Abstract
RAS-signaling mutations induce the myelomonocytic differentiation and proliferation of hematopoietic stem and progenitor cells. Moreover, they are important players in the development of myeloid neoplasias. RAF kinase inhibitor protein (RKIP) is a negative regulator of RAS-signaling. As RKIP loss has recently been described in RAS-mutated myelomonocytic acute myeloid leukemia, we now aimed to analyze its role in myelomonocytic differentiation and RAS-driven leukemogenesis. Therefore, we initially analyzed RKIP expression during human and murine hematopoietic differentiation and observed that it is high in hematopoietic stem and progenitor cells and lymphoid cells but decreases in cells belonging to the myeloid lineage. By employing short hairpin RNA knockdown experiments in CD34+ umbilical cord blood cells and the undifferentiated acute myeloid leukemia cell line HL-60, we show that RKIP loss is indeed functionally involved in myelomonocytic lineage commitment and drives the myelomonocytic differentiation of hematopoietic stem and progenitor cells. These results could be confirmed in vivo, where Rkip deletion induced a myelomonocytic differentiation bias in mice by amplifying the effects of granulocyte macrophage-colony-stimulating factor. We further show that RKIP is of relevance for RAS-driven myelomonocytic leukemogenesis by demonstrating that Rkip deletion aggravates the development of a myeloproliferative disease in NrasG12D-mutated mice. Mechanistically, we demonstrate that RKIP loss increases the activity of the RAS-MAPK/ERK signaling module. Finally, we prove the clinical relevance of these findings by showing that RKIP loss is a frequent event in chronic myelomonocytic leukemia, and that it co-occurs with RAS-signaling mutations. Taken together, these data establish RKIP as novel player in RAS-driven myeloid leukemogenesis.
Introduction
Activating RAS-signaling mutations comprise sequence variants within the RAS oncogenes themselves, but also can affect upstream activators and regulators of RAS-signaling cascades. Among others, these include CBL, PTPN11, NF1 as well as a wide range of receptor tyrosine kinases with relevance for physiological and malignant hematopoiesis. RAS-signaling mutations skew hematopoiesis into the myelomonocytic lineage and ultimately drive the proliferation of these cells.1 Mechanistically, they constitutively activate downstream signaling cascades, including the RAS-mitogen activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) and Phosphoinositide 3-kinase/AKT pathways.2 While this already causes myelomonocytic lineage commitment and increased proliferation of hematopoietic stem and progenitor cells (HSPC) per se,1,3,4 it also increases the sensitivity to granulocyte macrophage-colony-stimulating factor (GM-CSF),5 which augments these biological effects even further. Importantly, both increased myelomonocytic lineage commitment and proliferation are considered as key steps in the pathogenesis of myelomonocytic leukemias. Indeed, RAS-signaling mutations are essential players within the development of these malignancies and cause a myeloproliferative disease (MPD) with hyper-proliferation of the monocytic and granulocytic lineages in mice.6–9 In agreement with these data, RAS-signaling mutations are frequently detected in myeloid neoplasias. More than 10-20% of acute myeloid leukemia (AML) cases exhibit either NRAS or KRAS mutations, respectively.10–12 A myeloid neoplasia with particular dependence on aberrant RAS-signaling is chronic myelomonocytic leukemia (CMML), an aggressive malignancy characterized by increased myelomonocytic differentiation and proliferation. Indeed, more than 40% of CMML patients exhibit one or more mutations in the RAS-signaling genes.13,14 Recently, it was shown that the extent of RAS-signaling activation in myeloid neoplasias is not only determined by the presence of mutations, but also by the aberrant expression profiles of one or more of its regulators. This can also be of relevance for RAS-driven myeloid leukemogenesis, as shown for the RAS-signaling inhibitor SPRY2, which demonstrates decreased expression levels in TET2-mutated patients.15
RAF kinase inhibitor protein (RKIP) is a negative regulator of various intracellular signaling modules, including the RAS-MAPK/ERK and nuclear factor-κB pathways.16,17 A somatic loss of RKIP expression has been described in a variety of solid cancers and a metastasis-suppressor function could be shown in vitro and in vivo.18,19 We have previously shown that a leukemia-specific loss of RKIP occurs in patients with therapy-related AML with a predisposing germline mutation in CRAF.20 On a functional level, RKIP drives the oncogenic potential of mutant CRAF, thereby contributing to leukemogenesis in these patients. Subsequently, we could show that RKIP loss is of relevance for other subtypes of AML as well.12,21,22 It occurs in up to 20% of AML cases and contributes to leukemogenesis by increasing the proliferation of AML cells.12,21 In agreement with the data from therapy-related AML (t-AML) patients with CRAF germline mutations, RKIP loss is correlated with RAS-signaling mutations and increased the leukemogenic potential of mutant RAS in a series of in vitro assays. Interestingly, we observed that RKIP loss is also correlated with myelomonocytic and monocytic AML phenotypes, which suggests that RKIP might play a role in myelomonocytic differentiation as well.12,20,23
In this study, we aimed to clarify a connection between RKIP and myeloid skewing of hematopoiesis in more detail and demonstrate that RKIP loss contributes to myelomonocytic lineage commitment of HSPC in vitro and in vivo. We further show relevance of RKIP for RAS-driven myelomonocytic leukemogenesis, by demon strating that Rkip deletion aggravates myelomonocytic MPD development in NrasG12D-mutated mice. Mechanistically, we show that RKIP loss potentiates the RAS-induced activation of the RAS-MAPK/ERK signaling cascade. Finally, we prove the clinical relevance of these findings by showing that RKIP loss is a frequent event in primary CMML patient samples and frequently co-occurs with RAS-signaling mutations. These data establish RKIP as a novel player in RAS-driven myeloid leukemogenesis.
Methods
Primary patient samples and cell lines
Chronic myelomonocytic leukemia patient samples were collected at the Division of Hematology, Medical University of Graz, Austria, as well as in the Austrian Biodatabase for CMML. All samples were processed and stored as described in detail in the Online Supplementary Methods. Healthy CD34+ HSPC were collected from umbilical cord blood specimens (EasySep, STEMCELL Technologies) according to the manufacturerś instructions and processed as described before.24 Peripheral blood samples from healthy donors were used to collect CD14+ monocytes (MACS, Miltenyi Biotec), B lymphocytes and granulocytes (LymphoprepTM, STEMCELL Technologies and human B Lymphocyte enrichment set, BD biosciences) according to the manufacturer’s protocol. 293T, NB4 and HL-60 cell lines were obtained from the German National Resource Center for Biological Material (DSMZ, Braunschweig, Germany). Low passage stocks were frozen and cells were always passaged for less than six months after resuscitation. Additionally, cells were screened by variable number of tandem repeat profiling (VNTR) for authenticity.21 Lentiviral transduction of cell lines and primary HSPC were performed as previously described.12,21,22
Mouse experiments
All mouse experiments were performed on a C57BL/6 strain background. Survival analyses were based on groups of at least eleven animals, all other experiments comprised at least three animals. Genotyping was performed using tail tips as previously described.19,25 Mice with complete deletion of Rkip (Rkip−/−) as well as their controls (Rkip+/+) were obtained from Professor John Sedivy (Brown University, Providence, RI, USA). Mx1-Cre mice were obtained from Dr. Karen Blyth (Cancer Research UK Beatson Institute, Glasgow, UK), Nras-LSLG12D (JAX stock #008304; hereafter referred to as Nras) from The Jackson Laboratory (Bar Harbor, ME, USA).26 Mx1-Cre and Nras animals were kept in a heterozygous situation and crossed to Rkip−/− and Rkip+/+ mice to obtain Mx1-Cre/Nras/Rkip−/− and Mx1-Cre/Nras/Rkip+/+ genotypes, respectively. Detailed procedures of mouse analysis are presented in the Online Supplementary Methods.
Immunoblot analysis, real time quantitative polymerase chain reaction, next-generation sequencing, flow cytometry and in vitro differentiation assays
These assays were extensively described previously11,21,22,27–30 and are presented in detail in the Online Supplementary Methods.
Database retrieval and statistical analyses
Microarray expression data for RKIP expression in murine hematopoietic cell compartments were downloaded via the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession numbers GSE27787,31 GSE5677,32 GSE2781633 and GSE20377.34 For the statistical analysis of in vitro and in vivo experiments, paired and unpaired Student’s t-tests, respectively, were employed. For comparisons in primary patient samples, we used the Wilcoxon-Mann-Whitney test for continuous variables and the Fisher’s Exact test for dichotomous variables. The effects of RKIP expression on survival were tested by the log-rank test. SPSS version 22.0 (SPSS Inc.) was employed for these calculations. All tests were two-sided and P<0.050 was considered statistically significant.
Study approval
The study was reviewed and approved by the institutional review board (28-481 ex 15/16) and conducted in accordance with the Declaration of Helsinki. Mouse experiments were approved by the Federal Ministry for Science, Research and Economy (GZ: BMWF-66.010/0050-II/3b/2013).
Results
RAF kinase inhibitor protein expression is high in hematopoietic stem and progenitor cells and lymphoid cells but low in cells belonging to the myeloid lineage
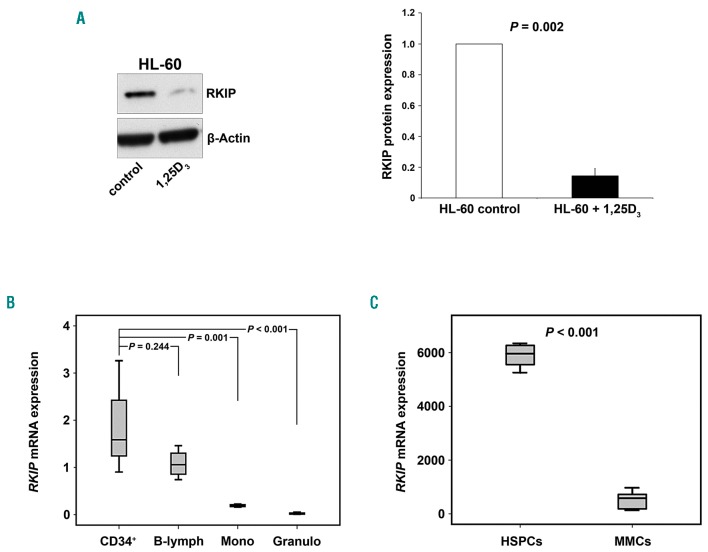
Hypothesizing that RKIP loss is a driver for myelomonocytic lineage commitment, one would assume that cells with myelomonocytic differentiation would demonstrate lower RKIP expression levels. In a first step, we therefore employed an HL-60 in vitro differentiation model. HL-60 is an undifferentiated AML cell line and can be forced into the myelomonocytic lineage by addition of 1,25 dihydroxyvitamin D3.35 Interestingly, we observed that 1,25D3-induced differentiation in these cells was accompanied by a significant decrease in the amount of RKIP protein (P=0.002) (Figure 1A). We then aimed to delineate RKIP expression in human hematopoiesis by studying CD34+ HSPC derived from three umbilical cord blood specimens on the one hand, as well as mature lymphocytes, granulocytes and monocytes from four healthy individuals on the other. Due to the restricted cell numbers available, we studied RKIP mRNA by the means of quantitative-polymerase chain reaction (qPCR) in these experiments. However, we have previously shown that decreased expression of RKIP at the protein level is accompanied by its downregulation at the mRNA level as well.12,20–22 In accordance with HL-60 data, RKIP was prominently expressed in healthy CD34+ HSPC (Figure 1B). RKIP expression levels in lymphocytes were similar to those observed in HSPC (P=0.244), while they were significantly reduced in monocytes (P=0.001) and granulocytes (P<0.001).
Figure 1.
RAF kinase inhibitor protein (RKIP) expression is low in cells belonging to the myeloid lineage. (A) Immature HL-60 acute myeloid leukemia (AML) cells were treated with 100nM 1,25-dihydroxyvitamin D3 (1,25D3) for 48 hours to induce myeloid differentiation. Immunoblot analysis demonstrates a decrease in RKIP protein expression in 1,25D3 treated cells. The graph represents the mean of three independent experiments ±Standard Deviation; expression values are given as x-fold expression of the HL-60 control. Statistical significance was evaluated using Student’s t-test. (B) Box plots showing RKIP mRNA expression, studied via quantitative polymerase chain reaction in CD34+ hematopoietic stem and progenitor cells (HSPC) (CD34+, n=3), lymphocytes (B-lymph, n=4), monocytes (Mono, n=4) and granulocytes (Granulo, n=4) from healthy donors. In comparison to HSPC, RKIP mRNA expression is significantly reduced in monocytes and granulocytes, while no significant difference was observed for lymphocytes. Graphs denote the x-fold RKIP expression levels of NB4 cells, which were used as a calibrator and arbitrarily set to a value of 1. P-values were calculated using Student’s t-test. (C) Box plots showing that the expression of RKIP mRNA in differentiated cells belonging to the myeloid lineage (MMC) is also decreased in mice. These data were generated by re-analysis of a previously published murine microarray gene expression profiling dataset.31 HSPC included long-term hematopoietic stem cells (lin−, Sca+, kit+, CD34−), short-term hematopoietic stem cells (lin−, Sca+, kit+, CD34+), LSK (lin−, Sca+, kit+) and hematopoietic progenitor cells (lin−). Myelomonocytic cells (MMC) included Gr-1+ neutrophils and Mac-1+ monocytes/macrophages. Statistical significance was calculated by Student’s t-test.
Finally, we sought to confirm these findings in a murine setting. We therefore performed a database retrieval via the Gene Expression Omnibus (GEO) database and re-analyzed RKIP mRNA in a previous publication of Konuma et al.31 (GEO data set GDS3997) who performed microarray analyses in different hematopoietic cell and progenitor compartments in C57BL/6 mice. In agreement with our findings in HL-60 and in healthy human individuals, RKIP mRNA levels were high in HSPC and lymphocytes but significantly reduced in more differentiated cells of the myeloid lineage (P<0.001) (Figure 1C). Of note, by focusing on specific HSPC compartments within GDS3997 and three additional datasets,31–34 we observed that RKIP expression remains high until the granulocyte-macrophage-progenitor (GMP) stage and decreases during the terminal stages of myelomonocytic differentiation (Online Supplementary Figure S1).
Knockdown of RAF kinase inhibitor protein amplifies the myelomonocytic differentiation of hematopoietic stem and progenitor cells
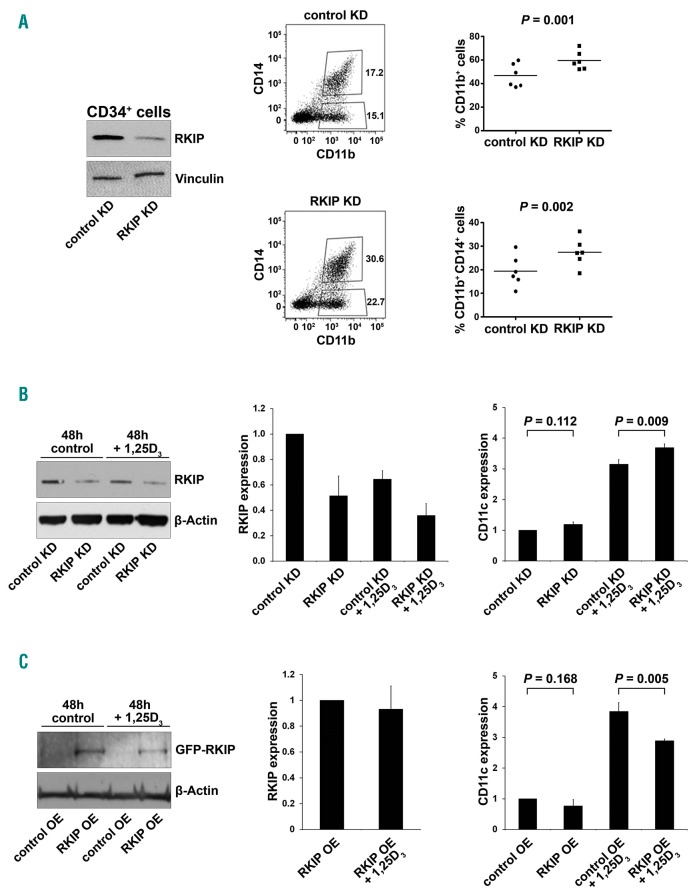
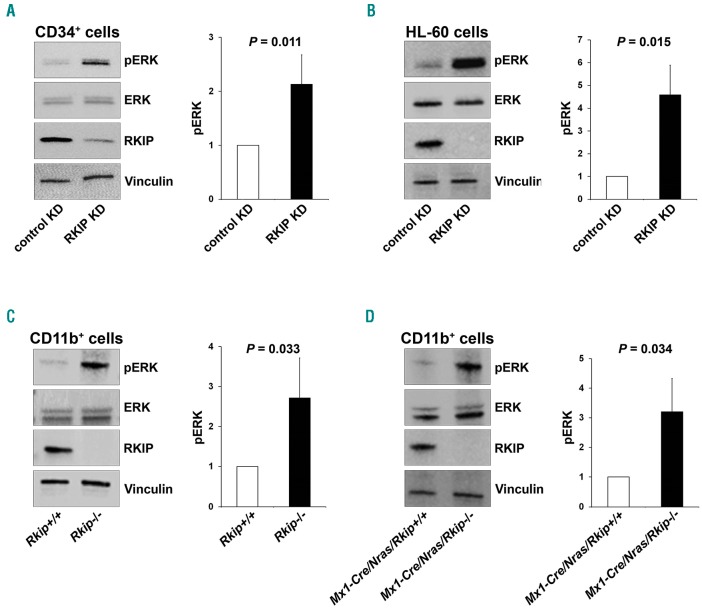
Having shown that RKIP levels are high in HSPC and reduced in differentiated myeloid cells, we next aimed to investigate whether RKIP expression levels are of functional relevance for the myeloid lineage commitment of HSPC. Therefore, we performed lentiviral knockdown of RKIP in CD34+ human HSPC isolated from umbilical cord blood specimens (Figure 2A). Subsequently, cells were treated with a granulocyte-macrophage colony-stimulating factor (GM-CSF)/flt-3 ligand (FL)/stem cell factor (SCF)/tumor necrosis factor-α (TNFα) cytokine mix to induce myelomonocytic differentiation.29 Five days later, the expression of myelomonocytic surface markers CD11b and CD14 was assessed. In these experiments, myelomonocytic differentiation was significantly increased in HSPC transduced with the RKIP shRNA (HSPC RKIP KD) as compared to HSPC control KD (P=0.001 for CD11b+ cells and P=0.002 for CD11b+ CD14+ cells) (Figure 2A), confirming a role of RKIP loss in myeloid differentiation of HSPC. RKIP KD as a single event without the addition of cytokines was insufficient to induce myelomonocytic differentiation (data not shown). Of note, RKIP is a negative regulator of RAS-MAPK/ERK signaling. To evaluate whether RKIP KD indeed enhances RAS-MAPK/ERK signaling in HSPC, we studied the phosphorylation of ERK (pERK) in the conditions mentioned above. Indeed, HSPC with RKIP KD displayed increased pERK levels (Figure 6A), suggesting that the RKIP-induced aggravation of differentiation might be mediated via activation of the RAS-MAPK/ERK pathway.
Figure 2.
RAF kinase inhibitor protein (RKIP) is functionally involved in myeloid lineage differentiation. (A) RKIP shRNA knockdown (KD) in CD34+ human hematopoi-etic stem and progenitor cells (HSPC) increased the myelomonocytic differentiation induced by a granulocyte-macrophage colony-stimulating factor (GM-CSF)/flt-3 ligand (FL)/stem cell factor (SCF)/tumor necrosis factor-α (TNFα) cytokine mix. (Left) Immunoblot showing the successful knockdown of RKIP. Control KD denotes control transduced cells. Representative flow cytometry plots showing increased expression of the myelomonocytic surface markers CD11b and CD14 in CD34+ HSPC with RKIP KD shown in the middle. (Right) Results of all six experiments performed; median is also shown. Statistical significance was calculated by Student’s t-test. Of note, RKIP KD as a single event without the addition of cytokines was insufficient to induce differentiation (data not shown). (B) RKIP KD in HL-60 AML cells increased 1,25D3-induced myelomonocytic differentiation, as assessed by flow cytometric expression of CD11c. (Left) Immunoblot showing the successful knockdown of RKIP; (right) mean CD11c expression of three independent experiments ±Standard Deviation (SD); expression values are given as x-fold expression of HL-60 control KD cells, statistical significance was evaluated by Student’s t-test. (C) RKIP overexpression (RKIP OE) in HL-60 AML cells reduced 1,25D3 induced myelomonocytic differentiation, as assessed by flow cytometric expression of CD11c. HL-60 cells were transduced with eGFP-C1-6xG-hRKIP (RKIP OE) or empty vector (control OE). (Left) Immunoblot showing successful RKIP overexpression; (right) mean CD11c expression of three independent experiments ±SD. Expression values are given as x-fold expression of HL-60 control OE cells. Statistical significance was evaluated by Student’s t-test. Note that HL-60 cells were treated with 10nM 1,25D3.
Figure 6.
RAF kinase inhibitor protein (RKIP) regulates RAS-MAPK/ERK signaling in the myeloid system. (A) RKIP shRNA knockdown (KD) in CD34+ human hematopoietic stem and progenitor cells (HSPC) increased RAS-MAPK/ERK signaling, as measured by the phosphorylation of ERK (pERK). (Left) A representative immunoblot. Graph represents the mean of three independent experiments ±Standard Deviation (SD); pERK intensity is given as x-fold change to the CD34+ control KD. (B) RKIP shRNA KD in HL-60 increased pERK levels as well. (Left) Representative immunoblot. Graph represents the mean of three independent experiments ±SD; pERK intensity is given as x-fold change to the HL-60 control KD. (C) Rkip deletion enhanced the activity of RAS-MAPK/ERK signaling in CD11b+ cells isolated from the bone marrow of mice. (Left) A representative immunoblot. Graph represents the mean of three independent experiments ±SD; pERK intensity is given as x-fold change to the Rkip+/+ control genotype. (D) Rkip deletion also increased the activity of RAS-MAPK/ERK signaling in CD11b+ cells isolated from Nras-mutated mice. A representative immunoblot and the graph is presented as described above, Mx1-Cre/Nras/Rkip+/+ were used as control group. Mice experiments were performed using n=3 mice for each genotype. Statistical significance was evaluated by Student’s t-test in all cases.
To further corroborate the role of RKIP in myelomonocytic differentiation, we again employed the 1,25D3-based HL-60 differentiation model and performed additional RKIP modulation by transfection of RKIP siRNA and over-expression constructs, respectively (Online Supplementary Figure S2). Initially, we thereby sought to confirm the role of RKIP on RAS-MAPK/ERK signaling and therefore studied the expression of pERK. In agreement with the results from healthy CD34+ HSPC, RKIP KD thereby increased pERK levels, which corroborates the role of RKIP as a regulator of RAS-MAPK/ERK signaling (Figure 6B). To study the role of RKIP on myelomonocytic differentiation, we assessed the CD11c surface expression in these experiments, which was previously established as the most suitable marker for this HL-60-based experimental approach.36 1,25D3-treated HL-60 cells harboring RKIP knockdown thereby demonstrated an increased potential to differentiate into the myeloid lineage as assessed by an increase in the expression of CD11c (P=0.009) (Figure 2B). The opposite effect was observed after RKIP overexpression (P=0.005) (Figure 2C). Finally, we studied the effect of RKIP modulation without additional 1,25D3 incubation. As seen for healthy HSPC, myelomonocytic differentiation could not be induced in this scenario (P=0.112 for RKIP knockdown and P=0.168 for RKIP overexpression).
Rkip deletion contributes to the development of a myelomonocytic-lineage-biased hematopoietic system in a murine in vivo model
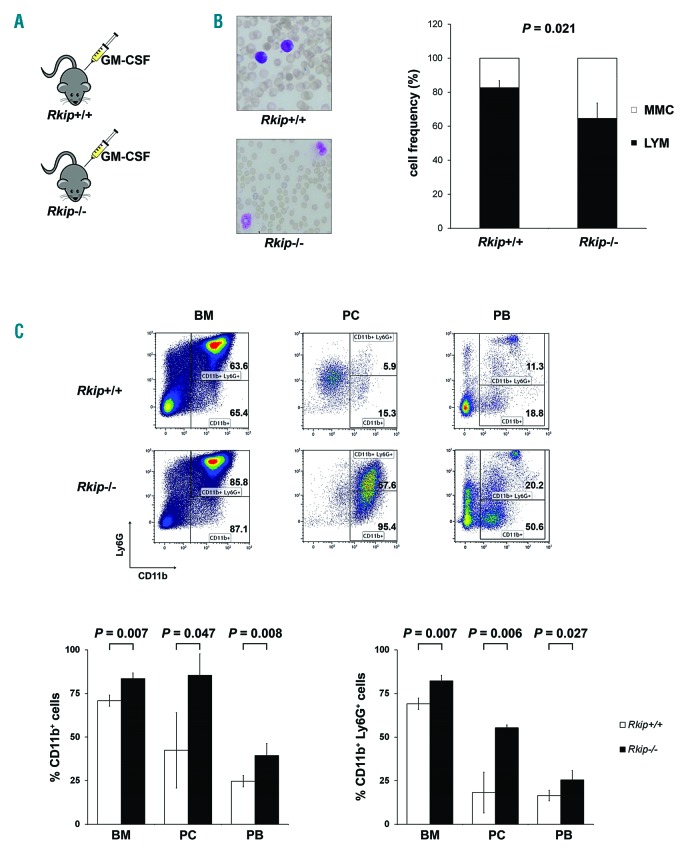
After demonstrating the functional involvement of RKIP loss in myelomonocytic differentiation in vitro, we focused on its effects in vivo. To do this, we analyzed the hematopoietic system in a murine model with a complete deletion of Rkip (Rkip−/−) (Online Supplementary Table S1 and Online Supplementary Figures S3 and S4). In agreement with the in vitro data, RKIP loss as a single event thereby proved insufficient to increase the amount of myelomonocytic cells. Rkip deletion alone further proved insufficient to increase the proliferation of specific HSPC compartments (Online Supplementary Figure S4). As our in vitro data already suggested that RKIP acts as a modulator for the sensitivity to extracellular inducers of differentiation, we aimed to challenge the hematopoietic system of Rkip−/−mice by the additional intraperitoneal injection of GM-CSF in a next step (Figure 3A). Indeed, Rkip−/− mice demonstrated an increased percentage of CD11b+ as well as CD11b+ Ly6G+ myelomonocytic cells in bone marrow, peritoneal cavity cells and peripheral blood in these experiments (Figure 3B and C). As seen in the in vitro experiments mentioned above, Rkip deletion also enhanced RAS-MAPK/ERK signaling, as evidenced by increased pERK levels in Rkip−/−mice (Figure 6C). Taken together, our data indicate that RKIP loss enhances RAS-MAPK/ERK signaling on the one hand and increases GM-CSF-induced myelomonocytic lineage commitment and differentiation of HSPC on the other. Furthermore, they suggest that RKIP exerts its role in terminal myelomonocytic differentiation by acting as a rheostat that modulates the sensitivity to external stimuli, such as 1,25D3 and growth factors, respectively.
Figure 3.
Rkip deletion causes increased myeloid lineage commitment in a murine model by increasing the sensitivity to granulocyte-macrophage colony-stimulating factor (GM-CSF). (A) To assess the effects of GM-CSF in vivo, four mice with a deletion of Rkip (Rkip−/−), as well as four control mice (Rkip+/+) were injected intraperitoneally with 500ng GM-CSF twice a day for four days and were analyzed on day 5. (B) Cytological analysis of peripheral blood smears from these mice revealed a significant increase in the number of myelomonocytic cells (MMC) in Rkip−/− animals. A representative peripheral blood smear picture is shown. Lymphoid cells are shown as LYM. Graphs show the average ±Standard Deviation (SD). Statistical significance was evaluated by Student’s t-test. (C) Representative flow cyto-metric plots of the mice presented in (B) showing an increased percentage of CD11b+ and CD11b+ Ly6G+ cells in the bone marrow (BM), peritoneal cavity (PC), and peripheral blood (PB) of Rkip−/− mice. Graphs show the average ±SD. Statistical significance was evaluated by Student’s t-test. RKIP: RAF kinase inhibitor protein.
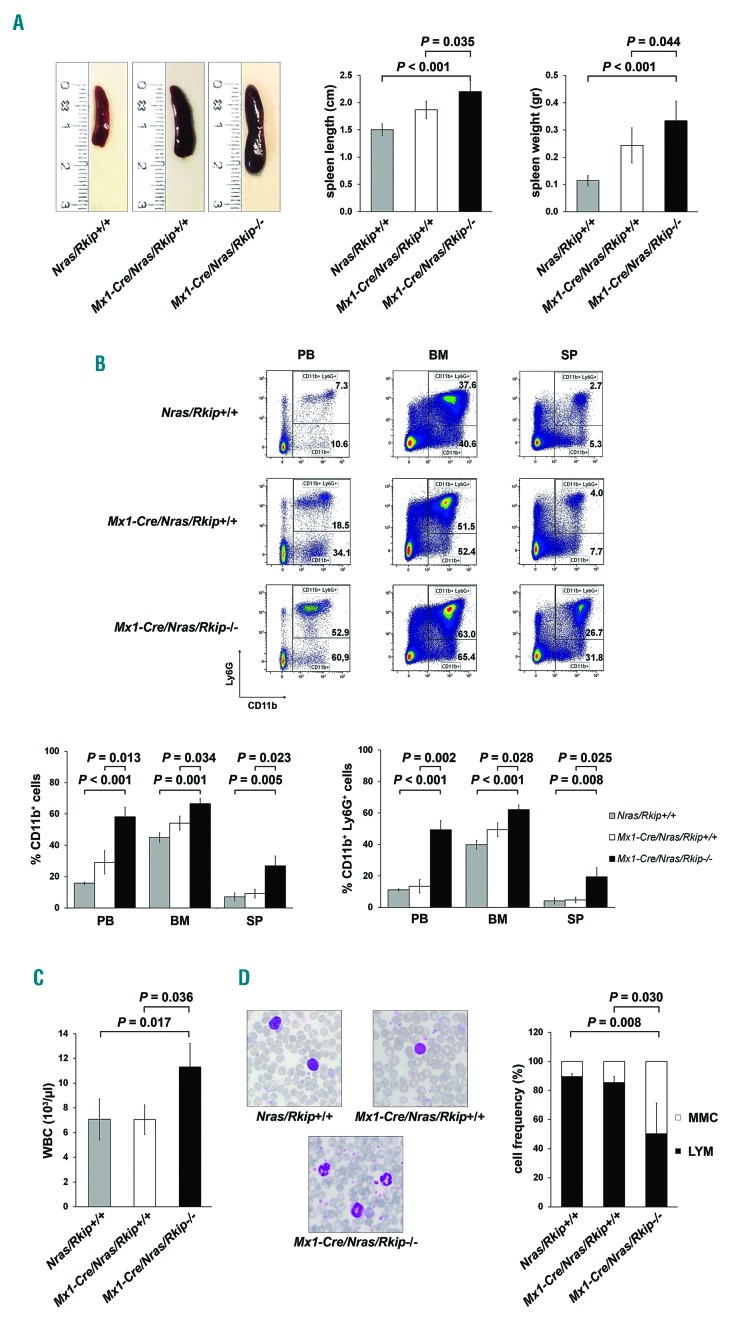
Rkip deletion aggravates myeloproliferation and the development of a myelomonocytic myeloproliferative disease in Ras-mutated mice
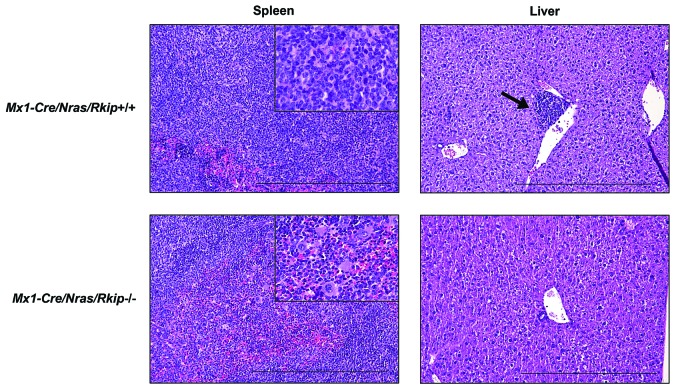
Increased myelomonocytic lineage commitment is a key step in myeloid leukemogenesis. However, Rkip−/− mice failed to develop myeloid neoplasias in our study (data not shown). As the functional assays delineating the role of RKIP in myelomonocytic differentiation suggested that RKIP rather acts as an amplifier of activated GM-CSF/RAS signaling, we next crossed Rkip−/− mice with Mx1-Cre/Nras-mutated animals (Online Supplementary Figure S5). The Mx1-Cre/Nras was chosen because: (i) RKIP loss and RAS-signaling mutations have previously been shown to co-occur in AML;12,20,22 and (ii) RKIP loss has been demonstrated to potentiate the oncogenic effects of RAS-signaling mutations in functional in vitro assays.12,20 Interestingly, Mx1-Cre/Nras mice on a pure C57BL/6 background develop a myeloproliferation that preferentially affects the myelomonocytic lineages. However, previously published data have demonstrated that these mice ultimately succumb to histiocytic sarcomas (HS) and only randomly develop a full blown MPD.8 In the current study, we elec-tively analyzed mice at an age of six months after the first pIpC injection and thereby observed that myeloproliferation was aggravated in Mx1-Cre/Nras/Rkip−/− mice, who consistently demonstrated splenomegaly, as well as increased myeloid infiltration of bone marrow, spleen and liver as compared to Nras/Rkip+/+ and to Mx1-Cre/Nras/Rkip+/+ animals (Figures 4A and B, and 5). In addi tion, only the Mx1-Cre/Nras/Rkip−/− genotype exhibited a full blown MPD, as evidenced by an accompanying leuko-cytosis in the peripheral blood (Figure 4C and D, and Online Supplementary Table S2).37 We then studied a potential involvement of the RAS-MAPK/ERK pathway in this process. As seen in the experiments described above, Rkip deletion again enhanced the activation of ERK. Importantly, these effects on RAS-MAPK/ERK signaling were visible both in the absence of mitogens and following GM-CSF stimulation (Figure 6D and Online Supplementary Figure S6). In agreement with data from Li et al.,8 HS were detected in all Nras-mutated mice studied; however, the phenotype was mitigated in the Mx1-Cre/Nras/Rkip−/− animals. Transformation into secondary AML did not occur in any of the mice, as assessed by morphological and flow cytometric evaluation of peripheral blood, bone marrow and spleen (data not shown). Interestingly, although MPD development was aggravated in Mx1-Cre/Nras/Rkip−/− animals, the median survival was similar between Mx1-Cre/Nras/Rkip−/− and Mx1-Cre/Nras/Rkip+/+ mice (P=0.339) (Online Supplementary Figure S7). Histopathological examination of moribund Mx1-Cre/Nras/Rkip+/+ mice thereby revealed that these mice suffered from extensive HS (Online Supplementary Table S3 and Online Supplementary Figure S8). In agreement with the data from 6-month old mice, the Mx1-Cre/Nras/Rkip−/− mice had a mitigated HS phenotype but an increased myeloproliferation/MPD occurrence. Future studies, using models without the predisposition to HS, will, therefore, be necessary to unambiguously delineate the effect of Rkip on the survival of Ras-driven MPD.
Figure 4.
Deletion of Rkip aggravates myeloproliferation and myeloproliferative disease (MPD) development in Nras-mutated mice. An Nras driven mouse model of myeloproliferation (Mx1-Cre/Nras) was used to study the effects of Rkip-/- on Ras-driven myeloproliferation and MPD development. Mice were electively killed at an age of six months after the first pIpC injection, Mx1-Cre/Nras/Rkip−/− mice (n=3) were compared to control mice (Mx1-Cre/Nras/Rkip+/+; n=3) as well as to mice without Mx1-Cre (Nras/Rkip+/+; n=4). (A) Representative images of spleens from Nras/Rkip+/+, Mx1-Cre/Nras/Rkip+/+ and Mx1-Cre/Nras/Rkip−/− mice as well as bar graphs of spleen lengths and weights, showing splenomegaly in animals with Rkip deletion. (B) Representative flow cytometric plots showing an increase in the percentage of CD11b+ and CD11b+ Ly6G+ myelomonocytic cells in peripheral blood (PB), bone marrow (BM) and spleen (SP) of Mx1-Cre/Nras/Rkip−/−mice when these were compared to Nras/Rkip+/+ animals as well as when compared to Mx1-Cre/Nras/Rkip+/+ littermates. (C) Peripheral blood counts demonstrate an increased number of white blood cells (WBC) in Mx1-Cre/Nras/Rkip−/− mice. (D) The leukocytosis in Mx1-Cre/Nras/Rkip−/−mice is caused by an increased number of myelomonocytic cells (MMC), as shown in the representative PB smear and as shown in the flow cyto-metric analyses shown above. Graphs show the average ± Standard Deviation. Statistical significance was calculated by Student’s t-test. RKIP: RAF kinase inhibitor protein.
Figure 5.
Induction of myeloproliferation in Nras-mutated mice with Rkip deletion coincides with a mitigation of histiocytic sarcoma development. Representative Hematoxylin & Eosin stained sections of spleen and liver of the mouse genotypes as indicated. Mice were electively killed at an age of six months after the first pIpC injection. As in the flow cytometric analyses, animals with Rkip deletion demonstrated increased myeloproliferation (spleen, bottom left and insert bottom left showing multiple megakaryocytes) as compared to the Rkip+/+ mice (spleen top left, and insert top left showing almost exclusively histiocytic sarcoma). The formation of histiocytic sarcomas was mitigated in the Rkip deleted genotypes as also clearly seen in the liver sample (bottom right, no infiltrate) compared to the Rkip+/+ liver showing infiltration by histiocytic sarcoma (top right, arrow). The black bar denotes a distance of 500 mm. RKIP: RAF kinase inhibitor protein.
Taken together, these data indicate that RKIP aggravates the effects of mutated Nras on RAS-MAPK/ERK signaling on the one hand, as well as on myeloproliferation and MPD development on the other.
RAF kinase inhibitor protein loss is frequently observed in primary chronic myelomonocytic leukemia patient specimens and co-occurs with RAS-signaling mutations
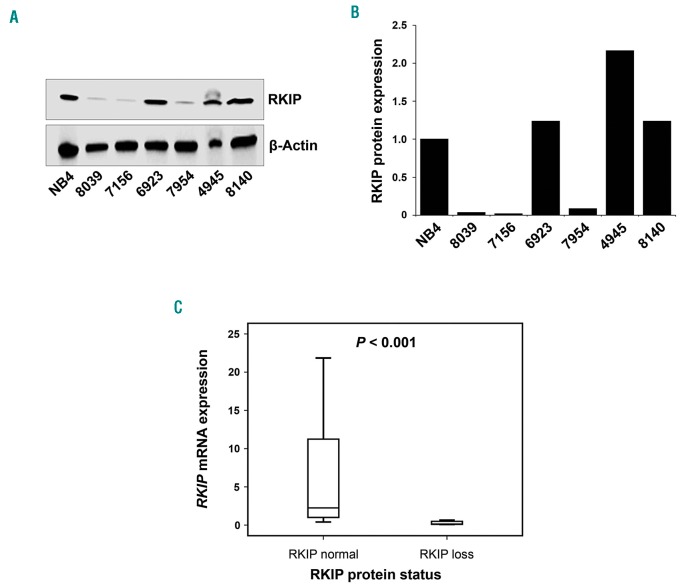
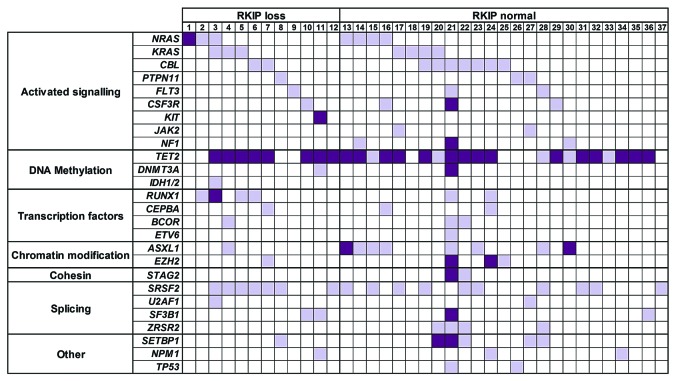
Finally, we aimed to delineate the clinical relevance of these findings and therefore analyzed a cohort of 41 primary CMML patients’ specimens for RKIP protein expression by immunoblot (Figure 7A and B; for details of clinical characteristics as well as treatment regimens administered see Online Supplementary Table S4). We chose this disease because increased myelomonocytic lineage commitment and pathological RAS-signaling are seminal steps in its pathogenesis. RKIP protein loss was defined as previously reported12,22 and could be detected in 12 of 41 (29.3%) of CMML patients’ samples. Interestingly, RKIP loss at the protein level also correlated with decreased expression of its mRNA (P<0.001) (Figure 7C). This is in agreement with data from AML, where RKIP loss has been shown to be caused by increased expression of miR-23a,21,22 and suggests that a similar mechanism might be present in CMML as well. Most interestingly, however, we observed that RKIP loss correlated with a more pronounced myelomonocytic phenotype, as assessed by the percentage of myelomonocytic cells (MMC; monocytes and granulocytes) in the peripheral blood (86% vs. 76%, P=0.030) (Online Supplementary Table S4). This is in agreement with our functional data and with previous findings from AML.12 To investigate the molecular landscape of CMML patients with RKIP loss, we performed next-generation sequencing (NGS) covering 39 genes with recurrent mutations in myeloid neoplasms (Figure 8). In total, these analyses could be performed in 37 of 41 patients. All together, we discovered 186 mutations in 37 of 37 (100%) of the patients. Thirty-three of 37 had more than one mutation with a median of four variants per sample (range 1-32). In agreement with previous studies,38 the most frequently affected genes were TET2 (75.7%), SRSF2 (46.0%), CBL (24.3%), and ASXL1 (24.3%). Mutations in NRAS and KRAS affected 13 of 37 (35.1%) of the patients (Online Supplementary Table S5). This high frequency was even increased, when mutations affecting RAS-signaling were grouped (including NRAS, KRAS, CBL, PTPN11, FLT3, CSF3R, KIT, JAK2, and NF1). Twenty-nine of 37 (78.4%) of CMML patients’ specimens showed one or more mutations within these genes. Most interestingly, however, almost all patients with RKIP loss (11 of 12, 91.7%) exhibited one or more mutations in RAS-signaling genes. Together with the previously published data from AML, this indicates that RKIP loss and RAS-signaling mutations co-occur in myeloid neoplasms, which proves the clinical relevance of the functional in vivo data mentioned above. Of note, RAS-signaling mutations were also frequent in patients with normal RKIP expression (18 of 25, 72%; comparison to patients with RKIP loss, P=0.232), which suggests that RKIP loss is not the only second genetic hit that interacts with RAS-signaling mutations in myeloid leukemogenesis. Indeed, such interactions have previously been shown for a variety of genetic aberrations, including aberrant expression of members of the dual specificity phosphatase (DUSP) and SPROUTY (Spry) families, as well as for mutations in ASXL1 and TET2.15,39–41 Finally, overall survival (OS) was similar between patients with and without RKIP loss (P=0.913) (Online Supplementary Figure S9). It must be noted, however, that these analyses are limited by the fact that these patients were managed with different treatment modalities, which ranged from best supportive care to high-dose chemotherapy. Subgroup analyses focusing on uniformly treated patients only could not be carried out due to the small sample size of this cohort.
Figure 7.
Loss of RAF kinase inhibitor protein (RKIP) is a frequent event in primary chronic myelomonocytic leukemia (CMML) patients’ samples. (A) Loss of RKIP at protein level was observed in 12 of 41 (29.3%) of cases. A representative immunoblot is shown, with RKIP protein loss being present in patients 8039, 7156 and 7954. (B) Graphs showing the x-fold change in RKIP protein expression as compared with NB4 acute myeloid leukemia (AML) cells, which were chosen as a calibrator due to their physiological RKIP expression levels.12 The expression of NB4 was arbitrarily set to 1. (C) Box plots illustrating RKIP mRNA levels in CMML patients with and without RKIP loss at the protein level. NB4 AML cells served as calibrator and statistical significance was calculated by the Wilcoxon-Mann-Whitney test.
Figure 8.
RAS-signaling mutations are frequent in chronic myelomonocytic leukemia (CMML) patients with RAF kinase inhibitor protein (RKIP) loss. Thirty-nine genes with recurrent mutations in myeloid neoplasias were screened for mutations by the means of next-generation sequencing.11 The heatmap shows the distribution and number of mutations observed in each CMML patient (n=37). Every column describes one CMML patient specimen, defined as either “RKIP loss” or “RKIP normal” according to the immunoblot results. Light purple indicates the presence of one mutation; dark purple is used to demonstrate the presence of multiple mutations. Genes without sequence variations in any of the patients are not shown.
Discussion
Hematopoietic stem and progenitor cells have the potential to differentiate into both myeloid and lymphoid hematopoietic cells. Although tight control and regulation programs are in place to maintain this system in homeostasis, skewing of hematopoiesis into the myeloid lineage can be achieved by a multitude of genetic aberrations. This includes RAS-signaling mutations,1 which drive the myelomonocytic lineage commitment by increasing the sensitivity of intracellular signaling cascades to extracellular growth factors, such as GM-CSF.5,42 In this study, we hypothesized that a loss of the RAS-signaling inhibitor RKIP plays a role in myelomonocytic differentiation as well. Therefore, we initially analyzed RKIP expression during human and murine hematopoietic differentiation and observed that it is high in HSPC and lymphoid cells but decreases in cells belonging to the myeloid lineage. By modulating RKIP expression in healthy HSPC and undifferentiated AML cell lines, we could further show that loss of RKIP expression is an important driver of myelomonocytic lineage commitment. This could be corroborated in subsequent in vivo studies, where we did show that RKIP loss increases the activation of RAS-MAPK/ERK signaling, and consequently, the GM-CSF-induced myelomonocytic differentiation of HSPC. Of note, we observed that RKIP exerts its role in myelomonocytic lineage commitment of HSPC by acting as an amplifier of GM-CSF signaling rather than inducing the differentiation process on its own. This has previously been shown for other alterations affecting RAS-signaling as well43–45 and further highlights the importance of physiological and pathological GM-CSF/RAS-sig-naling regulation in hematopoiesis.
Increased myelomonocytic lineage commitment has also been proposed to be an essential pre-phase of myeloid neoplasms.46 Indeed, a role of RKIP in myeloid leukemogenesis has been suggested previously, as its somatic loss of expression was described as a frequent event in AML.12,20–22 In line with our functional data presented above, it thereby correlated with myelomonocytic AML phenotypes.12 In the current study, we further strengthen these data by demonstrating that RKIP loss is indeed of functional relevance for the development of myelomonocytic leukemias. Again, it acted as an amplifier of pathologic RAS-signaling, as it aggravated the activity of the RAS-MAPK/ERK pathway as well as the development of a myelomonocytic MPD in mice that carry a somatically inducible mutation in Nras within the hematopoietic system. These data are further strengthened by our analysis of 41 primary CMML patients’ specimens, where we observed that RKIP loss occurs in almost 30% of cases on the one hand, and that it co-occurs with RAS-signaling mutations on the other. The data are, therefore, in agreement with previous studies of our group, where we did observe a clinical correlation and a functional synergism between RAS-signaling mutations and RKIP loss in different subtypes of AML.12,20,22 They are also in agreement with previous observations, where RAS-driven leukemogenesis could be significantly aggravated by additional inactivation of RAS-MAPK/ERK signaling inhibitors belonging to the dual specificity phosphatase (DUSP) and SPROUTY (Spry) families.15,39,41 Together with the previously shown aggravation of RAS-induced myeloid leukemogenesis by mutations in ASXL1 and TET2,15,40 respectively, these data indicate that activated RAS-signaling in human leukemias is far more complex than initially believed and cannot be explained by the occurrence of RAS-signaling mutations alone.
Finally, our data might also be of relevance for the future development of targeted treatment approaches in myeloid neoplasias, particularly for those aiming to inhibit specific signal transduction cascades. This is based on our observation that both the signaling and leukemogenic effects of RAS mutations can be influenced by aberrant expression of RAS-signaling regulator proteins. So far, development of these agents has often been hindered by the fact they showed disappointing efficacy in clinical trials, even though the results from pre-clinical models had been promising. An example for such a history of drug development are MEK-inhibitors, which efficiently attenuate Ras-driven MPD in mice, but show disappointing results in clinical trials of myeloid malignancies.47 Among others, one reason for this is the fact that the monogenic pre-clinical model does not adequately reflect the situation in myeloid neoplasia patients, who usually exhibit a complex network of co-occurring and interacting genetic aberrations within their neoplastic clone. Therefore, more detailed knowledge of the co-occurrence of mutational and non-mutational aberrations in patients’ specimens, as well as the functional consequences thereof, might not only help to extend our knowledge about the pathogenesis of this aggressive malignancy, but also to more specifically select patients that might profit from targeted therapies directed at cellular signaling. One successful example of this approach is the recent observation that sensitivity to MEK inhibitors in Nras-mutated mice can be increased by the co-occurrence of Tet2 deletion and decreased Spry2 expression levels.15 With these data, the authors identified a specific subgroup of RAS-mutated patients that will be the best candidates for MEK-directed therapy. The fact that simultaneous occurrence of RAS mutations and RKIP loss potentiated RAS-MAPK/ERK signaling as well, might identify another group of patients with particular sensitivity to this therapeutic approach. Future studies will, therefore, be warranted to specifically test this hypothesis.
In conclusion, we show that the RAS-signaling regulator RKIP plays a central role in myelomonocytic lineage commitment of HSPC. We further show its relevance for myelomonocytic leukemogenesis by demonstrating that Rkip deletion enhances RAS-MAPK/ERK signaling and aggravates the development of a myelomonocytic MPD in Nras-mutated mice. Finally, we prove the clinical relevance of these findings by showing that RKIP loss is a frequent event in primary CMML patients’ samples and frequently co-occurs with RAS-signaling mutations. These data establish RKIP as a novel player in RAS-driven myeloid leukemogenesis.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. John Sedivy for providing Rkip−/− mice.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/2/375
Funding
This study was supported by research funding from the Austrian Science Fund (grant P26619-B19 to A. Zebisch) and from the Science Foundation Ireland (grant 14/IA/2395 to W. Kolch). Work in the laboratories of A. Zebisch, A. Wölfler, and H. Sill is further supported by Leukämiehilfe Steiermark. PhD candidate V. Caraffini received funding from the Austrian Science Fund (grant P26619-B19 to A. Zebisch) and was trained within the frame of the PhD Program Molecular Medicine of the Medical University of Graz. PhD candidate J.L. Berg received funding from the Medical University of Graz within the PhD Program Molecular Medicine.
This work was supported by Biobank Graz.
References
- 1.Fatrai S, Van Gosliga D, Han L, Daenen SMGJ, Vellenga E, Schuringa JJ. KRASG12V enhances proliferation and initiates myelomonocytic differentiation in human stem/progenitor cells via intrinsic and extrinsic pathways. J Biol Chem. 2011;286(8):6061–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. [DOI] [PubMed] [Google Scholar]
- 3.Geest CR, Coffer PJ. MAPK signaling pathways in the regulation of hematopoiesis. J Leukoc Biol. 2009;86237–250. [DOI] [PubMed] [Google Scholar]
- 4.Zebisch A, Czernilofsky A, Keri G, Smigelskaite J, Sill H, Troppmair J. Signaling Through RAS-RAF-MEK-ERK: from Basics to Bedside. Curr Med Chem. 2007;14(5):601–623. [DOI] [PubMed] [Google Scholar]
- 5.Van Meter MEM, Díaz-Flores E, Archard JA, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109(9):3945–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun BS, Tuveson DA, Kong N, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101(2):597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan IT, Kutok JL, Williams IR, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113(4):528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Haigis KM, McDaniel A, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011; 117(6):2022–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Kong G, Liu Y, et al. Nras(G12D/+) promotes leukemogenesis by aberrantly regulating hematopoietic stem cell functions. Blood. 2013;121(26):5203–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashofer K, Gornicec M, Lind K, et al. Detection of prognostically relevant mutations and translocations in myeloid sarcoma by next generation sequencing. Leuk Lymphoma. 2018;59(2):501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zebisch A, Wölfler A, Fried I, et al. Frequent loss of RAF kinase inhibitor protein expression in acute myeloid leukemia. Leukemia. 2012;26(8):1842–1849. [DOI] [PubMed] [Google Scholar]
- 13.Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6(2):e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geissler K, Jäger E, Barna A, et al. Chronic myelomonocytic leukemia patients with RAS pathway mutations show high in vitro myeloid colony formation in the absence of exogenous growth factors. Leukemia. 2016;30(11):2280–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunimoto H, Meydan C, Nazir A, et al. Cooperative Epigenetic Remodeling by TET2 Loss and NRAS Mutation Drives Myeloid Transformation and MEK Inhibitor Sensitivity. Cancer Cell. 2018;33(1):44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeung K, Seitz T, Li S, et al. Suppression of Raf-1 kinase activity and MAP kinase sig nalling by RKIP. Nature. 1999; 401(6749):173–177. [DOI] [PubMed] [Google Scholar]
- 17.Al-Mulla F, Bitar MS, Taqi Z, Yeung KC. RKIP: Much more than Raf Kinase inhibitory protein. J Cell Physiol. 2013;228(8):1688–1702. [DOI] [PubMed] [Google Scholar]
- 18.Al-Mulla F, Hagan S, Behbehani AI, et al. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol. 2006;24(36):5672–5679. [DOI] [PubMed] [Google Scholar]
- 19.Escara-Wilke J, Keller JM, Ignatoski KMW, et al. Raf kinase inhibitor protein (RKIP) deficiency decreases latency of tumorigenesis and increases metastasis in a murine genetic model of prostate cancer. Prostate. 2015;75(3):292–302. [DOI] [PubMed] [Google Scholar]
- 20.Zebisch A, Haller M, Hiden K, et al. Loss of RAF kinase inhibitor protein is a somatic event in the pathogenesis of therapy-related acute myeloid leukemias with C-RAF germline mutations. Leukemia. 2009; 23(6):1049–1053. [DOI] [PubMed] [Google Scholar]
- 21.Hatzl S, Geiger O, Kuepper MK, et al. Increased expression of miR-23a mediates a loss of expression in the RAF kinase inhibitor protein RKIP. Cancer Res. 2016; 76(12):3644–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caraffini V, Perfler B, Berg JL, et al. Loss of RKIP is a frequent event in myeloid sarcoma and promotes leukemic tissue infiltration. Blood. 2018;131(7):826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zebisch A, Staber PB, Delavar A, et al. Two Transforming C-RAF Germ-Line Mutations Identified in Patients with Therapy-Related Acute Myeloid Leukemia. Cancer Res. 2006;66(7):3401–3408. [DOI] [PubMed] [Google Scholar]
- 24.Taschner S, Koesters C, Platzer B, et al. Down-regulation of RXR expression is essential for neutrophil development from granulocyte-monocyte progenitor. Blood. 2007;109(3):971–979. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Liu Y, Li Z, et al. Endogenous oncogenic NRAS mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood. 2010;116(26):5991–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal R, Lind K, Heitzer E, et al. Somatic TP53 mutations characterize preleukemic stem cells in acute myeloid leukemia. Blood. 2017;129(18):2587–2591. [DOI] [PubMed] [Google Scholar]
- 28.Gaksch L, Kashofer K, Heitzer E, et al. Residual disease detection using targeted parallel sequencing predicts relapse in cytogenetically normal acute myeloid leukemia. Am J Hematol. 2018;93(1):23–30. [DOI] [PubMed] [Google Scholar]
- 29.Caux C, Massacrier C, Dubois B, et al. Respective involvement of TGF- and IL-4 in the development of Langerhans cells and non-Langerhans dendritic cells from CD34+progenitors. J Leukoc Biol. 1999; 66(5):781–791. [DOI] [PubMed] [Google Scholar]
- 30.Zebisch A, Lal R, Müller M, et al. Acute myeloid leukemia with TP53 germ line mutations. Blood. 2016;128(18):2270–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konuma T, Nakamura S, Miyagi S, et al. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp Hematol. 2011;39(6):697–709. [DOI] [PubMed] [Google Scholar]
- 32.Sung L-Y, Gao S, Shen H, et al. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006; 38(11):1323–1328. [DOI] [PubMed] [Google Scholar]
- 33.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/-catenin Pathway Is Required for the Development of Leukemia Stem Cells in AML. Science. 2010;327(5973):1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White SL, Belov L, Barber N, Hodgkin PD, Christopherson RI. Immunophenotypic changes induced on human HL60 leukaemia cells by 1 ,25-dihydroxyvitamin D3and 12-O-tetradecanoyl phorbol-13-acetate. Leuk Res. 2005;29(10):1141–1151. [DOI] [PubMed] [Google Scholar]
- 36.Kim K, Seoh JY, Cho SJ. Phenotypic and Functional Analysis of HL-60 Cells Used in Opsonophagocytic-Killing Assay for Streptococcus pneumoniae. J Korean Med Sci. 2015;30(2):145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):238–245. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimi A, Balasis ME, Vedder A, et al. Robust patient-derived xenografts of MDS/MPN overlap syndromes capture the unique characteristics of CMML and JMML. Blood. 2017;130(4):397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Z, Chen CC, Rillahan CD, et al. Cooperative loss of RAS feedback regulation drives myeloid leukemogenesis. Nat Genet. 2015;47(5):539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Wahab O, Adli M, Lafave LM, et al. ASXL1 Mutations Promote Myeloid Transformation Through Loss of PRC2-Mediated Gene Repression. Cancer Cell. 2012;22(2):180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geiger O, Hatzl S, Kashofer K, et al. Deletion of SPRY4 is a frequent event in secondary acute myeloid leukemia. Ann Hematol. 2015;94(11):1923–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Studzinski GP. Activation of extracellular signal-regulated kinases (ERKs) defines the first phase of 1,25-dihydroxyvi-tamin d3-induced differentiation of HL60 cells. J Cell Biochem. 2001;80(4):471–482. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Studzinski GP. Kinase Suppressor of RAS (KSR) Amplifies the Differentiation Signal Provided by Low Concentrations 1,25-Dihydroxyvitamin D3. J Cell Physiol. 2004;198(3):333–342. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Zhao Y, Kauss MA, Spindel S, Lian H. Akt regulates vitamin D3-induced leukemia cell functional differentiation via Raf/MEK/ERK MAPK signaling. Eur J Cell. Biol 2009;88(2):103–115. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Studzinski GP. Oncoprotein Cot1 represses kinase suppressors of Ras1/2 and 1,25-dihydroxyvitamin D3-induced differentiation of human acute myeloid leukemia cells. J Cell Physiol. 2011; 226(5):1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mason CC, Khorashad JS, Tantravahi SK, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016;30(4):906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CC, Shah NP. The role of kinase inhibitors in the treatment of patients with acute myeloid leukemia. Am Soc Clin Oncol Educ Book. 2013;313–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.