Abstract
Consumption of probiotics contributes to a healthy microbiome of the GIT leading to many health benefits. They also contribute to the modulation of the immune system and are becoming popular for the treatment of a number of immune and inflammatory diseases. The main objective of this study was to evaluate anti-inflammatory and modulatory properties of Streptococcus thermophilus. We used peripheral blood mononuclear cells from healthy donors and assessed modifications in the mRNA expression of their genes related to innate and adaptive immune system. Our results showed strong immune modulatory effects of S. thermophilus 285 to human peripheral blood mononuclear cells with an array of anti-inflammatory properties. S. thermophilus 285 reduced mRNA expression in a number of inflammatory immune mediators and markers, and upregulated a few of immune markers. S. thermophilus is used in the dairy industry, survives during cold storage, tolerates well upon ingesting, and their consumption may have beneficial effects with potential implications in inflammatory and autoimmune disorders.
1. Introduction
The human body and, in particular, the gastrointestinal tract (GIT) hosts a variety of microbial populations referred to collectively as the microbiome [1]. The microbiome of the GIT plays a key role in the maintenance of a healthy immune system [1, 2], and disruptions to the microbiome composition can lead to serious effects on health [3–5]. In order to maintain a healthy microbiome, regular ingestion of probiotic supplements, or the ingestion of fermented dairy products/capsules has been suggested. These practices have led to various improved health outcomes, ranging from enhanced overall human wellbeing to the treatment of infections, constipation, diarrhoea etc [1].
The majority of probiotics belong to the lactic acid bacteria (LAB) family; gram positive lactic acid producing microorganisms that include several genera such as bifidobacteria, lactobacilli streptococci and enterococci [1]. The small intestine and the colon are highly enriched with these microorganisms [6–8], which are routinely supplemented in foods as live strains due to their beneficial effects on human health [1, 2, 8–13]. Streptococcus species such as exopolysaccharide-producing strains of Streptococcus thermophilus (ST) [12, 14, 15] are among those consumed. These characteristics of S. thermophiles enable them to be used in fermented milk products (i.e. yogurt) including flavoring of dairy, and is recognized as the next most important species after Lactococcus lactis [16, 17]. ST and L. brevis synergistically display well established health benefits, and S. thermophilus is one of the bacteria in the VSL#3 probiotic mixture, which has long been broadly applied in the treatment of inflammatory conditions [18, 19]. In addition, probiotics interact with the immune system leading to immunomodulation and anti-inflammatory properties [4, 20, 21].
The ‘hygiene hypothesis’ suggests that the positive trend in the incidence of immune-related disorders can been attributed to intestinal dysbiosis, resulting in immune dysfunction (ie. asthma, eczema, allergies and autoimmune diseases). Use of probiotic bacteria can increase abundance and concurrently modulate immune cells, including B, T helper (Th)-1, Th-2, Th-17 and regulatory T (Treg) cells. This in turn, directly influences human health and modulates pathologies of immune/autoimmune diseases [1, 2, 13]. In fact, we previously noted that S. thermophilus 1342, S. thermophilus 1275 and S. thermophilus 285 modulate the U937 monocyte cell line. Specifically, we showed that interleukin (IL)-4, IL-10, GM-CSF and CXCL8 production were increased, and, cell surface marker expression CD11c, CD86, C206, CD209, MHC-1 were upregulated [1]. In another study, S. thermophilus 1275 and Bifidobacterium longum BL536 demonstrated increased levels of transforming growth factor (TGF)-beta (a key factor in the differentiation of Treg and T-helper Th)-17 cells by bulk peripheral blood mononuclear cell (PBMC) cultures [22]. Primary macrophages co-cultured with ST bacteria stimulate production of anti-inflammatory IL-10 and pro-inflammatory IL-12 cytokines [23].
Peripheral blood mononuclear cells (PBMC) isolated from whole blood constitute a wide range of diverse immune cells that play vital roles in balancing immune homeostasis and keeping human health in check [24, 25]. These cells are crucial components of the innate and adaptive immune system, defend the body against bacterial, viral and parasitic infections, as well as destroying foreign antigens and cancer cells [25]. PBMC are predominantly made up of lymphocytes (~70–90%), monocytes (~10–20%) and other cells such as dendritic cells comprise less than 1–2% [26]. In spite of variations in the fraction of subtypes of immune cells within the total PBMC isolated from different samples [26], isolation, characterization and molecular studies of these cells have benefited medical research [27].
Herein, we describe changes in the expression of genes associated with innate and adaptive immunity including cytokines, chemokines and immune cell marker expression by human PBMC following exposure to live S. thermophilus 285 bacteria.
2. Material and methods
2.1. Bacterial strains
Pure bacterial cultures of S. thermophilus 285 were obtained from Victoria University culture collection (Werribee, VIC, Australia). Stock cultures were stored in cryobeads at −80°C. Prior to each experiment the cultures were propagated in M17 broth (Oxoid, Denmark) with 20 g/L lactose and incubated at 37°C under aerobic conditions. Bacteria were also cultured in M17 agar (1.5% w/v agar) with 20 g/L lactose (Oxoid, Denmark), to assess characteristics, morphology, purity and gram-positive confirmation [1].
2.2. Preparation of live bacterial suspensions
Media were prepared and autoclaved at 121°C for 15 minutes (mins) prior to experiments. Bacterial cultures were grown 3 times in M17 broth with 20 g/L lactose, at 37°C aerobically for 18 hours (hr) with a 1% inoculum transfer rate [28]. Cultures grow optimally at 37–42°C for 24 hrs [15]. The growth period of cultures were consistent at 18 hr (at the end of the exponential growth phase) and before stationary growth phase to prevent cell lysis. Bacteria were harvested during stationary growth phase on the day of experiment, centrifuged (6000×g) for 15 min at 4°C, followed by two washes with Dulbecco’s phosphate-buffered saline (DPBS) (Invitrogen, Pty Ltd. Australia) and resuspended in the Roswell Park Memorial Institute (RPMI) 1640 culture media. These samples constituted the live-cell suspensions.
2.3. Enumeration of bacterial cells
Bacterial strains were scraped from M17 agar and transferred into Dulbecco’s PBS (Invitrogen, Pty Ltd. Australia) adjusted to a final concentration of 108 colony forming units (cfu)/ml by measuring the optical density at 600 nm, and washed two times with PBS and resuspended in RPMI 1640 prior to co-culturing with PBMC [1].
2.4. Isolation, culture, and stimulation of PBMC
2.4.1. Isolation of PBMC using Ficoll-Paque
PBMC isolation from whole blood was via Ficoll-Paque density gradient centrifugation [9]. Three buffy coats were collected from the Australian Red Cross Blood Bank on the day of experiment (Victoria University human research ethics). Calcium and magnesium free PBS, pH7.2, (Invitrogen, Pty Ltd. Australia) was used after adding 2 mM EDTA and 2% heat-inactivated fetal bovine serum (FBS) (Invitrogen, Pty Ltd. Australia); PBS buffer. SEPMATE tubes (50 ml) with inner inserts (STEMCELL technology, Canada) were used to isolate PBMC following Ficoll-Paque density gradient protocol [29, 30]. PBMCs were washed, counted and the required number of PBMC were co-cultured with S. thermophilus 285 and the remaining PBMC were stored in freeze mix and transferred into liquid nitrogen for future use.
2.4.2. Stimulation of PBMC with S. thermophilus 285
PBMC (3x 107 cells) were resuspended in RPMI 1640 media supplemented with 10% heat-inactivated FBS (Invitrogen, Pty Ltd. Australia), 1% antibiotic-antimycotic solution and 2 mM L-glutamine in cell culture flasks, and 3x108 S. thermophilus 285 bacteria were added. PBMC with RPMI media without the addition of ST285 bacteria were used as a control and incubated at 37°C, 5% CO2 for 24 hrs [1]. We previously demonstrated that 24 hrs co-culture was optimal for stimulation of U937 monocyte/macrophage cell line, and all incubations described herein were for 24 hrs [1]. PBMCs were snap frozen post incubation and stored at -80°C prior to RNA extraction.
2.5. RNA extraction from PBMC
Total RNA was extracted from stimulated PBMCs using the RNeasy® mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, cells were centrifuged and harvested, supernatants were removed and RNA extracted from each cell pellet and resuspended in lysis buffer supplemented with β-mercaptoethanol to disrupt the cells. PBMC were lysed and each cell lysate passed through the supplied Qia-shredder columns to homogenize and was subsequently mixed with equal volume of 70% ethanol. Cell lysates were transferred onto RNeasy mini-spin columns and DNA was removed using DNase digestion/ treatment using RNase-Free DNase Set (Qiagen, Hilden, Germany.) The RNA Integrity Number (RIN) of all RNA samples were measured using an Agilent 2100 Bioanalyzer and Agilent RNA 6000 nano kit (Agilent Technologies, Santa Clara, CA, USA); with a minimum RIN of 7.5 used as the criterion for inclusion in gene expression analysis. The concentration of each individual RNA sample was measured using a Qubit RNA BR Assay (Invitrogen) in triplicate. Several blood samples were collected for PBMC isolation, treatment and extraction of RNA and only RNA samples with the highest RIN numbers (all above 8) were included for PCR.
2.6. Assessing changes in the expression of genes associated with innate and adaptive immunity
Aliquots of each RNA sample were reverse-transcribed to make complementary DNA (cDNA) using RT2 first strand kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the ‘Human Innate and Adaptive immune Response’ kit (Qiagen, Hilden, Germany) to evaluate gene/mRNA expression. The relative expression profiles of treated PBMC samples were analyzed in comparison with untreated PBMC cultured in RPMI using Thermo-cycler (Biorad, Melbourne Australia). The RT2 qPCR Primer innate and adaptive immune response arrays target a set of 84 innate and adaptive immune-related genes and five housekeeping genes, an RT control, a positive PCR control, and a human genomic DNA contamination control. The levels of the expression of these genes were calculated using the Qiagen web-based software (Qiagen, Germany) and then calculated the fold changes and analyzed data manually to compare results. Differential expression (up and down regulation) of the genes were identified using the criteria of a > 2.0-fold increase/decrease in gene expression in treated PBMCs in comparison with those genes in control PBMC cultures.
2.7. Data analysis
The Delta-Delta CT (ΔΔCT) was used to calculate fold-changes [31]. Fold-regulation represents fold-change results in a biologically meaningful way. In our RT2 profiler PCR array results, fold-change values greater than one, indicate a positive (or an up-) regulation, in fact in upregulated genes, the fold-regulation is equal to the fold-change. Fold-change values less than one specifies a negative (or a down) regulation, and in this case, the fold-regulation is the negative inverse of the fold-change [32–34]. Data related to changes in the expression of the genes were analyzed by ΔΔCT method using Qiagen RT2 profiler data analysis webportal that utilises the delta delta CT method in determining fold-changes. The raw CT values were uploaded to the Qiagen data analysis webportal with the lower limit of detection set for 35 cycles and 3 internal controls: PCR array reproducibility, RT efficiency and genomic DNA contamination were assessed to ensure all arrays successfully passed all of these control checks. Normalization of the raw data was performed using the included housekeeping genes (HKG) panel. Then using the ΔΔCT method, both housekeeping gene references and untreated/ controls were assessed to calculate relative expression of mRNA.
2.8. Statistical analysis
The p values are calculated based on a Student’s t-test of the Triplicate 2^ (- Delta CT) [(2^-ΔCT)] values for each gene in the treatment group vs. the control group [32, 33, 35, 36].
3. Results
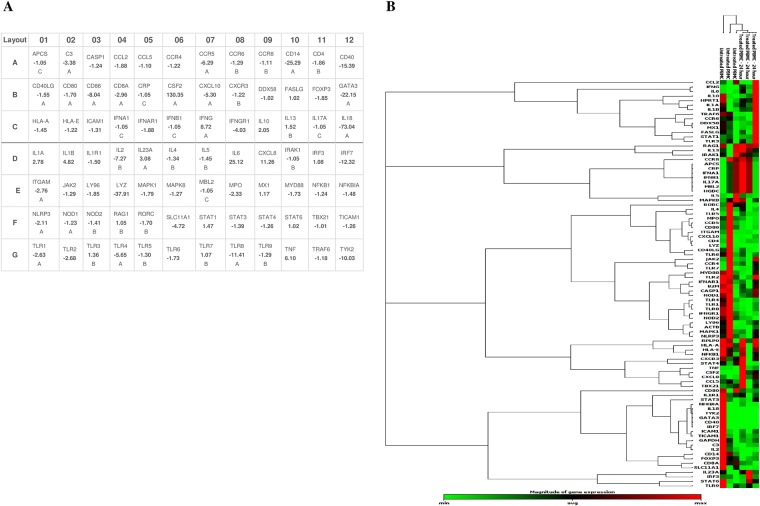
Among 84 genes assessed, 31 genes were significantly altered > 2.0 fold up/down in PBMC samples (n = 3) following exposure to S. thermophilus 285 compared to control PBMC (Fig 1, Table in S1 Table and S1 Fig).
Fig 1. Effects of co-culturing S. thermophilus 285 with PBMCs (n = 3) on gene/RNA expression compared to control PBMCs after 24 hrs.
(A) All 84 genes are shown including those with significant high up/down regulated genes (more than 2-fold) and those with no significant change (less than 2-fold). The housekeeping genes (HKG) panel and other genes used for normalization of the raw data are not presented. Letter A specifies the gene’s average threshold cycle to be reasonably high (> 30) in either the treated samples or the controls and relatively low (< 30) in the other/opposite sample. Thus, in case of presenting fold changes with letter A, the estimate fold change may be an underestimate. Letter B suggests a reasonably high (> 30) gene’s average threshold cycle that means a low level of average expression of relevant gene, in both test/treated samples and untreated control samples, and the p-value for the fold-change might be either relatively high (p > 0.05). Thus, in case of presenting fold changes with letter B, the estimate fold change may be slightly overestimate or unavailable. Letter C indicates that that gene’s average threshold cycle is either not determined or greater than the defined default 35 cut-off value, in both test/treated samples and control samples, suggesting that its expression was not detectable, resulting in the fold-change values being un-interpretable [86, 87] [88]. (B) Presentation of data as a hierarchical clusters of average gene/RNA expressions of PBMC (n = 3) co-cultured with S. thermophilus 285, compared to control. Green represents down regulated genes to red represents upregulated genes.
3.1. S. thermophilus 285 alters cytokine gene expression levels of PBMC
3.1.1. Interleukin mRNA expression levels
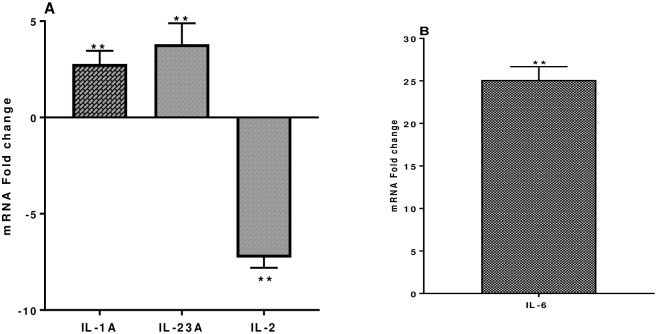
IL-1α and IL-6 are secreted by dendritic cells (DC), B cells and macrophages (MQ) are involved in acute phase responses, B cell maturation, macrophage differentiation, promote Th2 differentiation and inhibit Th1 polarization. IL-1α is upregulated 2.78 ± 0.6 fold and IL-6 25.12 ± 0.61 fold (Fig 2). IL- 23α is secreted by CD4+ T cells and aids in the stimulation of Th17 cells together with IL-6. IL-23α is highly upregulated 3.8 ± 1.0 fold (Fig 2). IL-2 has an array of functions it activates T cell proliferation and increases or decreases inflammatory responses. IL-2 is downregulated 7.27 ± 0.53 fold (Fig 2). IL-17A a pro-inflammatory cytokine secreted by Th17 cells, was not altered following PBMC co-cultured with S. thermophilus 285.
Fig 2. (A) IL-1α, IL-23α and IL-2 and (B) IL-6, mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where ** p < 0.04.
3.1.2. Th1/Th2 mRNA expression levels
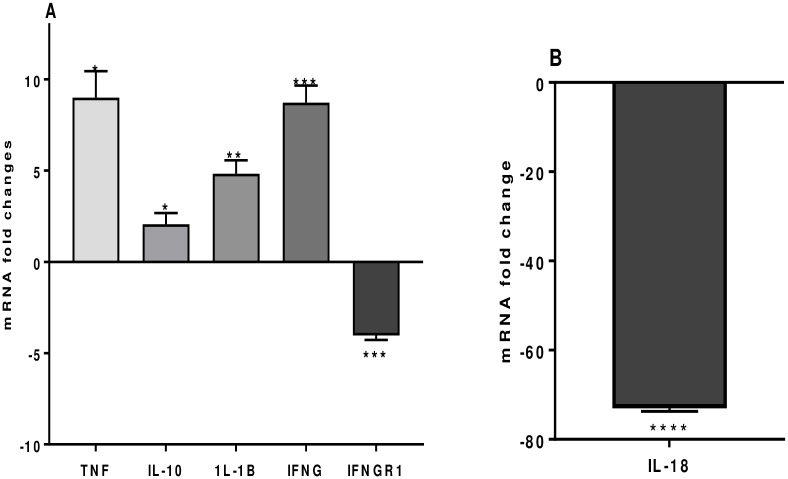
IFNγ, a Th1 cytokine important in the defense against bacterial infection is upregulated 8.73 ± 0.94 fold. Likewise, the Th1 cytokine IL-1β is upregulated 4.82 ± 0.74 fold (Fig 3). Of interest, IL-18 a Th1 inducing pro-inflammatory cytokine was vastly downregulated (75 ± 0.66 fold), in addition, IFNγR1, a transmembrane protein which interacts with IFNγ, is also downregulated 4.03 ± 0.25 fold (Fig 3). Tumor-necrosis factor-alpha (TNFα), important in the defense against bacterial infections, and in acute phase reactions is upregulated 6.10 ± 1.4 fold (Fig 3). IL-10, an anti-inflammatory cytokine secreted by Th2 and Treg cells is upregulated 2.05 ± 0.52 fold (Fig 3). Gene expressions of other cytokines, IFNB1, IL-4, IL-5 and IL-13 are not significantly altered.
Fig 3. A) TNF-α, IL-10, IL-1β, IFN-γ, and IFN- γ–R and (B) IL-18, mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
(The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where * p < 0.05, ** p < 0.04, *** p < 0.02 and **** p < 0.01.
3.2. S. thermophilus 285 alters chemokine gene expression levels of PBMC
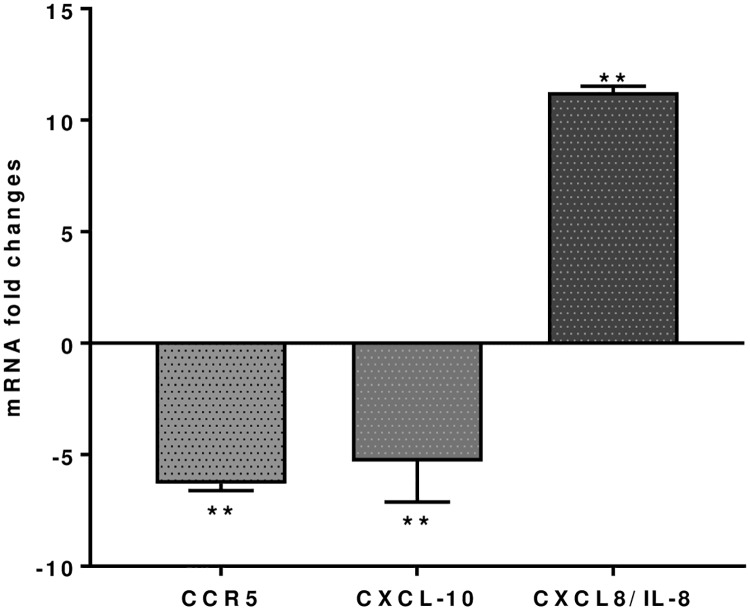
Chemokine (CXCL8, IL-8) is important in the innate immune system, it stimulates chemotaxis and is upregulated 11.26 ± 0.27 fold following S. thermophilus 285 co-culture with PBMC cells. However, CCR5 and CXCL10 (INP10) are down regulated 6.29 ± 0.32 and 5.30 ±1.8 fold respectively (Fig 4). No significant differences are noted for gene expressions of other chemokines, including CCL2 (MCP-1), CCL5 (RANTES), CCL8, CCR4, CCR8, CXCR3, CCL2, IFNA1.
Fig 4. CCR5, CXCL10 and CXCL8 (IL-8), mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where ** p < 0.04.
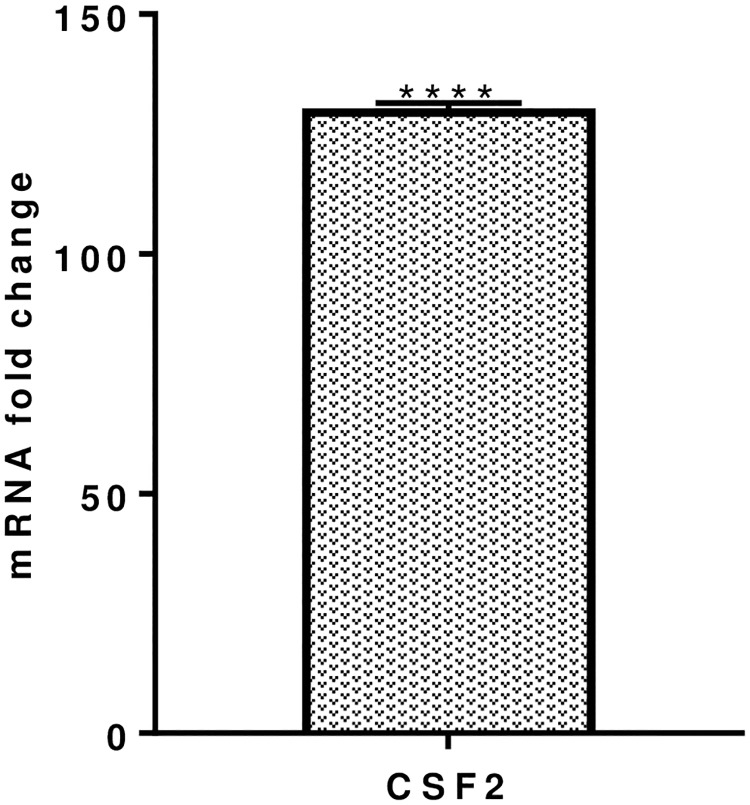
3.3. Colony stimulating factor mRNA expression levels
Colony-stimulating factor (CSF)-2, secreted by MQs, NK cells and T cells, enables cell proliferation and differentiation and is significantly increased by 130.35 ± 1.0 fold (Fig 5) after co-culturing PBMC with S. thermophilus 285 bacteria.
Fig 5. CSF-2, mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where **** p < 0.01.
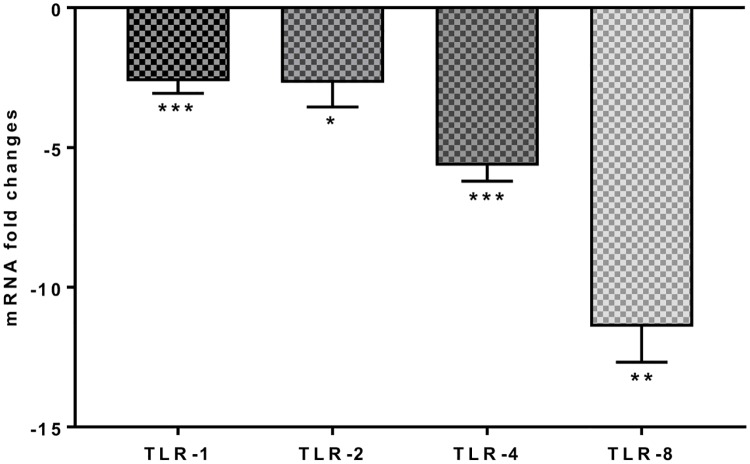
3.4. S. thermophilus 285 alters Toll like receptor gene expression levels of PBMC
TLR (toll like receptor)-1, TLR-2, TLR-4 and TLR-8 are part of the innate immune response and involved in the defense response to bacteria. PBMC co-cultured with S. thermophilus 285 induced downregulation of TLRs at varying levels; TLR-1 (-2.63 ± 0.43), TLR-2 (-2.69 ± 0.8 fold), TLR-4 (-5.65 ± 0.56 fold), TLR-8 (-11.41 ± 1.27 fold) (Fig 6). However, changes to other pattern recognition receptors such as, TLR-3, TLR-5, TLR-6, TLR-9 were not significant.
Fig 6. TLR-1, TLR-2, TLR-4 and TLR-8, mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where * p < 0.05, ** p < 0.04 and *** p < 0.02.
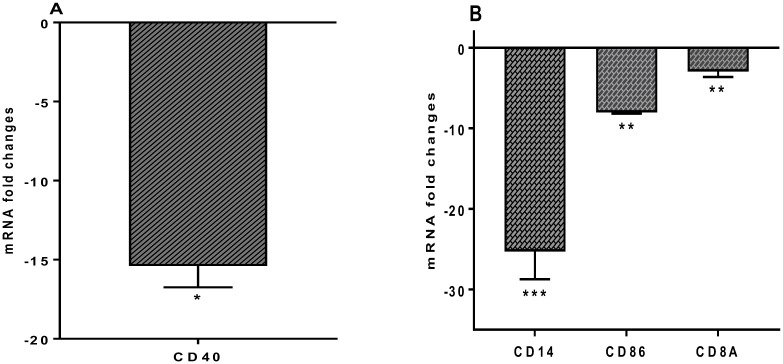
3.5. Cell surface markers CD14, CD40, CD86 mRNA expression levels
Expression of the monocyte cell surface markers CD14, CD40 and CD86 significantly downregulated -25.29 ± 3.46, -15.39 ± 1.36, -8.04 ± 0.14 fold, respectively (Fig 7). Expression of the CD8A gene, which is involved in adaptive immunity and in response to defense against viruses, was downregulated by -2.96 ± 0.68 fold (Fig 7). Expression of CD4, CD80, FOXP3, STAT3, CD40LG (TNFSF5), HLA-A, HLA-E and RORC genes do not show significant changes.
Fig 7. (A) CD40 and (B) CD14, CD86 and CD8A, mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where * p < 0.05, ** p < 0.04 and *** p < 0.02.
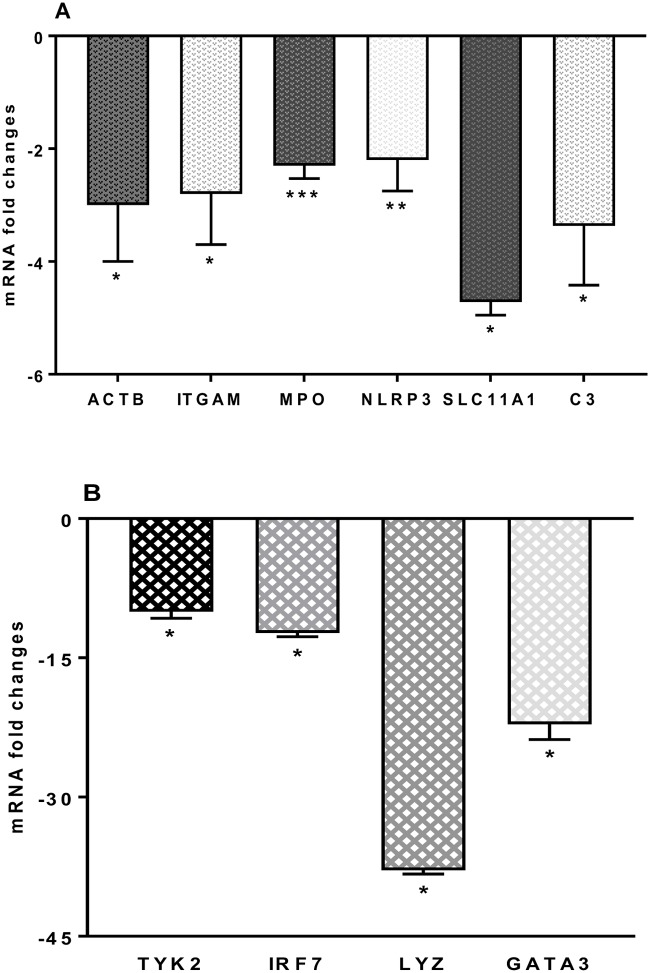
3.6. Changes to other innate and adaptive molecules, mRNA expression levels
Changes to other genes were also noted following S. thermophilus 285 co-culture with PBMC. ACTB (-3.01 ± 1.0) fold, ITGAM (-2.76 ± 0.9) were both downregulated. Downregulated genes were noted to the following: MPO (2.33 ± 0.2), NLRP3 (2.11 ± 0.6), SLC11A1 (4.72 ± 0.23) and complement component (C)-3 (3.38 ± 1.5), TYK2 (10.03 ± 0.7), IRF7 (12.32 ± 0.4), LYZ (37.91 ± 0.4) and GATA3 (22.15 ± 1.64) (Fig 8). Other immune markers including FASLG (TNFSF6), CRP, IFNAR1, JAK2, IL-1R1, MAPK8 (JNK1), IRF3, MBL2, NFKB1, MX1, ICAM1, MBL2, MYD88, NOD1 (CARD4), NOD2, DDX58 (RIG-I), RAG1 and TICAM1 (TRIF) showed no significant mRNA gene changes in the levels of their expression.
Fig 8. (A) ACTB, CCR5, ITGAM, MPO, NLRP3, SLC11A1, and C3 and (B) TYK2, IRF7, LYZ and GATA3, mRNA fold change following 24 h co-culture of S. thermophilus 285 with PBMCs (n = 3), compared to control PBMC.
The innate and adaptive RT2 gene profiler arrays were used to determine changes in gene expression. Symbols represent p value for Tukey Test (One way ANOVA) where * p < 0.05, ** p < 0.04 and *** p < 0.02.
4. Discussion
Our previous publications illustrated immune modulatory effects of S. thermophilus 285, S. thermophilus 1275 and S. thermophilus 1342 on U937 monocytic cell line and human monocytes by using secreted cytokines for bioplex assays, as well as flow cytometry of immune cell surface marker changes. The current study, aimed to get a more comprehensive overview of the data, by undertaking an in depth gene array analysis of the effects of probiotics to human PBMC.
4.1. S. thermophilus 285 promotes Th2 polarization
IL-1α secreted by peripheral blood DC and B cells induces Th2 differentiation and inhibits Th1 polarization [37], is significantly upregulated. Similarly, Enterococcus faecium NCIMB 10415 was shown to upregulate IL-1α in porcine jejunal epithelial cells (IPEC-J2) in vitro, [38]. IL-6 produced by Th2 cells is increased in the presence of S. thermophilus 285 by PBMC which was also shown previously to be upregulated by pro-monocyte cell line U937 [1]. Others have shown that PBMC co-cultured with S. thermophilus 1275 also increases IL-6 [39]. Likewise, mixed probiotics of S. thermophilus, Lactobacillus (L.) rhamnosus, L. casei, L. acidophilus, B. longum and B. bifidum stimulated PBMC to produce IL-6 [40, 41]. Our study shows that IL-1α and IL-6 are increased, highlighting the role of S. thermophilus 285 in stimulation of immune responses involved in acute phase; B cell maturation, macrophage differentiation, promotion of Th2 differentiation and inhibiting Th1 polarization.
IL-10 is an anti-inflammatory cytokine secreted by Th2 and Treg cells and co-culture of S. thermophilus 285 with PBMC increased expression of IL-10. Cultured PBMC with other live S. thermophilus strain (S. thermophilus 1275) also showed increased IL-10 [22, 39, 42–45]. Similarly, in a study using mixed probiotic cultures (S. thermophilus, L. rhamnosus, L. casei, L. acidophilus, B. longum and B. bifidum) high levels of IL-10 were stimulated by PBMC [40]. Conversely, in a study using B. breve and S. thermophilus combined to stimulate PBMC, IL-10 was only increased in the presence of B. breve, whereas exposing PBMC to S. thermophilus reduced the IL-10 level [46]. We also previously noted that monocyte cell line (U937), co-cultured with S. thermophilus 1342 stimulated production of high levels of IL-10 [1].
IL-18 is involved in the initiation of severe inflammatory responses, indicating the role of IL-18 in inflammatory and autoimmune disorders. Co-culture of PBMC with S. thermophilus 285 significantly downregulated IL-18 which indicates an anti-inflammatory role for S. thermophilus 285 bacteria. Likewise, a mixture of Lactobacilli species (L. rhamnosus, L. paracasei, and L. plantarum) was shown to supress the secretion of pro-inflammatory IL-18 gene by undifferentiated IPEC-1 intestinal porcine epithelial cell line [47], highlighting supportive role of lactobacilli probiotics in functioning against inflammation and suppression of immune response activities. However, other studies with other probiotics such as, L. rhamnosus E509, L. rhamnosus GG E522 (ATCC 53103), L. bulgaricus E585 and S. pyogenes serotype T1IH32030, increased IL-18 production by human PBMC [48]. Hence, different probiotic strains induce different cytokine profiles.
IL-2 is involved in signalling of immune responses and activates proliferation of lymphocytes. We note downregulation of IL-2 gene expression in PBMC after exposure to ST285. IL-23 known to activate Th17 cells was upregulated although IL-17, the key pro-inflammatory cytokine secreted by Th17 cells was not altered. Upregulation of IL-1α, IL-6, IL-10, and downregulation of IL-2, IL-18 and an absence of change in IL-17A (despite increase in IL-23α) designates ST285 to possess anti-inflammatory effects on human PBMC.
4.2 S. thermophilus 285 stimulates expression of cytokines involved in the defence against bacteria
IFN-γ is an adaptive immunity cytokine secreted by Th1 cells in the defense response to microbes and viruses. IFN-γ is predominantly secreted by NK, NKT cells as part of the innate immune response, and by CD4 Th1 and CD8+ T cells of the adaptive immune response [49]. S. thermophilus 285 upregulated IFN-γ gene expression by human PBMCs. This is similar to studies of a combination of probiotic strains including S. thermophilus, Lactobacillus, Bifidobacterium, Propionibacterium, E. coli and Leuconostoc [50], where upregulation of IFN-γ mRNA expression by PBMC was noted [50]. Likewise, co-cultures of pooled PBMC with ST1275 also induced upregulation of IFN-γ [39]. We previously noted that monocyte cell line co-cultured with S. thermophilus 1342, S. thermophilus 1275 or S. thermophilus 285 strains induced high levels of IFN-γ secretion [1]. In a study with Lactobacilli (L. rhamnosus E509, L. rhamnosus GG E522 (ATCC 53103) and L. bulgaricus E585), and streptococci (S. pyogenesserotype T1 IH32030), IFN-γ was produced by human PBMC [48].
IL-1β secretion by monocytes is involved in regulating immune and inflammatory responses to bacterial infections and injury, hence its role in innate immunity [51]. IL-1β is upregulated by S. thermophilus 285 co-cultured with PBMC, which is in accord with studies of PBMC co-cultured with mixed probiotics (S. thermophilus, L. rhamnosus, L. casei, L. acidophilus, B. longum and B. bifidum) [40]. We previously noted in monocyte cell line co-cultured with three different strains of S. thermophilus, only S. thermophilus 1342 stimulated production of high levels of IL-1β whereas, S. thermophilus 1275 and S. thermophilus 285 did not induce IL-1β cytokine [1]. A mixture of Lactobacilli strains (L. rhamnosus, L. paracasei, and L. plantarum) co-cultured with intestinal porcine epithelial cell line (IPEC-1) also upregulated IL-1β gene expression [47]. Similarly, the combination of L. casei Shirota, L. rhamnosus GG, L. plantarum NCIMB 8826 and L. reuteri NCIMB 11951, B. bifidum MF 20/5 and B. longum SP 07/3 co-cultured with PBMC, significantly augmented IL-1β production [41].
TNFα plays a key role in the defense against bacterial infections. It is a pro-inflammatory cytokine, which also supports recruitment and activation of T and B cells to promote an adaptive immune response. We previously demonstrated high levels of TNFα secretion by U937 monocyte cell line in the presence of S. thermophilus 1342, S. thermophilus 1275 and S. thermophilus 285 [1]. Likewise, our current findings show that ST285 co-cultured with PBMC results in upregulation of TNFα. However, in a study using B. breve and ST together to stimulate PBMC, TNF-α secretion was inhibited [46]. In addition, a mixture of strains of probiotics (L. casei Shirota, L. rhamnosus GG, L. plantarum NCIMB 8826 and L. reuteri NCIMB 11951, B. bifidum MF 20/5 and B. longum SP 07/3) co-cultured with PBMC, significantly increased the production of TNFα [41]. In another study of human PBMCs exposed to different probiotics (L. mesenteroides ssp. cremoris PIA2 (DSM 18892) S. pyogenes serotype T1M1, S. thermophilus THS, E. coli (DH5α), L. rhamnosus Lc705 (DSM 7061), L. lactis ssp. cremoris ARH74 (DSM 18891), L. rhamnosus GG (ATCC 53103), L. helveticus Lb 161, L. helveticus 1129, B. longum 1/10, B. breve Bb99 (DSM 13692), B. animalis ssp. lactis Bb12, and Propionibacterium (P.) freudenreichii ssp. shermanii JS (DSM 7067)), all induced TNF-α mRNA expression [50]. Given that IFNγ, IL-1β and TNFα are upregulated by PBMC following co-culture with S. thermophilus 285 this suggests that S. thermophilus 285 induces powerful defense against invading pathogens and could be beneficial against virus infection and tumours.
The upregulation of IFNγ, IL-1β and TNFα coupled with a significant decrease in IFNγ receptor and IL-18 shows an antagonising effect of S. thermophilus 285 inflammatory responses and leading to an overall anti-inflammatory profile.
4.3. S. thermophilus 285 activates mRNA expression of CXCL8 and downregulates CCR5 and CXCL10
IL-8, also known as CXCL8 is an important chemokine of the innate immune system, involved in the recrutiment of neutrophils and other granulocytes as the first line of defense [52]. S. thermophilus 1342, S. thermophilus 1275 and S. thermophilus 285 were previously shown to activate U937 monocyte cell line to produce high levels of IL-8 [1]. The probiotic L. paracasei DG also increases expression of IL-8 to the human monocyte cell line, THP-1 [53]. Likewise, short chain fatty acids, produced by probiotic bacteria, also stimulate IL-8 secretion and mRNA levels to the human epithelial cell line HT-29 [11]. These studies are in accord to our current findings that S. thermophilus 285 upregulates CXCL8 production by human PBMC.
C-C chemokine receptor type 5 (CCR5, CD195) is involved in Th1 immune responses and its gene expression is downregulated by PBMC following S. thermophilus 285 co-culture. However, in mice prolonged feeding with VSL#3 probiotic mixture shows significant gene upregulation of CCR5 [54]. Differences could be attributed to one probiotic strain applied and varying effects of the strain (S. thermophilus) used in current study versus a mixture of different strains and species used in mice VSL#3 (L. delbruekii Bulgaricus, L. casei, L. plantarum, L. acidophilus, B. breve, B. longum, B. infantis and S. thermophilus).
CXC motif chemokine 10 (CXCL10), or IFN-γ-induced protein-10 (IP-10), is secreted by a number of cell types (endothelial cells, monocytes and fibroblasts). Few roles have been ascribed to CXCL10 including chemo-attraction of NK cells, monocytes/macrophages, T cells and DCs, favouring adhesion of T cells to endothelial cells, anti-cancer/tumour action, and preventing angiogenesis and bone marrow colony development. CXCL10 is downregulated in PBMC culture following S. thermophilus 285 exposure. Conversely, monocyte-derived DCs co-cultured with B. breve Bb99, L. lactics subsp. cremoris ARH74 and S. thermophilus THS increased expression of CXCL10 and S. thermophilus was the most efficient probiotic in the induction of CXCL10 [23]. Additionally, microarray results of the intestines of mice prolonged administrated with VSL#3 probiotic mixture in healthy mice showed differential effects on intestinal immune parameters, including upregulation of CXC10 which contrasts with our findings [54]. The difference are most likely due to cell types, as well as bacterial strains in our study (PBMC co-cultured with S. thermophilus 285 bacteria) compared to using mouse cells exposed to three strains (B. breve Bb99, L. lactics subsp. cremoris ARH74 and S. thermophilus THS) in the other study. Also in the latter experiments, it is quite predictable to observe different results in mice intestine administered with VSL#3 due to different cells involved in mice study in contrast to PBMC cell population.
In summary, increased expression of IL-8 on its own could singularly be indicative of inflammation, but in the context of all other upregulated anti-inflammatory cytokine and mediators found in this study, this may not be interpreted as an inflammatory effect. IL-8 upregulation might also be interpreted as requirement for the initial stimulatory effect of S. thermophilus 285 to switch on the immune responses by initiating innate immunity, which by the progress of immune response, expression of CCR5 (which in turn influences Th1 immune responses), as well as CXCL10 (induced by IFNγ) are reduced by S. thermophilus 285. This might be suggestive of modulation of immune responses by S. thermophilus 285 to keep the adaptive immune responses in check.
4.4. S. thermophilus 285 significantly upregulates mRNA expression level of colony stimulating factor
CSF (GM-CSF) is secreted by machrophages, NK cells and T cells, enables cell proliferation and differentiation, stimulates the production of various immune cells, in particular it increases the production of machrophages which are important in fighting againts infections. CSF-2, is vastly increased (130 fold) by PBMC co-cultured with S. thermophilus 285 which is in alignment to our previous data whereby S. thermophilus 1275, S. thermophilus 1342 and S. thermophilus 285 induced U937 monocyte cell line to secrete high levels of GM-CSF with ST285 being the highest inducer [1]. Likewise, another study used RT2 Profiler PCR Arrays for mouse cytokines and chemokines to demonstrate that L. rhamnosus GR-1 (GR-1) induced high levels of granulocyte CSF (G-CSF) mRNA (60-fold) to bone marrow-derived mouse macrophages [55]. Likewise, PBMC co-cultured with B. infantis 52486 significantly increases GM-CSF [56].
GM-CSF is generally accepted as an inflammatory cytokine, its inflammatory activity is primarily associated with its role as granulocytes and macrophages growth and differentiation factor. GM-CSF-mediated inflammation has also been associated with certain types of autoimmune diseases such as rheumatoid arthritis and multiple sclerosis. However, in many instances GM-CSF plays anti-inflammatory/regulatory roles; GM-CSF can modulate differentiation of DC to render them into tolerogenic DCs, which, can stimulate anti-inflammatory Treg cells [57]. In addition, either of pro-inflammatory or regulatory effects of GM-CSF appears to be dependent on the amount of CSF and the presence of other relevant cytokines in the context of an immune response. There is also evidence that G-CSF induces expansion of IL-10-producing cells [58]. Our results show very high overexpression of CSF, which might be suggestive of anti-inflammatory effect of S. thermophilus 285 on PBMC.
4.5. S. thermophilus 285 downregulates mRNA expression levels of toll-like receptors
Toll-like receptors (TLRs) recognize pathogen-associated molecular patterns (PAMPs) that are expressed on infectious bacteria and mediate the production of cytokines necessary for the development of effective immunity [59]. TLRs recognize pathogens and activate the innate immune responses. TLR-1, TLR-2, TLR-4 and TLR-8 are part of the innate immune response and are involved in defense against bacteria. Co-culturing S. thermophilus 285 with human PBMC downregulated the expression of TLR. Similarly, E. coli K88 and mycotoxin zearalenone (ZEA) infection of IPEC-1 epithelial cell line was protected in the presence of mixed Lactobacillus strains (L. acidofilus ID11692, L. plantarum ID1253 and L. paracasei ID13239) by downregulating TLR-1, TLR-2 and TLR-4 gene expression [60].
TLRs are critical in bacterial recognition and host defence, such as lipo-teichionic acid (LTA) and lipo-polysaccharide (LPS) from Gram-positive and Gram-negative bacteria respectively [61, 62]. Activation of some of these molecules and mediators like TLR (especially TLR-2 and TLR-4) arbitrates to pro-inflammatory actions and further defensive functions of innate immunity [63–65]. The TLR-2 and TLR-4 activation and expression by LPS (pathogens) is known as one of the most important mechanisms by which the immune system controls reactions to bacteria in particular in the activation phase, therefore, over-expression of TLR-2 and TLR-4 during any bacterial infection could cause an elevated inflammatory response in the body. While early activation of TLRs expression is reported in response to bacterial LPS from pathogenic Salmonella typhimurium [61] as well as E. coli infection in bovine intestinal epithelial cells [66], our results show tolerance as a result of co-culturing PBMC with S. thermophilus 285 by down regulation of TLRs genes.
Downregulated mRNA expression of TLRs genes, specifically TLR-1, TLR-2, TLR-4 and TLR-8 indicates anti-inflammatory characteristics for S. thermophilus 285. Given that TLR-1, TLR-2, TLR-4 and TLR-8 are members of the innate immune response and play key roles in the defense against bacteria, downregulation of TLRs could be suggestive of a protective mechanism to keep S. thermophilus 285 safe by tolerance towards S. thermophilus 285. Perhaps designing experiments that allow different incubation period, as well as adding pathogenic bacteria to the co-cultured S. thermophilus 285-PBMC can help to illustrate if lesser co-culture time and/or presence of pathogens can result in a shift towards upregulation of TLRs instead.
4.6. S. thermophilus 285 downregulates cell surface markers CD14, CD40, CD86
CD14, CD40 CD86 are expressed on the cell surface of monocytes, macrophages and DC. CD14 is expressed on the surface of monocytes and primarily binds to bacterial constituents [67–69]. We previously showed that U937 monocyte cell line exposed to S. thermophilus 1342, S. thermophilus 1275 or S. thermophilus 285 enhanced expression of CD14 after 24 and 48 hrs, and S. thermophilus 285 was the most potent at 48 hrs [1]. However, in bulk PBMC cultures, CD14 expression was significantly downregulated in the presence of S. thermophilus 285, which is in accordance with downregulation of TRLs in particular TRL-4. In other studies, the combination of 3 probiotics (L. acidophilus, L. delbrueckii ssp. bulgaricus and B. bifidum) stimulated increased expression of cell surface markers, CD14, CD80 and MHC class II [1]. E. coli Nissle 1917, widely used as a probiotic for the treatment of inflammatory bowel disorders, expresses a K5 capsule important in E. coli mediating interactions with intestinal epithelial cells and chemokine expression. E. coli Nissle 1917 has been shown to induce mRNA expression of CD14 by intestinal Caco-2 cells [70].
CD40 is a costimulatory protein on antigen presenting cells and is essential for their activation. CD40 is a key mediator in a wide range of inflammatory and immune responses and its gene expression was downregulated by PBMC in the presence of S. thermophilus 285. In previous experiments with U937 monocyte cell line, co-culture with S. thermophilus 1342, S. thermophilus 1275 or S. thermophilus 285, resulted in small increase in CD40 [1].
CD86 (B7-2) is expressed on APCs and delivers co-stimulatory signals required for the activation and survival of T cells. CD86 plays the role of the ligand for T cells external CD28, and CTLA-4 (CD28) in regulation and cell to cell dis-association. CD86 acts in conjunction with CD80 to prime Th cells, delivering opposing functions on Treg cells through CTLA-4 and T cell surface CD28 protein. Expression of CD86 by PBMC is downregulated significantly, suggesting an anti-inflammatory profile following exposure to S. thermophilus 285. S. thermophilus bacteria promote CD86 expression required for T cell activation and the maintenance of immune responses, CD86 downregulation by S. thermophilus 285 suggests a regulating and damping effect of S. thermophilus 285 on PBMC, being interpreted as immunomodulation of adaptive immunity [71]. We previously noted using U937 monocyte cell line in the presence of S. thermophilus 1342, S. thermophilus 1275 and S. thermophilus ST285 increased expression of CD86 [1]. Similarly, L. plantarum WCFS1 and L. fermentum GR1485 upregulate CD86 on monocytes, conversely, L. rhamnosus and L. delbruekii reduced its expression [72].
Additionally, monocytes isolated from PBMC and differentiated into immature DCs by GM-CSF and IL-4, and co-cultured with B. breve Bb99, L. lactis subsp. cremoris ARH74 and S. thermophilus THS also increase CD86 expression [23]. Another study used bone marrow-derived DCs from DQ8-transgenic mice and co-culture with L. plantarum and L. paracasei and B. lactis increases CD86 differentially with the highest CD86 being noted in co-administration of L. plantarum and L. paracasei [73]. The contrast between these studies to the findings herein could be due to the differences in the nature of studies; we co-cultured PBMC with S. thermophilus 285 bacteria only and the other studies used mouse bone marrow-derived DCs co-cultured with three different probiotics leading to predictable differences.
Given the downregulation of cell surface markers and their roles in immunity, CD14 (involved in innate immunity), CD40 (involved in innate immunity), and CD86 (T cell activation), following S. thermophilus 285 co-culture is suggestive of an anti-inflammatory anti-activation profile for S. thermophilus 285. In addition, as all these cell surface markers are interlinked with defence against bacteria either through innate or adaptive immune responses, downregulation of these markers could be suggestive of S. thermophilus 285 initiating self-tolerance via regulating immune responses, which in turn modulates the immune responses too.
4.7. S. thermophilus 285 differentially downregulates mRNA expression level of other innate and adaptive immune response markers and chemokines
Complement component 3 (C3) is associated with complement cascades in immune responses by enhancing antibody function, phagocytosis and stimulation of inflammation [74–76]. GATA3 transcriptome is also important in both humoral immunity and inflammatory responses. Downregulation of C3 gene expression and significant reduced expression of GATA3 transcriptome by PBMC co-cultured with S. thermophilus 285 in noted. Similarly, lipoteichoic acid (p-LTA) extracted from L. plantarum K8 inhibits C3 mRNA in vitro and in vivo. In human clinical studies, blocking GATA3 is able to control allergy responses, inflammatory diseases and asthma [77]. C3 and GATA3 downregulation suggests that S. thermophilus 285 is able to lower inflammation (C3), as well as being a viable candidate for further pre-clinical and clinical studies for the management of such diseases.
Interferon regulatory factor (IRF) 7, integrin alpha M (ITGAM), Lysozyme (LYZ) and NALP3 are other innate immune response factors. IRF7, a member of IRF family and present on monocytes, macrophages, granulocytes, and NK cells, and expressed predominantly in macrophages (a component of the inflammasome) [78]. IRF7 plays a role in the transcriptional activation of virus-inducible cellular genes, including the type I interferon genes. ITGAM is involved in a number of inflammatory responses (i.e. cell-mediated cytotoxicity, phagocytosis, and chemotaxis). LYZ acts as an antimicrobial enzyme present in neutrophils and macrophages. IRF7 gene regulation decreased considerably along with ITGAM gene expression, which is downregulated when PBMC are co-cultured with S. thermophilus 285. NALP3 and LYZ are downregulated markedly in co-culture of PBMC with S. thermophilus 285. However, in a previous study, we showed significant upregulation of CD11b (ITGAM) by monocytic U937 cells when co-cultured with S. thermophilus 1342, S. thermophilus 1275 and S. thermophilus 285 bacteria [1]. S. thermophilus 285-induced downregulation of IRF7, ITGAM, NALP3 and LYZ in PBMC, suggestive of an anti-inflammatory effect of S. thermophilus 285 on PBMC as well.
Non-receptor tyrosine-protein kinase (TYK2) is an enzyme [7] that contributes to adaptive immune responses due to its implication in IFNα, IL-6, IL-10 and IL-12 signaling, also involved in transducing signals of IL-6, IL-10 and IL-23. TYK2 gene expression is significantly downregulated in PBMC co-cultured with S. thermophilus 285, supporting an anti-inflammatory profile for S. thermophilus 285. In addition, myeloperoxidase (MPO), an enzyme abundantly expressed by neutrophils and promotes inflammation, is also involved in autoimmune disorders (multiple sclerosis, rheumatoid arthritis) [79, 80]. A decreased expression of MPO has been suggested to manage these autoimmune disorders by decreasing the inflammatory state. S. thermophilus 285 co-cultured with PBMC decreased the expression of MPO, suggestive of an anti-inflammatory benefit of S. thermophilus 285.
IFNAR1 is a type I membrane protein which is a receptor for IFNα and IFNβ involved in defence against viruses. IFNAR1 signaling is associated with pro-inflammatory cytokine production [81]. In fact, IFNAR1 knockout mice show decreased pro-inflammatory cytokiens and chemokines [81]. IFNAR1 is significantly downregulated by PBMC following co-culture with ST285, supporting an anti-inflammatory role of S. thermophilus 285. In addition, SLC11A1 involved in T cell activation, is involved in inflammatory disorders such as autoimmune type 1 diabetes [82, 83], is downregulated by PBMC in the presence of S. thermophilus 285. Furthermore, the Beta-actin (ACTB) which stimulates eNOS and increase nitric oxide (NO) [84] involved in immunity and inflammation [85], is downregulated by PBMC following co-culture with S. thermophilus 285.
We determined the immune modulatory effects of S. thermophilus 285 to human PBMC and show that it has an array of anti-inflammatory immune-modulatory properties. S. thermophilus 285 decreases mRNA expression IL-18, IFNγR1, CCR5, CXCL10, TLR-1, TLR-2, TLR-4, TLR-8, CD14, CD40, CD86, C3, GATA3, ITGAM, IRF7, NLP3, LYZ, TYK2, IFNR1, and upregulates IL-1α, IL-1β, IL-6, IL-8, IL-10, IL-23, IFNγ, TNFα, CSF-2. No changes to mRNA expression are noted with IFNA1, IFNB1, IL-4, IL-5, IL-13, CCL2, CCL5, CCL8, CCR4, CCR8, CXCR3, TLR-3, TLR-5, TLR-6, TLR-9, CD4, CD80, FOXP3, STAT3, CD40LG, HLA-A, HLA-E, RORC. The data demonstrates a predominant anti-inflammatory profile exhibited by S. thermophilus 285, and further work is required to determine its effects in inflammatory disease models in vitro and in vivo, such as multiple sclerosis, inflammatory bowel disease and allergies. Future investigations using RNA-Seq and Western blots are some of the next logical steps to confirm and further investigate these results.
5. Conclusion
Probiotics are beneficial microorganism with immunomodulatory properties, which aid the maintenance of a healthy immune system. S. thermophilus is often used in fermented dairy products such as cheeses and yogurts and is believed to potentially have health benefits. We determined the immune modulatory effects of S. thermophilus 285 to human peripheral blood mononuclear cells and show that it has an array of anti-inflammatory immune-modulatory properties. S. thermophilus 285 decreases mRNA expression IL-18, IFN receptor, CCR5, CXCL10, TLR-1, TLR-2, TLR-4, TLR-8, CD14, CD40, CD86, C3, GATA3, ITGAM, IRF7, NLP3, LYZ, TYK2, IFNR1, and upregulates IL-1α, IL-1β, IL-6, IL-8, IL-10, IL-23, IFN-γ, TNF-α, CSF-2. No changes to mRNA expression were noted with IFNA1, IFNB1, IL-4, IL-5, IL-13, CCL2, CCL5, CCL8, CCR4, CCR8, CXCR3, TLR-3, TLR-5, TLR-6, TLR-9, CD4, CD80, FOXP3, STAT3, CD40LG, HLA-A, HLA-E, RORC.
Supporting information
(DOCX)
Green represents down regulated genes to red represents upregulated genes.
(DOCX)
Acknowledgments
All authors were supported by VU Research and in particular, Institute for Health and Sport and the Institute for Sustainable Industries and Liveable Cities at Victoria University.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Dargahi N, Johnson J, Donkor O, Vasiljevic T, Apostolopoulos V. Immunomodulatory effects of Streptococcus thermophilus on U937 monocyte cell cultures. Journal of Functional Foods. 2018;49:241–9. [Google Scholar]
- 2.Dargahi N, Katsara M, Tselios T, Androutsou ME, De Courten M, Matsoukas J, et al. Multiple sclerosis: Immunopathology and treatment update. Brain Sciences. 2017;7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen H, Drømtorp SM, Axelsson L, Grimmer S. Immunomodulation of Monocytes by Probiotic and Selected Lactic Acid Bacteria. Probiotics and Antimicrobial Proteins. 2015;7(1):14–23. 10.1007/s12602-014-9174-2 [DOI] [PubMed] [Google Scholar]
- 4.Stagg J, Divisekera U, Duret H, Sparwasser T, Teng MWL, Darcy PK, et al. CD73-deficient mice have increased antitumor immunity and are resistant to experimental metastasis. Cancer Research. 2011;71(8):2892–900. 10.1158/0008-5472.CAN-10-4246 [DOI] [PubMed] [Google Scholar]
- 5.Stagg AJ, Hart AL, Knight SC, Kamm MA. Interactions between dendritic cells and bacteria in the regulation of intestinal immunity. Best Practice and Research: Clinical Gastroenterology. 2004;18(2):255–70. 10.1016/j.bpg.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 6.Michałkiewicz J, Krotkiewski M, Gackowska L, Wyszomirska-Gołda M, Helmin-Basa A, Dzierzanowska D, et al. Immunomodulatory Effects of Lactic Acid Bacteria on Human Peripheral Blood Mononuclear Cells. Microbial Ecology in Health and Disease. 2003;15(4):185–92. [Google Scholar]
- 7.Maassen CBM, Van Holten-Neelen C, Balk F, Heijne Den Bak-Glashouwer MJ, Leer RJ, Laman JD, et al. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine. 2000;18(23):2613–23. 10.1016/s0264-410x(99)00378-3 [DOI] [PubMed] [Google Scholar]
- 8.Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frøkiær H, Ferlazzo G. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. International Immunology. 2007;19(12):1319–27. 10.1093/intimm/dxm103 [DOI] [PubMed] [Google Scholar]
- 9.Asarat M, Apostolopoulos V, Vasiljevic T, Donkor O. Short-chain fatty acids produced by synbiotic mixtures in skim milk differentially regulate proliferation and cytokine production in peripheral blood mononuclear cells. International Journal of Food Sciences and Nutrition. 2015;66(7):755–65. 10.3109/09637486.2015.1088935 [DOI] [PubMed] [Google Scholar]
- 10.Asarat M, Apostolopoulos V, Vasiljevic T, Donkor O. Short-chain fatty acids regulate cytokines and Th17/treg cells in human peripheral blood mononuclear cells in vitro. Immunological Investigations. 2016;45(3):205–22. 10.3109/08820139.2015.1122613 [DOI] [PubMed] [Google Scholar]
- 11.Asarat M, Vasiljevic T, Apostolopoulos V, Donkor O. Short-Chain Fatty Acids Regulate Secretion of IL-8 from Human Intestinal Epithelial Cell Lines in vitro. Immunological Investigations. 2015;44(7):678–93. 10.3109/08820139.2015.1085389 [DOI] [PubMed] [Google Scholar]
- 12.Salazar N, Prieto A, Leal JA, Mayo B, Bada-Gancedo JC, de los Reyes-Gavilán CG, et al. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. Journal of Dairy Science. 2009;92(9):4158–68. 10.3168/jds.2009-2126 [DOI] [PubMed] [Google Scholar]
- 13.Dargahi N, Johnson J, Donkor O, Vasiljevic T, Apostolopoulos V. Immunomodulatory effects of probiotics: Can they be used to treat allergies and autoimmune diseases? Maturitas. 2019;119:25–38. 10.1016/j.maturitas.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 14.Di Caro S, Tao H, Grillo A, Elia C, Gasbarrini G, Sepulveda AR, et al. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Digestive and Liver Disease. 2005;37(5):320–9. 10.1016/j.dld.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 15.Purwandari U, Vasiljevic T. Rheological properties of fermented milk produced by a single exopolysaccharide producing Streptococcus thermophilus strain in the presence of added calcium and sucrose. International Journal of Dairy Technology. 2009;62(3):411–21. [Google Scholar]
- 16.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiology Reviews. 2005;29(3 SPEC. ISS.):435–63. [DOI] [PubMed] [Google Scholar]
- 17.Uriot O, Denis S, Junjua M, Roussel Y, Dary-Mourot A, Blanquet-Diot S. Streptococcus thermophilus: From yogurt starter to a new promising probiotic candidate? Journal of Functional Foods. 2017;37:74–89. [Google Scholar]
- 18.Mennigen R, Nolte K, Rijcken E, Utech M, Loeffler B, Senninger N, et al. Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. American Journal of Physiology—Gastrointestinal and Liver Physiology. 2009;296(5):G1140–G9. 10.1152/ajpgi.90534.2008 [DOI] [PubMed] [Google Scholar]
- 19.Dai C, Zheng CQ, Meng FJ, Zhou Z, Sang LX, Jiang M. VSL#3 probiotics exerts the anti-inflammatory activity via PI3k/Akt and NF-κB pathway in rat model of DSS-induced colitis. Molecular and Cellular Biochemistry. 2013;374(1–2):1–11. 10.1007/s11010-012-1488-3 [DOI] [PubMed] [Google Scholar]
- 20.Han GK, Kim NR, Min GG, Jung ML, Seung YL, Mi YK, et al. Lipoteichoic acid isolated from Lactobacillus plantarum inhibits lipopolysaccharide-induced TNF-α production in THP-1 cells and endotoxin shock in mice. Journal of Immunology. 2008;180(4):2553–61. [DOI] [PubMed] [Google Scholar]
- 21.Vliagoftis H, Kouranos VD, Betsi GI, Falagas ME. Probiotics for the treatment of allergic rhinitis and asthma: systematic review of randomized controlled trials. Annals of Allergy, Asthma & Immunology. 2008;101(6):570–9. [DOI] [PubMed] [Google Scholar]
- 22.Donkor ON, Ravikumar M, Proudfoot O, Day SL, Apostolopoulos V, Paukovics G, et al. Cytokine profile and induction of T helper type 17 and regulatory T cells by human peripheral mononuclear cells after microbial exposure. Clinical and Experimental Immunology. 2012;167(2):282–95. 10.1111/j.1365-2249.2011.04496.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latvala S, Pietilä TE, Veckman V, Kekkonen RA, Tynkkynen S, Korpela R, et al. Potentially probiotic bacteria induce efficient maturation but differential cytokine production in human monocyte-derived dendritic cells. World Journal of Gastroenterology. 2008;14(36):5570–83. 10.3748/wjg.14.5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaczorowski KJ, Shekhar K, Nkulikiyimfura D, Dekker CL, Maecker H, Davis MM, et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(30):E6097–E106. 10.1073/pnas.1705065114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green AC, Rudolph-Stringer V, Chantry AD, Wu JY, Purton LE. Mesenchymal lineage cells and their importance in B lymphocyte niches. Bone. 2019;119:42–56. 10.1016/j.bone.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito S, Shiozaki A, Nakashima A, Sakai M, Sasaki Y. The role of the immune system in preeclampsia. Molecular Aspects of Medicine. 2007;28(2):192–209. 10.1016/j.mam.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 27.Corkum CP, Ings DP, Burgess C, Karwowska S, Kroll W, Michalak TI. Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT™) and standard density gradient. BMC Immunology. 2015;16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husson-Kao C, Mengaud J, Cesselin B, Van Sinderen D, Benbadis L, Chapot-Chartier MP. The Streptococcus thermophilus autolytic phenotype results from a leaky prophage. Applied and Environmental Microbiology. 2000;66(2):558–65. 10.1128/aem.66.2.558-565.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betsou F, Gaignaux A, Ammerlaan W, Norris PJ, Stone M. Biospecimen Science of Blood for Peripheral Blood Mononuclear Cell (PBMC) Functional Applications. Current Pathobiology Reports. 2019;7(2):17–27. [Google Scholar]
- 30.Grievink HW, Luisman T, Kluft C, Moerland M, Malone KE. Comparison of Three Isolation Techniques for Human Peripheral Blood Mononuclear Cells: Cell Recovery and Viability, Population Composition, and Cell Functionality. Biopreservation and Biobanking. 2016;14(5):410–5. 10.1089/bio.2015.0104 [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Zhang T, Xiu H-H, Liu J-X, Ma Y, Xu K-Q, Huang W-Q. Protective effect of aspirin-triggered resolvin D1 on hepatic ischemia/reperfusion injury in rats: The role of miR-146b. International Immunopharmacology. 2017;51:140–7. 10.1016/j.intimp.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 33.Yang Z, Zhong L, Zhong S, Xian R, Yuan B. miR-203 protects microglia mediated brain injury by regulating inflammatory responses via feedback to MyD88 in ischemia. Molecular Immunology. 2015;65(2):293–301. 10.1016/j.molimm.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 34.Souza BM, Preisser TM, Pereira VB, Zurita-Turk M, Castro CP, Cunha VP, et al. Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microbial Cell Factories. 2016;15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.dos Santos GG, Reinders J, Ouwehand K, Rustemeyer T, Scheper RJ, Gibbs S. Progress on the development of human in vitro dendritic cell based assays for assessment of the sensitizing potential of a compound. Toxicology and Applied Pharmacology. 2009;236(3):372–82. 10.1016/j.taap.2009.02.004 [DOI] [PubMed] [Google Scholar]
- 36.Biasin V, Wygrecka M, Marsh LM, Becker-Pauly C, Brcic L, Ghanim B, et al. Meprin β contributes to collagen deposition in lung fibrosis. Scientific Reports. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, Shapira I, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7119–24. 10.1073/pnas.0902745106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kern M, Günzel D, Aschenbach JR, Tedin K, Bondzio A, Lodemann U. Altered Cytokine Expression and Barrier Properties after In Vitro Infection of Porcine Epithelial Cells with Enterotoxigenic Escherichia coli and Probiotic Enterococcus faecium. Mediators of Inflammation. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donkor ON, Henriksson A, Vasiljevic T, Shah NP. Proteolytic activity of dairy lactic acid bacteria and probiotics as determinant of growth and in vitro angiotensin-converting enzyme inhibitory activity in fermented milk. Lait. 2007;87(1):21–38. [Google Scholar]
- 40.Djaldetti M, Bessler H. Probiotic strains modulate cytokine production and the immune interplay between human peripheral blood mononucear cells and colon cancer cells. FEMS Microbiology Letters. 2017;364(3). [DOI] [PubMed] [Google Scholar]
- 41.Dong H, Rowland I, Yaqoob P. Comparative effects of six probiotic strains on immune function in vitro. British Journal of Nutrition. 2012;108(3):459–70. 10.1017/S0007114511005824 [DOI] [PubMed] [Google Scholar]
- 42.Donkor ON, Henriksson A, Vasiljevic T, Shah NP. Effect of acidification on the activity of probiotics in yoghurt during cold storage. International Dairy Journal. 2006;16(10):1181–9. [Google Scholar]
- 43.Donkor ON, Shah NP, Apostolopoulos V, Vasiljevic T. Development of allergic responses related to microorganisms exposure in early life. International Dairy Journal. 2010;20(6):373–85. [Google Scholar]
- 44.Donkor ON, Stojanovska L, Ginn P, Ashton J, Vasiljevic T. Germinated grains—Sources of bioactive compounds. Food Chemistry. 2012;135(3):950–9. 10.1016/j.foodchem.2012.05.058 [DOI] [PubMed] [Google Scholar]
- 45.Donkor ON, Tsangalis D, Shah NP. Viability of probiotic bacteria and concentrations of organic acids in commercial yoghurts during refrigerated storage. Food Australia. 2007;59(4):121–6. [Google Scholar]
- 46.Ménard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53(6):821–8. 10.1136/gut.2003.026252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taranu I, Marin DE, Braicu C, Pistol GC, Sorescu I, Pruteanu LL, et al. In vitro transcriptome response to a mixture of lactobacilli strains in intestinal porcine epithelial cell line. International Journal of Molecular Sciences. 2018;19(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miettinen M, Matikainen S, Vuopio-Varkila J, Pirhonen J, Varkila K, Kurimoto M, et al. Lactobacilli and streptococci induce interleukin-12 (IL-12), IL-18, and gamma interferon production in human peripheral blood mononuclear cells. Infection and Immunity. 1998;66(12):6058–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoenborn JR, Wilson CB. Regulation of Interferon-γ During Innate and Adaptive Immune Responses. Advances in Immunology 2007. p. 41–101. 10.1016/S0065-2776(07)96002-2 [DOI] [PubMed] [Google Scholar]
- 50.Kekkonen RA, Kajasto E, Miettinen M, Veckman V, Korpela R, Julkunen I. Probiotic Leuconostoc mesenteroides ssp. cremoris and Streptococcus thermophilus induce IL-12 and IFN-γ production. World Journal of Gastroenterology. 2008;14(8):1192–203. 10.3748/wjg.14.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1beta secretion. Cytokine & growth factor reviews. 2011;22(4):189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baggiolini M, Clark-Lewis I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Letters. 1992;307(1):97–101. 10.1016/0014-5793(92)80909-z [DOI] [PubMed] [Google Scholar]
- 53.Balzaretti S, Taverniti V, Guglielmetti S, Fiore W, Minuzzo M, Ngo HN, et al. A novel rhamnose-rich hetero-exopolysaccharide isolated from Lactobacillus paracasei DG activates THP-1 human monocytic cells. Applied and Environmental Microbiology. 2017;83(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mariman R, Tielen F, Koning F, Nagelkerken L. The probiotic mixture VSL#3 has differential effects on intestinal immune parameters in healthy female BALB/c and C57BL/6 mice. Journal of Nutrition. 2015;145(6):1354–61. 10.3945/jn.114.199729 [DOI] [PubMed] [Google Scholar]
- 55.Meshkibaf S, Fritz J, Gottschalk M, Kim SO. Preferential production of G-CSF by a protein-like Lactobacillus rhamnosus GR-1 secretory factor through activating TLR2-dependent signaling events without activation of JNKs. BMC Microbiology. 2015;15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.You J, Yaqoob P. Evidence of immunomodulatory effects of a novel probiotic, Bifidobacterium longum bv. infantis CCUG 52486. FEMS Immunology and Medical Microbiology. 2012;66(3):353–62. 10.1111/j.1574-695X.2012.01014.x [DOI] [PubMed] [Google Scholar]
- 57.Bhattacharya P, Budnick I, Singh M, Thiruppathi M, Alharshawi K, Elshabrawy H, et al. Dual Role of GM-CSF as a Pro-Inflammatory and a Regulatory Cytokine: Implications for Immune Therapy. Journal of Interferon and Cytokine Research. 2015;35(8):585–99. 10.1089/jir.2014.0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malashchenko VV, Meniailo ME, Shmarov VA, Gazatova ND, Melashchenko OB, Goncharov AG, et al. Direct anti-inflammatory effects of granulocyte colony-stimulating factor (G-CSF) on activation and functional properties of human T cell subpopulations in vitro. Cellular Immunology. 2018;325:23–32. 10.1016/j.cellimm.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 59.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nature Immunology. 2010;11(5):373–84. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 60.Taranu I, Marin DE, Pistol GC, Motiu M, Pelinescu D. Induction of pro-inflammatory gene expression by Escherichia coli and mycotoxin zearalenone contamination and protection by a Lactobacillus mixture in porcine IPEC-1 cells. Toxicon. 2015;97:53–63. 10.1016/j.toxicon.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 61.Arce C, Ramírez-Boo M, Lucena C, Garrido JJ. Innate immune activation of swine intestinal epithelial cell lines (IPEC-J2 and IPI-2I) in response to LPS from Salmonella typhimurium. Comparative Immunology, Microbiology and Infectious Diseases. 2010;33(2):161–74. 10.1016/j.cimid.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 62.Kajikawa A, Nordone SK, Zhang L, Stoeker LL, LaVoy AS, Klaenhammer TR, et al. Dissimilar properties of two recombinant Lactobacillus acidophilus strains displaying Salmonella FliC with different anchoring motifs. Applied and Environmental Microbiology. 2011;77(18):6587–96. 10.1128/AEM.05153-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugitharini V, Shahana P, Prema A, Berla Thangam E. TLR2 and TLR4 co-activation utilizes distinct signaling pathways for the production of Th1/Th2/Th17 cytokines in neonatal immune cells. Cytokine. 2016;85:191–200. 10.1016/j.cyto.2016.06.024 [DOI] [PubMed] [Google Scholar]
- 64.Islam MA, Pröll M, Hölker M, Tholen E, Tesfaye D, Looft C, et al. Alveolar macrophage phagocytic activity is enhanced with LPS priming, and combined stimulation of LPS and lipoteichoic acid synergistically induce pro-inflammatory cytokines in pigs. Innate Immunity. 2013;19(6):631–43. 10.1177/1753425913477166 [DOI] [PubMed] [Google Scholar]
- 65.Sugitharini V, Pavani K, Prema A, Berla Thangam E. TLR-mediated inflammatory response to neonatal pathogens and co-infection in neonatal immune cells. Cytokine. 2014;69(2):211–7. 10.1016/j.cyto.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 66.Takanashi N, Tomosada Y, Villena J, Murata K, Takahashi T, Chiba E, et al. Advanced application of bovine intestinal epithelial cell line for evaluating regulatory effect of lactobacilli against heat-killed enterotoxigenic Escherichia coli-mediated inflammation. BMC Microbiology. 2013;13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bron PA, Tomita S, Mercenier A, Kleerebezem M. Cell surface-associated compounds of probiotic lactobacilli sustain the strain-specificity dogma. Current Opinion in Microbiology. 2013;16(3):262–9. 10.1016/j.mib.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 68.van Baarlen P, Wells JM, Kleerebezem M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends in Immunology. 2013;34(5):208–15. 10.1016/j.it.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 69.Lee IC, Tomita S, Kleerebezem M, Bron PA. The quest for probiotic effector molecules—Unraveling strain specificity at the molecular level. Pharmacological Research. 2013;69(1):61–74. 10.1016/j.phrs.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 70.Hafez M, Hayes K, Goldrick M, Grencis RK, Roberts IS. The K5 capsule of Escherichia coli strain nissle 1917 is important in stimulating expression of toll-like receptor 5, CD14, MyD88, and TRIF together with the induction of interleukin-8 expression via the mitogen-activated protein kinase pathway in epithelial cells. Infection and Immunity. 2010;78(5):2153–62. 10.1128/IAI.01406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fleischer J, Soeth E, Reiling N, Grage-Griebenow E, Flad HD, Ernst M. Differential expression and function of CD80 (B7-1) and CD86 (B7-2) on human peripheral blood monocytes. Immunology. 1996;89(4):592–8. 10.1046/j.1365-2567.1996.d01-785.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hajebi A, Motevalian SA, Rahimi-Movaghar A, Sharifi V, Amin-Esmaeili M, Radgoodarzi R, et al. Major anxiety disorders in Iran: Prevalence, sociodemographic correlates and service utilization. BMC Psychiatry. 2018;18(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Arienzo R, Maurano F, Lavermicocca P, Ricca E, Rossi M. Modulation of the immune response by probiotic strains in a mouse model of gluten sensitivity. Cytokine. 2009;48(3):254–9. 10.1016/j.cyto.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 74.Appledorn DM, McBride A, Seregin S, Scott JM, Schuldt N, Kiang A, et al. Complex interactions with several arms of the complement system dictate innate and humoral immunity to adenoviral vectors. Gene Therapy. 2008;15(24):1606–17. 10.1038/gt.2008.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunological Reviews. 2001;180:35–48. 10.1034/j.1600-065x.2001.1800103.x [DOI] [PubMed] [Google Scholar]
- 76.Rus H, Cudrici C, Niculescu F. The role of the complement system in innate immunity. Immunologic Research. 2005;33(2):103–12. 10.1385/IR:33:2:103 [DOI] [PubMed] [Google Scholar]
- 77.Maneechotesuwan K, Yao X, Ito K, Jazrawi E, Usmani OS, Adcock IM, et al. Suppression of GATA-3 nuclear import and phosphorylation: A novel mechanism of corticosteroid action in allergic disease. PLoS Medicine. 2009;6(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu X, Du Y, Cai C, Cai B, Zhu M, Xing C, et al. Inflammasome activation negatively regulates MyD88-IRF7 type I IFN signaling and anti-malaria immunity. Nature Communications. 2018;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strzepa A, Pritchard KA, Dittel BN. Myeloperoxidase: A new player in autoimmunity. Cellular Immunology. 2017;317:1–8. 10.1016/j.cellimm.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Papayannopoulos V, Zychlinsky A. NETs: a new strategy for using old weapons. Trends in Immunology. 2009;30(11):513–21. 10.1016/j.it.2009.07.011 [DOI] [PubMed] [Google Scholar]
- 81.Goritzka M, Durant LR, Pereira C, Salek-Ardakani S, Openshaw PJ, Johansson C. Alpha/beta interferon receptor signaling amplifies early proinflammatory cytokine production in the lung during respiratory syncytial virus infection. Journal of Virology. 2014;88(11):6128–36. 10.1128/JVI.00333-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thayer TC, Wilson SB, Mathews CE. Use of nonobese diabetic mice to understand human type 1 diabetes. Endocrinology and Metabolism Clinics of North America. 2010;39(3):541–61. 10.1016/j.ecl.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dai YD, Marrero IG, Gros P, Zaghouani H, Wicker LS, Sercarz EE. Slcllal enhances the autoimmune diabetogenic T-cell response by altering processing and presentation of pancreatic islet antigens. Diabetes. 2009;58(1):156–64. 10.2337/db07-1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kondrikov D, Fonseca FV, Elms S, Fulton D, Black SM, Block ER, et al. β-actin association with endothelial nitric-oxide synthase modulates nitric oxide and superoxide generation from the enzyme. Journal of Biological Chemistry. 2010;285(7):4319–27. 10.1074/jbc.M109.063172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Butterworth RF. Neuroinflammation in acute liver failure: Mechanisms and novel therapeutic targets. Neurochemistry International. 2011;59(6):830–6. 10.1016/j.neuint.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 86.Goad J, Ko Y-A, Kumar M, Syed SM, Tanwar PS. Differential Wnt signaling activity limits epithelial gland development to the anti-mesometrial side of the mouse uterus. Developmental Biology. 2017;423(2):138–51. 10.1016/j.ydbio.2017.01.015 [DOI] [PubMed] [Google Scholar]
- 87.Gaston JD, Bischel LL, Fitzgerald LA, Cusick KD, Ringeisen BR, Pirlo RK. Gene Expression Changes in Long-Term in Vitro Human Blood-Brain Barrier Models and Their Dependence on a Transwell Scaffold Materia. Journal of Healthcare Engineering. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abubaker J, Tiss A, Abu-Farha M, Al-Ghimlas F, Al-Khairi I, Baturcam E, et al. DNAJB3/HSP-40 Cochaperone Is Downregulated in Obese Humans and Is Restored by Physical Exercise. PLoS ONE. 2013;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]