs.
Left: a representative image of a dendritic segment taken with two-photon imaging with orientation preference (color) measured through the contralateral eye with several callosal-recipient and non-callosal-recipient spines (shape).
Right: the relationship of distance between spines in a pair and orientation preference is calculated using four pairing conditions: all orientation-selective spines (n = 3622 spine pairs, 146 dendrites, 12 mice), callosal-recipient with callosal-recipient spines (n = 322 spine pairs, 102 dendrites, 12 mice), non-callosal-recipient with non-callosal-recipient spines (n = 2152 spine pairs, 139 dendrites, 12 mice), and non-callosal-recipient with callosal-recipient spines (n = 658 spine pairs, 102 dendrites, 12 mice). Callosal-recipient spines are more likely to cluster with non-callosal spines with a similar orientation preference. Error bars represent SEM. *p < 0.001, permutation test.
Anatomical Evidence of Functional Clustering
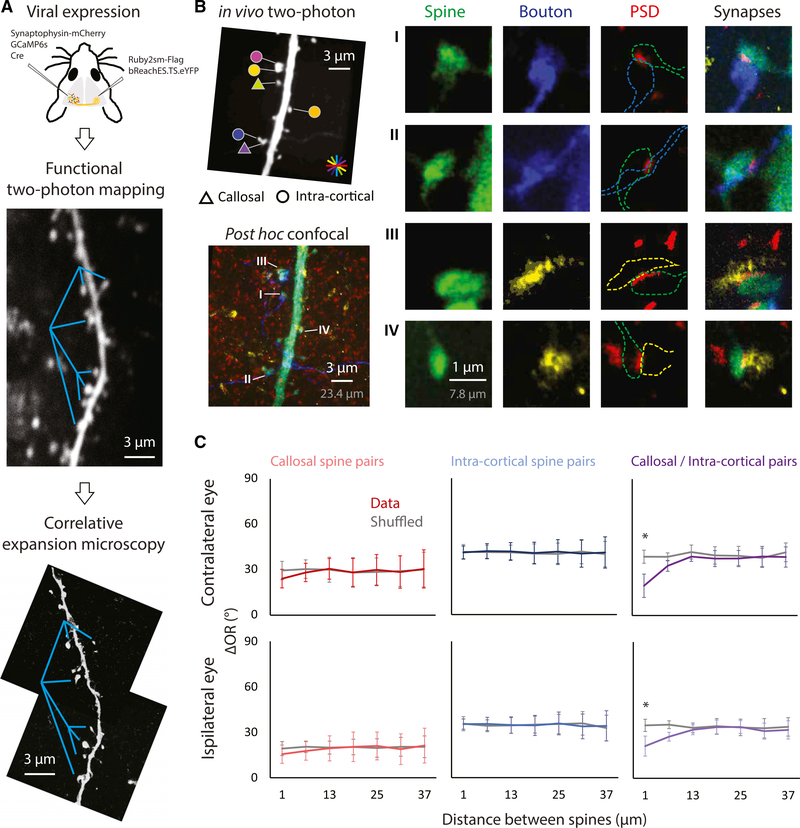
To confirm the properties of callosal-recipient spines defined by optogenetic stimulation, we turned to a post hoc correlative anatomy approach that would allow us to identify dendritic spines that exhibit the morphological criteria consistent with synaptic contact with callosal axons. We applied an advanced version of expansion microscopy, X10 ExM (Truckenbrodt et al., 2018), using viral transfection with the structural marker Ruby2sm-Flag to label callosal axon terminals and viral transfection with synaptophysin-mCherry to label intra-cortical axon terminals. After functional mapping with conventional two-photon microscopy, tangential brain slices were collected and immunocytochemistry was performed to identify in vivo imaged layer-2/3 dendrites (anti-GFP), postsynaptic densities (anti-Homer1), and callosal (anti-Flag) and intra-cortical axon terminals (anti-mCherry). The tissue, consisting of targeted dendritic branches, was embedded in the hydrogel, digested, and physically expanded approximately 8 times. This enabled imaging resolution of ~30 nm with conventional confocal microscopy to visualize synaptic contacts (Figures 3A, 3B, S3A, and S3B). Synaptic contacts were defined by proximity (<150 nm) between the signal of presynaptic terminals and the signal of the postsynaptic density of the given spine (Dani et al., 2010). We found that ~10% of spines were monosynaptic callosal recipient and ~65% of spines received synaptic inputs from intra-cortical axon terminals. Consistent with high-throughput CRASM, anatomically defined callosal-recipient spines displayed a strong ipsilateral dominance (Figure S3C), a high response probability to the laser stimulation in CRASM (Figure S3D), and a higher likelihood of clustering with intra-cortical spines with a similar orientation preference, a clustering effect that was not observed within either callosal or intra-cortical groups (Figure 3C).
Figure 3. Functional Clustering Between the Callosal and Intra-Cortical Spine Pairs Supported with Anatomical Evidence of Monosynaptic Connections.
(A) The structural markers for callosal (Ruby2sm-Flag) and intra-cortical (synaptophysin-mCherry) axonal terminals were expressed through a viral approach. The same dendritic segment that underwent functional mapping was reimaged with confocal microscopy post-tissue expansion. Cyan lines, references for comparison.(B) Left: the pictured dendritic segment was imaged after histological staining. Fluorophores targeted callosal axons (blue), intra-cortical boutons (yellow), dendritic spines (green), and postsynaptic densities (red). Right: four example spines receiving callosal (top two) or intra-cortical (bottom two) synaptic inputs. Scale bars are provided both in physical size post-expansion (gray) and back-calculation with expansion factor (white).(C) The relationship of the distance between spines in a pair and orientation preference is analyzed in a similar way as in Figure 2. Consistent with results from high-throughput CRASM, callosal spines are more likely to cluster with the intra-cortical spines with similar orientation preference (n = 131 callosal spine pairs; n = 923 intra-cortical spine pairs; n = 298 callosal/intra-cortical pairs, 52 dendrites, 6 mice). Error bars represent SEM. *p < 0.001, permutation test.
Large-Scale Dendritic Organization of Callosal Inputs
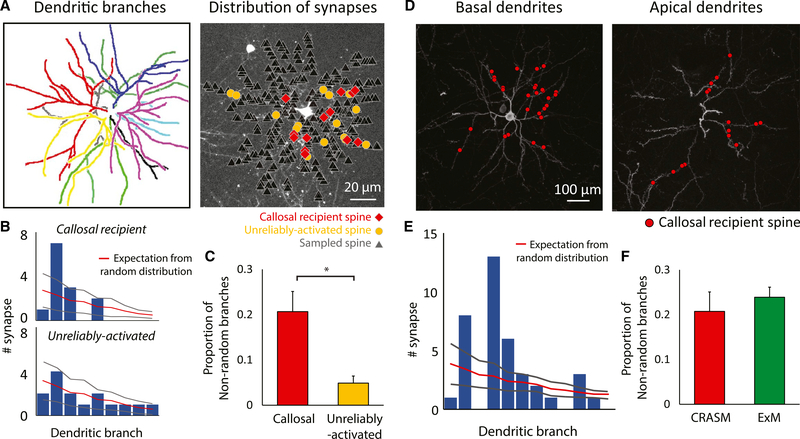
Two recent studies have noted larger spatial-scale non-uniformities in the distribution of excitatory and inhibitory synaptic inputs within the dendritic field of single pyramidal neurons in the hippocampi of mice (Druckmann et al., 2014; Bloss et al., 2016). Thus, we sought to determine whether callosal inputs are randomly distributed throughout the dendritic field or biased to particular dendritic branches. After comparing the actual synapses defined by optogenetic stimulation, we found that callosal inputs on dendrites were biased toward specific branches, an effect that was nearly absent in the population of unreliably-activated spines (Figures 4A–4C). We then applied the same analysis to the anatomically defined callosal-recipient spines and found a similar bias (Figures 4D–4F). Consistent with the previous demonstration of a strong ipsilateral bias in ocular dominance for callosal inputs, the dendritic branches with a callosal bias exhibited significant ipsilateral bias compared with other branches (P < 0.001), but were not significantly different in average orientation preference (P = 0.721) or orientation tuning index (P = 0.367, Wilcoxon rank sum test). In addition, we tested whether callosal spines exhibited a differential distribution in their distance from the soma or in their dendritic branch order and found no evidence for differential distributions (Figures S4A and S4B). Finally, we tested whether those branches that had a higher density of callosal inputs had different rules for fine-scale functional clustering. Limiting the cluster analysis to the 20% of branches that exhibited a higher density, we found that the same rules applied: functional clustering was present in callosal-recipient/non-callosal-recipient spine pairs but absent in both callosal/callosal and non-callosal/non-callosal pairs (Figure S4C).
Figure 4. Non-random Distribution of Callosal Inputs on Dendrites.
(A) Left: different primary dendritic branches labeled by color. Right: callosal-recipient (red) and unreliably activated (yellow) spines derived from optogenetic synaptic mapping are highlighted amongst the total spines recorded in the imaging experiment with CRASM.(B) Comparing the experimentally derived distribution of spines on dendritic branches to the random distribution of the example cell. The bar plot shows the number of synapses on each dendritic branch (sorted according to their surface area from high to low), while the overlaid red line indicates the number of synapses expected in a Poisson distribution. Gray lines indicate ±2 SD.(C) The proportion of dendritic branches receiving a significantly biased number of reliably activated inputs (callosal) is significantly higher than ones receiving a significantly biased number of unreliably activated inputs. Error bars represent SEM. *, P=0.00812, Wilcoxon rank sum test; n = 12 cells, 12 mice.(D) Dendritic distribution of callosal synapses in the basal dendrites (left) and apical dendrites (right) defined by expansion microscopy.(E) Comparison of the experimentally derived distribution of callosal-recipient spines on dendritic branches to the random distribution. Gray lines indicate ±2 SD.(F) Results from anatomical mapping using expansion microscopy are not statistically different from results using optogenetic synaptic mapping. Error bars represent SEM. p = 0.545, Wilcoxon rank sum test; n = 8 cells, 4 mice for ExM experiment.
DISCUSSION
We have identified the source of synaptic inputs to individual functionally characterized dendritic spines in the murine visual cortex using two new approaches: (1) a high-throughput method coupling in vivo 3D AOD imaging with in vivo CRASM and (2) a correlative approach combining conventional two-photon in vivo imaging and anatomical post hoc synapse identification with X10 ExM. Using both of these approaches, we found evidence for orientation-specific functional clustering in the fine-scale spatial arrangement of callosal inputs relative to non-callosal inputs. At a coarser scale, we observed that callosal inputs tend to bias themselves toward a small subset of branches but widely distribute within the branches. These results suggest that both the coarse- and fine-scale spatial arrangements of synaptic inputs within the dendritic field play an important role in the integration of inputs from different sources.
The spatial-length constant of ~5 μm has emerged as a fundamental distance for several different aspects of dendritic function, including functional synaptic clustering (lacaruso et al., 2017; Scholl et al., 2017), the synchronization of spontaneous activity (Kleindienst et al., 2011; Takahashi et al., 2012; Winnubst et al., 2015), and intradendritic plasticity signaling mechanisms (Larkum and Nevian, 2008; Nishiyama and Yasuda, 2015). While these studies emphasize a common distance for physiological and molecular synaptic interactions, the source of the synaptic inputs that engage this dendritic structure has remained unclear. Functionally clustered spines could receive their input from repeated synaptic contacts arising from a single axon, or single synaptic contacts arising from different axons originating in the same or different brain regions. Recent studies in the murine visual cortex found that single axons forming multiple synapses on the postsynaptic cell’s dendrites are frequently observed, but the spatially clustered synapses are not specifically favored over widespread spacing (Kasthuri et al., 2015; Lee et al., 2016). Thus, functional clustering emerging from the innervation of a single axon is not commonplace in V1. While it might be possible that different axons from a single source might participate in functional clustering (Lee et al., 2016), our results suggest that this is not a common occurrence, at least for callosal inputs. Instead, we find that synaptic clusters reflect the interaction of functionally similar inputs from different sources: distinct inter- and intra-cortical networks that are biased to the ipsilateral or contralateral eye, respectively. In this framework, functional clustering could then nonlinearly amplify the impact of excitatory postsynaptic potentials (Polsky et al., 2004; Losonczy et al., 2008; Branco et al., 2010) and generate robust responses to binocular inputs that exhibit orientation-matched features. Callosal connections are thought to contribute to gain modulation of sensitivity (Wunderle et al., 2015) and many other visual functions, such as re-establishing the continuity of the visual field across the vertical meridian (Schmidt et al., 2010), anticipating shape and motion across the visual field (Peiker et al., 2013), and enhancing binocular response (Conde-Ocazionez et al., 2018). Our results suggest that the fine-scale spatial arrangement of synaptic inputs within the dendritic field could be crucial for the visual-field integrative function of visual callosal connections.
It is unclear whether the underlying developmental mechanism responsible for this network interaction is contingent on functional clustering, but results from previous studies suggest that an activity-dependent mechanism is likely. The formation of interhemispheric axonal projections in rodent cortical regions is severely disrupted by inhibiting projection-neuron activity (Mizuno et al., 2007), inhibiting target-neuron activity (Mizuno et al., 2010), blocking unilateral sensory input (Innocenti and Frost, 1979; Olavarria et al., 1987), or interrupting the balanced activity between both hemispheres (Suárez et al., 2014). Thus, one could speculate that Hebbian mechanisms allow this late-arriving interhemispheric projection (Mizuno et al., 2007) to preferentially strengthen synaptic inputs to spines that are located near intra-cortical spines with similar orientation preferences.
In conclusion, we developed two new approaches to bridge the gap of knowledge between functional dendritic organization and network integration. Consistent results were derived with a high-throughput all-physiological CRASM method and a correlative anatomical method. These procedures can be applied to probe the functional synaptic organization of diverse neuronal networks innervating a single neuron and to potentially understand the role of dendritic computation in neural-circuit integration.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Fitzpatrick (david.fitzpatrick@mpfi.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All procedures were approved by the Max Planck Florida Institute for Neuroscience Institutional Animal Care and Use Committee and adhered to the standards of the National Institutes of Health. Female mice (Mus musculus, Jackson Laboratory) were used in all experiments. Ages of animals ranged from 3 to 12-weeks. No a priori sample size estimation was performed but sample sizes are similar to other studies which performed in vivo physiology followed by ex vivo histology.
METHOD DETAILS
Virus Injection
Mice (N = 17) were anesthetized and maintained with isoflurane (1%–3%) delivered in O2. Atropine (0.2 mg/kg, SC) was administered to reduce secretions and a 1:1 mixture of lidocaine and bupivacaine (<0.1 ml) was administered subcutaneously in the scalp. Animals were placed on a feedback-controlled heating pad to maintain internal temperature at 37°C. Under sterile surgical conditions, a small craniotomy (0.8 mm) was made over the binocular V1 region. AAV2/1.hSyn.Cre (Penn Vector Core) was diluted (1:100,000) in phosphate-buffered saline (Sigma) and mixed with AAV2/1.CAG.Flex.GCaMP6s (Penn Vector Core) to sparsely express GCaMP6s in layer 2/3 cortical neurons. Beveled glass micropipettes (10–20 μm outer diameter, Drummond Scientific Company) were lowered into the brain, and 390–650 nl of virus were injected over 10 min (Nanoject II, Drummond Scientific Company) at 200–400 μm below the pia. AAV2.CamKIIa.bReaChES.TS.eYFP (UNC Virus Vector Core) was injected into the contralateral binocular V1 region. For the expansion microscopy experiments (N = 6), structural labeling of the callosal axons was achieved by co-expressing AAV1.CAG.Ruby2sm- Flag (Penn Vector Core) with bReaChES by mixing 1:1 viruses. To label axonal boutons from local cortical neurons, AAV8.2-hEF1a-synaptophysin-mCherry (Gene Delivery Technology Core) was co-injected with GCaMP6s virus. To ascertain whether we could reliably and strongly activate callosal boutons in the contralateral hemisphere, we co-expressed GCaMP6s (AAV1.Syn.GCaMP6s) with bReaChES in a separate set of animals (N = 3) and imaged the calcium activity of boutons to determine the optimal parameters (Figure S1D–G). To prevent adherence of scar tissue to the arachnoid membrane, we filled each craniotomy with a small amount of bone wax and closed the scalp incision with 6–0 Ethilon sutures.
Cranial Window and Fiber-Optic Implantation
Viruses (above) were allowed two to three weeks of expression prior to craniotomy and fiber-optic implantation. Anesthesia was induced with 1%–3% isoflurane. Atropine (0.2 mg/kg, SQ) and a 1:1 mixture of lidocaine and bupivacaine were administered. Animals were placed on a feedback-controlled heating pad to maintain an internal temperature of 36 to 38°C. Isoflurane was delivered between 1 and 3% throughout the surgical procedure to maintain a surgical plane of anesthesia. Breathing patterns, external temperature, and internal temperature were continuously monitored during the procedure and the subsequent imaging session. The scalp was retracted and a custom titanium headplate was adhered to the skull with C&B Metabond (Parkell). A 3.5–4.0 mm craniotomy was performed with disposable Miltex biopsy punches (3 mm diameter, Patterson) over the viral injection site. One to two pieces of commercial coverglass (3 mm, #1 thickness, Warner Instruments) or custom coverglass (3 mm diameter, 0.7mm thickness, Warner Instruments) were adhered to a larger coverglass (5 mm diameter, #1.5 thickness, Electron Microscopy Sciences) using optical adhesive (# 71, Norland Products) and placed onto the brain to gently compress the underlying cortex and dampen biological motion during imaging. The cranial window was sealed with Vetbond (3M). Fiber-optic implantation in contralateral V1 involved cutting an optic fiber (high OH, 400 μm core, 0.39 NA, Thorlabs) with a diamond knife followed by insertion into the 1.25 mm diameter of a ceramic ferrule (Thorlabs) with a 440 μm bore. The optic fiber was adhered to the ferrule with epoxy (Gorilla Glue Company). Both ends of the optic fiber were finely ground with sandpaper and grinding puck (Thorlabs). The optic fiber was implanted perpendicular to the surface of V1 that had been previously injected with bReaChES. Dental cement (C&B Metabond) was applied to adhere the cranial window and optic fiber to the surface of the skull. Eyes were lubricated hourly with Silicon Oil AP150 Wacker (Sigma-Aldrich). Upon completion of the surgical procedure, anesthesia was maintained with isoflurane (0.5%−2%).
Two-Photon Imaging
In vivo ChR2-assisted synaptic mapping (CRASM) was accomplished through the use of the commercial two-photon AOD microscope, which was operated with the software MES (Femtonics Ltd) and coupled to a Chameleon Ultra II laser (COHERENT). The laser was tuned to λ=910 nm in order to excite GCaMP6s. Both signals were collected through a water-immersion objective (16x, CFI75, Nikon Instruments). This microscope allowed for 3D DRIFT AO random-access scanning, by quickly drifting the focal spot in 3D in an arbitrary direction and speed (Szalay et al., 2016). We first took a reference Z-stack of the targeted neuron, and manually drew an ROI along the dendritic branches of a layer-2/3 neuron in V1. Using the aforementioned Z-stack, we selected guide points along the dendritic branch to manually draw the ROI and fit the 3D trajectory. A 3D ribbon scan, following either a transverse or longitudinal drifting pattern, measured the selected branch with optimized speed (12–30 Hz) and spatial resolution (10–14.3 pixels/μm). Motion correction was sometimes performed using MES.
For the correlative ExM experiment, imaging was performed with a Bergamo II microscope (Thorlabs) running Scanimage 4 (Pologruto et al., 2003; Janelia Research Campus) with 910 nm excitation provided by an Insight DS+ laser (Spectraphysics). Average excitation power at the exit of the objective (16x, CFI75, Nikon Instruments) ranged from 10 to 40 mW. Individual neurons in layer 2/3 in binocular V1 were selected for imaging based on several criteria: visible dendritic spines, nuclear exclusion, orientation-tuned responses, and a lack of large blood vessels obscuring the dendritic field (Wilson et al., 2016). Images of dendritic segments were acquired at 15Hz (512×512, resolution: 8.42–12.2 pixels/μm) and images of soma were acquired at 15 Hz (512×512, resolution: 4.2–5.9 pixels/μm). Z-stacks of individual cells were acquired prior to dendritic imaging by averaging 50 frames per plane using 1–2 μm z-steps. Multiple dendrites, both basal and apical, across multiple depths were imaged on individual cells. This imaging method only allows visualization of a fraction of spines on dendrites in horizontal planes. Two-photon frame triggers from Scanimage were synchronized with stimulus information using Spike2 (CED). Throughout the experiment, dendrites were carefully monitored for indications of photodamage.
Laser and Visual Stimuli
For the CRASM experiment, laser illumination (MGL-F-593.5, Changchun New Industries Optoelectronics Technology) was delivered (593.5 nm, 40 mW, 0.5 s ON, 3 s OFF for 9 trials) to V1 during two-photon imaging. Orange laser stimulation was triggered by scripts written in Psychopy (Peirce, 2007). Laser output was measured from the optic fiber tip, and maintained at no more than 60 mW during experiments to prevent cell damage. We chose the laser power that insured reliable and homogeneous activation. The gray screen in front of the mice served to reduce retinal sensitivity by light adaptation, which effectively eliminates retinal activation by laser stimulation (Danskin et al., 2015). Control experiments were conducted by imaging a population of V1 neurons during laser stimulation in animals either expressing bReaChES or lacking expression of bReaChES.
Visual stimuli were displayed on an LED monitor (29 cm × 51 cm, height × width) with a resolution of 1920 × 1080 pixels. The monitor was placed centrally in front of the animal at a distance of 21 cm from the eyes to cover about 100 degrees in azimuth and 70 degrees in elevation. The refresh rate of the monitor was 120 Hz, and the mean luminance for gray background was 54 cd/m2. Stimuli were generated using Psychopy (Peirce, 2007). Monocular orientation selectivity of somata and dendritic spines were assessed by presenting full-field square wave gratings (contrast 100%, spatial frequency (SF) 0.06 cycles per degree (CPD), and temporal frequency (TF) 2 Hz, stimulus duration 2 s, drifting in both directions in 4 different orientations (0–135°, spaced at 45°)) to each eye individually. Typically, 8 stimulus trials were presented along with blank stimulus trials (random order) with 1 s inter-stimulus intervals.
Fixation and Immunostaining
Upon completion of imaging, isoflurane was raised to 5% and 0.2 ml Euthasol was delivered IP. The animal was transcardially perfused with 20 ml of 0.9% NaCl in 0.1M phosphate buffer (PB) (w/v) and then 50 ml of 4% paraformaldehyde in 0.1 M PB. The brain was removed and placed in 4% PFAfor 2–3 hr. A vibratome (Leica VT1200S) was used to cut tangential sections (40–100 mm thick) of the imaged hemisphere. Tangential sections were cut parallel to the in vivo cranial window surface to aid in post hoc identification of dendrites previously imaged in vivo. Slices were rinsed once in PB and incubated in blocking buffer (2% normal goat serum, Jackson ImmunoResearch, Cat # 017–000-121,0.3M NaCl, 1% Bovine Serum Albumin, Sigma, Cat#A2153, 0.3% Triton X-100, in 0.1M PB). After washing three times in PB, slices were incubated for 10–12 hr in blocking buffer containing primary antibodies (Rat anti DYKDDDDK Tag at 1:500, Aligent Technologies 200474; Chicken anti GFP at 1:1000, Aves Lab, Inc GFP1020; Rabbit anti Homer at 1:500, Synaptic Systems 160 003; Rabbit anti RFP at 1:1000, Rockland Antibodies 600–401-379; mouse anti RFP at 1:1000, Abcam AB65856). After washing three times in PBS, slices were incubated for 8–10 hr in blocking buffer containing either 1:500 (Alexa 488 goat anti chicken Molecular Probes, ab1500169; Alexa 568 goat anti rat or goat anti mouse, Molecular Probes, A11077 and A11031) or 1:250 secondary antibodies (CF405M goat anti rabbit, Biotium 20181; CF633 goat anti mouse, Biotium 20121; CF647 goat anti rat, Biotium 20283). Slices were washed three times using 1X PBS, then acutely mounted to a slide using 0.1M PB for identifying previously imaged dendrites using a fluorescence microscope (Olympus BX53). Slices were either mounted to a slide using SlowFade Gold (Thermofisher Scientific) or continuously processed for tissue expansion.
In some cases, enhancement of the far red channel was needed post-digestion. After washing three times in 0.1M PBS, slices were treated with 0.3% H2O2 in PBS for 10 min followed by additional 0.1M PBS washes and blocking buffer containing 4 drops/ml of avidin (Avidin/Biotin blocking kit Vector Labs SP-2001) for 1 hr. After a brief rinse, 4 drops/ml of biotin were added to blocking buffer containing primary antibody rat anti DYKDDDDK Tag antibody at 1:500 for 10–12 hr. After washing briefly in PBS, slices were incubated for 2 hr in biotinylated goat anti rat at 1:1000, Vector Labs BA-9400. After polymerization and digestion, sections were washed three times in PBS followed by incubation in streptavidin Alexa 647 for 2 hr at room temperature.
Expansion Microscopy
10 ml of gelling solution was prepared by dissolving 33% (w/w) of N,N-dimethylacrylamide (DMAA) and sodium acrylate (SA) monomers at a molar ratio of 4:1 (DMAA:SA) in ddH2O. The solution was then bubbled for 20–40 min at room temperature with N2 to remove molecular oxygen from the solution and. 0.3–0.5 ml of the gelation solution was added to each sample in the 24-well tissue culture dish for 10 min while rocking. After exchanging the new solution, initiator potassium persulfate (KPS) at 0.4 molar% and 8 μl of N,N,N′,N′-tetramethyl-ethane-1,2-diamine (TEMED) relative was added to the gelation solution to enable the polymerization. Tissue with solution was quickly moved to the chamber made with 0.2 mm iSpacer (SunJin Lab) and coverslips at room temperature for at least 6 hr.
The polymerized gels were detached from the coverslip and placed into digestion buffer (50 mM Tris buffer, 0.8 M guanidinium chloride, 8 U/ml proteinase K, 0.5% Triton-X 100, pH 8.0). Digestion was carried out at 40–50°C in a humidified chamber for at least 10 h. The digested samples were then placed into 10-cm tissue culture dish (USA Scientific) filled with ddH2O for expansion. After replacement 3–4 times with 30–40 min per expansion step, final expansion of ~8× can be consistently achieved. The expansion factor for each experiment was determined by direct comparison of physical distances between blood vessels (averaged across 5 pairs) in pre-expansion and post-expansion images. This allowed the conventional confocal imaging to resolve a structure smaller than ~35 nm pre-expansion. Synaptic contacts were defined by the distance (<150 μm) between the half maximum point of presynaptic terminal signal (Flag/mCherry) and the peak of the postsynaptic density signal (Homer1) within a GFP labeled spine, measured along the axis orthogonal to the synaptic cleft.
Confocal Imaging
All expansion microscopy imaging was performed using a Leica SP5 Resonant confocal microscope with a Leica HCX PL APO 63×/1.3 NA objective or HC APO L 63×/0.90 long working distance (2.2 mm) objective. Images were acquired using 405, 488, 561, and 633 nm laser lines with emission channels optimized for each fluorophore (CF405, Alexa 488, Alexa 568, and CF647) while minimizing cross-talk between channels. Images were acquired at 256×256 or 512×512 with sampling resolution ranging from 0.15 to 0.3 μm per pixel. Z-stacks were acquired using 0.5 μm steps and the confocal pinhole was set to 1 AU. Confocal z-stacks were aligned to in vivo images using an affine transform applied in ImageJ. Colocalization was performed manually. The imaging data was analyzed using custom-written MATLAB (MathWorks, Natick) scripts, and ImageJ (NIH).
ANALYSIS
Images were corrected for in-plane motion using a correlation-based approach (MATLAB). ROI drawing was performed in ImageJ (Schindelin et al., 2012). For somata and spines, ROIs were circular or drawn manually. For dendrites, polygonal ROIs were drawn spanning the extent of a short, contiguous dendritic segment. Spine distances on dendritic segments were measured by reconstructing dendritic arbors using Simple Neurite Tracer (Schindelin et al., 2012). Fluorescence time-courses were computed as the mean of all pixels within the ROI at each time point and were extracted using Miji (Sage et al., 2012, ImageJ User & Developer Conference). Fluorescence time-courses were then synchronized with either laser or visual stimulation, and evoked responses were computed as changes in fluorescence relative to baseline fluorescence. Dendritic-spine fluorescence signals were sometimes contaminated by regenerative dendritic events. We used a subtraction procedure to isolate spine signals: (1) subtracting stimulus artifacts from the background, (2) performing a robust fit (MATLAB) of the spine signal against the dendritic signal for stimulus-evoked data and (3) subtracting a scaled version of the dendritic signal, where the scaling factor equals the slope from the robust fit (Wilson et al., 2016). Following subtraction, dendritic spines correlated with dendritic signals (r > 0.50) were excluded from analysis (17%).
In the CRASM experiment, callosal-recipient dendritic spines were identified as those reliably responding to laser stimulation on more than 80% of trials. Spines that never responded to laser stimulation were classified as non-callosal-recipient spines, and the remaining population of less-reliably activated spines were considered unreliably-activated spines. Orientation selectivity for individual neurons and spines was quantified by a previously reported method (Lee et al., 2016). For computing tuning properties, the fluorescence signal was calculated as ΔF/F=(F-F0)/F0, where F0 is the baseline fluorescence signal averaged over the 1 s period immediately before the start of visual stimulation and F is the fluorescence signal averaged over the first 1.5 s period after the start of the visual stimulation. Orientation tuning curves were obtained by calculating the mean fluorescence signal (ΔF/F) for each orientation, and then fitting a Gaussian curve to the resulting data. Dendritic spines were considered to be visually responsive if the maximum stimulus-related fluorescence response (ΔF/F) to any orientation was greater than 5% on average, and also greater than 2 standard deviations (SD) above the mean baseline fluorescence. In addition, we required that spines respond at least 2 SD above baseline on at least 20% of the trials tested. Spines were considered to be orientation selective if they were visually responsive and also met the following criteria: (1) well fit by the Gaussian function (r > 0.7, p < 0.05), and (2) tuning index (TI) > 0.2
where μpref denotes the mean response to the preferred orientation and μortho equals mean response to the orthogonal orientation. The preferred orientation and orthogonal orientation were calculated from Gaussian curve fitting. Cells and spines used for analysis in this paper were required to have orientation selective responses for individual eyes. Ocular dominance (OD) was defined as Responseipsi / (Responseipsi + Responsecontra).
To characterize the statistical significance of functional clustering, we compared the relationship between pair-wise spine distance and the difference in orientation preference. In the shuffled data, preferred orientation of spines was randomly permuted within the dendritic field of each cell. This procedure was repeated 10,000 times to derive the P values for each binned inter-spine distance. To provide more conservative statistics, the analysis for CRASM experiment was repeated by randomly permuting the preferred orientation of spines within different dendrites. Only dendritic branches with at least 4 orientation selective spines were included in the analysis. Functional clustering was defined as the dendritic distance when the experimentally derived Δorientation was less than that of the shuffled results.
We applied the same branch-level analysis previously described (Druckmann et al., 2014) to characterize branch-wise structural synaptic connectivity. For the purpose of our analysis on input homogeneity, we categorized the branches into separate primary basal dendritic branches directly connecting to the soma, and separate apical tuft dendritic branches from the main trunk. The experimentally derived number of synaptic contacts on individual branches was compared to the number generated by random distribution (100 repetitions), based on surface area of the relevant dendritic arbor. The fraction of branches significantly different from chance was calculated for both CRASM and anatomical methods.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed in MATLAB. We used two-sided non-parametric Wilcoxon ranked sum for paired results. A permutation test was applied to compare the experimentally derived data to shuffled data in the analysis for functional clustering. Correlations were calculated as Pearson’s correlation coefficient. Exact sample sizes are included in the text or figure legends. No estimates of statistical power were performed prior to experiments; animal numbers were minimized to conform to ethical guidelines while accurately measuring parameters of animal physiology. Statistical analyses were performed in MATLAB and significance was defined as p < 0.05.
DATA AND SOFTWARE AVAILABILITY
Data are available from the lead author upon reasonable request. Software Data are available from the lead author upon reasonable request.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti DYKDDDDK Tag (Flag) | Aligent Technologies | Cat # 200474; RRID: AB_10596510 |
| Chicken anti GFP | Aves Labs, Inc | Cat# GFP1020; RRID: AB_10000240 |
| Rabbit anti Homer1 | Synaptic Systems | Cat# 160 003; RRID: AB_887730 |
| Mouse anti RFP | Abcam | Cat# AB65856; RRID: AB_1141717 |
| Rabbit anti RFP | Rockland Antibodies | Cat# 600–401–379; RRID: AB_2209751 |
| Alexa 488 goat anti chicken | Abcam | Cat# ab1500169; RRID: AB_2636803 |
| CF405M goat anti rabbit | Biotium | Cat# 20181; RRID: AB_10561325 |
| Alexa 568 goat anti mouse | Molecular Probes | Cat# A11031; RRID: AB_144696 |
| Alexa 568 goat anti rat | Molecular Probes | Cat# A11077; RRID: AB_141874 |
| CF633 goat anti mouse | Biotium | Cat# 20121; RRID: AB_10854245 |
| CF647 goat anti rat | Biotium | Cat# 20283; RRID: AB_10852692 |
| Biotinylated goat anti rat | Vector Labs | Cat# BA-9400; RRID: AB_2336202 |
| Steptavidin Alexa 647 | Invitrogen | Cat#S21374; RRID: AB_2336066 |
| Alexa 568 donkey anti-rat | Abcam | Abcam cat # AB175475; RRID: AB_2636887 |
| Bacterial and Virus Strains | ||
| AAV2/1.hSyn.Cre | UPenn Vector Core Upenn | cat # AV-1-PV2676 |
| AAV2/1.CAG.FLEX.GcaMP6s | Upenn Vector Core Upenn | cat # AV-1-PV2818 |
| AAV1.Syn.GcaMP6s | Upenn Vector Core Upenn | cat # AV-1-PV2824 |
| AAV1.CAG.Ruby2sm-Flag | Upenn Vector Core Upenn | cat # AV-1-PV3509 |
| AAV8.2-hEF1a- synaptophysin-mCherry | Gene Delivery Technology Core | cat # AAV RN1 |
| AAV2.CamKIIa.bReaChES.TS.eYFP | UNC Virus Vector Core | (hSyn)Red Shifted Optical Excitation |
| Biological Samples | ||
| Mouse | Charles River | C57BL/6NCrl |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Avidin/biotin blocking kit | Vector Labs | Cat# SP2001; RRID: AB_2336231 |
| Sodium Acrylate | Sigma | Cat# 408220 |
| N,N’- dimethylacrylamide | Sigma | Cat# 274135 |
| Acrylamide | Sigma | Cat# A9099 |
| Potassium persulfate | Sigma | Cat# 216224 |
| N,N,N’, N’- tetramethyl-ethane-1,2-diamine (TEMED) | Sigma | Cat# T7024 |
| Acryloyl-X, SE, 6-((acryloyl)amino)hexanoic Acid, Succinimidyl Ester | Thermo Fisher | Cat# A20770 |
| Experimental Models: Organisms/Strains | ||
| Mouse | Charles River | C57BL/6NCrl |
| Software and Algorithms | ||
| Miji | Sage et al., 2012, ImageJ User & Developer Conference | https://imagej.net/Miji |
| PsychoPy | Peirce, 2007 | http://www.psychopy.org |
| FIJI/ImageJ | NIH | http://fiji.sc |
| MATLAB | MathWorks | https://ch.mathworks.com/products/matlab.html |
Highlights.
Callosal/non-callosal inputs with similar orientation preference cluster together
Correlative imaging supports callosal and intra-cortical functional clustering
Callosal inputs are biased toward specific dendritic branches
ACKNOWLEDGMENTS
We are grateful to Femtonics for assisting with the application of AOD microscopy, to Dominique Ouimet-Erlacher for providing surgical expertise and animal care, and to the Fitzpatrick lab and the Max Planck Florida Institute for Neuroscience for their support. We thank Jung Ho Hyun, Kanghoon Jung, Patrick Hannan, and Hyung-Bae Kwon for sharing their experience with in vivo optogenetic stimulation. This work was supported by grants from the National Institutes of Health (2R01 EY006821-28), The Celia Lipton Farris and Victor W. Farris Foundation, and the Max Planck Florida Institute for Neuroscience.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.neuron.2018.12.005.
REFERENCES
- Bloss EB, Cembrowski MS, Karsh B, Colonell J, Fetter RD, and Spruston N (2016). Structured Dendritic Inhibition Supports Branch- Selective Integration in CA1 Pyramidal Cells. Neuron 89, 1016–1030. [DOI] [PubMed] [Google Scholar]
- Branco T, Clark BA, and Häusser M (2010). Dendritic discrimination of temporal input sequences in cortical neurons. Science 329, 1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri C, Restani L, and Caleo M (2010). Callosal contribution to ocular dominance in rat primary visual cortex. Eur. J. Neurosci. 32, 1163–1169. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Ocazionez SA, Jungen C, Wunderle T, Eriksson D, Neuenschwander S, and Schmidt KE (2018). Callosal Influence on Visual Receptive Fields Has an Ocular, an Orientation-and Direction Bias. Front. Syst. Neurosci 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, and Zhuang X (2010). Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danskin B, Denman D, Valley M, Ollerenshaw D, Williams D, Groblewski P, Reid C, Olsen S, Blanche T, and Waters J (2015). Optogenetics in Mice Performing a Visual Discrimination Task: Measurement and Suppression of Retinal Activation and the Resulting Behavioral Artifact. PLoS ONE 10, e0144760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S, Feng L, Lee B, Yook C, Zhao T, Magee JC, and Kim J (2014). Structured synaptic connectivity between hippocampal regions. Neuron 81, 629–640. [DOI] [PubMed] [Google Scholar]
- Iacaruso MF, Gasler IT, and Hofer SB (2017). Synaptic organization of visual space in primary visual cortex. Nature 547, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti GM, and Frost DO (1979). Effects of visual experience on the maturation of the efferent system to the corpus callosum. Nature 280, 231–234. [DOI] [PubMed] [Google Scholar]
- Jia H, Rochefort NL, Chen X, and Konnerth A (2010). Dendritic organization of sensory input to cortical neurons in vivo. Nature 464, 1307–1312. [DOI] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, et al. (2015). Saturated Reconstruction of a Volume of Neocortex. Cell 162, 648–661. [DOI] [PubMed] [Google Scholar]
- Kleindienst T, Winnubst J, Roth-Alpermann C, Bonhoeffer T, and Lohmann C (2011). Activity-dependent clustering of functional synaptic inputs on developing hippocampal dendrites. Neuron 72, 1012–1024. [DOI] [PubMed] [Google Scholar]
- Larkum ME, and Nevian T (2008). Synaptic clustering by dendritic signalling mechanisms. Curr. Opin. Neurobiol 18, 321–331. [DOI] [PubMed] [Google Scholar]
- Lee WC, Bonin V, Reed M, Graham BJ, Hood G, Glattfelder K, and Reid RC (2016). Anatomy and function of an excitatory network in the visual cortex. Nature 532, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczy A, Makara JK, and Magee JC (2008). Compartmentalized dendritic plasticity and input feature storage in neurons. Nature 452, 436–441. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Hirano T, and Tagawa Y (2007). Evidence for activity-dependent cortical wiring: formation of interhemispheric connections in neonatal mouse visual cortex requires projection neuron activity. J. Neurosci 27, 6760–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Hirano T, and Tagawa Y (2010). Pre-synaptic and post-synaptic neuronal activity supports the axon development of callosal projection neurons during different post-natal periods in the mouse cerebral cortex. Eur. J. Neurosci 31, 410–424. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, and Yasuda R (2015). Biochemical Computation for Spine Structural Plasticity. Neuron 87, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olavarria J, Malach R, and Van Sluyters RC (1987). Development of visual callosal connections in neonatally enucleated rats. J. Comp. Neurol 260, 321–348. [DOI] [PubMed] [Google Scholar]
- Peiker C, Wunderle T, Eriksson D, Schmidt A, and Schmidt KE (2013). An updated midline rule: visual callosal connections anticipate shape and motion in ongoing activity across the hemispheres. J. Neurosci 33, 18036–18046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JW (2007). PsychoPy-Psychophysics software in Python. J. Neurosci. Methods 162, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, and Svoboda K (2007). Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci 10, 663–668. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, and Svoboda K (2003). ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng Online 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polsky A, Mel BW, and Schiller J (2004). Computational subunits in thin dendrites of pyramidal cells. Nat. Neurosci 7, 621–627. [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Sankaran S, Marshel JH, Kim CK, Ferenczi E, Lee SY, Berndt A, Ramakrishnan C, Jaffe A, Lo M, et al. (2015). Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restani L, Cerri C, Pietrasanta M, Gianfranceschi L, Maffei L, and Caleo M (2009). Functional masking of deprived eye responses by callosal input during ocular dominance plasticity. Neuron 64, 707–718. [DOI] [PubMed] [Google Scholar]
- Schindelin J, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KE, Lomber SG, and Innocenti GM (2010). Specificity of neuronal responses in primary visual cortex is modulated by interhemispheric corticocortical input. Cereb. Cortex 20, 2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl B, Wilson DE, and Fitzpatrick D (2017). Local Order within Global Disorder: Synaptic Architecture of Visual Space. Neuron 96, 1127–1138 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez R, Fenlon LR, Marek R, Avitan L, Sah P, Goodhill GJ, and Richards LJ (2014). Balanced interhemispheric cortical activity is required for correct targeting of the corpus callosum. Neuron 82, 1289–1298. [DOI] [PubMed] [Google Scholar]
- Szalay G, Judák L, Katona G, Ócsai K, Juhász G, Veress M, Szadai Z, Fehér A, Tompa T, Chiovini B, et al. (2016). Fast 3D Imaging of Spine, Dendritic, and Neuronal Assemblies in Behaving Animals. Neuron 92, 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kitamura K, Matsuo N, Mayford M, Kano M, Matsuki N, and Ikegaya Y (2012). Locally synchronized synaptic inputs. Science 335, 353–356. [DOI] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, et al. (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol 34, 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckenbrodt S, Maidorn M, Crzan D, Wildhagen H, Kabatas S, and Rizzoli SO (2018). X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Rep. 19, e45836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, Whitney DE, Scholl B, and Fitzpatrick D (2016). Orientation selectivity and the functional clustering of synaptic inputs in primary visual cortex. Nat. Neurosci 19, 1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnubst J, Cheyne JE, Niculescu D, and Lohmann C (2015). Spontaneous Activity Drives Local Synaptic Plasticity In Vivo. Neuron 87, 399–410. [DOI] [PubMed] [Google Scholar]
- Wunderle T, Eriksson D, Peiker C, and Schmidt KE (2015). Input and output gain modulation by the lateral interhemispheric network in early visual cortex. J. Neurosci 35, 7682–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Liu M, and Cang J (2013). Sublinear binocular integration preserves orientation selectivity in mouse visual cortex. Nat. Commun 4, 2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the lead author upon reasonable request. Software Data are available from the lead author upon reasonable request.