Abstract
The gut microbiota plays a key role in the development of chronic inflammatory liver disease. The gut-liver axis involves inflammatory cells, cytokines, and other molecules that cause liver deterioration. Dysbiosis is important in understanding several liver diseases, especially in relation to the development of autoimmune liver disease. The aim of this review is to provide a current overview of alterations in the gut and oral microbiota associated with autoimmune liver diseases.
Keywords: gut microbiota, oral microbiota, autoimmune hepatitis, primary biliary cholangitis
Introduction
Primary biliary cholangitis (PBC) and autoimmune hepatitis (AIH) are classically viewed as distinct autoimmune liver diseases. Autoimmune liver diseases are thought to be triggered by environmental factors in genetically susceptible individuals. Genome-wide association and murine model studies have expanded our knowledge of autoimmune liver diseases, however, the pathogenesis of disease remains obscure. Recently culture-independent techniques have revolutionized knowledge of the gut and oral microbiota. This review provides an overview of gut and oral microbiota in autoimmune liver diseases.
Gut-Liver axis
The gut and liver are closely related. The liver is significantly affected by the gut and its contents, as 70% of the blood supplied to the liver is collected from intestinal circulation through the portal venous system. Both the gut and liver play a pivotal role in the absorption and metabolism of various nutrients and drugs. Abnormal bile acid homeostasis may lead to diarrhea and bacterial overgrowth1). Bacterial overgrowth, increased intestinal permeability, failure to inactivate endotoxins, and activated innate immunity all contribute to the pathological state of bacterial translocation.
Gut and oral microbiota
The gut microbiota collectively refers to the 100 trillion bacteria, with an estimated mass of 1-2 kg, that inhabit the human gastrointestinal tract. This very diverse ecosystem comprises over 2,000 distinct species and has a collective genome containing 150-fold more genes than the human genome2). The metabolic activity of the gut microbiota provides benefits to human health by supplying essential nutrients and maximizing the efficiency of energy harvest from ingested food. However, the microbiota also contains numerous potential opportunistic pathogens. Moreover, gut microbial products activate Toll-like receptors (TLRs) and induce inflammation that defines disease. Maintaining the homeostasis of the gut microbiota has thus necessitated the development of a specialized mucosal immune system3). Characterization of the microbiota has been performed by culture and biochemical typing, which have been gold standards for the identification of bacterial species. Recently, however, advances in culture-independent techniques have revolutionized our knowledge about the gut microbiota. These techniques, based on sequence divergences in the small subunit ribosomal ribonucleic acid (16S rRNA), allow the demonstration of gut microbial diversity, providing qualitative and quantitative information on bacterial species as well as alterations in the gut microbiota in healthy and diseased states4).
The oral cavity is a large reservoir of bacteria of more than 700 species or phylotypes and is profoundly relevant to host health and disease5-7). The role of the oral and gut microbiota in the pathogenesis of immune-related diseases has been highlighted in autoimmune diseases, such as autoimmune encephalomyelitis, rheumatoid arthritis, and inflammatory bowel disease8-14). A previous report revealed evidence of pervasive changes in the immune system-microbiota interface in the saliva of patients with cirrhosis that were similar to those found in stool15).
Gut and oral microbiota in AIH
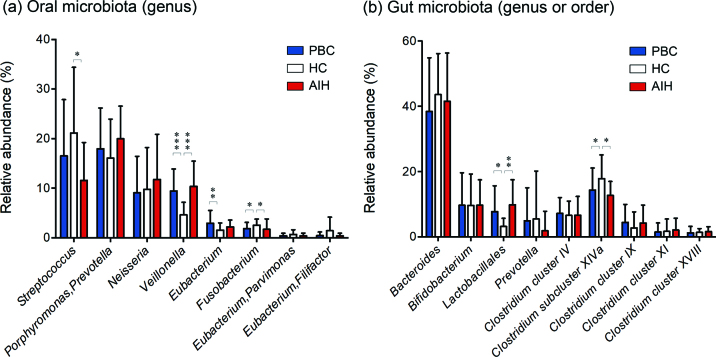
AIH manifests as chronic liver inflammation of an unknown cause. It generally affects young to middle-aged females and is associated with the presence of autoantibodies and hypergammaglobulinemia16). However, the pathogenesis of this disease remains obscure. It is increasingly recognized that the composition of the gut microbiota plays a critical role in influencing the predisposition to AIH17-21) (Table 1). In addition, an increase in the abundance of Veillonella dispar was associated with disease severity, and the metagenomic profile suggests that this bacterium may be involved in lipopolysaccharide (LPS) biosynthesis and amino acid metabolism20). Lin R et al. reported that increased intestinal permeability and bacterial translocation occurred in AIH, wihich associated with disease severity17). However, direct evaluation of the oral microbiome has not been performed in AIH. Our data indicated a significant increase in the abundance of the genus Veillonella in the oral microbiota of patients with AIH, and the abundance of the genus Veillonella was positively correlated with the levels of proinflammatory cytokines, such as IL-1β, IL-6, IL-8, and IL-12p70 in the saliva of patients with AIH21) (Figure 1a). However, it is unknown whether the inflammatory state in the oral cavity of AIH patients is the cause or consequence of imbalances in the salivary microbiota and whether the oral cavity or the gut immune response exerts greater influence on the observed dysbiosis of the oral microbiota. In our study, changes in the gut microbiota composition in AIH were characterized by an increase in abundance of the order Lactobacillales and a decreased abundance of the genus Clostridium subcluster XIVa21) (Figure 1b). Interestingly, our study suggested that the relative abundance of Lactobacillales in feces was positively correlated with the relative abundance of Veillonella in saliva from patients with AIH (Figure 2). Lactobacillus species had relatively large effect sizes in Behcet’s disease microbiota, consistent with the inductive effect of Lactobacillus on systemic inflammation. Animal studies using germ-free mice reported that some bacterial species independently promoted arthritis by activating Th17 cells22,23). Dysbiosis of the oral microbiota is directly and/or indirectly related to the gut microbiota and may be correlated with disease onset.
Table 1.
Changes in the gut and oral microbiota associated with autoimmune liver diseases
| Gut microbiota | |||
| Disease | Comparison | Genus (or Order) | Author/year |
| AIH (n=24) | Healthy vs AIH |
Lactobacillus ↓
Bifidobacterium ↓ |
Lin/201517) |
| AIH (n=17) | Healthy vs AIH |
Lactobacillales ↑
Clostridium subcluster XIVa ↓ |
Abe/201821) |
| AIH (n=99) | Healthy vs AIH |
Veillonella ↑, Lactobacillus ↑, Streptococcus ↑,
Klebsiella ↑, Oscillospira ↓, Parabacteroides ↓, Coprococcus ↓ |
Wei/201920) |
| PBC (n=79) | Healthy vs PBC |
Enterobacteriaceae ↑ Klebsiella ↑,
Haemophilus ↑ Veillonella ↑, Clostridium ↑ Lactobacillus ↑, Streptococcus ↑, Pseudomonas ↑, Bacteroidetes ↓, Sutterella ↓ Oscillospira ↓, Faecalibacterium ↓ |
Tang/201726) |
| PBC (n=39) | Healthy vs PBC |
Lactobacillales ↑
Clostridium subcluster XIVa ↓ |
Abe/201821) |
| Oral microbiota | |||
| Disease | Comparison | Genus (or Order) | Author/year |
| AIH (n=17) | Healthy vs AIH |
Veillonella ↑, Streptococcus ↓,
Fusobacterium ↓ |
Abe/201821) |
| PBC (n=39) | Healthy vs PBC |
Veillonella ↑, Eubacterium ↑
Fusobacterium ↓ |
Abe/201821) |
Fig. 1.
Analysis of the gut and oral microbiota of the PBC, AIH and HC groups based on the T-RFLP profiles.
(a) Mean genus abundance of oral microbiota in the PBC, AIH and HC groups. The plotted values are the mean abundance values of the 8 abundant genera in each group. (b) Mean genus or order abundance of gut microbiota in the PBC, AIH and HC groups. The plotted values are the mean abundance values of the 8 abundant genera and 1 abundant order in each group. The results are expressed as the means ± SDs. Differences were compared using the Mann-Whitney U test; *P<0.05, **P<0.01, ***P<0.0005.
Fig. 2.

Correlation between the gut microbiota and oral microbiota in AIH patients.
The relative abundance of Lactobacillales in feces was positively correlated with the relative abundance of Veillonella in saliva from patients with AIH. The P-value was calculated with the Spearman rank correlation test. P<0.05 was considered significant.
Gut and oral microbiota in PBC
PBC is a progressive autoimmune liver disease characterized by portal inflammation, immune-mediated destruction of the intrahepatic bile ducts, and the presence of highly specific anti-mitochondrial antibodies in serum24,25). Tang R et al. reported that the 16S rRNA gene was analyzed longitudinally by next-generation sequencing (NGS) in PBC patients before treatment with ursodeoxycholic acid (UDCA) using stools of 114 healthy subjects matched in age, gender and body mass index with 79 patients with naive PBC. The number and diversity of observed operational taxonomic units (OTUs) were significantly reduced in the naive PBC patients. The results of principal component analysis by calculating the UniFrac distance showed that the distribution was significantly different between the naive patients with PBC and the healthy subjects. The exploration cohort and the verification cohort had similar results26). Moreover, to investigate the changes in specific gut microbiota components in naive PBC patients, the relative abundance of bacteria at the genus level was compared with that of the healthy subjects. At the genus level, there were changes in the relative abundances of 12 bacteria in naïve patients with PBC compared to healthy subjects. Eight bacteria (Haemophilus, Veillonella, Clostridium, Lactobacillus, Streptococcus, Pseudomonas, Klebsiella, and Enterobacteriaceae) had increased relative abundances, and four (Sutterella, Oscillospira, Faecalibacterium, and Bacteroides) had decreased relative abundances (Table 1).
Our study indicated that there was a significantly higher frequency of bacteria in the genus Veillonella and the genus Eubacterium and a lower frequency of bacteria in the genus Fusobacterium in the PBC group than in the healthy subjects (Figure 1a). Moreover, the changes in the gut microbiota composition in PBC were characterized by an increase in the abundance of the order Lactobacillales and by a decrease in the abundance of the genus Clostridium subcluster XIVa (Figure 1b). The relative abundance of Streptococcus was negatively correlated with the levels of IL-1β, IL-4, IL-6, IL-7, IL-8, IL-12p70, IL-17, G-CSF, IFN-γ and TNF-α, while the relative abundances of Veillonella and Prevotella/Porphyromonas were positively correlated with the IgA level in the saliva of patients with PBC. Moreover, the relative abundances of Neisseria and Eubacterium/Filifactor were positively correlated with the salivary cytokine levels in patients with PBC21).
In 37 patients with naive PBC, UDCA was orally administered for 6 months, and changes in the gut microbiota were evaluated before and after treatment. The abundances of Haemophilus, Veillonella, Streptococcus, and Pseudomonas decreased after UDCA treatment, and those of Sutterella, Oscillospira, and Bacteroides increased after UDCA treatment in PBC patients26).
Conclusion
These findings suggest that the gut and oral microbiota may play different roles in the pathophysiology of AIH and PBC. Further studies of the establishment and modification of the gut and oral microbiota structure may contribute to the development of a therapeutic strategy for patients with autoimmune liver disease.
Conflict of interest disclosure
The authors declare no conflicts of interest associated with this manuscript.
References
- 1.Rai R, Saraswat VA, Dhiman RK. Gut microbiota: its role in hepatic encephalopathy. J Clin Exp Hepatol, 5: 29-36, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464: 59-65, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol, 10: 159-169, 2010. [DOI] [PubMed] [Google Scholar]
- 4.Fraher MH, O’Toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol, 9: 312-22, 2012. [DOI] [PubMed] [Google Scholar]
- 5.Avila M, Ojcius DM and Yilmaz, O. The oral microbiota: living with a permanent guest, DNA Cell Biol, 28: 405-411, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewhirst, FE, Chen, T, Izard, J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J. Bacteriol, 192: 5002-5017, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis MA, Zenobia C. and Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe, 10: 302-306, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One, 10(9): e0137429, 2015. doi: 10.1371/journal.pone.0137429. PMID: 26367776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A, 114(40): 10719-10724, 2017. doi: 10.1073/pnas.1711233114. PMID: 28893994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A, 114(40): 10713-10718, 2017. doi: 10.1073/pnas.1711235114. PMID: 28893978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med, 21(8): 895-905, 2015. doi: 10.1038/nm.3914. PMID: 26214836 [DOI] [PubMed] [Google Scholar]
- 12.Phillips R. Rheumatoid arthritis: Microbiome reflects status of RA and response to therapy. Nat Rev Rheumatol, 11(9): 502, 2015. doi: 10.1038/nrrheum.2015.109. PMID: 26241185 [DOI] [PubMed] [Google Scholar]
- 13.Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res, 21(1): 15-25, 2014. doi: 10.1093/dnares/dst037. PMID: 24013298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science, 358(6361): 359-365, 2017. doi: 10.1126/science.aan4526. PMID: 29051379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology, 62(4): 1260-1271, 2015. doi: 10.1002/hep.27819. PMID: 25820757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krawitt EL. Autoimmune hepatitis. N Engl J Med, 334: 897-903, 1996. [DOI] [PubMed] [Google Scholar]
- 17.Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol, 8(5): 5153-5160, 2015. PMID: 26191211 [PMC free article] [PubMed] [Google Scholar]
- 18.Yuksel M, Wang Y, Tai N, Peng J, Guo J, Beland K, et al. A novel “humanized mouse” model for autoimmune hepatitis and the association of gut microbiota with liver inflammation. Hepatology, 62(5): 1536-1550, 2015. doi: 10.1002/hep.27998. PMID: 26185095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czaja AJ. Factoring the intestinal microbiome into the pathogenesis of autoimmune hepatitis. World J Gastroenterol, 22(42): 9257-9278, 2016. Review. PMID: 27895415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, Xiao X, Lian M, Li B, Chen Y, Zhang J, Li Y, Huang B, Li Y, Cao Q, Fan Z, Chen X, Fang JY, Gershwin ME, Tang R, Ma X. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2019. pii: gutjnl-2018-317836. doi: 10.1136/gutjnl-2018-317836. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Abe K, Takahashi A, Fujita M, Imaizumi H, Hayashi M, Okai K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS One, 13(7): e0198757, 2018. Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity, 32: 815-827, 2010. doi: 10.1016/j.immuni.2010.06.001. PMID: 20620945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu J, Kubota T, Takada E, Takai K, Fujiwara N, Arimitsu N, et al. Bifidobacteria Abundance-Featured Gut Microbiota Compositional Change in Patients with Behcet’s Disease. PLoS One, 11: e0153746, 2016. doi: 10.1371/journal.pone.0153746. eCollection 2016. PMID: 27105322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med, 353: 1261-1273, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Selmi C, Bowlus CL, Gershwin ME, Coppel RL. Primary biliary cirrhosis. Lancet, 377: 1600-1609, 2011. [DOI] [PubMed] [Google Scholar]
- 26. Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, Yang F, Miao Q, Xiao X, Zhang H, Lian M, Jiang X, Zhang J, Cao Q, Fan Z, Wu M, Qiu D, Fang JY, Ansari A, Gershwin ME, Ma X. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut, 67: 534-541, 2018. [DOI] [PubMed] [Google Scholar]