Abstract
On Earth all life is exposed to dramatic changes in the environment over the course of the day; consequently, organisms have evolved strategies to both adapt to and anticipate these 24-hr oscillations. As a result time-of-day is major regulator of mammalian physiology and processes including transcription, signaling, metabolism and muscle contraction all of which oscillate over the course of the day. In particular, the heart is subject to wide fluctuations in energetic demand throughout the day as a result of waking, physical activity, and food intake patterns. Daily rhythms in cardiovascular function ensures that increased delivery of oxygen, nutrients, and endocrine factors to organs during the active period, as well as removal of metabolic by-products, are in balance. Failure to maintain these physiologic rhythms invariably has pathologic consequences. The current review highlights rhythms that underpin cardiac physiology. More specifically, we summarize the key aspects of cardiac physiology that oscillates over the course of the day, and discuss potential mechanisms that regulate these 24-hr rhythms.
Keywords: Chronobiology, Heart, Mitochondrial Quality Control, Post-translational Modification, Redox Biology
INTRODUCTION
Life on Earth is subjected to dramatic fluctuations in the environment over the course of a normal day. These include, but are not limited to, changes in light intensity, humidity, and temperature, as well as the presence/absence of natural predators and prey. It is therefore not surprising that organisms have evolved strategies not only to adapt to 24-hr oscillations in environmental parameters, but also to anticipate them. This concept is readily illustrated at the level of behaviors, such as sleep/wake and fasting/feeding cycles. Given known influences that behaviors and environmental factors have on biological systems, it is not surprising that time-of-day has emerged as a significant modulator of essentially all aspects of mammalian physiology (1, 2). The cardiovascular system serves as an excellent example. Associated with the psychological and physical stresses of waking, blood pressure and heart rate increase steeply; these parameters remain elevated throughout the awake period, but decrease again during the sleep period in health individuals (3). In doing so, daily rhythms in cardiovascular function ensures that supply and demand remain balanced (e.g., increased delivery of oxygen, nutrients, and endocrine factors to organs during the active period, as well as removal of metabolic by-products) (4–6). Various additional cellular processes (e.g., transcription, translation, cellular signaling, metabolism) and organ physiology (e.g., hormone release, neurological activity, muscular contraction) also oscillate over the course of the day (2). Underscoring their fundamental importance, failure to maintain these physiologic rhythms invariably has pathologic consequences (7). The current review article, focusing on cardiac physiology, has two primary goals: 1) summarize the extent to which cardiac physiology oscillates over the course of the day; and 2) discuss recent insights regarding putative mechanisms orchestrating 24-hr rhythms in cardiac physiology.
It is important to appreciate the concept that cells/organs/organisms are not only capable of adapting to, but can also anticipate stimuli/stresses. With regards to adaptation, a ‘stimulus-response’ concept is typically considered, wherein a cell/organ/organism responds upon challenge with a stimulus/stress (both acutely and chronically, depending on the nature of the stimulus/stress and/or its duration). Such a concept is often utilized as an explanation for time-of-day-dependent oscillations in cardiovascular parameters. Indeed, associated with the steep rise in blood pressure and heart rate in the early hours of the morning, are increases in multiple neurohumoral factors that are known to augment these parameters (e.g., norepinephrine, epinephrine) (8–10). It is therefore not surprising that classically, 24-hr oscillations in heart rate and blood pressure were considered to be largely mediated by daily fluctuations in neurohumoral factors and secondary to behaviors such as sleep/wake and fasting/feeding cycles. However, in recent years studies in both humans and animal models have challenged this concept. For example, when healthy individuals are subject to a light/dark cycle of 28-hrs, termed forced dyssynchrony, 24-hr oscillations persist for distinct cardiovascular parameters, including heart rate (11). Such observations suggest that daily rhythms in heart rate are not simply an adaptation to environmental and/or behavioral rhythms. Instead, an intrinsic timekeeping mechanism, namely the circadian clock, must be present within organisms, to drive 24-hr rhythms in distinct biological processes.
Circadian clocks are cell autonomous molecular mechanisms comprised of a series of positive and negative feedback loops, with a free running period of approximately 24-hrs (2). Circadian clocks confer the selective advantage of anticipation, allowing a cell/organ/organism to prepare for a stimulus/stress prior to its onset, thereby facilitating a temporally appropriate response. At the core of the mammalian mechanism are two transcription factors, CLOCK (Circadian Locomotor Output Cycles Kaput) and BMAL1 (Brain and Muscle ARNT-Like 1), which, upon heterodimerization, bind to E-boxes in the promotor of target genes, invariably leading to induction (12, 13). Target genes include negative feedback components, such as period (PER1/2/3), cryptochrome (CRY1/2) and Rev-erb (REV/ERBα/β) isoforms; PER/CRY heterodimer complexes repress CLOCK/BMAL1 transcriptional activity through physical interactions, while REV-ERBα/β represses BMAL1 at a transcriptional level (14–16). The CLOCK/BMAL1 heterodimer influences expression of numerous additional target genes, some of which are integral to the clock mechanism, whereas others, termed clock controlled/output genes (CCGs), have the potential to modulate cellular function in a time-of-day-dependent manner. Circadian clocks have been described in all mammalian cells, including cardiomyocytes (2, 17). It has been estimated that between 3%–16% of an organ’s transcriptome is circadian regulated, impacting a diverse array of biological processes (18). Studies, using mouse models in which the circadian clock mechanism has been genetically disrupted only in cardiomyocytes, suggest that up to 10% of the cardiac transcriptome is regulated in some manner by the cardiomyocyte circadian clock; known functions of the encoded proteins range from transcription, signal transduction, cellular constituent turnover, transport, and metabolism (19). All of these processes influence contractile function of the heart, suggesting that the cardiomyocyte circadian clock enables the heart to anticipate day-night differences in contractile demand through pre-emptive modulation of critical cellular processes.
It is apparent, therefore, that both extrinsic (i.e., neurohumoral factors) and intrinsic (i.e., circadian clock) mechanisms have the potential to contribute towards daily fluctuations in cardiac physiology, with both contributing at multiple levels. If considered as independent factors, the effects of neurohumoral factors and circadian clocks would be additive. However, neurohumoral factors are extracellular stimuli, for which, target tissue responsiveness is modulated by circadian clocks. In the case of the heart, the cardiomyocyte circadian clock has been shown to influence responsiveness to hormones (e.g., epinephrine, insulin) and nutrients (e.g., fatty acids) (20–23). Circadian clocks therefore amplify or dampen the responsiveness of the heart to neurohumoral stimuli, depending on the time-of-day. Moreover, the timing of the circadian clock mechanism needs to reset/entrain; for the heart, important entrainment factors (termed zeitgebers) include neurohumoral factors such as cortisol and norephinephrine (17, 24). It is also noteworthy that the levels of neurohumoral stimuli are themselves often under circadian clock control (6). For example, daily fluctuations in plasma cortisol levels are dependent on the circadian clock within the adrenal cortex (25). Taken together, a complex interplay between neurohumoral and circadian clock influences likely contribute towards time-of-day-dependent oscillations in cardiac physiology.
Daily Oscillations in Cardiac Processes
As discussed above, day-night differences in cardiac physiology have been reported at multiple levels. Therefore the purpose of the following subsections is to highlight what is known regarding circadian control of these processes, and their potential importance in cardiac physiology.
1. Cardiac Function
Electrophysiology:
In healthy subjects, time-of-day-dependent oscillations have been reported for multiple cardiac function parameters, including contractility and electrophysiology. The latter includes the R-R interval (which has an inverse relationship with heart rate), QT interval, and heart rate variability (HRV) (26, 27). In humans, both R-R and QT intervals are increased during the night, whereas HRV is increased in the morning (26–28). Classically, diurnal variations in these parameters have been explained by fluctuations in autonomic and sympathetic tone; however, during forced dyssynchrony, 24-hr rhythms in several of these electrophysiologic parameters persist, suggesting a potential contribution of the intrinsic timekeeping mechanism (11). Time-of-day-dependent rhythms in heart rate and electrophysiologic parameters have also been described in various animal models, ranging from dogs to rodents (29, 30). Interestingly, ex vivo perfused mouse hearts tend to exhibit increased heart rates in the middle of the active phase (compared to the sleep phase) (20). Moreover, 24-hr oscillations in spontaneous beating has been reported in cultured neonatal rat cardiomyocytes (31). Such observations suggest that daily rhythms in heart rate may be mediated, in part, by intrinsic circadian clocks. Use of genetically modified mouse models have helped to reinforce this concept. Both germline BMAL1 knockout and CLOCKΔ19 mutant (a dominant negative mutant of CLOCK, due to deletion of the transactivation domain) mice exhibit a pronounced attenuation of day-night differences in heart rate; in both cases, the mesor (daily average) is lower (i.e., bradycardia) (32). Both germline BMAL1 knockout and CLOCKΔ19 mutant mice exhibit alterations in behaviors and systemic factors, which could contribute to the observed heart rate phenotype. To avoid these systemic perturbations, cell-type specific models have been employed. Cardiomyocyte-specific CLOCKΔ19 mutant (CCM) mice exhibit a reduced amplitude of day-night differences in heart rate and bradycardia (20). However, inducible cardiomyocyte-specific BMAL1 knockout mice exhibit normal day-night differences in heart rate (29). Collectively, these observations suggest that circadian regulation of extra-cardiac influences (such as autonomic tone) potentially play a dominant role in mediating time-of-day-dependent oscillations in heart rate, while the exact contribution of the cardiomyocyte circadian clock remains somewhat less clear.
Multiple ion channel subunits have been reported to fluctuate in atria and/or ventricles, in a time-of-day-dependent manner, including integral components of K+ (KCHIP2, KCNA5, KCND2, KCNH2, and KCNK3), Na+ (SCN5a), and Ca2+ (CACNA1D) channels (29, 33–37). Daily rhythms in distinct ion channel components are associated with predicted day-night differences in electrophysiological properties of the heart. For example, elevated cardiac KCNA5 protein levels during the active period is associated with increased steady state current (IKur) density in rat myocytes (37). In many instances, rhythms in these ion channel subunits are lost following disruption of circadian clocks and/or autonomic blockade (19, 20, 29, 33–37). For example, CACNA1D, encoding for a subunit in L-type voltage gated ion channels, is rhythmic at both mRNA and protein expression levels in wild-type hearts, but not CCM hearts (20). Another subunit of this channel, CACNA1C, has also appears to be regulated by the cardiomyocyte clock, not only at the level of expression, but also post-translationally via the PI3K/AKT signaling axis, which is also under clock control (19, 38). Together, these studies illustrate that the electrophysiologic properties of the heart are markedly different depending on the time-of-day, potentially contributing towards daily oscillations in heart rate.
Contractility:
Similar to heart rate, contractility of the heart exhibits time-of-day-dependent oscillations at multiple levels. Echocardiographic studies in humans suggest that over a 24-hr period, although systolic function does not exhibit fluctuations in healthy subjects, multiple diastolic parameters do, such as relaxation time, which peaks in the early hours of the morning (39, 40). When the intrinsic contractile properties of the heart are investigated, through the use of isolated perfused mouse and rat hearts, parameters including cardiac output and rate pressure product are elevated during the active period; this was most striking for contractile reserve; when ex vivo mouse hearts are challenged with high workloads, cardiac output and rate pressure product were almost 2-fold higher during the active phase (compared to the sleep phase) (20, 41). Moreover, genetic disruption of the cardiomyocyte circadian clock abolishes these time-of-day-dependent rhythms in contractile function of the heart ex vivo, underscoring mediation by this intrinsic mechanism (20). Contractility of the heart is influenced by numerous mechanisms, including metabolism, cell signaling, Ca2+ homeostasis, and myofilament composition, all of which are under strict circadian control (5, 42). β-Adrenergic signaling is a well-established positive ionotrope, in large part through protein kinase A (PKA) activation and PKA activity in the heart is increased at the beginning of the active period, associated with increased phospholambam phosphorylation and SERCA2a activity (43). In contrast, calcineurin activity increases in the heart at the beginning of the sleep phase, thereby reversing the actions of PKA (43). Both PKA and calcineurin appear to be directly regulated by the cardiomyocyte circadian clock (19, 20), suggesting that both extrinsic and intrinsic mechanisms contribute towards time-of-day-dependent rhythms in this ionotropic signaling axis. Contractile protein components also exhibit significant day-night differences in the heart, at multiple levels. For example, the calcium sensitivity of myosin ATPase activity is augmented during the active period, and this day-night difference is absent following genetic disruption of the cardiomyocyte circadian clock (44). Numerous myofilament components also fluctuate over the course of the day, including myosin binding protein C, desmin, tropomyosin, troponins I and T, as well as titin cap (44). The extent to which daily oscillations in myofilament proteins, and/or posttranslational modifications of contractile components, plays a significant role in day-night differences of cardiac contractility remains an important, yet unanswered question.
2. Transcription, Translation, and Post-Translational Modifications
Transcription:
Hypothesis generating approaches designed to determine potential molecular underpinnings of day-night differences in cardiac function have yielded several important concepts. The first is that a surprisingly large number of mRNA species fluctuate in the heart over the course of a 24-hr period. For example, when mice are housed under standard 12h:12h light:dark cycles, approximately 13% of genes expressed in the heart exhibit statistically significant changes over the course of the day (45). Two major expression patterns were observed for these genes: cosine (single daily peak) and biphasic (two peaks per day) (45). In the latter case, the two peaks were often observed at the light-dark transitions. Bioinformatics approaches revealed that genes exhibiting time-of-day-dependent variations in hearts clustered into diverse biological functions, ranging from cellular growth and remodeling, to transcription, translation, mitochondrial function, and cellular signaling (45). When mice were housed in constant darkness, thereby eliminating the acute effects of light stimulation, approximately 6% of the cardiac transcriptome was identified as circadian regulated (18), which was similar to the estimate (8%) reported by Storch et al (46). Again, two primary patterns of oscillating genes were identified: cosine and biphasic (peaking at times when light:dark transitions would have had occurred if mice had not been housed in constant darkness) (18). Interestingly, the heart exhibited temporal gene expression patterns that were markedly distinct to those observed in other tissues such as kidney, aorta, skeletal muscle, white adipose tissue, or brown adipose tissue. As one example, the time-of-day-dependent oscillation of vascular endothelial growth factor alpha is essentially antiphase in heart compared to brown adipose tissue (18). Collectively, these studies highlight that the heart exhibits extensive, divergent, and unique daily fluctuations in its transcriptome.
Many questions have arisen following temporal characterization of the cardiac transcriptome, including: 1) what mechanism(s) orchestrate time-of-day-dependent fluctuations in the cardiac transcriptome; and 2) do daily changes in mRNA species translate to fluctuations in corresponding proteins and/or cardiac function? With regards to the first question, both extrinsic and intrinsic factors have the potential to mediate temporal control of the cardiac transcriptome. A plethora of neurohumoral factors are known to impact gene expression in the heart, ranging from endocrine factors (e.g., glucocorticoids, insulin, epinephrine) to nutrients (e.g., fatty acids, glucose, vitamins) and neural stimulation (e.g., norepinephrine). Many of these extrinsic stimuli fluctuate over the course of the day. For example, norepinephrine, which peaks in the early hours of the morning, can acutely increase expression of multiple immediate early genes in the heart (47). Insulin, which fluctuates with fasting/feeding cycles, also influences expression of numerous genes in the heart involved in cellular growth and metabolism, in part through regulation of FOXO transcription factors (48). Fatty acids, which increase during periods of fasting and/or following a high fat meal, impact gene expression through numerous mechanisms, including direct binding to nuclear receptors (e.g., PPAR isoforms) (49). Similarly, cortisol, which peaks in the early hours of the morning, binds to nuclear (glucocorticoid) receptors, modulating expression of target genes (50). It is therefore not surprising that the cardiac transcriptome fluctuates dramatically over the course of the day. However, intrinsic circadian clocks also modulate cardiac gene expression in a time-of-day-dependent manner; indeed, the classic mammalian mechanism is composed of a series of transcription factors (51). In mice, selective overexpression of a dominant negative CLOCK mutant protein (CCM), or deletion of BMAL1 (CBK), specifically in cardiomyocytes, results in a temporal suspension of the circadian clock in the heart at the beginning of the sleep phase (19, 20). Interestingly, the majority of genes that exhibit 24-hr oscillations in expression in normal murine hearts also become temporally suspended in CCM and CBK hearts, indicating that as much as 10% of the cardiac transcriptome is regulated in some manner by the cardiomyocyte circadian clock (19, 20). Moreover, gene ontology analysis suggested that clock regulated genes in the heart are involved in processes such as metabolism, cellular signaling, and electrophysiology. What is less clear is which of these genes are directly regulated by a circadian clock component (e.g., the CLOCK/BMAL1 heterodimer), as opposed to secondarily (e.g., through clock-controlled transcription factors, such as KLF15) (52). A number of bioinformatic filters have been applied to CCM and CBK transcriptomic data, in an attempt to identify direct target genes of the CLOCK/BMAL1 heterodimer; only a small number of target genes were identified from this stringent analysis, many of which are known to influence cardiac metabolism (see subsection on ‘Metabolism‘) (19). However, the contribution of other circadian clock components (e.g., REV-ERB isoforms) remains less clear. Interestingly, agonists to REV-ERBα/β have recently been reported to dramatically impact gene expression in the heart (53); however, whether these effects are direct (i.e., activation of REV-ERBα/β in the heart) or indirect (e.g., agonist-induced changes in neurohumoral factors) is currently unknown.
Translation and Protein Turnover:
In order to influence cardiac function over the course of the day, mRNA fluctuations must translate to meaningful alterations in protein levels and subsequently function. Moreover, the levels of some proteins may fluctuate independently to changes in mRNA. The latter was highlighted in the liver, where combined transcriptomic and proteomic approaches revealed that only approximately 50% of protein expression oscillations could be explained by alterations in gene expression (54). Time-of-day-dependent proteomic (and transcriptomic) analyses have been performed for the heart; when proteomic data were compared to previously published transcriptomic analyses, it appeared that only a sub-set of oscillating proteins could be readily explained by fluctuations in reciprocal mRNA species (44). Collectively, these observations suggest that at the proteomic level, the heart is markedly different during the day versus night, and the likelihood exists that daily rhythms in the cardiac proteome are driven by a combination of transcriptional and protein turnover events.
Protein turnover is considered the culmination of protein synthesis (i.e., translation) and degradation (through, for example, the proteasome and/or autophagy), both of which are circadian regulated in various tissues, including the heart. Cardiac protein synthesis is lowest in the middle of the active period, and peaks at the beginning of the sleep phase; these rhythms are associated with concomitant oscillations in the activation status of the mTOR/S6/4EBP1 signaling axis, an established mechanism promoting translation initiation (22). Extrinsic modulators of protein synthesis and the mTOR/S6/4EBP1 signaling axis thus might be considered as key drivers for time-of-day-dependent rhythms, such as insulin or IGF1 (55); however, a number of key observations suggest that this is not the case. For example, in ad libitum fed mice, plasma insulin levels are highest towards the beginning of the active period, a time at which protein synthesis is low, while plasma IGF1 levels do not fluctuate over the course of the day (56, 57). Moreover, during continuous fasting, despite chronically low plasma insulin levels, the mTOR/S6/4EBP1 signaling axis in the heart remains active at the wake-to-sleep phase transition (58); similar observations have been reported in skeletal muscle (59). Such observations cannot discount the possibility that locally generated IGF1 acts in a paracrine/autocrine fashion, which persists during fasting. On the other hand, genetic disruption of the cardiomyocyte circadian clock results in loss of time-of-day-dependent oscillations in both cardiac protein synthesis, as well as the mTOR/S6/4EBP1 signaling axis, both of which are chronically activated in CBK and CCM hearts (22). Furthermore, rapamycin (an established mTOR inhibitor) normalizes rates of protein synthesis in CBK hearts (22). Collectively, these observations highlight a critical role for the cardiomyocyte circadian clock as a mediator of time-of-day-dependent rhythms in cardiac protein synthesis.
Damaged and short-lived proteins are targeted for degradation via the ubiquitin-proteasome system (UPS). This includes circadian clock components, which oscillate in an UPS-dependent manner (60–62). Consistent with an obligate relationship between circadian clock and the UPS, several E3-ligases and proteasome subunits are directly regulated by cell autonomous circadian clocks (54, 63). Although circadian control of the UPS system has not been investigated directly in the heart, transcriptomic analyses have revealed that a number of UPS components exhibit time-of-day-dependent oscillations; these include various proteasome subunits, such as PSMB9, PSMD5, and PSMD10 (19). Proteins, as well as other cellular constituents, are also targeted for degradation via autophagic pathways. Evidence has accumulated suggesting circadian regulation of autophagy in multiple organs, including the heart, as discussed in more detail below.
Posttranslational Modifications:
The post translational modification (PTM) of proteins is an important mechanism responsible for fine-tuning protein function, localization and stability and appreciable evidence has accumulated, highlighting that PTM of proteins exhibit time-of-day-dependent oscillations in multiple tissues, including the heart. Many studies have focused on core circadian clock components, but such changes extend far beyond the clock mechanism. These PTMs include phosphorylation, acetylation, O-GlcNAcylation, ubiquitination, SUMOylation, ADP-ribosylation, methylation and S-nitrosation (Table 1A) (64–68). Here, we have focused on phosphorylation, acetylation and O-GlcNAcylation, which together represent key connections between the circadian clock and cardiac processes, such as signaling and metabolism.
Table 1A:
Known post translational modifications of the mammalian circadian clock
| Protein | Known PTMs |
|---|---|
| CLOCK | P, O-GlcNAc, ADPrib, Me, |
| BMAL1 | P, O-GlcNAc, Ac, Ub, S-Ni, SUMO |
| PER1/2 | P, O-GlcNAc, Ac, Ub, Me |
| CRY1/2 | P, Ub |
| REV-ERBα/β | P, Ub |
Information from Stojkovic et al (2014), and PhopshositePlus (https://www.phosphosite.org/)
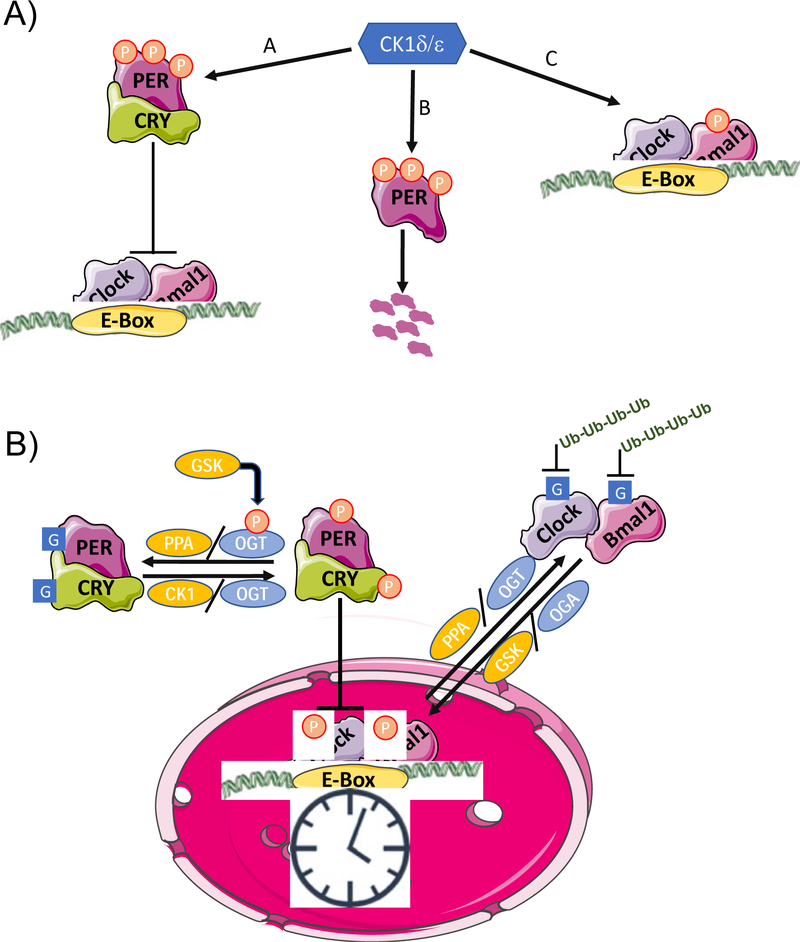
Phosphorylation is the most widely studied circadian-regulated PTM. In the liver, over 20,000 phosphorylation sites have been reported to oscillate in a time dependent manner, including those on core clock proteins (69). Interestingly, the magnitude of changes in amplitude of phosphorylation levels are substantially greater than those in transcript or protein levels; the phase distributions between peaks in phosphorylation and protein levels were also divergent (69). Moreover, different kinase pathways, responsible for regulating different physiological pathways, were partitioned into day and night phases. Overall, these findings provide strong evidence demonstrating the importance of phosphorylation in regulating biological functions over the course of the day. One such function includes the circadian clock itself. In mammals, casein kinase (CK) 1δ and 1ε play a central role in the regulation of core clock components (68). PER1/2 proteins, which repress CLOCK and BMAL1, are phosphorylated by CK1δ/ε, regulating their nuclear translocation and ubiquitin-mediated degradation (68). In doing so, CK1δ/ε is critical for maintaining the ~24 hour periodicity of the clock (68). This is evident by the fact that the tau mutant hamster, which has a point mutation in CK1ε, possesses a clock with a free running period of 20 hours (70). The complexity surrounding CK1δ/ε-mediated PER1/2 phosphorylation is demonstrated by the fact that in some cells it promotes cytoplasmic accumulation, whereas in other it facilitates nuclear translocation (Figure 1A) (68).
Figure 1.
A) Functions of Caesin Kinase (CK) 1d/e in the mammalian clock. A) Interaction with CRY stabilizes PER, allowing it be phosphorylated by leading to its nuclear translocation and repression of Clock/Bmal1 transcription; B) CK1δ/ε phosphorylation of PER in the cytosol leads to proteasomal degradation; C) CK1 δ/ε phosphorylation of Bmal1 increases its transcriptional activity. B) Cross-talk between phosphorylation (P) and O-GlcNAcylation (G) in the regulation of the circadian clock. GSK3β phosphorylation of OGT results in its activation, leading to O-GlcNAcylation of PER and CRY, which prevents their phosphorylation thereby repressing their effects on CLOCK/BMAL1 transcription. O-GlcNAcylation of CLOCK also represses the effects of PER. Phosphorylation and O-GlcNAcylation also play a role in regulating the ubiquitination and subsequent degradation of CLOCK and BMAL1
CRY1/2, repressor of CLOCK/BMAL1 transactivation, is also a known target of AMPK; AMPK-dependent phosphorylation of CRY1/2 promotes its proteasomal degradation (67). AMPK has also been shown to phosphorylate CK1ɛ, increasing its activity and subsequently decreasing PER2 stability (71). Following phosphorylation by dual-specificity tyrosine-phosphorylation-regulated kinase (DYRK1A), CRY2 (but not CRY1) is phosphorylated by GSK3β, which accelerates its degradation. GSK3β also phosphorylates REV-ERBα, a negative regulator of BMAL1, increasing its stability (67, 72). Importantly, both AMPK and GSK3β have been shown to exhibit circadian rhythms in the heart, which likely plays a role not only in control of the cardiomyocyte circadian clock, but also as modulators of cardiac physiology.
Although protein levels of BMAL1 and CLOCK exhibit relatively modest time-of-day dependent fluctuations in many tissues relative to mRNA levels, both demonstrate more robust rhythms in phosphorylation levels, which is important for regulation of their stability and consequently their transcriptional activity (66). BMAL1 phosphorylation by CKIδ/ε occurs on a highly conserved Ser90 residue, required for dimerization with CLOCK; consequently, mutation of this site to alanine prevents phosphorylation and results in disruption of circadian clock function (66). Multiple PTMs are dependent on BMAL1:CLOCK heterodimerization and/or Ser90 phosphorylation (66). Moreover, the number of kinases reported to phosphorylate BMAL1 is growing, and include ERK, JNK, GSK3β, cyclin-dependent kinase 5 (Cdk5), PKCγ, and calmodulin-dependent protein kinase II (CaMKII) (66, 73). In addition to BMAL1, CLOCK is phosphorylated by both GSK3β and Cdk5. In general, phosphorylation deficient forms of both BMAL1 and CLOCK exhibit increased stability or reduced activities (74). The cyclic control of clock components, as well as downstream processes, through phosphorylation, requires not only kinases, but also phosphatases; protein phosphatase-1 (PP1) and PP2A are the best characterized in terms of circadian regulation. In the case of PP1, previous studies suggest that activity of this phosphatase likely oscillates over the course of the day in the heart, secondary to circadian regulation of inhibitor-1 (an established inhibitor of PP1). Interestingly, the protein phosphatase 2 regulatory subunit B gamma (PPP2R2C) gene has been linked to cross talk between the circadian clock and heart disease (75). The interactions between kinases/phosphatases with the mammalian circadian clock are summarized in Table 1B.
Table 1B:
Interactions between kinases and phosphatases and the mammalian circadian clock
| Kinase/Phosphatase | Clock Target |
|---|---|
| PP1 | PER1/2 |
| PP5 | CK1 |
| CK1 | PER1/2, CRY1/2 |
| CK2 | PER1/2, BMAL1 |
| GSK3b | CRY1/2,BMAL1, CLOCK, REV-ERBα/β |
| AMPK | CRY1/2 |
Based on Figure 1 from Reischl and Kramer (2011)
The role of acetylation in regulating the circadian clock was first recognized when rhythms in H3 histone acetylation in mouse liver were shown to accompany CLOCK-BMAL1 transcriptional activity, and that CLOCK exhibited histone acetyltransferase activity (76). In addition to specific histone targets, CLOCK also acetylates BMAL1, and BMAL1 acetylation demonstrates time-of-day dependent rhythms. Importantly, BMAL1 acetylation is required for normal clock function and is necessary to inactivate BMAL1 activity via the recruitment of CRY proteins to the CLOCK-BMAL1 complex (67). SIRT1, an NAD+-dependent deacetylase, antagonizes CLOCK-mediated acetylation; SIRT1 directly interacts with CLOCK in a circadian-dependent manner, and is responsible for deacetylation of PER2, which stimulates PER2 degradation. Conversely, knockdown of SIRT1 increases PER2 acetylation and its stability (77). The precise mechanism by which acetylation stabilizes PER2 is not known; however, the fact that it occurs on lysine residues that can also be ubiquitinated could be a factor. In the mouse heart, rhythms in the acetylation of the PER1/2 promoter preceded those of PER1/2 mRNA, consistent with regulation of the cardiomyocyte circadian clock by acetylation (78).
The activity of SIRT1 is dependent on NAD+, and nicotinamide phosphoribosyltransferase (NAMPT) is the rate limiting step in the NAD+ salvage pathway. NAMPT itself is directly regulated by the circadian clock, resulting in rhythms in both NAD+ levels, and SIRT1 activity (79–81). The importance of this pathway in circadian regulation is demonstrated by the fact that chronically elevated NAD+ levels, due to systemic KO of the NAD hydrolase CD38, resulted in disruption of circadian behavior and dysregulation of peripheral clock genes (82). Pharmacological inhibition of NAMPT also altered the circadian expression of PER2, resulting in an earlier onset of its peak expression and increased BMAL1 acetylation, similar to that seen in response to loss of SIRT1 (81). The circadian rhythms of protein acetylation in the heart has not been examined fully; however, as in other tissues, the heart exhibits time-of-day-dependent changes in both NAMPT and NAD+ levels. Moreover, NAMPT and NAD+ levels are both chronically low in CCM hearts (56, 83). Contrary to what might be anticipated, CCM hearts exhibit chronically low protein acetylation, suggesting that other HATs and HDACs are likely clock controlled in the heart. Given the importance of NAD+, SIRT1, and acetylation in regulation of cardiac processes, this axis likely represents an important link between the cardiomyocyte circadian clock and cardiac physiology.
Another link between regulation of cardiac physiology and the circadian clock is modification of serine and threonine residues proteins by O-linked-N-acetylglucosamine (O-GlcNAc) (Figure 1B). O-GlcNAc is a reversible PTM, which is formed by the metabolism of UDP-N-acetylglucosamine (UDP-GlcNAc) by O-GlcNAc transferase; O-GlcNAc is removed from proteins by O-GlcNAcase. UDP-GlcNAc is synthesized by the hexosamine biosynthesis pathway (HBP), which requires fructose-6-phosphate, glutamine, acetyl-CoA and UTP; consequently, O-GlcNAc modifications are typically consider to be nutrient regulated PTMs (84). Durgan et al. first reported a link between protein O-GlcNAcylation and the cardiomyocyte circadian clock, when they observed that O-GlcNAc levels and OGT protein had a time-of-day-dependent rhythm in WT hearts, that was absent in CCM hearts (83). While OGA levels did not show any rhythmic changes, it was chronically repressed in CCM hearts. Both BMAL1 and PER1 were shown to be O-GlcNAcylated; BMAL1 O-GlcNAcylation peaked in the middle of the dark phase (83). Subsequent studies in Drosophila further substantiated a link between O-GlcNAc and the circadian clock, demonstrating that changes in OGT expression altered the length of circadian rhythms, secondary to PER O-GlcNAcylation (85). O-GlcNAcylation of BMAL1 and CLOCK were found to inhibit their ubiquitination, thereby increasing their stability (86, 87), while changes in OGT expression resulted in alterations in BMAL1/CLOCK target genes (86). Another study identified specific sites of O-GlcNAcylation on PER, and showed that O-GlcNAc modification of Ser492 attenuated the interaction between PER and CLOCK (88). Interestingly, O-GlcNAcylation of PER2 occurs in a region that is known to regulate sleep phase in humans, and competes with phosphorylation in the same region (89).
While REV-ERBα has not been shown to be an O-GlcNAc target, it interacts directly with OGT in both the cytosol and nucleus, where it stabilizes OGT protein levels (90). As a result of influencing OGT stability, REV-ERBα indirectly regulates cellular O-GlcNAc levels; consequently, changes in REV-ERBα protein levels over the course of the day could represent a mechanism leading to the rhythmic changes in O-GlcNAc observed in the heart (83). In cell culture, REV-ERBα, OGT and O-GlcNAc levels all peaked 24 hours following serum shock and REV-ERBα was found to regulate O-GlcNAc levels of over 200 proteins (90). One of those proteins was Akt, and REV-ERBα influenced both basal and insulin stimulated Akt phosphorylation. These studies suggest that REV-ERBα could be an important link between time-of-day-dependent changes in cardiac O-GlcNAc levels (83) and insulin sensitivity (22).
3. Metabolism, Mitochondria, and Redox Biology
Metabolism:
To be able to continuously adapt to changing workload demands throughout the day, the heart requires a high level of metabolic flexibility. Consequently, the heart is capable of using a wide array of substrates for energy production, including glucose, fatty acids, lactate, pyruvate, ketone bodies, and distinct amino acids (91). Both substrate availability and hormone levels fluctuate with fasting/feeding cycles, which undoubtedly contribute towards daily rhythms in cardiac metabolism. Changes in physical activity across sleep/wake cycles also impacts cardiac metabolism. However in addition to these extrinsic factors, there is substantial evidence that the cardiomyocyte circadian clock plays a significant role in regulating myocardial metabolism. For example, rhythms in glycogen and protein synthesis persist even during prolonged fasting, suggesting that feeding behavior is not the main driving factor (91). Moreover, time-of-day dependent metabolic oscillations persist in in vitro models, such as the isolated perfused heart (91). The importance of these intrinsic factors is underscored by observations that genetic disruption of CLOCK and/or BMAL1 abolishes time-of-day dependent rhythms in myocardial glucose and fatty acid metabolism (20, 21, 91). In the case of fatty acid metabolism, peroxisome proliferator-activated receptors (PPARs) play an important role in transcriptional regulation of cardiac metabolism. Consistent with this at the beginning of the active phase, there is a coordinated regulation of PPARα modulators, such that PPARα target genes, which regulate fatty acid metabolism are induced in the heart (23). Fasting, which increases circulating fatty acids, induces transcriptional activation of fatty acid metabolism in a PPARα dependent manner in the heart and this is markedly attenuated in CCM hearts (23). CCM hearts exhibit a sustained increase in fatty acid oxidation compared to control littermate hearts independent of the time of day, while triglyceride synthesis is chronically low. Similar metabolic inflexibility of CCM hearts was observed during chronic alteration of fatty acid availability, induced either by diabetes or high fat feeding (56, 92). Although AMPK mediated phosphorylation of ACCβ is a key mechanism for increasing fatty acid oxidation it is not appear involved in clock-dependent regulation of cardiac fatty acid metabolism (56). However, PTM of carboxylases, such as ACCβ, with biotin, is essential for enzymatic activity (93) and recent studies have shown that biotinylation was regulated by the circadian clock in the heart (94). Biotinylation levels are decreased in CBK hearts and ACCβ, biotinylation was also decreased, consistent with higher fatty acid oxidation rates. Moreover, dietary supplementation with biotin, normalized ACCβ biotinylation and reduced fatty acid oxidation to control levels (94).
Although AMPK is not responsible for time of day dependent changes in fatty acid oxidation, AMPK phosphorylation and activity peak in the middle of the active phase in the heart, which is associated with an increase in glucose metabolism; these rhythms are absent in CCM hearts (83, 92). NAD+ represents an key link between metabolism and circadian clocks (95) and as discussed earlier in the heart, NAMPT, which is involved in NAD+ biosynthesis exhibits a diurnal rhythm (83) which is rhythm is absent in CCM hearts. Circadian regulation of cardiomyocyte NAD+ levels could have wide ranging effects given its central role in regulating numerous metabolic processes (83).
Akt phosphorylation, a key component in insulin signaling, is dependent on PI3K; Pik3r1, encoding for the p85α subunit of PI3K, exhibits time-of-day-dependent rhythms in the heart, which are absent in both CCM and CBK hearts (19). Disruption of the cardiomyocyte clock resulted in augmented phosphorylation of Akt at both ser473 and thr308 in response to insulin treatment, which was most pronounced at the beginning of the active period (22). Surprisingly, despite higher Akt phosphorylation, insulin-mediated glycolysis and glucose oxidation was attenuated in CBK hearts, suggesting a disconnect between Akt phosphorylation and subsequent activation of GLUT4 translocation (22). Insulin-mediated phosphorylation of AS160, a downstream target of Akt responsible for regulating GLUT4 trafficking and glucose metabolism, was also blunted in CBK hearts. Similarly, insulin-mediated phosphorylation of GSK3β (another Akt target), was attenuated throughout out the day in both CBK and CCM hearts. Collectively, these observation raised the possibility that increased phosphatase activity might be counteracting the effects of Akt (22). PP1 expression, which regulates both AS160 and GSK3β phosphorylation is not affected by disruption of the circadian clock in heart; however, inhibitor-1 (I-1), which represses PP1 activity, was ~50% lower in both CBK and CCM hearts. I-1 mRNA and protein levels also exhibited time of day dependent changes, which were again significantly blunted in CBK hearts. These data suggest that disruption of the cardiomyocyte circadian clock leads to impaired insulin mediated glucose metabolism due to chronic activation of PP1, resulting from lower I-1 levels (22).
In contrast to carbohydrate and lipid metabolism, limited information on circadian regulation of ketone body metabolism in the heart is available. Circulating ketone body concentrations can vary widely in response to different physiological states, from approximately 50μM under normal conditions to the low millimolar range in response to fasting (and even higher in pathological conditions such as diabetic ketoacidosis) (96). Ketone bodies are readily oxidized by the heart in direct proportion to their availability, and are used preferentially over carbohydrates and fatty acids for energy production (96). There are reports of diurnal variations in circulating ketone bodies in humans (97, 98) and rats (99). In rats these rhythms were maintained during 24 hours of fasting (99), which suggests that they could be regulated at least in part by circadian clocks. In the heart, β-hydroxybutyrate dehydrogenase 1 (BDH1), which plays a key role in oxidation of ketone bodies (96) is markedly repressed in CCM and CBK hearts, consistent with it being regulated by the cardiomyocyte circadian clock (19). This was associated with a >90% decrease in activity and >60% decrease in β-hydroxybutyrate oxidation (19). The growing evidence that ketone body metabolism is increased in the failing heart (100), suggests that studies to better understand the mechanisms underlying the circadian clock regulation of cardiac ketone body metabolism are needed.
Our understanding of time-of-day-dependent regulation of amino acid metabolism in the heart is not well developed; however, there are rhythmic changes in amino acids levels, as well as the expression of enzymes that regulated amino acid metabolism (20, 45, 46, 101, 102). Glutamine synthetase, which plays a key role in cardiac glutamine metabolism, exhibits a clear change over the course of day at both the gene and protein level, which was repressed in CCM hearts (83). In addition, oxidation of the branched chain amino acid leucine was attenuated in CBK hearts (94); whereas leucine incorporation into proteins was higher in CBK hearts. This observation is consistent with studies reporting that time-of-day dependent rhythms in protein synthesis are mediated by the cardiomyocyte circadian clock (22, 103). The sub-section on ‘Translation and Protein Turnover‘ describes in greater detail the mechanisms by which circadian clocks impact protein synthesis (and degradation).
Mitochondria:
A key cornerstone of cardiac physiology is mitochondria function. Mitochondria are essential not only for ATP synthesis, but also for calcium homeostasis, redox signaling, and metabolite provision. Circadian regulation of mitochondrial function has been reported in a number of cells (e.g., fibroblasts) and tissues (e.g., liver). However, less is known for the heart. Time-of-day-dependent regulation of cardiac mitochondrial function was first suggested in 2001, when rat myocardial oxygen consumption was found to be the higher in the middle of the night (compared to in the middle of the day) (104). To date, we are unaware of a study that has directly assessed mitochondrial function in the heart at different times of the day. However, we have recently measured the activity of mitochondrial complexes in mouse hearts at 6-hr intervals over a 24-hr period, revealing that complex I activity is highest at the beginning of the active phase (unpublished observations). More is known about the impact that genetic disruption of the cardiomyocyte circadian clock has on cardiac mitochondrial parameters. For example, oxygen consumption has been measured in isolated subsarcolemmal (SSM) and intramyofibrillar (IFM) mitochondria from CCM and control littermate hearts. In SSM, but not IFM, state 3 respiration was decreased in CCM hearts, with glutamate, pyruvate, palmitoycarnitine, palmitoyl-CoA or acetyl carnitine as substrates (but not succinate, DHQ, or octanoyl-CoA). This was in the absence of mitochondrial DNA content, number, citrate synthase activity, gross morphology, or volume density between CCM and control hearts (20). Kohsaka et al (105) have reported a more severe mitochondrial phenotype in CBK hearts, which exhibit decreased mitochondrial protein content, diminished complex I and IV activities, as well as more divergent mitochondrial sizes.
As discussed in previous sections, multiple omics approaches support the concept of circadian regulation of mitochondrial function (18, 46, 101, 106–108). Microarray studies performed on extra-cardiac tissues (e.g., SCN, liver) isolated from mice housed in constant darkness, revealed that many genes related to mitochondrial function and metabolism, oscillate over the course of the day, including those encoding electron transport chain proteins (Ndufa2, Ndufb5, Ndufc1, Ndufv2, Cox7b, 4a, 6a1, 6c, 7a3, 7c and Atp5g2, 5l, and 5c1) (18, 108). A similar microarray study investigating the heart reported that, of the 462 genes oscillating, 5 were mitochondrial, including a mitochondrial uncoupling protein, a membrane carrier and a subunit of cytochrome c oxidase (46). When transcriptomic studies are performed in hearts from animals as diverse as mice to primates and humans, genes involved in ATP generation, oxidative metabolism, and mitochondrial function, are consistently identified as oscillating over the course of the day (101, 106, 107). Using genetically modified mouse models with circadian gene disruption, transcripts encoding distinct components of electron transport chain complexes, such as Ndufs7, Sdhc, Cox7b, Atp5g2 and Uqcrc1, are chronically repressed in the heart (and oscillate in control hearts) (105). mRNAs encoding for the mitochondrial biogenesis protein PGC1α, as well as fusion proteins (Mfn2 and Opa1), are also decreased in CBK hearts (105). Interestingly, Mfn2, Opa1, and Drp1 have all been shown to be circadian regulated in the liver, and that liver cell mitochondria are enlarged/swollen in hepatocyte-specific Bmal1 knockout mice (109). Taken together, these observations suggest that cardiac mitochondrial function and morphology likely fluctuate over the course of the day, through processes governed by cardiomyocyte circadian clock. Despite these studies, the extent to which mitochondrial function is regulated by circadian clock in the heart remains unclear.
Cellular Redox Status:
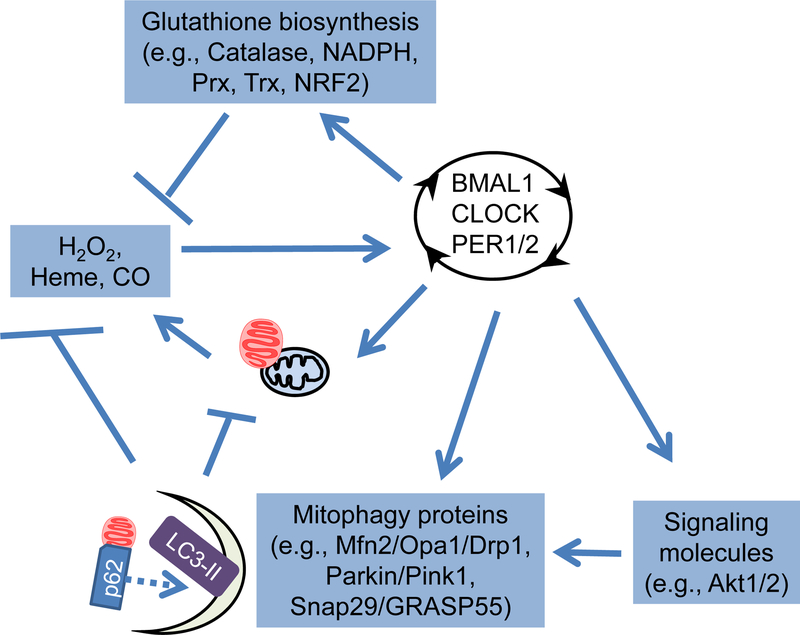
Dysregulation of redox status has been shown to affect cardiac health and response to stress (110–112). Consequently, it is important to understand whether redox signaling in the heart is circadian dependent; moreover, redox signaling may modulate circadian clock function (Figure 2). It is also possible that antioxidant defenses may also be circadian regulated and could mediate time-of-day dependent responses of the heart to stress. Significant progress has been made towards a better understanding of the cross talk between redox biology and the circadian clock (113, 114). As early as the 1980s, glutathione and lipid peroxidation products were found to be circadian regulated in rat blood, liver, stomach, kidney and heart, with glutathione levels higher in the dark phase (115). In human red blood cells kept in complete darkness, the oxidized/hyperoxidized PRX-SO2/3 dimer peaks at circadian time 16, which was associated with circadian regulation of hemoglobin autoxidation, NADH and NADPH (116). Although peroxiredoxin dimer oscillations are transcription independent in red blood cells, these oscillations are lengthened in embryonic fibroblast cells from mCry1/2−/−mice compare to those from wildtype mice, indicating that both transcription dependent and independent redox oscillations exist (116). Peroxiredoxin oscillations also occur in synchronized mouse 3T3 cells, the unicellular pico-eukaryotic alga, heads of the fruit fly D. Melanogaster, and the filamentous fungus N. crassa, suggesting that this is a highly conserved mechanism (116–118). Importantly, peroxiredoxin oscillations are also observed in the heart (119) supporting the concept of time-of-day dependent changes in cardiac redox status.
Figure 2. Crosstalk between circadian clock components and reactive species.
Both reactive species and antioxidants are circadian regulated. Moreover, the activities of circadian clock components can also be modulated by reactive species. To what extent this cross talk occurs in the heart requires full elucidation.
In the rat heart, higher levels of lipid peroxidation and lower levels of selenium-dependent glutathione peroxidase activity have been reported during the dark phase (120). Moreover, microarray analysis demonstrated that several isoforms of glutathione transferase are repressed in CBK hearts (19). In aortas of germline BMAL1 knockout mice, superoxide is increased, and BH4 and BH2 levels are imbalanced (121). Both L-NAME (NOS inhibitor) and BH4 (eNOS cofactor) decreased superoxide in BMAL1 knockout mice, suggesting an uncoupling of eNOS activity (121). The enzymes involved in biopterin metabolism and eNOS coupling, GTPCH-1 and DHFR, have also been shown to be circadian regulated in aortas (121). In human aortic smooth muscle and endothelial cells, NADPH oxidase 4 (NOX4) gene expression is circadian regulated; genetic disruption of BMAL1 induced NOX4 gene expression both in vivo and in vitro (122). These observations provide strong support for circadian clock dependent regulation of redox signaling in the heart and vascular system.
Circadian clock transcription factor function is also influenced by reactive oxygen species. For example, exposing a cell line from zebrafish embryos to H2O2, resulted in elevated zCry1a and zPer2 gene expression, and also induced zCry1a, zPer2, zPer1 and zCat gene oscillation (123). Interestingly the rhythms in catalase activity were anti-phase relative to zPer2 and zCry1a expression; ectopic expression of zCat resulted in attenuated light induction of zCry1a and zPer2 gene expression (123). The binding and regulation of heme seems to be a common feature shared among NPAS2, mPer2, Rev-erbα and β suggesting a possible mechanisms for reactive species-mediated regulation of circadian rhythms (124–128). Moreover, NADPH is circadian regulated not only in the human red blood cells, but also in human osteosarcoma (U2OS) cells (116, 129); inhibition of the pentose phosphate pathway (PPP; critical for NADPH generation) increased BMAL1 reporter gene expression (129). Moreover, inhibition of PPP in organotypic slice culture of the suprachiasmatic nucleus (SCN) and the liver lengthened the oscillation of Per2:Luc gene expression (129). Taken together, significant cross talk between redox homeostasis and the circadian clock exists (Figure 2); however, to what extent this cross talk occurs in the heart, and how their perturbation may contribute to heart diseases, remain to be determined.
Mitochondrial Quality Control:
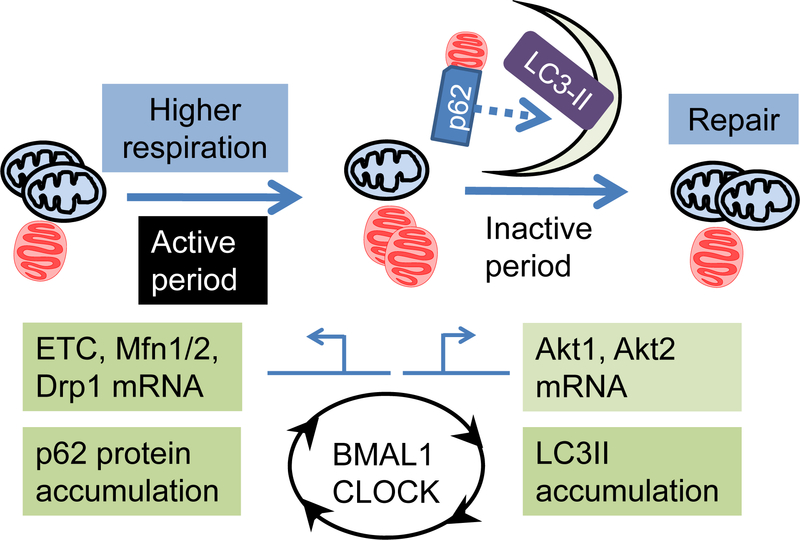
Given the importance of mitochondria for cellular functions, it is essential that there are mechanisms to ensure mitochondrial quality control. This is especially important in the heart, since oxidative phosphorylation is the main source of energy in the mammalian heart (130), and ~30% of a cardiomyocytes volume consists of mitochondria (131). Using pulse chase as the primary method of assessment, it was estimated that the half-life of heart mitochondria in rodents was ~16 days (132). Despite their long half-life, it is essential that damaged mitochondria are replaced (i.e., degraded and synthesized). Since there is a strong circadian regulation of mitochondrial protein levels, and likely a circadian regulation of mitochondrial function, there is a strong possibility that cardiac mitochondria are more likely replaced at specific times of the day. A key pathway for removal of damaged mitochondria involves mitophagy (a specialized form of autophagy). Diurnal changes of autophagy were observed as early as in the 1970s in kidney tubules of normal rats (133). Circadian regulation of autophagy has also been investigated in other tissues, including the liver and the heart. The use of both candidate testing and hypothesis generating studies suggest that genes and proteins known to be involved in mitochondrial dynamics and mitophagy may be circadian controlled in the heart (19, 22, 58). Since autophagy is sensitive to nutrient deprivation, it has been proposed that diurnal changes of autophagy might be secondary to feeding/fasting cycles (134). However, recent studies in the liver revealed that autophagy rhythms are dependent on both C/EBP (i.e., extrinsic nutrient response) and BMAL1 (intrinsic timekeeping mechanism) (135). Regarding circadian regulation of mitophagy, Drp1 phosphorylation (a fission protein which promotes mitophagy) (136) is circadian regulated in fibroblasts (137). Moreover, mitophagy proteins PINK1 and Bnip3 are BMAL1-regulated in the liver (109). Less is known regarding circadian control of mitophagy in the heart. Studies in mice revealed that autophagy appears to increase approximately 4-hrs into the sleep period, and greater fasting-induced autophagy during the sleep phase, again suggesting involvement of a non-nutrient signal (22, 58) (Figure 3). Clearly, additional studies are required to elucidate the extent to which mitochondrial quality control is regulated by the cardiomyocyte circadian clock.
Figure 3. Circadian regulation of genes influencing mitochondrial function and morphology.
Mitochondrial respiration generally peaks early in the active period, while autophagy/mitophagy peaks during inactive period, presumably ensuring mitochondrial remodeling and quality control. Oscillation of mRNAs encoding mitochondrial electron transport chain (ETC) proteins, fission protein Drp1, and Akt1/2, as well and autophagy adaptor protein sequestosome1/p62 are controlled by BMAL1 and CLOCK in the heart. However, whether mitophagic flux is higher in the inactive phase remains unclear. How regulation of mitochondrial function, dynamics and quality control by circadian clock contributes to cardiac physiology and pathology is an important question to be address.
Summary, Implications, and Future Perspectives
As summarized and discussed in this review, great advances to improve our understanding of circadian regulation of cardiac physiology have been made. The heart is a drastically distinct organ during the day, compared to the night, in terms of transcriptional, translational, and post-translational regulation of proteins, as well as regulation of metabolism, redox status and mitochondrial structure/function. An important topic not discussed here is that accumulating evidence indicates that disruption of circadian governance over cardiac processes invariably leads to pathological consequences, highlighting an urgent need to expand, enhance, substantiate, and improve our understanding of the fundamental biology of circadian regulation of cardiac function. Specific area of future study may include: 1) determining the exact mechanisms driving daily fluctuations in cardiac processes and/or whether perturbation of these mechanisms play a causal role in the etiology of common cardiac diseases; 2) improving our understanding of circadian control of organelles (e.g., lysosomes, mitochondria) in the heart; and 3) unveiling the complex interplay between extrinsic (i.e., neurohumoral) and intrinsic (i.e., circadian clock) influences contributing towards circadian control of cardiac physiology.
ACKNOWLEDGEMENTS
This work was supported by the National Heart, Lung, and Blood Institute (R01HL142216; JZ, JCC, MEY).
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Edery I 2000. Circadian rhythms in a nutshell. Physiol Genomics 3: 59–74 [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. 2008. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9: 764–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degaute JP, van de Borne P, Linkowski P, Van Cauter E. 1991. Quantitative analysis of the 24-hour blood pressure and heart rate patterns in young men. Hypertension 18: 199–210 [DOI] [PubMed] [Google Scholar]
- 4.Bass J, Takahashi JS. 2010. Circadian integration of metabolism and energetics. Science 330: 1349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martino TA, Young ME. 2015. Influence of the cardiomyocyte circadian clock on cardiac physiology and pathophysiology. J Biol Rhythms 30: 183–205 [DOI] [PubMed] [Google Scholar]
- 6.Gamble KL, Berry R, Frank SJ, Young ME. 2014. Circadian clock control of endocrine factors. Nat Rev Endocrinol 10: 466–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smolensky MH, Hermida RC, Reinberg A, Sackett-Lundeen L, Portaluppi F. 2016. Circadian disruption: New clinical perspective of disease pathology and basis for chronotherapeutic intervention. Chronobiol Int 33: 1101–19 [DOI] [PubMed] [Google Scholar]
- 8.Prinz PN, Halter J, Benedetti C, Raskind M. 1979. Circadian variation of plasma catecholamines in young and old men: relation to rapid eye movement and slow wave sleep. J Clin Endocrinol Metab 49: 300–4 [DOI] [PubMed] [Google Scholar]
- 9.Richards AM, Nicholls MG, Espiner EA, Ikram H, Cullens M, Hinton D. 1986. Diurnal patterns of blood pressure, heart rate and vasoactive hormones in normal man. Clin Exp Hypertens A 8: 153–66 [DOI] [PubMed] [Google Scholar]
- 10.Turton MB, Deegan T. 1974. Circadian variations of plasma catecholamine, cortisol and immunoreactive insulin concentrations in supine subjects. Clin Chim Acta 55: 389–97 [DOI] [PubMed] [Google Scholar]
- 11.Scheer FA, Hu K, Evoniuk H, Kelly EE, Malhotra A, et al. 2010. Impact of the human circadian system, exercise, and their interaction on cardiovascular function. Proc Natl Acad Sci U S A 107: 20541–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gekakis N, Staknis D, Nguyen H, Davis F, Wilsbacher L, et al. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564–69 [DOI] [PubMed] [Google Scholar]
- 13.Hogenesch J, Gu Y, Jain S, Bradfield C. 1998. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Aad Sci U.S.A 95: 5474–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kume K, Zylka M, Sriram S, Shearman L, Weaver D, et al. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- 15.Shearman L, Sriram S, Weaver D, Maywood E, Chaves I, et al. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288: 1013–19 [DOI] [PubMed] [Google Scholar]
- 16.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, et al. 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–60 [DOI] [PubMed] [Google Scholar]
- 17.Durgan D, Hotze M, Tomlin T, Egbejimi O, Graveleau C, et al. 2005. The intrinsic circadian clock within the cardiomyocyte. Am J Physiol Heart Circ Physiol 289: H1530–H41 [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111: 16219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young ME, Brewer RA, Peliciari-Garcia RA, Collins HE, He L, et al. 2014. Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J. Biol. Rhythms 29: 257–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray M, Shaw C, Moore M, Garcia R, Zanquetta M, et al. 2008. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function; metabolism; and gene expression. Am J Physiol Heart Circ Physiol 294: H1036–H47 [DOI] [PubMed] [Google Scholar]
- 21.Durgan DJ, Tsai JY, Grenett MH, Pat BM, Ratcliffe WF, et al. 2011. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int 28: 187–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinnis GR, Tang Y, Brewer RA, Brahma MK, Stanley HL, et al. 2017. Genetic disruption of the cardiomyocyte circadian clock differentially influences insulin-mediated processes in the heart. J Mol Cell Cardiol 110: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durgan D, Trexler N, Egbejimi O, McElfresh T, Suk H, et al. 2006. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem 281: 24254–69 [DOI] [PubMed] [Google Scholar]
- 24.Young ME, Bray MS. 2007. Potential role for peripheral circadian clock dyssynchrony in the pathogenesis of cardiovascular dysfunction. Sleep Med 8: 656–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son GH, Chung S, Choe HK, Kim HD, Baik SM, et al. 2008. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci U S A 105: 20970–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong TQ Jr., Goldberger JJ, Parker M, Wang T, Kadish AH. 1995. Circadian variation in human ventricular refractoriness. Circulation 92: 1507–16 [DOI] [PubMed] [Google Scholar]
- 27.Boudreau P, Dumont G, Kin NM, Walker CD, Boivin DB. 2011. Correlation of heart rate variability and circadian markers in humans. Conf Proc IEEE Eng Med Biol Soc 2011: 681–2 [DOI] [PubMed] [Google Scholar]
- 28.Bexton RS, Vallin HO, Camm AJ. 1986. Diurnal variation of the QT interval--influence of the autonomic nervous system. Br Heart J 55: 253–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, et al. 2013. The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol 304: C954–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Hanafy MA, Killingsworth CR, Walcott GP, Young ME, Pogwizd SM. 2014. Morning surge of ventricular arrhythmias in a new arrhythmogenic canine model of chronic heart failure is associated with attenuation of time-of-day dependence of heart rate and autonomic adaptation, and reduced cardiac chaos. PLoS One 9: e105379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.du Pre BC, Dierickx P, Crnko S, Doevendans PA, Vos MA, et al. 2017. Neonatal rat cardiomyocytes as an in vitro model for circadian rhythms in the heart. J Mol Cell Cardiol 112: 58–63 [DOI] [PubMed] [Google Scholar]
- 32.Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, Fitzgerald GA. 2007. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci U S A 104: 3450–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong M, Watanabe E, Yamamoto N, Nagahata-Ishiguro M, Maemura K, et al. 2013. Circadian expressions of cardiac ion channel genes in mouse might be associated with the central clock in the SCN but not the peripheral clock in the heart. Biol Rhythm Res 44: 519–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder EA, Burgess DE, Zhang X, Lefta M, Smith JL, et al. 2015. The cardiomyocyte molecular clock regulates the circadian expression of Kcnh2 and contributes to ventricular repolarization. Heart Rhythm 12: 1306–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, et al. 2012. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 483: 96–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko ML, Shi L, Tsai JY, Young ME, Neuendorff N, et al. 2011. Cardiac-specific mutation of Clock alters the quantitative measurements of physical activities without changing behavioral circadian rhythms. J Biol Rhythms 26: 412–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita T, Sekiguchi A, Iwasaki YK, Sagara K, Iinuma H, et al. 2003. Circadian variation of cardiac K+ channel gene expression. Circulation 107: 1917–22 [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Zhu D, Yuan J, Han Z, Wang Y, et al. 2016. CLOCK-BMAL1 regulate the cardiac L-type calcium channel subunit CACNA1C through PI3K-Akt signaling pathway. Can J Physiol Pharmacol 94: 1023–32 [DOI] [PubMed] [Google Scholar]
- 39.Karabag T, Aydin M, Dogan SM, Sayin MR, Cetiner MA. 2011. The influence of circadian variations on echocardiographic parameters in healthy people. Echocardiography 28: 612–8 [DOI] [PubMed] [Google Scholar]
- 40.Voutilainen S, Kupari M, Hippelainen M, Karppinen K, Ventila M. 1996. Circadian variation of left ventricular diastolic function in healthy people. Heart 75: 35–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young M, Razeghi P, Cedars A, Guthrie P, Taegtmeyer H. 2001. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res 89: 1199–208 [DOI] [PubMed] [Google Scholar]
- 42.Young ME. 2009. Anticipating anticipation: pursuing identification of cardiomyocyte circadian clock function. J Appl Physiol 107: 1339–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachan N, Dey A, Rotter D, Grinsfelder DB, Battiprolu PK, et al. 2011. Sustained hemodynamic stress disrupts normal circadian rhythms in calcineurin-dependent signaling and protein phosphorylation in the heart. Circ Res 108: 437–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podobed P, Pyle WG, Ackloo S, Alibhai FJ, Tsimakouridze EV, et al. 2014. The day/night proteome in the murine heart. Am J Physiol Regul Integr Comp Physiol 307: R121–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martino T, Arab S, Straume M, Belsham DD, Tata N, et al. 2004. Day/night rhythms in gene expression of the normal murine heart. J Mol Med 82: 256–64 [DOI] [PubMed] [Google Scholar]
- 46.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, et al. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83 [DOI] [PubMed] [Google Scholar]
- 47.Hannan RD, Stennard FA, West AK. 1993. Expression of c-fos and related genes in the rat heart in response to norepinephrine. J Mol Cell Cardiol 25: 1137–48 [DOI] [PubMed] [Google Scholar]
- 48.Ni YG, Wang N, Cao DJ, Sachan N, Morris DJ, et al. 2007. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc Natl Acad Sci U S A 104: 20517–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barger P, Kelly D. 2000. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc Med 10: 238–45 [DOI] [PubMed] [Google Scholar]
- 50.Oakley RH, Cruz-Topete D, He B, Foley JF, Myers PH, et al. 2019. Cardiomyocyte glucocorticoid and mineralocorticoid receptors directly and antagonistically regulate heart disease in mice. Sci Signal 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi J 1993. Circadian-clock regulation of gene expression. Curr Opin Genet Dev 3: 301–09 [DOI] [PubMed] [Google Scholar]
- 52.Hsieh PN, Zhang L, Jain MK. 2018. Coordination of cardiac rhythmic output and circadian metabolic regulation in the heart. Cell Mol Life Sci 75: 403–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Zhang R, Tien CL, Chan RE, Sugi K, et al. 2017. REV-ERBalpha ameliorates heart failure through transcription repression. JCI Insight 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, et al. 2006. Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–15 [DOI] [PubMed] [Google Scholar]
- 55.Dorn GW 2nd, Force T. 2005. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest 115: 527–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peliciari-Garcia RA, Goel M, Aristorenas JA, Shah K, He L, et al. 2016. Altered myocardial metabolic adaptation to increased fatty acid availability in cardiomyocyte-specific CLOCK mutant mice. Biochim. Biophys. Acta 1860: 1579–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawai M, Delany AM, Green CB, Adamo ML, Rosen CJ. 2010. Nocturnin suppresses igf1 expression in bone by targeting the 3’ untranslated region of igf1 mRNA. Endocrinology 151: 4861–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brewer RA, Collins HE, Berry RD, Brahma MK, Tirado BA, et al. 2018. Temporal partitioning of adaptive responses of the murine heart to fasting. Life Sciences 197: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shavlakadze T, Anwari T, Soffe Z, Cozens G, Mark PJ, et al. 2013. Impact of fasting on the rhythmic expression of myogenic and metabolic factors in skeletal muscle of adult mice. Am J Physiol Cell Physiol 305: C26–35 [DOI] [PubMed] [Google Scholar]
- 60.Willis MS, Patterson C. 2013. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N Engl J Med 368: 455–64 [DOI] [PubMed] [Google Scholar]
- 61.Portbury AL, Ronnebaum SM, Zungu M, Patterson C, Willis MS. 2012. Back to your heart: ubiquitin proteasome system-regulated signal transduction. J Mol Cell Cardiol 52: 526–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schibler U 2009. The 2008 Pittendrigh/Aschoff lecture: peripheral phase coordination in the mammalian circadian timing system. J Biol Rhythms 24: 3–15 [DOI] [PubMed] [Google Scholar]
- 63.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. 2002. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol 12: 551–7 [DOI] [PubMed] [Google Scholar]
- 64.Stojkovic K, Wing SS, Cermakian N. 2014. A central role for ubiquitination within a circadian clock protein modification code. Front Mol Neurosci 7: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papazyan R, Zhang Y, Lazar MA. 2016. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 23: 1045–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamaru T, Takamatsu K. 2018. Circadian modification network of a core clock driver BMAL1 to harmonize physiology from brain to peripheral tissues. Neurochem Int 119: 11–16 [DOI] [PubMed] [Google Scholar]
- 67.Hirano A, Fu YH, Ptacek LJ. 2016. The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol 23: 1053–60 [DOI] [PubMed] [Google Scholar]
- 68.Gallego M, Virshup DM. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–48 [DOI] [PubMed] [Google Scholar]
- 69.Robles MS, Humphrey SJ, Mann M. 2017. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab 25: 118–27 [DOI] [PubMed] [Google Scholar]
- 70.Dey J, Carr AJ, Cagampang FR, Semikhodskii AS, Loudon AS, et al. 2005. The tau mutation in the Syrian hamster differentially reprograms the circadian clock in the SCN and peripheral tissues. J Biol Rhythms 20: 99–110 [DOI] [PubMed] [Google Scholar]
- 71.Jordan SD, Lamia KA. 2013. AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol 366: 163–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin L, Wang J, Klein PS, Lazar MA. 2006. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311: 1002–5 [DOI] [PubMed] [Google Scholar]
- 73.Reischl S, Kramer A. 2011. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett 585: 1393–9 [DOI] [PubMed] [Google Scholar]
- 74.Hiraishi T, Seo Y, Murakami M, Watari H. 1990. Detection of Biexponential Relaxation in Intracellular K in the Rat Heart by Double-Quantum 39K NMR. Journal of Magnetic Resonance 87: 169–73 [Google Scholar]
- 75.Yan X, Huang Y, Wu J. 2018. Identify Cross Talk Between Circadian Rhythm and Coronary Heart Disease by Multiple Correlation Analysis. J Comput Biol 25: 1312–27 [DOI] [PubMed] [Google Scholar]
- 76.Etchegaray JP, Lee C, Wade PA, Reppert SM. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–82 [DOI] [PubMed] [Google Scholar]
- 77.Wang RH, Zhao T, Cui K, Hu G, Chen Q, et al. 2016. Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging. Sci Rep 6: 28633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, et al. 2004. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem 279: 7091–7 [DOI] [PubMed] [Google Scholar]
- 79.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, et al. 2009. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wijnen H 2009. Circadian rhythms. A circadian loop asSIRTs itself. Science 324: 598–9 [DOI] [PubMed] [Google Scholar]
- 81.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. 2009. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. 2011. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 3: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, et al. 2011. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 286: 44606–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright JN, Collins HE, Wende AR, Chatham JC. 2017. O-GlcNAcylation and cardiovascular disease. Biochem Soc Trans 45: 545–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. 2012. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev 26: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, et al. 2013. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab 17: 303–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma YT, Luo H, Guan WJ, Zhang H, Chen C, et al. 2013. O-GlcNAcylation of BMAL1 regulates circadian rhythms in NIH3T3 fibroblasts. Biochem Biophys Res Commun 431: 382–7 [DOI] [PubMed] [Google Scholar]
- 88.Li YH, Liu X, Vanselow JT, Zheng H, Schlosser A, Chiu JC. 2019. O-GlcNAcylation of PERIOD regulates its interaction with CLOCK and timing of circadian transcriptional repression. PLoS Genet 15: e1007953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, et al. 2013. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab 17: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berthier A, Vinod M, Porez G, Steenackers A, Alexandre J, et al. 2018. Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV-ERBalpha complex. Proc Natl Acad Sci U S A 115: E11033–E42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chatham JC, Young ME. 2013. Regulation of myocardial metabolism by the cardiomyocyte circadian clock. J Mol Cell Cardiol 55: 139–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsai JY, Kienesberger PC, Pulinilkunnil T, Sailors MH, Durgan DJ, et al. 2009. Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem 285: 2918–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tong L 2013. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70: 863–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He L, Hamm JA, Reddy A, Sams D, Peliciari-Garcia RA, et al. 2016. Biotinylation: a novel posttranslational modification linking cell autonomous circadian clocks with metabolism. Am J Physiol Heart Circ Physiol 310: H1520–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Asher G, Schibler U. 2011. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–37 [DOI] [PubMed] [Google Scholar]
- 96.Cotter DG, Schugar RC, Crawford PA. 2013. Ketone body metabolism and cardiovascular disease. Am J Physiol Heart Circ Physiol 304: H1060–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hocking MD, Rayner PH, Nattrass M. 1986. Metabolic rhythms in adolescents with diabetes. Arch Dis Child 61: 124–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iwata S, Ozawa K, Shimahara Y, Mori K, Kobayashi N, et al. 1991. Diurnal fluctuations of arterial ketone body ratio in normal subjects and patients with liver dysfunction. Gastroenterology 100: 1371–8 [PubMed] [Google Scholar]
- 99.Ahlersova E, Ahlers I, Benninghausova Z, Toropila M, Datelinka I. 1985. Circadian rhythm of ketone bodies in the blood of fasting rats. Physiol Bohemoslov 34: 177–81 [PubMed] [Google Scholar]
- 100.Aubert G, Martin OJ, Horton JL, Lai L, Vega RB, et al. 2016. The Failing Heart Relies on Ketone Bodies as a Fuel. Circulation 133: 698–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang L, Prosdocimo DA, Bai X, Fu C, Zhang R, et al. 2015. KLF15 Establishes the Landscape of Diurnal Expression in the Heart. Cell Rep 13: 2368–75 [DOI] [PubMed] [Google Scholar]
- 102.Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. 2012. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A 109: 5541–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rau E, Meyer DK. 1975. A diurnal rhythm of incorporation of L-[3H] leucine in myocardium of the rat. Recent Adv Stud Cardiac Struct Metab 7: 105–10 [PubMed] [Google Scholar]
- 104.Young ME, Razeghi P, Cedars AM, Guthrie PH, Taegtmeyer H. 2001. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res 89: 1199–208 [DOI] [PubMed] [Google Scholar]
- 105.Kohsaka A, Das P, Hashimoto I, Nakao T, Deguchi Y, et al. 2014. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS. ONE 9: e112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, et al. 2018. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruben MD, Wu G, Smith DF, Schmidt RE, Francey LJ, et al. 2018. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci Transl Med 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–20 [DOI] [PubMed] [Google Scholar]
- 109.Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen NH, et al. 2015. Hepatic Bmal1 Regulates Rhythmic Mitochondrial Dynamics and Promotes Metabolic Fitness. Cell Metab 22: 709–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, et al. 2014. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515: 431–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Turko IV, Li L, Aulak KS, Stuehr DJ, Chang JY, Murad F. 2003. Protein tyrosine nitration in the mitochondria from diabetic mouse heart. Implications to dysfunctional mitochondria in diabetes. J Biol Chem 278: 33972–7 [DOI] [PubMed] [Google Scholar]
- 112.Liu H, Colavitti R, Rovira II, Finkel T. 2005. Redox-dependent transcriptional regulation. Circ. Res 97: 967–74 [DOI] [PubMed] [Google Scholar]
- 113.Peliciari-Garcia RA, Darley-Usmar V, Young ME. 2018. An overview of the emerging interface between cardiac metabolism, redox biology and the circadian clock. Free Radic Biol Med 119: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wende AR, Young ME, Chatham J, Zhang J, Rajasekaran NS, Darley-Usmar VM. 2016. Redox biology and the interface between bioenergetics, autophagy and circadian control of metabolism. Free Radic. Biol. Med 100: 94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Farooqui MY, Ahmed AE. 1984. Circadian periodicity of tissue glutathione and its relationship with lipid peroxidation in rats. Life Sci 34: 2413–8 [DOI] [PubMed] [Google Scholar]
- 116.O’Neill JS, Reddy AB. 2011. Circadian clocks in human red blood cells. Nature 469: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, et al. 2011. Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qipshidze N, Tyagi N, Metreveli N, Lominadze D, Tyagi SC. 2012. Autophagy mechanism of right ventricular remodeling in murine model of pulmonary artery constriction. Am. J. Physiol Heart Circ. Physiol 302: H688–H96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rhee SG, Kil IS. 2016. Mitochondrial H2O2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: Linking mitochondrial function to circadian rhythm. Free Radic Biol Med 99: 120–27 [DOI] [PubMed] [Google Scholar]
- 120.Lapenna D, De Gioia S, Mezzetti A, Porreca E, Ciofani G, et al. 1992. Circadian variations in antioxidant defences and lipid peroxidation in the rat heart. Free Radic Res Commun 17: 187–94 [DOI] [PubMed] [Google Scholar]
- 121.Anea CB, Cheng B, Sharma S, Kumar S, Caldwell RW, et al. 2012. Increased superoxide and endothelial NO synthase uncoupling in blood vessels of Bmal1-knockout mice. Circ Res 111: 1157–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anea CB, Zhang M, Chen F, Ali MI, Hart CM, et al. 2013. Circadian clock control of Nox4 and reactive oxygen species in the vasculature. PLoS One 8: e78626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hirayama J, Cho S, Sassone-Corsi P. 2007. Circadian control by the reduction/oxidation pathway: catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci U S A 104: 15747–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. 2002. NPAS2: a gas-responsive transcription factor. Science 298: 2385–7 [DOI] [PubMed] [Google Scholar]
- 125.Boehning D, Snyder SH. 2002. Circadian rhythms. Carbon monoxide and clocks. Science 298: 2339–40 [DOI] [PubMed] [Google Scholar]
- 126.Kaasik K, Lee CC. 2004. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430: 467–71 [DOI] [PubMed] [Google Scholar]
- 127.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, et al. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 14: 1207–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, et al. 2007. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–9 [DOI] [PubMed] [Google Scholar]
- 129.Rey G, Valekunja UK, Feeney KA, Wulund L, Milev NB, et al. 2016. The Pentose Phosphate Pathway Regulates the Circadian Clock. Cell Metab 24: 462–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duncker DJ, Bache RJ. 2008. Regulation of coronary blood flow during exercise. Physiol Rev 88: 1009–86 [DOI] [PubMed] [Google Scholar]
- 131.Schaper J, Meiser E, Stammler G. 1985. Ultrastructural morphometric analysis of myocardium from dogs, rats, hamsters, mice, and from human hearts. Circ. Res 56: 377–91 [DOI] [PubMed] [Google Scholar]
- 132.Menzies RA, Gold PH. 1971. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem 246: 2425–29 [PubMed] [Google Scholar]
- 133.Pfeifer U, Scheller H. 1975. A morphometric study of cellular autophagy including diurnal variations in kidney tubules of normal rats. J. Cell Biol 64: 608–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sachdeva UM, Thompson CB. 2008. Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy 4: 581–89 [DOI] [PubMed] [Google Scholar]
- 135.Ma D, Panda S, Lin JD. 2011. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J 30: 4642–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, et al. 2014. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J 33: 2798–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schmitt K, Grimm A, Dallmann R, Oettinghaus B, Restelli LM, et al. 2018. Circadian Control of DRP1 Activity Regulates Mitochondrial Dynamics and Bioenergetics. Cell Metab 27: 657–66 e5 [DOI] [PubMed] [Google Scholar]