Abstract
Readthrough therapy relies on the use of small molecules that enable premature termination codons in mRNA open reading frames to be misinterpreted by the translation machinery, thus allowing the generation of full-length, potentially functional proteins from mRNA carrying nonsense mutations. In patients with hemophilia A, nonsense mutations potentially sensitive to readthrough agents represent approximately 16% of the point mutations. The aim of this study was to measure the readthrough effect of different compounds and to analyze the influence of premature termination codon context in selected nonsense mutations causing hemophilia A. To this end, primary fibroblasts from three patients with hemophilia A caused by nonsense mutations (p.W1586X, p.Q1636X and p.R1960X) and Chinese hamster ovary (CHO) cells transfected with 12 different plasmids encoding mutated F8 (p.Q462X, p.Q1705X, p.Q1764X, p.W274X, p.W1726X, p.W2015X, p.W2131X, p.R1715X, p.R1822X, p.R1960X, p.R2071X and p.R2228X) were treated with gentamicin, geneticin, PTC124, RTC13 or RTC14. Responses were assessed by analyzing not only F8 mRNA expression and FVIII biosynthesis (FVIII antigen by ELISA, western blot and immunofluorescence) but also the FVIII activity (by chromogenic assay). In the patients’ fibroblasts, readthrough agents neither stabilized F8 mRNA nor increased FVIII protein or activity to detectable levels. In CHO cells, only in five of the 12 F8 variants, readthrough treatment increased both FVIII antigen and activity levels, which was associated with a reduction in intracellular accumulation of truncated forms and an increase in full-length proteins. These results provide experimental evidence of genetic context dependence of nonsense suppression by readthrough agents and of factors predicting responsiveness.
Introduction
Hemophilia A (HA) is an X-linked disorder caused by molecular defects in the coagulation factor VIII gene (F8). Nonsense mutations represent approximately 16% of point mutations leading HA (http://www.factorviii-db.org). These patients usually have severe HA and a high risk of FVIII inhibitor development.1 HA predisposes patients to recurrent bleeds, primarily into the joints and soft tissues, with the severity of the disease inversely correlated to FVIII:C activity. Therapy is based on the prevention of bleeding using FVIII replacement (either recombinant or plasma-derived), with an annual estimated cost of >150,000 euros in severely affected patients.2
Although major advances have been reported with gene therapy in both HA3 and hemophilia B (HB),4 it has yet to be approved for clinical use and will not, therefore, be commercially available for the majority of patients in the immediate future. Recently, readthrough therapy has emerged as a potential strategy for the treatment of hereditary diseases caused by nonsense mutations. It is based on the use of small molecules that cause ribosomes to ignore premature termination codons (PTC) during translation, thus allowing the generation of full-length, potentially functional, proteins from the mutant mRNA. PTC are estimated to occur in approximately 10-15% of the patients with monogenic disease (homo or heterozygotes), who may potentially benefit from readthrough agents (RTA). However, this approach has important limitations, including the toxicities of some of these compounds (which require long-term administration), the usually very low levels of readthrough that can be obtained, and the fact that an amino acid different from that originally encoded may be introduced at the PTC site, with potentially detrimental consequences in terms of protein function. Nonetheless, clinical trials or pilot studies of RTA have already been carried out in patients with cystic fibrosis,5,6 Duchenne muscular dystrophy7,8 or hemophilia,9 although often with inconclusive results.
The aminoglycosides gentamicin and geneticin, as well as non-aminoglycoside compounds, such as PTC124 (Ataluren),10 RTC13 and RTC14,11 have been used as RTA based on their ability to suppress PTC and to allow the generation of full-length proteins with at least partial functionality. However, the efficiency of RTA therapy varies according to not only the PTC type and sequence context,12–14 but also to the specific agent that is used, which can influence the binding of specific aminoacidyl-tRNA to the PTC and thus the amino acid added to the nascent peptide, with consequences for the functionality of the recoded protein.15 Here, we present an analysis of the effect of five RTA (gentamicin, geneticin, PTC124, RTC13 and RTC14) on F8 mRNA expressed in primary skin fibroblasts from three patients with HA as well as in a Chinese hamster ovary (CHO)-cell-based model of HA. Our aim was to assess the readthrough effect of these RTA on the FVIII activity, in addition to FVIII:Ag levels, and the influence of the molecular context, including type of stop codon, adjacent sequences, and the amino acid originally encoded by the wild-type (WT) protein at the mutated site.
Methods
Patients and isolation of skin fibroblasts
Four patients with HA caused by either nonsense mutations (p.W1568X, p.Q1636X and p.R1960X) or a missense mutation (p.R1960Q), diagnosed at the Hemophilia Unit of the Vall d’Hebron University Hospital and genetically characterized at the Congenital Coagulopathies Laboratory of the Blood and Tissue Bank of Catalonia (BST)16 were selected for this study. All participating patients and controls provided informed consent in accordance with the Declaration of Helsinki. The study was approved by our institutional Research Ethics Committee.
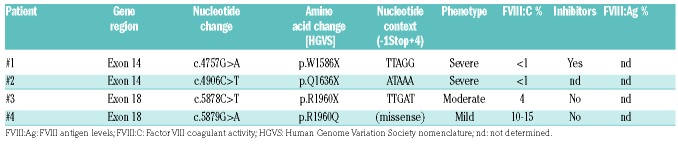
The genetic characteristics of each patient and their plasma FVIII:C activities at the time of diagnosis are summarized in Table 1.
Table 1.
Molecular and clinical data of patients with hemophilia A included in the study.
Generation of F8 variants harboring premature termination codon mutations
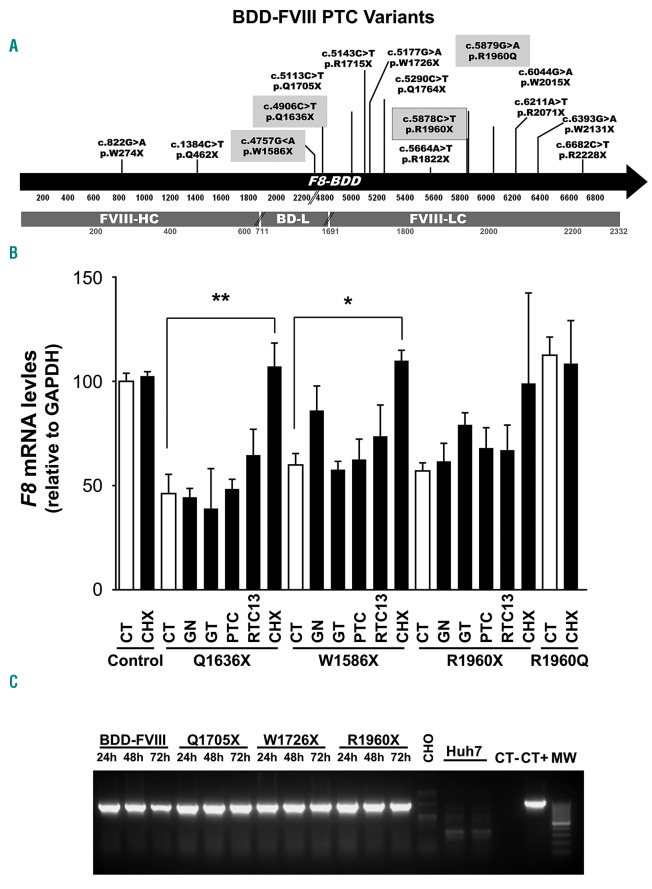
All F8 B-domain deleted (F8BDD) cDNA variants were designed according to our hypotheses and purchased from GeneArt (Thermo Fisher Scientific, Waltham, MA, USA). All the mutations studied are shown in Figure 1A.
Figure 1.
F8 mutations studied and detection of F8 mRNA levels. (A) Schematic representation of the distribution of premature termination codons (PTC) across the F8BDD cDNA generated by site-directed mutagenesis for the Chinese hamster ovary (CHO) model or naturally occurring in the hemophilia A (HA) patients. The black arrow represents the F8 cDNA (5’ to 3’), numbers below the arrow correspond to the nucleotide position, while the gray bar represents the BDD-FVIII protein, and numbers below correspond to the amino acid position according to the Human Genome Variation Society (HGVS) nomenclature. The distribution of mutations in F8 mRNA analyzed in CHO model, patients’ fibroblasts (in gray boxes) or both cellular models (black lined gray box) are also shown. BD-L: BDD-linker; FVIII-HC: heavy chain; FVIII-LC: light chain. (B) F8 mRNA levels detected by quantitative real-time polymerase chain reaction in the fibroblasts of HA-patients or a normal control. GAPDH: glyceraldehyde 3-phosphate dehydrogenase; Control: fibroblasts of a HB patient; Q1636X, W1586X and R1960X HA patients fibroblasts harboring these nonsense mutations; and R1960Q: HA patient fibroblasts harboring this missense mutation. CT: untreated cells; GN: geneticin 100 mg/mL; GT: gentamicin 100 mg/mL; PTC: PTC124 10 mM; RTC13: RTC13 10 mM; CHX: cycloheximide 1 mg/mL (n=3). (C) Time course of F8BDD mRNA levels detected by BDD-specific semi-quantitative polymerase chain reaction in the CHO-based HA-model. F8BDD: WT variant; p.Q1705X, p.W1726X and p.R1960X: F8BDD variants harboring PTC; CHO: non transfected CHO cells; Huh-7: human hepatocellular carcinoma cell line (a cell line that expresses high levels of endogenous F8 but used here as a negative control of the F8BDD-specific amplification); MW: 100-bp DNA ladder. P values: *P<0.05, **P<0.01 and *** P<0.001. (n=3).
Cell lines and Chinese hamster ovary-cellular model
Human hepatocarcinoma cell line Huh-7 was kindly provided by Dr. J. Quer (VHIR, Barcelona) while CHO-S cells were purchased from Thermo Fisher Scientific. Both cell lines were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, penicillin and streptomycin (all from Biowest, Nuaillé, France). For CHO transfection, cells were passed three times and seeded at 4-5 × 104 cells in 12-well plates to achieve a confluence of 50-80%. The day after, the cells were transfected with 1.5 mg of the WT or mutated F8BDD plasmids using 1.5 mL lipofectamine-LTX (Thermo Fisher Scientific) per well, following the manufacturer’s recommendations.
Readthrough agent treatment
Patient-derived skin fibroblasts (3.5x103 cells/cm2) were seeded in 6-well plates and grown to 70-80% confluence before RTA treatment, while CHO cells were treated after 8 hours of transfection. The cells were treated with gentamicin, geneticin (Thermo Fisher Scientific), PTC124 (Selleckchem Co., Houston, TX, USA), RTC13 or RTC14 (ID: 5735019 and ID: 5311257) (ChemBridge. San Diego, CA, USA) using a range of concentrations reported to induce PTC readthrough in vitro (50-100 mg/mL for gentamicin and geneticin, 10 mM for PTC124, RTC13 and RTC14).17,18
F8 mRNA analysis
Total RNA was extracted using the RNeasy mini kit followed by on-column DNase I treatment (Qiagen. Hilden, Germany). Single-stranded cDNA was generated with the high capacity cDNA reverse transcription kit (Thermo Fisher Scientific) using 500 ng of total RNA and random primers in a final volume of 25 mL, as previously described.19 The cDNA obtained was used to quantify F8 mRNA expression.
FVIII Ag levels of F8BDD variants
Human FVIII antigen levels (FVIII:Ag) were determined in culture supernatants using the IMUBIND® factor VIII ELISA kit (Sekisui Diagnostics, Lexington, MA, USA).
FVIII immunodetection of F8BDD variants
Chinese hamster ovary lines transfected with the F8BDD variants were stained with several anti-FVIII antibodies for the immunodetection of FVIII in cell homogenates (Online Supplementary Table S1).
FVIII activity of F8BDD variants
The FVIII activity (FVIII:C) of the supernatants of cultured fibroblasts or CHO cells transfected with F8BDD variants, treated or not with RTA, was assessed by the chromogenic method using the COAMATIC FVIII kit (Chromogenix, Werfen, Barcelona, Spain). The protocol was modified from Yatuv et al.20
Results
F8 mRNA levels after readthrough agent treatment
In the fibroblasts of HA-patients harboring nonsense mutations, F8 mRNA levels measured by quantitative real-time-polymerase chain reaction (qRT-PCR) were <60% (p.Q1636X: 46.23%±9.19; p.W1586X: 59.89%±5.55; p.R1960X: 57.09%±3,81) of those detected in control fibroblasts from healthy individuals or from the HA patients caused by the missense mutation. Treatment with the protein synthesis inhibitor cycloheximide, which also inhibits nonsense-mediated decay (NMD), restored the levels of PTC-containing transcripts to normal values, which suggested a role for NMD in our HA patients harboring nonsense mutations (Figure 1B). We then analyzed the ability of RTA to suppress PTC and stabilize PTC-containing mRNA, as reported in previous studies.21,22 Although some of the RTA increased F8 mRNA levels in the fibroblasts of HA patients, the differences were not statistically significant: p.W1586X: 59.89%±5.55 (CT) versus 86.01%±11.82 (100 mg geneticin/mL); p.Q1636X: 46.23%±9.19 (CT) versus 64.68%±12.41 (10 mM RTC13), and p.R1960X: 57.09%±3.82 (CT) versus 79.21%±5.66 (100 mg gentamicin/mL) (Figure 1B). A possible explanation for this could be that none of the nonsense mutations analyzed in patients’ fibroblasts met the PTC rule.
In addition, we used CHO cells, which is the most commonly used cell line for commercial production of FVIII,23 to analyze F8 mRNA levels to measure the effects of RTA. Time course qRT-PCR analysis using F8BDD-specific primers showed that the CHO cell model expressed high levels of F8 mRNA without any presence of NMD over time (Figure 1C).
Analysis of the human FVIII:Ag levels in the culture media
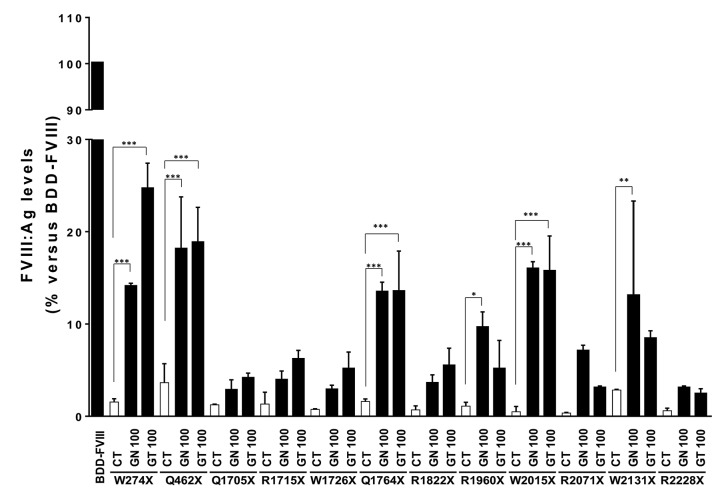
Although fibroblasts ectopically express F8 mRNA, they do not synthesize FVIII protein.24 For this reason, and because none of the nonsense mutations analyzed in patients’ fibroblasts met the PTC rule, all protein studies were performed in the CHO cellular model. To determine the ability of the already established RTAs geneticin and gentamicin to increase FVIII production, FVIII:Ag levels in the culture supernatants of the treated CHO cells were analyzed by ELISA (Table 2 and Figure 2). Compared to the controls (CT), these two drugs, each at 100 mg/mL, significantly increased the human FVIII:Ag levels of the variants p.W274X (CT=1.55 vs. 24.6% with gentamicin), p.Q462X (CT=3.65 vs. 18.75% with gentamicin), p.Q1764X (CT=1.6 vs. 13.45% with geneticin), p.W2015X (CT=0.5 vs. 15.9% with geneticin) and p.W2131X (CT=2.85 vs. 13% with geneticin). In the p.R1960X variants, FVIII:Ag levels increased significantly only in cells treated with 100 mg geneticin/mL (CT=1.1 vs. 9.55%). Smaller, non-significant, increases were observed for the p.Q1715X, p.R1715X, p.W1726X, p.R18822x, p.R2071X and p.R2228X variants.
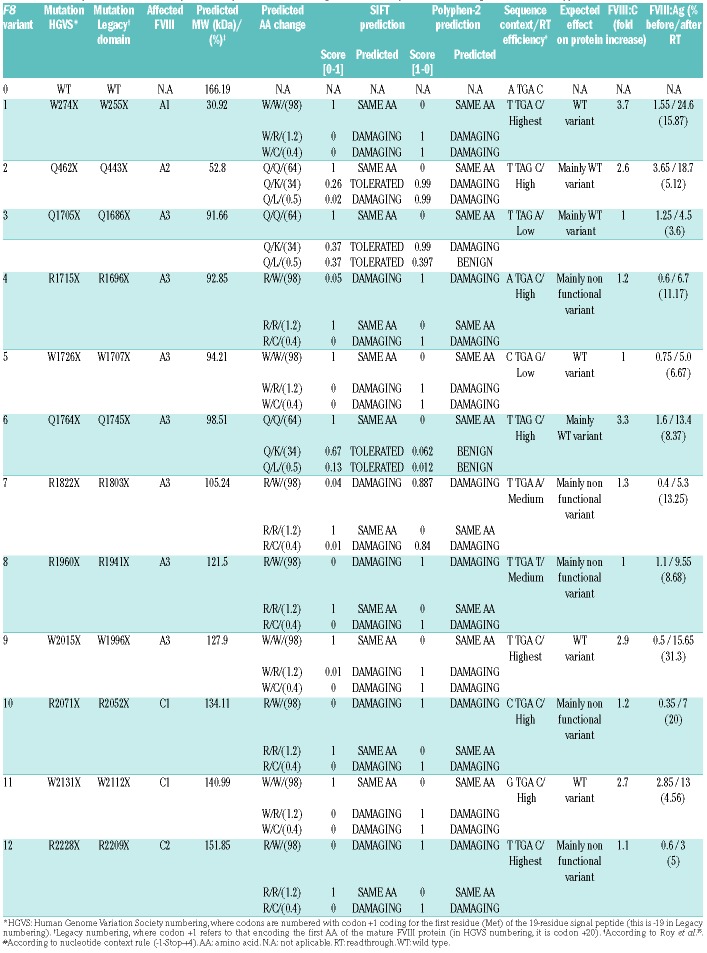
Table 2.
Summary of the in silico analysis of the predicted readthrough effect at the protein level using the SIFT and Polyphen-2 softwares.
Figure 2.
Readthrough agents (RTA) can increase FVIII antigen levels in the supernatants of some transfected Chinese hamster ovary (CHO) cells. Human FVIII:Ag levels analyzed by ELISA in the supernatants of CHO cells transiently transfected with the F8 variants, showing the increase in FVIII:Ag levels before and after RTA-treatment versus the F8 wild-type (WT) variant. BDD-FVIII: WT; CT: untreated controls; GN100: 100 mg geneticin/mL and GT100: 100 mg gentamicin/mL. *P<0.05; **P<0.01; ***P<0.001. (N=3).
Immunodetection of FVIII after readthrough agent treatment
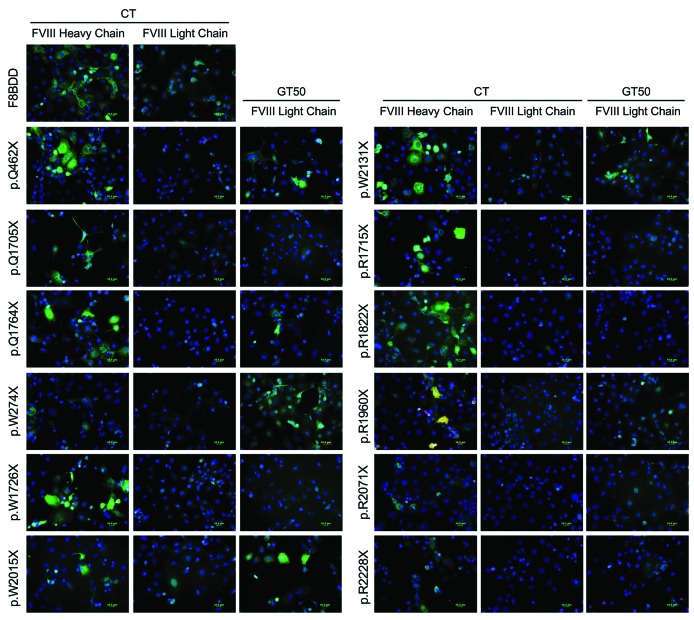
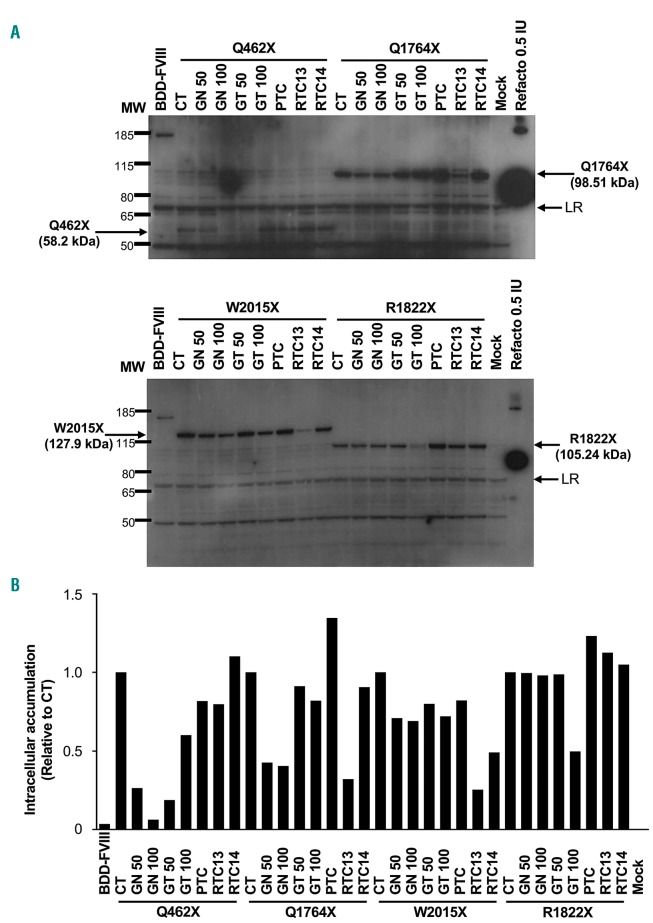
To establish the mechanism by which the previously used RTA increased FVIII production, protein levels were analyzed in transfected CHO cells by immunofluorescence using antibodies generated against either heavy (N-terminal) or light (C-terminal) chains (Online Supplementary Table S1). Before RTA treatment, light chain staining was detected only in cells transfected with the WT F8BDD variant (Figure 3). However, after treatment of the transfectants with 50 mg gentamicin/mL, staining for light chain was visible in cells transfected with variants p.W274X, p.Q462X, p.Q1764X, p.R1960X, p.W2015X, and p.W2131X (Figure 3). FVIII production in cell lysates was also analyzed, using an anti-FVIII antibody raised against FVIII heavy chain (Nt). Before RTA treatment, truncated FVIII accumulated intracellularly in cells transfected with the different constructs harboring PTC, which is, in addition to a faulty degradation, one of the possible consequences of the inability of FVIII to fold properly.23 In contrast, treatment with some of the RTA reduced the amount of intracellular accumulation (Figure 4 and Online Supplementary Figure S1). In the p.Q462X, the highest reduction in FVIII accumulation was observed with geneticin, at the two tested concentrations. The p.Q1764X and p.W2015X variants were also responsive to geneticin, but the highest reduction in intracellular accumulation of FVIII was achieved with RTC13. The p.R1822X showed a reduction in intracellular accumulation only with the higher concentration of gentamicin tested. Other variants also showed a qualitative reduction (p.Q1705X for geneticin and gentamicin; R1960X for geneticin, gentamicin but also PTC124, and p.W2131X for geneticin, and RTC13) or only a minimal qualitative reduction (p.W1726X and p.R2228X) in the intracellular accumulation of truncated FVIII (Online Supplementary Figure S1).
Figure 3.
Gentamicin increased FVIII biosynthesis in some transfected Chinese hamster ovary (CHO) cells. Representative images of the immunofluorescence detection of FVIII in transiently transfected CHO cells stained with antibodies against either FVIII heavy chain or FVIII light chain (both in green). Gentamicin treatment (GT50: 50 mg/mL) increased FVIII labeling using the anti-FVIII light chain antibody in CHO cells transiently transfected with variants harboring the p.W274X, p.Q462X, p.Q1764X, p.W2015X and p.W2131X premature termination codons.
Figure 4.
Readthrough agents (RTA) reduced intracellular accumulation of FVIII in transfected Chinese hamster ovary (CHO) cells. Representative images of the immunodetection of FVIII heavy chain in cell homogenates of transiently transfected CHO cells. (A) Western blot of CHO cells transfected with either wild-type (WT) BDD-FVIII (166.19 kDa), the p.Q462X (52.8 kDa), p.Q1764X (98.51 kDa) (top panel), p.W2015X (127.9 kDa), or the p.R1822X (105.24 kDa) variants (bottom panel). The intracellular accumulation of BDD-FVIII or the truncated proteins before and after RTA-treatment is shown. (B) Densitometry of the bands in (A). CT: Untreated cells; GN50: 50 mg geneticin/mL; GN100: 100 mg geneticin/mL; GT50: 50 mg gentamicin/mL; GT100: 100 mg gentamicin/mL; PTC: PTC124 10 mM; RTC13 and RTC14: 10 mM; Mock: untransfected cells; MW: Pageruler Plus prestained protein ladder; Refacto®: recombinant human BDD-FVIII (Pfizer); LR: loading reference used for densitometry normalization.
Rescue of FVIII:C activity after readthrough agent treatment
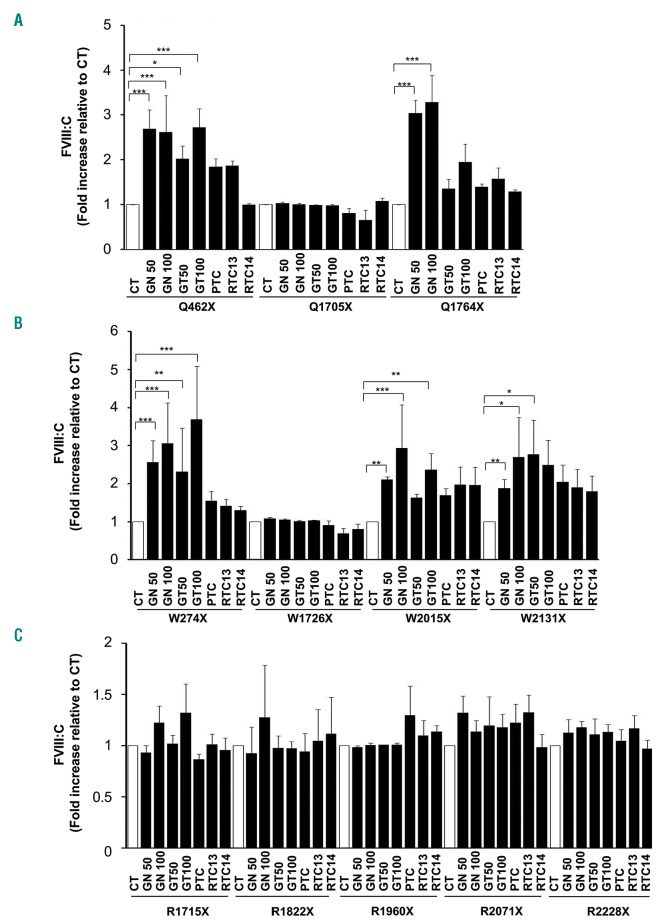
Finally, we focused in the RTA-mediated effect on the protein functionality rather than on the protein levels. To this end, we analyzed the FVIII:C activity by means of a chromogenic assay using not only the well established RTA geneticin and gentamicin, but also other RTA such as PTC, RTC13 and RTC14. RTA treatment resulted in a significant dose-dependent increase in FVIII:C in only five of the 12 variants (Table 2 and Figure 5). The strongest readthrough response was that of p.W274X, in which a 3.7-fold increase in activity was obtained after treatment with 100 mg gentamicin/mL. Smaller increases in FVIII:C activity were measured at a lower dose of gentamicin (50 mg/mL) or geneticin (50 and 100 mg/mL). Increases in FVIII:C after exposure to 100 mg geneticin/mL were also observed in the p.W2015X and p.W2131X variants (2.9-and 2.7-fold, respectively) and in the p.Q462X and p.Q1764X variants (up to 2.6- and 3.3-fold, respectively). These variants were also responsive, although to a lesser extent, to treatment with 100 mg gentamicin/mL but the increases in FVIII:C were smaller. Variants responsive to either gentamicin or geneticin showed a slight response to other RTA, particularly to PTC124 and RTC13, but these increases were not statistically significant.
Figure 5.
Readthrough agents (RTA) increase FVIII coagulant activity in the supernatants of transfected Chinese hamster ovary (CHO) cells. (A) Fold increase of FVIII:C (relative to untreated controls) in the supernatants of cultured CHO cells transiently transfected with variants harboring a QX premature termination codons (PTC) (see legend to Figure 1). (B) Fold increase of FVIII:C in the supernatants of cultured CHO cells transiently transfected with variants harboring a WX PTC (see legend to Figure 1). (C) Fold increase of FVIII:C in the supernatants of cultured CHO cells transiently transfected with variants harboring a RX PTC (see legend to Figure 1). CT: untreated controls; GN50: 50 mg geneticin/mL; GN100: 100 mg geneticin/mL; GT50: 50 μg gentamicin/mL; GT100: 100 mg gentamicin/mL; PTC: PTC124 10 mM; RTC13 and RTC14: 10 μM. *P<0.05; **P<0.01; ***P<0.001. (N=3).
In silico analysis of the premature termination codons
Variable responses to RTA-induced nonsense suppression related to the sequences adjacent to the PTC have been reported. We therefore analyzed the PTC type and the context effect in both the patients’ fibroblasts and the 12 F8BDD variants (Tables 1 and 2, and Online Supplementary Table S2). In patients’ fibroblasts responsive to RTA, we would expect at least a partial restoration of the F8 mRNA levels; however, none of the PTC sequence context filled the PTC rule, so we cannot expect any mRNA stabilization. In contrast, in the CHO model, we could evaluate the readthrough effect on the production of full-length protein and on the restoration of FVIII:C activity. The results suggested a TGA≥TAG hierarchy of PTC suppression sensitivity and a positive RTA readthrough effect on both FVIII:Ag levels and FVIII:C activity for sequences in which a C was located at position +4 and a T at position −1 as present in variants pW274X, pQ462X, pQ1764X and pW2015X. Furthermore, the pW2131X variant also showed responsiveness even when a G is present at position -1 instead of a T. In contrast, variants pR1715X, pR2071X and pR2228X, even containing a TGA and a T at position –1 and a C at position +4 were unable to significantly increase FVIII:C activity. Similarly, the pR1822X variant was also unresponsive to RTA, despite the fact that it contains a TGA and a T at position –1.
Although positions −5 (A, C>G) and +8 (G>C) were also reported to influence readthrough, our data show that T, G and A at position −5 as well as T or C at position +8 promoted PTC suppression, as measured by FVIII:C levels; other tested combinations may have positive effects measured by FVIII:Ag levels. Whether positive effects are associated with yet other nucleotide combinations could not be determined due to the limited number of F8 variants tested (Online Supplementary Table S2).
Discussion
The present study represents a comprehensive analysis of factors influencing the responsiveness to RTA of a series of nonsense mutations that are representative of those causing HA. Readthrough therapy is not expected to restore the synthesis and activity of the recoded proteins to curative levels in most patients harboring nonsense mutations. However, in the case of hemophilia, even a relatively small percentage of restoration may be very valuable. Thus, in patients with severe HA (<1% of FVIII:C), a slight increase in activity to 2-5% could significantly improve the bleeding phenotype as reported for FIX production in severe HB patients treated with gen-tamicin.9
Our study uses a CHO-based HA model in which NMD is absent, as cells were transfected with plasmid containing the F8 cDNA, but lacking the exon-exon junction complexes (EJC) which are necessary for PTC-containing mRNA to be targeted by NMD, thus providing a clean tool to amplify and detect eventual readthrough activity of RTA. While cDNA models do not mimic the in vivo situation (in which NMD is present), they can be used for assessing potential readthrough effects and to identify new RTA.10,11,17,25 In practice, to show a readthrough effect in vivo or in primary cells, RTA will probably have to be combined with NMD inhibition. Nonetheless, the CHO-based HA model allowed us to demonstrate that the effectiveness of readthough therapy is mutation-specific and that in vitro assays may be useful for classifying patients that can likely benefit from RTA.
It has recently become clear that different types of endothelial cells, including liver sinusoidal endothelial cells (LSEC), constitute the main source of FVIII production in humans,24,26–29 perhaps to a greater extent than hepatocytes. This may explain why, although ectopically expressed at the mRNA level, none of the fibroblasts from HA patients analyzed showed a significant stabilization of F8 mRNA levels. However, as the PTC sequence context does not fill the PTC rule, we cannot draw any conclusions as to the effectiveness of the RTA used. Furthermore, since fibroblasts do not produce FVIII,24 we could not expect to detect FVIII protein.
Despite the limitations of this study, which include the relatively low number of mutations analyzed, the intrinsic variability in the transfection efficiencies, and the low levels of FVIII:C achieved (in the low range of detection by FVIII:C assays, i.e. 0.5-3%), the observed increases in the FVIII:C activity in some mutations suggest a potential phenotypic improvement, from severe to moderate/mild HA. Thus, RTA treatment significantly rescued FVIII production in some of the F8BDD variants, as measured by FVIII:Ag levels. This increase was associated with a qualitative reduction in intracellular accumulation of FVIII observed in the western blots, and a concomitant increase in light-chain FVIII detection by immunofluorescence. Moreover, data obtained in the FVIII:C analyses highlighted the importance of the amino acid incorporated upon RTA treatment in the partial restoration of FVIII:C activity. In our study, after drug treatment, only variants that natively encoded tryptophan (Trp=W) or glutamine (Gln=Q) and had a cytosine at position +4, regardless of the PTC type, were able to significantly increase FVIII:C in the culture supernatants, while this activity was not increased in variants natively encoding arginine (Arg=R), even when a cytosine at position +4 was present, likely because the RTA may promote the incorporation of amino acids that do not necessarily restore FVIII functionality.
Nonsense suppression was observed in only five of the 12 transfected F8BDD variants (p.Q462X, p.Q1764X, p.W274X, p.W2015X and p.W2131X). This apparent discrepancy between protein levels and restored activity can be explained by the context-dependent ability of RTA to restore the full-length recoded protein13 and by the preferred aminoacyl tRNA that will recognize each PTC in RTA-treated cells.15 Accordingly, upon secretion, only full-length protein that has incorporated the native amino acid (or another one with similar chemical properties) is expected to have a greater chance of restoring maximum or partial levels of FVIII:C activity. Additionally, the phylogenetic conservation of this amino acid, or whether deleterious missense mutations have been described in this codon, are additional important factors to be considered.
Under our experimental conditions, the ability of RTA to increase C-terminal (light chain) positive staining or FVIII:Ag levels did not always correlate with the ability of RTA to restore FVIII:C activity. This suggests that the amino acid incorporated at the PTC and its structural similarity with the natively encoded amino acid can be critical for the restoration of FVIII:C, but not necessarily for that of FVIII:Ag. According to the PTC context rule,13 the hierarchy of RTA responsiveness is T-TGA-C>T-TAG-C> T-TAA-C; thus, a stronger RTA effect at TGA (opal) is expected in PTC originally encoding tryptophan and argi-nine. However, we found that PTC generated at triplets natively encoding arginine and fulfilling the PTC context rule led to production of a non-functional protein, although this situation can also result in a gain of function, as reported in a cellular model of readthrough in hemophilia B.30 Conversely, according to the PTC context rule, a PTC generated at triplets encoding glutamine is predicted to be less responsive to RTA treatment. However, in our study, RTA allowed partial restoration of FVIII:C levels in 2 of 3 recoded FVIII mutants originally encoding glutamine (pQ462X and pQ1764X). A similar result was reported in a study of cystic fibrosis in which patients harboring the Y122X mutation had a better than expected response to RTA, even though the PTC was caused by a TAA codon, followed by a C nucleotide at position +4, which was previously reported to have the lowest RTA sensitiveness.13 However, both the readthrough effect due to the presence of cytosine at position +4, and the incorporation of the natively encoded tyrosine might have been facilitated by the RTA treatment,15 which would account for the clinical benefit observed.
Our work points out the importance of not only evaluating RTA-mediated protein biosynthesis, but also the functional activity of the recoded protein generated. This is particularly important in studies of the potential clinical benefit of RTA therapy and the search for safer and more effective RTA. By taking into account the functionality of the restored protein generated by RTA, our cellular model, or other similar models, provide a method for testing the effectiveness of new RTA.31
Although we evaluated only a small number of F8BDD variants, and these results cannot be generalized, our analysis of the molecular environment of these PTC provides valuable information on additional factors involved in eventual responses to RTA treatment. From these results we can conclude that: 1) responsiveness is greater in PTC with the consensus sequence T-Stop-C; 2) in TGA (opal) than in TAG (amber) PTC; and 3) it will be higher when the mutated original amino acid is tryptophan or glutamine as opposed to arginine, in which, despite the efficient intracellular reduction of truncated protein and extracellular increase of FVIII:Ag levels, the increase observed in FVIII:C activity was minimal.
In conclusion, the results obtained with our model represent a proof-of-concept that RTA are capable of suppressing PTC in some nonsense mutations causing HA, albeit with different efficiencies and different functional consequences depending on the sequence context. This model, linked to a functional assay based on the determination of FVIII:C activity, and the application of the PTC rule described here, allowed for the first time the identification of five RTA-responsive nonsense mutations in HA and may facilitate the classification of nonsense mutations reported in the HA database into categories based on the predicted levels of responsiveness to RTA, thus promoting patient selection in readthrough therapy clinical trials.32
Supplementary Material
Acknowledgments
The authors thank the participating patients and their families. We also thank Yvonne Richaud-Patin and Dr. Angel Raya (CMRB, Spain) for their help with the fibroblast isolation.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/2/508
Funding
This project was supported by two grants from the Instituto de Salud Carlos III (PI11/03024 and PI11/03029) / e-Rare-2 (HEMO-iPS) that were cofinanced by the European Regional Development Fund (FEDER), and the Europe Aspire 2013 Award in Hemophilia Research.
References
- 1.Gouw SC, van den Berg HM, Oldenburg J, et al. F8 gene mutation type and inhibitor development in patients with severe hemophilia A: systematic review and meta-analysis. Blood. 2012;119(12):2922–2934. [DOI] [PubMed] [Google Scholar]
- 2.Escobar MA. Health economics in haemophilia: a review from the clinician’s perspective. Haemophilia. 2010;16:29–34. [DOI] [PubMed] [Google Scholar]
- 3.Rangarajan S, Walsh L, Lester W, et al. AAV5-Factor VIII Gene Transfer in Severe Hemophilia A. N Engl J Med. 2017; 377(26):2519–2530. [DOI] [PubMed] [Google Scholar]
- 4.George LA, Sullivan SK, Giermasz A, et al. Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant. N Engl J Med. 2017;377(23):2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linde L, Boelz S, Nissim-Rafinia M, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117(3):683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sermet-Gaudelus I, Renouil M, Fajac A, et al. In vitro prediction of stop-codon suppression by intravenous gentamicin in patients with cystic fibrosis: a pilot study. BMC Med. 2007;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel RS, Flanigan KM, Wong B, et al. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS One. 2013;8(12):e81302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushby K, Finkel R, Wong B, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014; 50(4):477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James PD, Raut S, Rivard GE, et al. Aminoglycoside suppression of nonsense mutations in severe hemophilia. Blood. 2005; 106(9):3043–3048. [DOI] [PubMed] [Google Scholar]
- 10.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007; 447(7140):87–91. [DOI] [PubMed] [Google Scholar]
- 11.Du L, Damoiseaux R, Nahas S, et al. Nonaminoglycoside compounds induce readthrough of nonsense mutations. J Exp Med. 2009;206(10):2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6(7):1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Floquet C, Hatin I, Rousset J-P, Bidou L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 2012; 8(3):e1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence. RNA Biol. 2015;12(9):950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy B, Leszyk JD, Mangus DA, Jacobson A. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc Natl Acad Sci U S A. 2015;112(10):3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal F, Farssac E, Altisent C, Puig L, Gallardo D. Rapid hemophilia A molecular diagnosis by a simple DNA sequencing procedure: identification of 14 novel mutations. Thromb Haemost. 2001;85(4):580–583. [PubMed] [Google Scholar]
- 17.Jung ME, Ku J-M, Du L, Hu H, Gatti RA. Synthesis and evaluation of compounds that induce readthrough of premature termination codons. Bioorg Med Chem Lett. 2011;21(19):5842–5848. [DOI] [PubMed] [Google Scholar]
- 18.Yu H, Liu X, Huang J, et al. Comparison of readthrough effects of aminoglycosides and PTC124 on rescuing nonsense mutations of HERG gene associated with long QT syndrome. Int J Mol Med. 2014; 33(3):729–735. [DOI] [PubMed] [Google Scholar]
- 19.Martorell L, Corrales I, Ramirez L, et al. Molecular characterization of ten F8 splicing mutations in RNA isolated from patient’s leucocytes: assessment of in silico prediction tools accuracy. Haemophilia. 2015;21(2):249–257. [DOI] [PubMed] [Google Scholar]
- 20.Yatuv R, Dayan I, Baru M. A modified chromogenic assay for the measurement of very low levels of factor VIII activity (FVIII:C). Haemophilia. 2006;12(3):253–257. [DOI] [PubMed] [Google Scholar]
- 21.Bedwell DM, Kaenjak A, Benos DJ, et al. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med. 1997;3(11):1280–1284. [DOI] [PubMed] [Google Scholar]
- 22.Keeling KM, Brooks DA, Hopwood JJ, et al. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10(3):291–299. [DOI] [PubMed] [Google Scholar]
- 23.Kumar SR. Industrial production of clotting factors: Challenges of expression, and choice of host cells. Biotechnol J. 2015;10(7):995–1004. [DOI] [PubMed] [Google Scholar]
- 24.Turner NA, Moake JL. Factor VIII Is Synthesized in Human Endothelial Cells, Packaged in Weibel-Palade Bodies and Secreted Bound to ULVWF Strings. PLoS One. 2015;10(10):e0140740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benhabiles H, Gonzalez-Hilarion S, Amand S, et al. Optimized approach for the identification of highly efficient correctors of nonsense mutations in human diseases. PLoS One. 2017;12(11):e0187930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollestelle MJ, Thinnes T, Crain K, et al. Tissue distribution of factor VIII gene expression in vivo--a closer look. Thromb. Haemost. 2001;86(3):855–861. [PubMed] [Google Scholar]
- 27.Wion KL, Kelly D, Summerfield JA, Tuddenham EG, Lawn RM. Distribution of factor VIII mRNA and antigen in human liver and other tissues. Nature. 1985;317(6039):726–729. [DOI] [PubMed] [Google Scholar]
- 28.Olgasi C, Talmon M, Merlin S, et al. Patient-Specific iPSC-Derived Endothelial Cells Provide Long-Term Phenotypic Correction of Hemophilia A. Stem Cell Rep. 2018;11(6):1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki Y, Kakisaka K, Matsumoto T, et al. Orthotopic liver transplantation for haemophilia A may not always lead to a phenotypic cure of haemophilia A: A case report. Haemophilia. 2018;24(6):e420–e422. [DOI] [PubMed] [Google Scholar]
- 30.Branchini A, Ferrarese M, Campioni M, et al. Specific factor IX mRNA and protein features favor drug-induced readthrough over recurrent nonsense mutations. Blood. 2017;129(16):2303–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baradaran-Heravi A, Balgi AD, Zimmerman C, et al. Novel small molecules potentiate premature termination codon readthrough by aminoglycosides. Nucleic Acids Res. 2016;44(14):6583–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmittgen TD, Zakrajsek BA, Mills AG, et al. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000; 285(2):194–204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.