Abstract
Objectives
Oncology has become more reliant on new testing methods and a greater use of electronic medical records, which provide a plethora of information available to physicians and researchers. However, to take advantage of vital clinical and research data for precision medicine, we must initially make an effort to create an infrastructure for the collection, storage, and utilization of this information with uniquely designed disease-specific registries that could support the collection of a large number of patients.
Materials and methods
In this study, we perform an in-depth analysis of a series of lung adenocarcinoma patients (n = 415) with genomic and clinical data in a recently created thoracic patient registry.
Results
Of the 415 patients with lung adenocarcinoma, 59% (n = 245) were female; the median age was 64 (range, 22–92) years with a median OS of 33.29 months (95% CI, 29.77–39.48). The most common actionable alterations were identified in EGFR (n = 177/415 [42.7%]), ALK (n = 28/377 [7.4%]), and BRAF V600E (n = 7/288 [2.4%]). There was also a discernible difference in survival for 222 patients, who had an actionable alteration, with a median OS of 39.8 months as compared to 193 wild-type patients with a median OS of 26.0 months (P<0.001). We identified an unprecedented number of actionable alterations [53.5% (222/415)], including distinct individual alteration rates, as compared with 15.0% and 22.3% in TCGA and GENIE respectively.
Conclusion
The use of patient registries, focused genomic panels and the appropriate use of clinical guidelines in community and academic settings may influence cohort selection for clinical trials and improve survival outcomes.
Introduction
In response to advances in genomic testing and the rapid integration of new drugs and publications, oncologists have been adapting the concept of precision medicine where evidence-based medicine guides treatment decisions for individuals [1]. However, more effort is required to translate these benefits into real-world durable survivals for patients. Therefore, in this pursuit, several organizations have implemented the utilization of guidelines and pathways to ensure that patients receive proper testing and are assigned to proper treatment options, which in theory should then translate into durable survival outcomes [2–6]. This spur towards personalized medicine is primarily driven by the advances in genomic testing, biomarker-driven therapy, and immunotherapy that have transformed the landscape of oncology care and have greatly improved outcomes for patients [7–12]. Next-generation sequencing (NGS) has been highly influential in its ability to identify genomic alterations that confer sensitivity to approved and investigational targeted therapies in patients suffering from a variety of advanced stage cancers. The application of molecular testing is transforming cancer into a diverse template of genomic alterations that drive oncogenesis [13].
In view of the vast clinical data offered by NGS in non-small cell lung cancer (NSCLC), City of Hope (COH) has established a concise and efficient patient registry (Registry of Hope) for the collection of genomic alterations and outcomes-focused clinical data. Here we present the results of genomic testing performed as part of routine clinical care and correlative analysis of 415 lung adenocarcinoma (LUAD) patients within the Thoracic Oncology Registry (THOR). We hypothesized that the application of broad genomic testing provides not only a comprehensive overview of clinical heterogeneity in lung cancer but may also guide the future of oncology care as more and more precision medicine therapeutics emerge. In this study, we also evaluate the genomic profile of our COH cohort as it compares to national testing results found in GENIE/TCGA databases.
Materials and methods
Patients
Patients with advanced LUAD (n = 415), were enrolled in this analysis and evaluated at COH from 2008 to 2016, the data was collected between 2016 and 2018 through retrospective chart review on patients who had LUAD diagnosis and molecular testing performed at the discretion of their primary clinical provider. All 415 patients had metastatic disease, with 89 percent of patients who presented with metastatic disease at the time of initial diagnosis while others later developed metastatic disease. THOR was used to perform this study and data was collected into the registry over time on eligible patients. Different NGS platforms were used as described in S1 Table. Patients were categorized by race/ethnicity according to what they had reported to their oncologist and the data was pulled from the electronic medical record. The categories included African American, American Indian or Alaska Native, Asian, White, Native Hawaiian or Other Pacific Island, Other, and Unknown/Declined to answer. Confounding variables, such as socioeconomic status, were not adjusted but the researchers felt that it was important to understand the diverse demographic makeup of the lung cancer patients and how it relates to their mutational status. The study was approved by the City of Hope institutional review board and in accord with an assurance filed with and approved by the Department of Health and Human Services at COH. This study was approved by the Institutional Review Board at COH under IRB 18217 and was conducted according to the Declaration of Helsinki. Data was de-identified and analyzed anonymously.
Genomic analysis
Patient genomic alteration data was collected manually from clinical genomic tests alongside the clinical information. The molecular testing results in this study were all performed at around the time of metastatic diagnosis and did not include multiple time points. Clinical actionability of genomic alterations was assessed for each patient and included genomic alterations that had US Food and Drug Administration (FDA) approved targeted therapy in NSCLC. The actionable genomic alterations in this study were defined as EGFR exon 19 deletions and L858R mutation, ALK rearrangement, ROS1 rearrangement, NTRK fusions, and BRAF V600E alterations based on FDA-approved therapy available. MET exon 14 splice site/deletion was also included as actionable based on FDA accelerated path to approval [14, 15]. These alterations were chosen because they are largely exclusive of one another. Any patient who had at least one of these six gene alterations was considered to have an actionable alteration and anyone whose genomic testing results did not identify any of the six gene alterations was considered wild-type. When individual genes were evaluated for survival and various statistical analyses the patients with alterations were only compared with patients who were tested negative for that specific gene. Tile plot maps were generated using seaborn library for Python (version 2.7.14) [16].
Comparison with TCGA and GENIE
For the alteration rate of the THOR data set, each gene that had an alteration was calculated as an individual patient divided by the number of patients who were tested for that gene. Genomic alteration data were retrieved from TCGA (study id = luad_tcga_pub) and GENIE (GENIE Cohort v4.1-public) [17, 18]. For each gene, only samples profiled in all molecular profiles including copy-number alteration, somatic mutations, and structural rearrangement were counted. Alteration rate for each gene was calculated by number of samples with at least one alteration divided by the number of profiled samples.
Statistical analysis
Patient characteristic parameters were evaluated using the χ2 test to test for association between characteristic values and age. OS was calculated from the date of diagnosis of metastatic disease to death or last follow-up visit. Patients who were thought to be alive at the end of the study were censored at the time of the last visit. Survival estimates for the studied patients were generated using the Kaplan-Meier method and the Cox proportional hazards models were used to estimate the hazard ratios. The analysis was performed using survival and survminer packages for R software (version 3.4.4) and SAS version 9.4 (SAS Institute, Cary, NC, United States). The assumption of proportional hazards was tested using goodness-of-fit tests, graphical methods and time-dependent variable methods. The extended Cox models using time-dependent variables were used to adjust the non-proportionality of variables. By adding interaction terms between time and the variables that violated the proportional hazards assumption, the models allow for the possible diverging survival curves over time.
Results
Patients
415 eligible patients with LUAD and tumor genotyping results were identified in THOR at the time of this study. The majority of patients were Stage IV (n = 369/415 [89%]) at the time of initial diagnosis and 46/415 (11%) patients were stage I-III at diagnosis who eventually recurred as metastatic disease. There were 281 patients who underwent broad-based genomic testing (more than 30 genes) using NGS and 134 patients were tested for a few genes in a small panel (less than 10 genes). 254 (61%) patients were never-smokers (n = 212) or had a smoking history of fewer than 10 pack-years (light smokers; n = 42) and 161 (39%) patients were smokers with a history of 10–29 pack-years (medium smokers; n = 74) or > = 30 pack-years (heavy smokers; n = 87). The overall median age at diagnosis was 64 (22–92) and the median OS was 33.29 months (95% CI, 29.77–39.48), with the majority of patients being female (n = 245 [59%]). Detailed patient characteristics noted in Table 1.
Table 1. Patient characteristics.
| Characteristic | Total, No. (%) | Age, No. (%) | P Value | Chi-square Statistic | |
|---|---|---|---|---|---|
| <70 | > = 70 | ||||
| Patients | 415 (100) | 289 (100) | 126 (100) | NA | NA |
| Sex | 0.341 | 0.906 | |||
| Female | 245 (59) | 175 (61) | 70 (56) | ||
| Male | 170 (41) | 114 (39) | 56 (44) | ||
| Race | 0.127 | 9.956 | |||
| African American | 10 (2) | 6 (2) | 4 (3) | ||
| American Indian or Alaska Native | 2 (0) | 1 (0) | 1 (1) | ||
| Asian | 136 (33) | 107 (37) | 29 (23) | ||
| White | 247 (60) | 161 (56) | 86 (68) | ||
| Native Hawaiian or Other Pacific Islander | 3 (1) | 3 (1) | 0 (0) | ||
| Other | 12 (3) | 8 (3) | 4 (3) | ||
| Unknown/Declined to Answer | 5 (1) | 3 (1) | 2 (2) | ||
| Ethnicity | 0.150 | 3.796 | |||
| Hispanic or Latino | 40 (10) | 28 (10) | 12 (10) | ||
| Not Hispanic or Latino | 369 (89) | 259 (90) | 110 (87) | ||
| Unknown/Declined to Answer | 6 (1) | 2 (1) | 4 (3) | ||
| Smoking Status | 0.036 | 8.550 | |||
| Never | 212 (51) | 159 (55) | 53 (42) | ||
| Light (<10 pack years) | 42 (10) | 31 (11) | 11 (9) | ||
| Medium (10–29 pack years) | 74 (18) | 44 (15) | 30 (24) | ||
| Heavy (> = 30 pack years) | 87 (21) | 55 (19) | 32 (25) | ||
| Stage | 0.004 | 13.595 | |||
| I | 15 (4) | 7 (2) | 8 (6) | ||
| II | 14 (3) | 5 (2) | 9 (7) | ||
| III | 17 (4) | 10 (3) | 7 (6) | ||
| IV | 369 (89) | 267 (92) | 102 (81) | ||
| EGFR (L858R/exon 19 deletion) * | 0.059 | 3.559 | |||
| Alteration | 177 (43) | 132 (46) | 45 (36) | ||
| Tested Negative | 238 (57) | 157 (54) | 81 (64) | ||
| ALK (Rearrangement)* | 0.001 | 10.349 | |||
| Positive | 28 (7) | 27 (10) | 1 (1) | ||
| Tested Negative | 349 (93) | 235 (90) | 114 (99) | ||
| ROS1 (Rearrangement)* | 0.233 | 1.422 | |||
| Positive | 3 (1) | 3 (2) | 0 (0) | ||
| Tested Negative | 254 (99) | 172 (98) | 82 (100) | ||
| BRAF (V600E)* | 0.319 | 0.995 | |||
| Positive | 7 (2) | 6 (3) | 1 (1) | ||
| Tested Negative | 281 (98) | 194 (97) | 90 (99) | ||
| MET (exon 14 splice-site/deletion)* | 0.022 | 5.228 | |||
| Positive | 7 (2) | 2 (1) | 5 (5) | ||
| Tested Negative | 280 (98) | 194 (99) | 86 (95) | ||
| KRAS | <0.001 | 12.410 | |||
| Positive | 97 (28) | 53 (22) | 44 (40) | ||
| Tested Negative | 255 (72) | 189 (78) | 66 (60) | ||
| TP53 | 0.306 | 1.046 | |||
| Positive | 140 (49) | 99 (52) | 41 (45) | ||
| Tested Negative | 143 (51) | 93 (48) | 50 (55) | ||
*Only patients who had genomic test results were counted for each gene. Total number of patients with ALK, ROS1, BRAF, MET, KRAS and TP53 tested were 377, 257, 288, 287, 352, and 283 respectively.
Genomic alterations
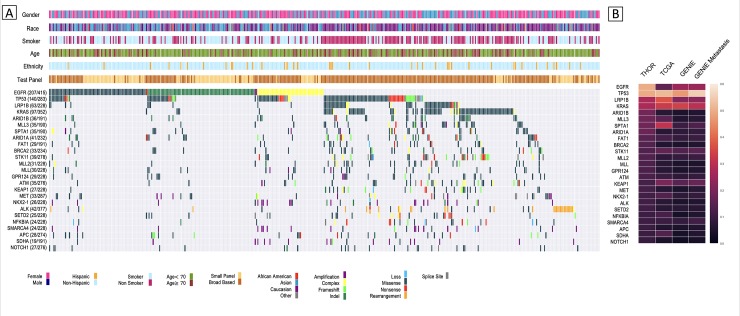
There were 323 different genes with evidence of genomic alteration in this group of patients. The most commonly occurring alterations in oncogenes were found in EGFR (n = 207/415 [50%]), KRAS (n = 97/352 [28%]), and ALK rearrangement (n = 28/377 [7%]), while the most commonly occurring tumor suppressor genes consisted of TP53 (n = 140/283 [49%]), LRP1B (n = 63/228 [28%]), and STK11 (n = 39/278 [14%]) (Fig 1A). The median number of genes altered in patients who underwent broad-based sequencing was 10. The most common actionable alterations were identified in EGFR L858R/exon 19 deletion (n = 177/415 [42.7%]), ALK rearrangement (n = 28/377 [7.4%]), ROS1 rearrangement (n = 3/257 [1.2%]), BRAF V600E (n = 7/288 [2.4%]), and MET exon 14 splice site/deletion (n = 7/287 [2.4%]). No NTRK fusion alterations were identified in our cohort. 86% (177/207) of patients with EGFR alterations had an actionable alteration while 14% (29/207) consisted of exon 18 mutations, exon 20 insertions and other EGFR alterations including amplifications or substitution variants of unknown significance. EGFR alterations were split into five subtypes with the majority of Asians (n = 51/99 [52%]) and whites (n = 46/95 [48%]) presenting with an exon 19 deletion. KRAS alterations predominated in whites (n = 77/97 [79%]) and patients with history of medium (n = 24/97 [25%]) or heavy smoking (n = 47/97 [48%]) (Table 2).
Fig 1. Landscape overview of molecular profiling in 415 THOR lung adenocarcinoma patients.
A) Tile plot of top 25 most prevalent genes and their demographic parameters sorted by genomic alteration rate and subtypes, including amplification, complex (more than one type of alteration), frameshift, indel (insertion/deletion), loss, missense, nonsense, rearrangement, and splice site. For each gene, the alteration rate was calculated by number of patients with alteration divided by the number of patients tested for this gene. B) Heatmap showing a comparison of the genomic alteration rates between THOR, TCGA, GENIE, and GENIE Metastatic for genes in 1A.
Table 2. Patients characteristic table for EGFR and KRAS alteration subtypes.
| Characteristic | EGFR, No. (%)* | KRAS, No. (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | exon 18 mutation | exon 19 deletion | exon 20 insertion | exon 21 mutation | Others | Total | G12 | G13 | Q61 | Others | |
| Patients | 207 (100) | 9 (4) | 106 (51) | 16 (8) | 72 (35) | 7 (3) | 97 (100) | 79 (81) | 7 (7) | 9 (9) | 2 (2) |
| Sex | |||||||||||
| Female | 130 (63) | 4 (44) | 65 (61) | 12 (75) | 47 (65) | 3 (43) | 49 (51) | 39 (49) | 5 (71) | 4 (44) | 1 (50) |
| Male | 77 (37) | 5 (56) | 41 (39) | 4 (25) | 25 (35) | 4 (57) | 48 (49) | 40 (51) | 2 (29) | 5 (56) | 1 (50) |
| Age | |||||||||||
| <70 | 155 (75) | 5 (56) | 84 (79) | 15 (94) | 48 (67) | 6 (86) | 53 (55) | 45 (57) | 3 (43) | 4 (44) | 1 (50) |
| > = 70 | 52 (25) | 4 (44) | 22 (21) | 1 (6) | 24 (33) | 1 (14) | 44 (45) | 34 (43) | 4 (57) | 5 (56) | 1 (50) |
| Race | |||||||||||
| African American | 4 (2) | 0 (0) | 3 (3) | 0 (0) | 1 (1) | 0 (0) | 4 (4) | 3 (4) | 0 (0) | 1 (11) | 0 (0) |
| American Indian or Alaska Native | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (2) | 2 (3) | 0 (0) | 0 (0) | 0 (0) |
| Asian | 99 (48) | 6 (67) | 51 (48) | 6 (38) | 37 (51) | 2 (29) | 11 (11) | 9 (11) | 1 (14) | 0 (0) | 1 (50) |
| White | 95 (46) | 3 (33) | 46 (43) | 10 (63) | 32 (44) | 4 (57) | 77 (79) | 62 (78) | 6 (86) | 8 (89) | 1 (50) |
| Native Hawaiian or Other Pacific Islander | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Other | 8 (4) | 0 (0) | 6 (6) | 0 (0) | 1 (1) | 1 (14) | 1 (1) | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Unknown/Declined to Answer | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 1 (1) | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Ethnicity | |||||||||||
| Hispanic or Latino | 17 (8) | 0 (0) | 13 (12) | 1 (6) | 2 (3) | 1 (14) | 11 (11) | 10 (13) | 0 (0) | 1 (11) | 0 (0) |
| Not Hispanic or Latino | 188 (91) | 9 (100) | 92 (87) | 15 (94) | 69 (96) | 6 (86) | 84 (87) | 68 (86) | 6 (86) | 8 (89) | 2 (100) |
| Unknown/Declined to Answer | 2 (1) | 0 (0) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | 2 (2) | 1 (1) | 1 (14) | 0 (0) | 0 (0) |
| Smoking Status | |||||||||||
| Never | 137 (66) | 5 (56) | 72 (68) | 9 (56) | 50 (69) | 3 (43) | 17 (18) | 14 (18) | 1 (14) | 1 (11) | 1 (50) |
| Light (<10 pack years) | 23 (11) | 1 (11) | 11 (10) | 3 (19) | 7 (10) | 2 (29) | 9 (9) | 7 (9) | 1 (14) | 1 (11) | 0 (0) |
| Medium (10–29 pack years) | 39 (19) | 3 (33) | 21 (20) | 3 (19) | 11 (15) | 1 (14) | 24 (25) | 19 (24) | 1 (14) | 3 (33) | 1 (50) |
| Heavy (> = 30 pack years) | 8 (4) | 0 (0) | 2 (2) | 1 (6) | 4 (6) | 1 (14) | 47 (48) | 39 (49) | 4 (57) | 4 (44) | 0 (0) |
*Three patients carried two subtypes of EGFR alteration (e.g., exon 19 deletion and exon 18 mutation) and were counted two times.
Comparison with TCGA and GENIE
While the gene alteration rates of LUAD patients from TCGA/GENIE were comparable [17, 18], evident differences were observed between the patients of THOR and these two public consortiums (Fig 1B). Importantly, a large proportion of patients had actionable alterations [53.5% (222/415)] with the highest rate of actionable EGFR mutations at 42.7% (177/415), as compared with 9.1% and 17.1% actionable rate in TCGA/GENIE respectively. In particular, our dataset had a significantly increased prevalence of EGFR (49.9%), BRCA2 (14.1%), and chromatin modifying genes (ARID1B [18.8%], ARID1A [17.7%], MLL2 [13.6%] and MLL [13.2%]) as compared to the other datasets (Table 3). Analysis of subgroups of patients in GENIE that were diagnosed with or without metastasis showed similar gene alteration rates of these genes, indicating the difference we observed between THOR and TCGA/GENIE patients was not directly linked to metastasis (Table 3).
Table 3. Genomic alteration rates of THOR referring to TCGA/GENIE/GENIE metastasis patients.
| Gene | Altered | Tested | THOR Alteration Rate (n = 415) | TCGA Alternation Rate (n = 507) | GENIE Alternation Rate (n = 6529) | GENIE Metastasis Alternation Rate (n = 2697) |
|---|---|---|---|---|---|---|
| EGFR | 207 | 415 | 50% | 16% | 25% | 25% |
| TP53 | 140 | 283 | 50% | 52% | 47% | 55% |
| LRP1B | 63 | 228 | 28% | 37% | 22% | 23% |
| KRAS | 97 | 352 | 28% | 33% | 34% | 30% |
| ARID1B | 36 | 191 | 19% | 7% | 5% | 6% |
| MLL3 | 35 | 190 | 18% | 15% | 6% | 7% |
| SPTA1 | 35 | 190 | 18% | 31% | 10% | 13% |
| ARID1A | 41 | 232 | 18% | 7% | 8% | 9% |
| FAT1 | 29 | 191 | 15% | 12% | 9% | 10% |
| BRCA2 | 33 | 234 | 14% | 6% | 5% | 6% |
| STK11 | 39 | 278 | 14% | 16% | 14% | 15% |
| MLL2 | 31 | 228 | 14% | 8% | 9% | 10% |
| MLL | 30 | 228 | 13% | 7% | 5% | 5% |
| GPR124 | 29 | 228 | 13% | 7% | 5% | 6% |
| ATM | 35 | 276 | 13% | 10% | 8% | 9% |
| KEAP1 | 27 | 228 | 12% | 20% | 15% | 17% |
| MET | 33 | 287 | 12% | 7% | 5% | 7% |
| NKX2-1 | 26 | 228 | 11% | 14% | 8% | 9% |
| ALK | 42 | 377 | 11% | 7% | 5% | 6% |
| SETD2 | 25 | 228 | 11% | 7% | 7% | 7% |
| NFKBIA | 24 | 228 | 11% | 12% | 4% | 6% |
| SMARCA4 | 24 | 228 | 11% | 10% | 9% | 11% |
| APC | 28 | 274 | 10% | 6% | 5% | 6% |
| SDHA | 19 | 191 | 10% | 14% | 4% | 6% |
| NOTCH1 | 27 | 276 | 10% | 5% | 3% | 4% |
Genomic alteration data were collected for 415 THOR lung adenocarcinoma patients, 507 TCGA Lung Adenocarcinoma patients (study id = luad_tcga_pan_can_atlas_2018), 6529 GENIE Lung Adenocarcinoma patients (GENIE Cohort v5.0-public, cancer type detailed = lung adenocarcinoma), and 2697 GENIE metastasis lung adenocarcinoma patients (GENIE Cohort v5.0-public, cancer type detailed = lung adenocarcinoma, sample type = Metastasis).
Survival
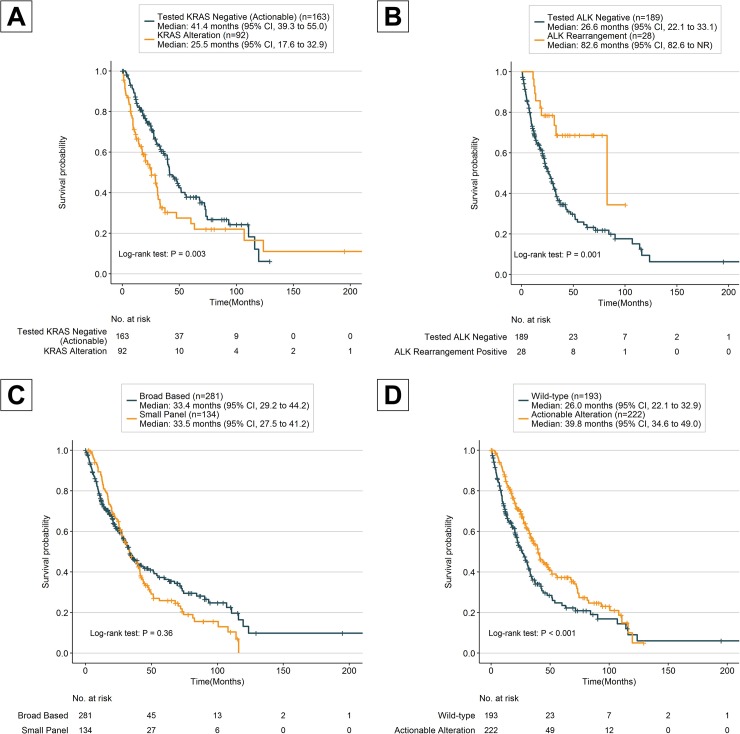
The median OS of all patients was 33.3 months (95% CI; 29.8–39.5) and female patients had a better median OS of 39.8 months (95% CI, 33.5–45.5 months; HR, 1.56; 95% CI, 1.21–2.00; Table 4) as compared to median OS of 27.4 months for male patients. The OS was also better for never/light smokers (<10 pack years) with a median OS of 39.5 months (95% CI, 32.9–45.5 months; HR, 1.42; 95% CI, 1.10–1.83; Table 4) as compared to medium/heavy smokers (> = 10 pack years) who had a median OS of 25.7 months (95% CI, 22.1–33.3 months). Two genes had violations of the proportional hazards assumption EGFR in actionable and KRAS in actionable, therefore, we added an interaction with time to account for changes in the HR. When comparing the median OS of KRAS positive patient (25.5 months; 95% CI, 17.6–32.9 months) with the median OS of actionable patient who had tested KRAS negative (41.1 months; 95% CI, 39.3–55.0 months), the difference was noticeable (Fig 2A), the HR changes from high risk of death in the KRAS positive patients initially to less risk over time (Table 4). Of the six actionable genes only ALK rearrangement patients had a difference in survival with a median OS of 82.6 months (95% CI, 82.6-NR; HR, 0.35; 95% CI, 0.17–0.68;) as compared to 26.6 months (95% CI, 22.1–33.1 months) for tested ALK rearrangement negative patients (Fig 2B). The genomic testing panel size had no discernible OS difference and broad-based sequencing had a median OS of 33.4 months (95% CI, 29.2–44.2 months) as compared to 33.5 months (95% CI, 27.5–41.2 months) for the small panel (Fig 2C). There was a discernible difference in survival for 222 patients, who had an actionable alteration (such as EGFR L858R/exon 19 deletion, ALK rearrangements, ROS1 rearrangements, BRAF V600E, NTRK fusions and MET exon 14 splice site/deletion), with a median OS of 39.8 months as compared to 193 patients who were wild-type with a median OS of 26.0 months (Fig 2D).
Table 4. Cox proportional hazard regression models for univariate survival.
| Risk Factor | Median Survival (95% CI), months | Hazard Ratio (95% CI) | P Value |
|---|---|---|---|
| Sex, Male vs Female | 27.4 (20.6–31.7) vs 39.8 (33.5–45.5) | 1.56 (1.21–2.00) | <0.001 |
| Age, > = 70 vs <70 | 29.0 (19.5–36.5) vs 36.3 (31.4–42.9) | 1.41 (1.08–1.85) | 0.012 |
| Smoking Status, Medium + Heavy vs Never + Light |
|||
| 25.7 (22.1–33.3) vs 39.5 (32.9–45.5) | 1.42 (1.10–1.83) | 0.006 | |
| EGFR (Actionable)ab | |||
| Positive vs Negative EGFR x time |
39 (32.6–43.8) vs 26 (22.1–32.9) | 0.46 (0.31–0.68) 1.02 (1.01–1.03) |
0.0001 0.0022 |
| ALK (Rearrangement)b | |||
| Positive vs Negative | 82.6 (82.6-NR) vs 26.6 (22.1–33.1) | 0.35 (0.17–0.68) | 0.002 |
| ROS1 (Rearrangement)b | |||
| Positive vs Negative | NR vs 26 (20.8–33.3) | - | - |
| BRAF (V600E)b | |||
| Positive vs Negative | 73.4 (11.6-NR) vs 28.0 (22.2–34.4) | 0.70 (0.26–1.91) | 0.487 |
| MET (exon 14 Splice-site/Deletion)b | |||
| Positive vs Negative | 17.2 (8.77-NR) vs 28.0 (22.2–34.4) | 0.59 (0.19–1.89) | 0.377 |
| KRAS in Actionablec | |||
| Positive vs Negative KRAS x time |
25.5 (17.6–32.9) vs 41.4 (39.3–55.0) | 2.80 (1.70–4.61) 0.98 (0.97–0.99) |
<0.0001 0.0065 |
| KRAS in Non-Actionabled | |||
| Positive vs Negative | 25.5 (17.6–32.9) vs 27.7 (22.2–42.9) | 1.06 (0.73–1.53) | 0.769 |
| TP53 in Actionablec | |||
| Positive vs Negative | 25.5 (20.6–34.9) vs 67.6 (39.5-NR) | 1.87 (1.12–3.12) | 0.017 |
| TP53 in Non-Actionabled | |||
| Positive vs Negative | 25.5 (20.6–34.9) vs 30.1 (21.6–54.0) | 1.13 (0.75–1.69) | 0.561 |
a. EGFR All patients had EGFR tested.
b. For each gene, patients carrying actionable alterations were compared with patients who were tested negative, with patients carrying actionable alterations from other genes excluded from its analysis.
c. For KRAS and TP53, patients who were tested positive with no actionable alteration were compared with patients who were tested negative but had actionable gene alterations.
d. For KRAS and TP53, patients who were tested positive were compared with patients who were tested negative and patients who had actionable gene alterations were excluded from both groups.
Fig 2. Kaplan-Meier estimates of overall survival.
A) Overall survival among KRAS altered and tested KRAS negative patients with actionable alteration. B) Overall survival among ALK rearrangement and tested ALK rearrangement negative patients. C) Overall survival among broad-based panel tested and small-panel tested patients. D) Overall survival among actionable altered and wild-type patients. P values comparing risk groups were calculated with the log-rank test.
Discussion
The actualization of tyrosine kinase inhibitors, monoclonal antibodies, and immunotherapy drugs has pushed the treatment of lung cancer forward, however, the reality remains that lung cancer is the leading cause of cancer deaths [19]. Genomic testing has become more crucial in oncology care and recently there have been several studies launched that aim to match targeted therapy to patients based on their omic profile [20–27]. It is now a standard recommendation that patients with advanced NSCLC undergo routine molecular testing for identification of certain known genomic abnormalities, most notably ALK rearrangements, EGFR mutations, BRAF V600E, ROS1 rearrangements, and NTRK fusions [5]. More so several inhibitors for various markers, such as ERBB2, MET, and RET are quickly being implemented in standard clinical care and several others await FDA approval [28–32]. Thus, this study aims to highlight a unique single-site perspective into the clinical heterogeneity found in our cohort of patients that is largely composed of clinically relevant markers that will benefit from recent advances in precision medicine and a cohort that is distinctly different from other databases [17, 18]. To achieve durable survivals for patients it vital to implement appropriate testing at diagnosis and upon progression, and guided treatment plans (both guidelines and pathways) through consolidation of clinical data into patient registries [33].
Precision genomics
In a recent study that compared the use of broad-based genomic sequencing versus small-panel testing, it was shown that there was no significant difference in OS [34]. While our data recapitulates that broad-based sequencing panels were not independently associated with better median OS, our results also show that patients with actionable alterations had a discernibly improved median OS as compared to wild-type patients who did not have an actionable alteration. Most notably ALK rearrangement patients had an improved OS as compared to tested ALK rearrangement negative (median OS 82.6 vs 26.6 months), which is suspected due to appropriate molecular testing at presentation and apt selection of therapy based on results [35, 36]. While Presley et al. include ALK in their small routine panel it should be noted that in a study evaluating the methods of ALK rearrangement testing it was found that comprehensive genomic profiling by NGS was able to identify a number of patients who had previously tested negative using the standard ALK fluorescence in situ hybridization (FISH) assay [37]. While there were several studies that showed improved PFS with targeted therapy which did not translate into OS benefit [7–12], more recent results in lung cancer have shown there is a durable survival benefit for patients who are properly treated with appropriate inhibitors as opposed to patients who do not receive targeted therapy [30, 38–41]. 53.5% (222/415) of patients presented with actionable alterations in our cohort had specific targeted therapies approved by the FDA. While many question the utility to targeted therapy due to low actionable alteration rates [42–44], this study shows that actionable alterations rates may vary demographically and by location presenting a challenge for improving outcomes that may be addressed through the implementation of clinical pathway guidelines and continuing genomic education [45].
Guided value-based medicine
Several studies have been performed that have determined the cost-effectiveness of NGS panels over single gene testing and in one study there was an improvement in PFS without additional health care costs to the patient [46–48]. In several meta-analyses of prospective clinical trials for solid tumors and hematological malignancies, personalized treatment strategies showed superior outcomes to those in control arms [49–54]. Therefore, the question becomes not whether genomic testing should be performed with a specific panel but whether the patient is treated accordingly based on all information available to improve their individual outcomes [54]. For this purpose, several associations including the National Comprehensive Cancer Network (NCCN) have issued guidelines which strongly emphasize not only testing for EGFR, ALK, ROS1, BRAF, and PD-L1 alterations but more importantly advise to conduct broader molecular profiling to identify rare mutational drivers [5, 6]. In practice, it was reported that the majority of oncologists (60%) in North America did not utilize genomic alteration results in their treatment decision making [55, 56].
To improve these statistics, the American Society of Clinical Oncology (ASCO) established in 2015 a task force on clinical pathways to improve treatment decision making through evidence-based clinical pathway guidelines [3]. Since then, ASCO revealed in a 2017 State of Cancer Care in America report a 42% increase from 2014 to 2016 of practices complying with a pathway program [57, 58]. Moreover, a recent study that evaluated 7 cancer programs, including COH (both academic and community), that employed clinical pathways demonstrated unprecedently high rates of molecular testing and concordant appropriate first-line treatment decision making based on actionable biomarkers [59]. Thusly, the end-goal for improvement of outcomes can only be achieved under the right circumstances where the appropriate testing is performed and the accurate treatment decisions are made.
THOR in comparison with TCGA and GENIE
While there is some overlap in the alteration rates between our cohort and the publicly available datasets (TCGA/GENIE), there are more differences in the comparable rates of occurrences. Our study identified a disproportionally high rate of EGFR alterations with 49.9% (207/415) and actionable alteration rate with 53.5%, which was notably higher than the TCGA/GENIE rates [17, 18]. However, these results are consistent with a large gene profiling study of NSCLC from the Icahn School of Medicine at Mount Sinai that was able to identify actionable alterations in 65% of the cases [60]. While it is estimated that the actionability rate for NSCLC is around 30%, the results of this single-site analysis warrant further consideration to understand the heterogeneity of lung cancer in different populations and how this may impact survival outcomes [17]. This may in part be due to referrals to COH for clinical care and potential clinical trials, but it cannot be discounted that the high rates of EGFR alterations may be due to a large number of Asian never-smokers at presentation. In fact, 72.8% (99/136) of Asians in our cohort presented with an EGFR alteration as compared with 50.0% EGFR incidence rate found in the GENIE dataset [17]. This large occurrence of EGFR in Asian never-smokers is consistent with another study that identified 75.3% of Asian never-smokers who harbored EGFR mutations [61].
Unexpectedly a number of chromatin modulating genes were irregularly prevalent in our cohort as compared with TCGA/GENIE. ARID1A alterations were highly expressed in our cohort alongside ARID1B and the two are known to be mutually exclusive within individual SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complexes [62]. However, ARID1A has been shown to be the strongest tumor suppressor gene amongst the ARID genes in various cancers [63–66] and ARID1A is known to have the highest alteration rate among the SWI/SNF subunit genes [67]. The dysfunction of these complexes is known to destabilize the lung cancer genome by affecting chromatin remodeling and also disrupt DNA repair [68–70]. Furthermore, the high prevalence of genes such as MLL, MLL2, and BRCA2 suggest that other chromatin remodeling motifs may be involved in tumorigenesis of these patients rather than an event specific to a particular tumor type [71, 72].
Conclusion
The limitation of current oncology practice is the speed of integration and adaptation to the rapidly advancing testing modalities and multiple therapeutic options that are garnering swift approval in various cancer types [73]. These efforts will require standardization of molecular testing modalities, cohesive guidelines, and pathways for precision medicine, and focused patient registries to enable cohort management and efficient clinical trial accrual. While our study was limited by the sample size and the demographics of our patients, it is still important to utilize these tools to guide physicians in determining which patient groups can benefit from clinical trials, tumor resistance or chemotherapy sensitivity. Review of the genomic data of our lung cancer patient cohorts in THOR showed significantly different incidences of actionable mutations as compared to the public datasets. Thus, instead of relying on generalized demographics and genomic results within the public datasets, the individual centers are required to perform their own assessments of their cohorts in order to have a proper analysis of their clinic population.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
For molecular markers, "0" means tested negative, and blank means not tested.
(XLSX)
Acknowledgments
We thank the clinical staff at City of Hope for their skill and dedication in helping the patients presented in this manuscript. The authors would like to acknowledge the American Association for Cancer Research and its support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of study authors. The results published here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Abbreviations
- NSCLC
Non-small cell lung cancer
- IRB
Institutional Review Board
- FISH
Fluorescent in situ hybridization
- LUAD
Lung adenocarcinoma
- NGS
Next-Generation Sequencing
- TKIs
tyrosine-kinase inhibitors
- FDA
U.S. Food and Drug Administration
- FFPE
formalin-fixed paraffin-embedded
- IHC
immunohistochemistry
- PCR
polymerase chain reaction
- PFS
progression-free survival
- THOR
Thoracic Oncology Registry
- OS
overall survival
- SWI/SNF
SWItch/Sucrose Non-Fermentable
- NCCN
National Comprehensive Cancer Network
- ASCO
American Society of Clinical Oncology
Data Availability
All relevant study data are within the manuscript and its Supporting Information files. Any additional data is available through TCGA (https://www.cancer.gov/tcga) and AACR GENIE (https://www.aacr.org/professionals/research/aacr-project-genie/aacr-project-genie-data/).
Funding Statement
Research reported in this publication included work performed in the Core facilities, Biostatistics Core and Center for Informatics Core, supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. Pilot Projects and Early Phase Clinical Research Support reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01CA218545 and U54CA209978. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The American Association for Cancer Research provided its financial and material support in the development of the AACR Project GENIE registry. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marignani P, Kim J, Greer W, Xu Z. P3.13–31 Creating a Precision Medicine Pipeline for Lung Cancers. Journal of Thoracic Oncology. 2018;13(10):S989 10.1016/j.jtho.2018.08.1872 [DOI] [Google Scholar]
- 2.Zon RT, Goss E, Vogel VG, Chlebowski RT, Jatoi I, Robson ME, et al. American Society of Clinical Oncology policy statement: the role of the oncologist in cancer prevention and risk assessment. J Clin Oncol. 2009;27(6):986–93. Epub 2008/12/17. 10.1200/JCO.2008.16.3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zon RT, Frame JN, Neuss MN, Page RD, Wollins DS, Stranne S, et al. American Society of Clinical Oncology Policy Statement on Clinical Pathways in Oncology. J Oncol Pract. 2016;12(3):261–6. Epub 2016/01/14. 10.1200/JOP.2015.009134 . [DOI] [PubMed] [Google Scholar]
- 4.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137(6):828–60. Epub 2013/04/05. 10.5858/arpa.2012-0720-OA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142(3):321–46. Epub 2018/01/23. 10.5858/arpa.2017-0388-CP . [DOI] [PubMed] [Google Scholar]
- 6.(NCCN). NcCN. NCCN Clinical practice guidelines in Oncology (NCCN Guidelines) for Non-small cell lung cancer. 2019 [cited 2019 January 4]. Available from: http://www.nccn.org.
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. Epub 2009/08/21. 10.1056/NEJMoa0810699 . [DOI] [PubMed] [Google Scholar]
- 8.Planchard D, Smit EF, Groen HJM, Mazieres J, Besse B, Helland A, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 2017;18(10):1307–16. Epub 2017/09/19. 10.1016/S1470-2045(17)30679-4 . [DOI] [PubMed] [Google Scholar]
- 9.Soria JC, Wu YL, Nakagawa K, Kim SW, Yang JJ, Ahn MJ, et al. Gefitinib plus chemotherapy versus placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): a phase 3 randomised trial. Lancet Oncol. 2015;16(8):990–8. Epub 2015/07/15. 10.1016/S1470-2045(15)00121-7 . [DOI] [PubMed] [Google Scholar]
- 10.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46. Epub 2012/01/31. 10.1016/S1470-2045(11)70393-X . [DOI] [PubMed] [Google Scholar]
- 11.Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–34. Epub 2013/07/03. 10.1200/JCO.2012.44.2806 . [DOI] [PubMed] [Google Scholar]
- 12.Le Tourneau C, Delord JP, Goncalves A, Gavoille C, Dubot C, Isambert N, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–34. Epub 2015/09/08. 10.1016/S1470-2045(15)00188-6 . [DOI] [PubMed] [Google Scholar]
- 13.Sabari JK, Santini F, Bergagnini I, Lai WV, Arbour KC, Drilon A. Changing the Therapeutic Landscape in Non-small Cell Lung Cancers: the Evolution of Comprehensive Molecular Profiling Improves Access to Therapy. Curr Oncol Rep. 2017;19(4):24 Epub 2017/03/18. 10.1007/s11912-017-0587-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PFIZER’S XALKORI® (CRIZOTINIB) RECEIVES FDA BREAKTHROUGH THERAPY DESIGNATION IN TWO NEW INDICATIONS 2018 [cited 2019 January 4]. Available from: https://investors.pfizer.com/investor-news/press-release-details/2018/Pfizers-XALKORI-crizotinib-Receives-FDA-Breakthrough-Therapy-Designation-in-Two-New-Indications/default.aspx.
- 15.Drilon AE, Camidge DR, Ou S-HI, Clark JW, Socinski MA, Weiss J, et al. Efficacy and safety of crizotinib in patients (pts) with advanced MET exon 14-altered non-small cell lung cancer (NSCLC). Journal of Clinical Oncology. 2016;34(15_suppl):108–. 10.1200/JCO.2016.34.15_suppl.108 [DOI] [Google Scholar]
- 16.Waskom M, Botvinnik O, O'Kane D, Hobson P, Ostblom J, Lukauskas S, et al. mwaskom/seaborn: v0.9.0 (July 2018). Zenodo. 2018. 10.5281/zenodo.1313201 [DOI] [Google Scholar]
- 17.AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discovery. 2017;7(8):818–31. 10.1158/2159-8290.CD-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Research N, Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543 10.1038/nature13385 https://www.nature.com/articles/nature13385#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. Epub 2018/01/10. 10.3322/caac.21442 . [DOI] [PubMed] [Google Scholar]
- 20.Gerber DE, Oxnard GR, Govindan R. ALCHEMIST: Bringing genomic discovery and targeted therapies to early-stage lung cancer. Clin Pharmacol Ther. 2015;97(5):447–50. Epub 2015/02/14. 10.1002/cpt.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coyne GO, Takebe N, Chen AP. Defining precision: The precision medicine initiative trials NCI-MPACT and NCI-MATCH. Curr Probl Cancer. 2017;41(3):182–93. Epub 2017/04/05. 10.1016/j.currproblcancer.2017.02.001 . [DOI] [PubMed] [Google Scholar]
- 22.Herbst RS, Gandara DR, Hirsch FR, Redman MW, LeBlanc M, Mack PC, et al. Lung Master Protocol (Lung-MAP)-A Biomarker-Driven Protocol for Accelerating Development of Therapies for Squamous Cell Lung Cancer: SWOG S1400. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21(7):1514–24. 10.1158/1078-0432.CCR-13-3473 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carey LA, Winer EP. I-SPY 2—Toward More Rapid Progress in Breast Cancer Treatment. N Engl J Med. 2016;375(1):83–4. Epub 2016/07/15. 10.1056/NEJMe1603691 . [DOI] [PubMed] [Google Scholar]
- 24.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba, II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. Epub 2014/05/23. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews A. ASCO and NCI Launch Largest Precision Medicine Trials Using Real-World Evidence. Am Health Drug Benefits. 2015;8(Spec Issue):37 Epub 2015/09/19. [PMC free article] [PubMed] [Google Scholar]
- 26.Seibel NL, Janeway K, Allen CE, Chi SN, Cho YJ, Glade Bender JL, et al. Pediatric oncology enters an era of precision medicine. Curr Probl Cancer. 2017;41(3):194–200. Epub 2017/03/28. 10.1016/j.currproblcancer.2017.01.002 . [DOI] [PubMed] [Google Scholar]
- 27.Takebe N, McShane L, Conley B. Biomarkers: exceptional responders-discovering predictive biomarkers. Nat Rev Clin Oncol. 2015;12(3):132–4. Epub 2015/02/18. 10.1038/nrclinonc.2015.19 . [DOI] [PubMed] [Google Scholar]
- 28.Ramalingam SS, Yang JC, Lee CK, Kurata T, Kim DW, John T, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2018;36(9):841–9. Epub 2017/08/26. 10.1200/JCO.2017.74.7576 . [DOI] [PubMed] [Google Scholar]
- 29.Shaw AT, Felip E, Bauer TM, Besse B, Navarro A, Postel-Vinay S, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 2017;18(12):1590–9. Epub 2017/10/28. 10.1016/S1470-2045(17)30680-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 2016;17(12):1653–60. Epub 2016/11/09. 10.1016/S1470-2045(16)30562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373(8):726–36. Epub 2015/08/20. 10.1056/NEJMoa1502309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuang JC, Stehr H, Liang Y, Das M, Huang J, Diehn M, et al. ERBB2-Mutated Metastatic Non-Small Cell Lung Cancer: Response and Resistance to Targeted Therapies. Journal of Thoracic Oncology. 2017;12(5):833–42. 10.1016/j.jtho.2017.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal A, Lewison G, Idir S, Peters M, Aldige C, Boerckel W, et al. The State of Lung Cancer Research: A Global Analysis. J Thorac Oncol. 2016;11(7):1040–50. 10.1016/j.jtho.2016.03.010 . [DOI] [PubMed] [Google Scholar]
- 34.Presley CJ, Tang D, Soulos PR, Chiang AC, Longtine JA, Adelson KB, et al. Association of Broad-Based Genomic Sequencing With Survival Among Patients With Advanced Non-Small Cell Lung Cancer in the Community Oncology Setting. JAMA. 2018;320(5):469–77. Epub 2018/08/09. 10.1001/jama.2018.9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, Chae KJ, Yoon SH, Kim M, Keam B, Kim TM, et al. Repeat biopsy of patients with acquired resistance to EGFR TKIs: implications of biopsy-related factors on T790M mutation detection. Eur Radiol. 2018;28(2):861–8. Epub 2017/08/09. 10.1007/s00330-017-5006-6 . [DOI] [PubMed] [Google Scholar]
- 36.Itchins M, Chia PL, Hayes SA, Howell VM, Gill AJ, Cooper WA, et al. Treatment of ALK-rearranged non-small cell lung cancer: A review of the landscape and approach to emerging patterns of treatment resistance in the Australian context. Asia Pac J Clin Oncol. 2017;13 Suppl 3:3–13. Epub 2017/08/11. 10.1111/ajco.12754 . [DOI] [PubMed] [Google Scholar]
- 37.Ali SM, Hensing T, Schrock AB, Allen J, Sanford E, Gowen K, et al. Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-Responsive ALK-Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization. Oncologist. 2016;21(6):762–70. Epub 2016/06/02. 10.1634/theoncologist.2015-0497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aisner D, Sholl LM, Berry LD, Haura EB, Ramalingam SS, Glisson BS, et al. Effect of expanded genomic testing in lung adenocarcinoma (LUCA) on survival benefit: The Lung Cancer Mutation Consortium II (LCMC II) experience. Journal of Clinical Oncology. 2016;34(15_suppl):11510–. 10.1200/JCO.2016.34.15_suppl.11510 [DOI] [Google Scholar]
- 39.Zhao H, Fan Y, Ma S, Song X, Han B, Cheng Y, et al. Final Overall Survival Results from a Phase III, Randomized, Placebo-Controlled, Parallel-Group Study of Gefitinib Versus Placebo as Maintenance Therapy in Patients with Locally Advanced or Metastatic Non–Small-Cell Lung Cancer (INFORM; C-TONG 0804). Journal of Thoracic Oncology . 2015;10(4):655–64. 10.1097/JTO.0000000000000445 [DOI] [PubMed] [Google Scholar]
- 40.Hida T, Nokihara H, Kondo M, Kim YH, Azuma K, Seto T, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): an open-label, randomised phase 3 trial. Lancet. 2017;390(10089):29–39. Epub 2017/05/16. 10.1016/S0140-6736(17)30565-2 . [DOI] [PubMed] [Google Scholar]
- 41.Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5(8):842–9. Epub 2015/05/15. 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tannock IF, Hickman JA. Limits to Personalized Cancer Medicine. N Engl J Med. 2016;375(13):1289–94. Epub 2016/09/30. 10.1056/NEJMsb1607705 . [DOI] [PubMed] [Google Scholar]
- 43.Prasad V. Perspective: The precision-oncology illusion. Nature. 2016;537(7619):S63 Epub 2016/09/08. 10.1038/537S63a . [DOI] [PubMed] [Google Scholar]
- 44.Joyner MJ, Paneth N, Ioannidis JP. What Happens When Underperforming Big Ideas in Research Become Entrenched? JAMA. 2016;316(13):1355–6. Epub 2016/07/29. 10.1001/jama.2016.11076 . [DOI] [PubMed] [Google Scholar]
- 45.Gutierrez ME, Choi K, Lanman RB, Licitra EJ, Skrzypczak SM, Pe Benito R, et al. Genomic Profiling of Advanced Non–Small Cell Lung Cancer in Community Settings: Gaps and Opportunities. Clinical Lung Cancer. 2017;18(6):651–9. 10.1016/j.cllc.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Bare LA, Bender RA, Sninsky JJ, Wilson LS, Devlin JJ, et al. Cost Effectiveness of Sequencing 34 Cancer-Associated Genes as an Aid for Treatment Selection in Patients with Metastatic Melanoma. Mol Diagn Ther. 2015;19(3):169–77. Epub 2015/05/01. 10.1007/s40291-015-0140-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallego CJ, Shirts BH, Bennette CS, Guzauskas G, Amendola LM, Horike-Pyne M, et al. Next-Generation Sequencing Panels for the Diagnosis of Colorectal Cancer and Polyposis Syndromes: A Cost-Effectiveness Analysis. J Clin Oncol. 2015;33(18):2084–91. Epub 2015/05/06. 10.1200/JCO.2014.59.3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haslem DS, Van Norman SB, Fulde G, Knighton AJ, Belnap T, Butler AM, et al. A Retrospective Analysis of Precision Medicine Outcomes in Patients With Advanced Cancer Reveals Improved Progression-Free Survival Without Increased Health Care Costs. J Oncol Pract. 2017;13(2):e108–e19. Epub 2016/09/08. 10.1200/JOP.2016.011486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong J, Pan K, Fakih M, Pal S, Salgia R. Value-based genomics. Oncotarget. 2018;9(21):15792–815. Epub 2018/04/13. 10.18632/oncotarget.24353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janku F, Berry DA, Gong J, Parsons HA, Stewart DJ, Kurzrock R. Outcomes of phase II clinical trials with single-agent therapies in advanced/metastatic non-small cell lung cancer published between 2000 and 2009. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(22):6356–63. Epub 2012/09/28. 10.1158/1078-0432.CCR-12-0178 . [DOI] [PubMed] [Google Scholar]
- 51.Jardim DL, Schwaederle M, Wei C, Lee JJ, Hong DS, Eggermont AM, et al. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. J Natl Cancer Inst. 2015;107(11). Epub 2015/09/18. 10.1093/jnci/djv253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33(32):3817–25. Epub 2015/08/26. 10.1200/JCO.2015.61.5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwaederle M, Zhao M, Lee JJ, Lazar V, Leyland-Jones B, Schilsky RL, et al. Association of Biomarker-Based Treatment Strategies With Response Rates and Progression-Free Survival in Refractory Malignant Neoplasms: A Meta-analysis. JAMA Oncol. 2016;2(11):1452–9. Epub 2016/06/09. 10.1001/jamaoncol.2016.2129 . [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Zhu D, Lu B, Liu G, Xu T. P2.01–102 Comprehensive Next-Generation Sequencing Guided Targeted Therapies Improve Clinical Outcomes of Lung Cancer Patients. Journal of Thoracic Oncology. 2018;13(10):S704 10.1016/j.jtho.2018.08.1157 [DOI] [Google Scholar]
- 55.Lim C, Tsao MS, Le LW, Shepherd FA, Feld R, Burkes RL, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26(7):1415–21. Epub 2015/04/30. 10.1093/annonc/mdv208 . [DOI] [PubMed] [Google Scholar]
- 56.Spicer J, Tischer B, Peters M. EGFR mutation testing and oncologist treatment choice in advanced NSCLC: global trends and differences. Ann Oncol. 2015;26(suppl 1):i57–161. [Google Scholar]
- 57.Oncology ASoC. The State of Cancer Care in America, 2016: A Report by the American Society of Clinical Oncology. Journal of Oncology Practice. 2016;12(4):339–83. 10.1200/JOP.2015.010462 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oncology TASoC. The State of Cancer Care in America, 2017: A Report by the American Society of Clinical Oncology. Journal of Oncology Practice. 2017;13(4):e353–e94. 10.1200/JOP.2016.020743 . [DOI] [PubMed] [Google Scholar]
- 59.Mason C, Ellis PG, Lokay K, Barry A, Dickson N, Page R, et al. Patterns of Biomarker Testing Rates and Appropriate Use of Targeted Therapy in the First-Line, Metastatic Non-Small Cell Lung Cancer Treatment Setting. J Clin Pathways. 4(1):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li SD, Ma M, Li H, Waluszko A, Sidorenko T, Schadt EE, et al. Cancer gene profiling in non-small cell lung cancers reveals activating mutations in JAK2 and JAK3 with therapeutic implications. Genome Med. 2017;9(1):89 Epub 2017/10/31. 10.1186/s13073-017-0478-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li C, Fang R, Sun Y, Han X, Li F, Gao B, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One. 2011;6(11):e28204 Epub 2011/12/06. 10.1371/journal.pone.0028204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20(3):251–4. Epub 2014/02/25. 10.1038/nm.3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aso T, Uozaki H, Morita S, Kumagai A, Watanabe M. Loss of ARID1A, ARID1B, and ARID2 Expression During Progression of Gastric Cancer. Anticancer Res. 2015;35(12):6819–27. Epub 2015/12/08. . [PubMed] [Google Scholar]
- 64.Cornen S, Adelaide J, Bertucci F, Finetti P, Guille A, Birnbaum DJ, et al. Mutations and deletions of ARID1A in breast tumors. Oncogene. 2012;31(38):4255–6. Epub 2012/01/18. 10.1038/onc.2011.598 . [DOI] [PubMed] [Google Scholar]
- 65.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2(10):899–905. Epub 2012/08/11. 10.1158/2159-8290.CD-12-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Streppel MM, Lata S, DelaBastide M, Montgomery EA, Wang JS, Canto MI, et al. Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus. Oncogene. 2014;33(3):347–57. Epub 2013/01/16. 10.1038/onc.2012.586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45(6):592–601. Epub 2013/05/07. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang HT, Chen SM, Pan LB, Yao J, Ma HT. Loss of function of SWI/SNF chromatin remodeling genes leads to genome instability of human lung cancer. Oncol Rep. 2015;33(1):283–91. Epub 2014/11/06. 10.3892/or.2014.3584 . [DOI] [PubMed] [Google Scholar]
- 69.Yoshimoto T, Matsubara D, Nakano T, Tamura T, Endo S, Sugiyama Y, et al. Frequent loss of the expression of multiple subunits of the SWI/SNF complex in large cell carcinoma and pleomorphic carcinoma of the lung. Pathol Int. 2015;65(11):595–602. Epub 2015/09/09. 10.1111/pin.12350 . [DOI] [PubMed] [Google Scholar]
- 70.Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014;74(9):2465–75. Epub 2014/05/03. 10.1158/0008-5472.CAN-13-3608 . [DOI] [PubMed] [Google Scholar]
- 71.Natarajan TG, Kallakury BV, Sheehan CE, Bartlett MB, Ganesan N, Preet A, et al. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell Int. 2010;10:13 Epub 2010/05/04. 10.1186/1475-2867-10-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daniel DC. Highlight: BRCA1 and BRCA2 proteins in breast cancer. Microsc Res Tech. 2002;59(1):68–83. Epub 2002/09/21. 10.1002/jemt.10178 . [DOI] [PubMed] [Google Scholar]
- 73.Yuan Y, Yost SE, Yuan YC, Solomon NM, Mambetsariev I, Pal S, et al. Genomic mutation-driven metastatic breast cancer therapy: a single center experience. Oncotarget. 2017;8(16):26414–23. Epub 2017/01/07. 10.18632/oncotarget.14476 [DOI] [PMC free article] [PubMed] [Google Scholar]