Abstract
Reduced TGF-β signaling is associated with osteoarthritis (OA). TGF-β is thought to act as a chondroprotective agent and provide anabolic cues to cartilage, thus acting as an OA suppressor in young, healthy cartilage. A potential approach to treating OA is to identify factors that act downstream of TGF-β’s anabolic pathway and target those factors to promote cartilage regeneration or repair. The aims of the present study were to (1) develop a scaffoldless tissue-engineered cartilage model with reduced TGF-β signaling and disrupted cartilage formation and (2) validate the system for identifying downstream effectors of TGF-β that promote cartilage formation. Sox9 was used to validate the model since Sox9 is known to promote cartilage formation and since TGF-β regulates Sox9 activity. Primary bovine articular chondrocytes were grown in Transwell supports to form cartilage tissue. An Alk5/TGF-β Type I receptor inhibitor, SB431542, was used to attenuate TGF-β signaling, and an adenovirus encoding FLAG-Sox9 was used to drive expression of Sox9 in the in vitro-generated cartilage. SB431542-treated tissues exhibited reduced cartilage formation, including reduced thicknesses and reduced proteoglycan staining compared to control tissue. Expression of FLAG-Sox9 in SB431542-treated cartilage allowed formation of cartilage despite antagonism of the TGF-β receptor. In summary, we developed a three-dimensional in vitro cartilage model with attenuated TGF-β signaling. Sox9 was used to validate the model for identification of anabolic agents that counteract loss of TGF-β signaling. This model has the potential to identify additional anabolic factors that could be used to repair or regenerate damaged cartilage.
Keywords: Osteoarthritis, cartilage, anabolic, TGF-β, Sox9, scaffoldless tissue engineering
Background
Transforming growth factor beta (TGF-β) signaling exhibits changes during aging that are associated with OA in humans 1, 2. In young, healthy cartilage, TGF-β signals through the TGF-β type I receptor (ALK5) and Smad2/3 to elicit an anabolic signal. Loss of this signal is associated with OA. Genetic variations in the SMAD3 gene are associated with OA susceptibility1, and OA cartilage exhibits reduced expression of the TGF-β type II receptor (TGFβR2). It was previously shown that mice expressing a dominant-negative mutation of TGFβR2 (DNIIR) 3–5 and mice with disruption of the Smad3 gene (Smad3ex8/ex8) 6 exhibit OA symptoms, establishing TGF-β as an OA suppressor in young, healthy cartilage. In aged cartilage, however, ALK5 levels are reduced, and expression of an alternate receptor, Acvrl1 (ALK1), which uses Smad1/5 and elicits a catabolic signal, is increased 7, 8. Thus, TGF-β has biphasic effects on the progression of OA, promoting cartilage formation in young tissue and promoting degradation in aged tissue. This study focuses on ways to determine the effectors of TGF-β’s anabolic pathway, which are potential drug targets for preventing OA and repairing or regenerating damaged cartilage.
To accelerate studies on cartilage signaling pathways, in vitro tissue-engineered cartilage models can be used instead of animal models for initial tests. For example, in vitro tissue-engineered cartilage models have been used to study cytokine signaling pathways 9, 10 and mechanical trauma signaling pathways 11. Studying the TGF-β signaling pathway in a tissue-engineered cartilage model could help rapidly determine the effectors of TGF-β’s anabolic pathway and therefore may accelerate identification of potentially druggable targets for promoting cartilage formation. Such a model could also eventually be used to screen for molecules capable of regenerating or repairing cartilage. One specific approach is to develop a tissue-engineered cartilage model with reduced TGF-β signaling and test which downstream effectors of TGF-β promote cartilage formation in this context.
Extensive work has been performed in the area of cartilage tissue engineering. Methods resulting in tissue histologically resembling cartilage without synthetic scaffolds embedded in the final tissue product have been developed. As previously described, scaffoldless tissue engineering “refers to any platform that does not require cell seeding or adherence within an exogenous, three-dimensional (3D) material” 12. In previous work, scaffoldless tissue engineering involved seeding cells onto a two-dimensional (2D) substrate upon which the cells produced extracellular matrix and formed tissue that remained on top of the substrate 13–17. For example, agarose molds and Transwell supports were used as substrates upon which cartilage-like tissues developed 13–17. These methods are easy to use because they involve off-the-shelf components, such as agarose or Transwell supports, on which cells can be seeded and cultivated.
To validate a scaffoldless tissue-engineered cartilage model with reduced TGF-β signaling, a molecule regulated by TGF-β known to promote cartilage formation should be tested. Sox9 is a transcription factor that is expressed in mature articular cartilage. Sox9 gene therapies have been shown to repair osteochondral defects in rabbit knee joints 18 and restore extracellular matrix in human OA cartilage 19. Also, we previously showed that Sox9 activity is regulated by TGF-β through Smad3. Furthermore, Sox9 is required for TGF-β-mediated regulation of genes important for cartilage function 20, 21.
The aims of the present study were to (1) develop a scaffoldless tissue-engineered cartilage model with reduced TGF-β signaling and disrupted cartilage formation and (2) validate the system for identifying downstream effectors of TGF-β that promote cartilage formation and compensate for low TGF-β signaling. We used an inhibitor of the TGF-β type I receptor ALK5, SB431542, in scaffoldless cultures to block TGF-β-mediated cartilage formation. The resulting cartilage had reduced tissue thickness and reduced proteoglycan staining. An adenovirus encoding FLAG-Sox9 (Ad-Sox9) was used to drive expression of Sox9 in the cultures. Expression of Sox9 in the SB431542-treated tissue promoted cartilage formation, validating the model for identification of anabolic factors that function in absence of TGF-β signaling.
Methods
Isolation of primary bovine chondrocytes
Cartilage was harvested from bovine metacarpophalangeal joints, minced, and washed. The cartilage was placed in PBS containing 2 mg/mL collagenase D (Roche, # 11088882001) and 1% penicillin-streptomycin on an orbital shaker at 37˚C for 36 hours. The digested tissue was filtered to isolate chondrocytes. Chondrocytes from five different cows were pooled together for each “n”.
Adenoviruses and TGF-β inhibitor
Adenovirus encoding for a wild-type, FLAG-tagged Sox9 protein and an enhanced green fluorescent protein (eGFP) reporter (Ad-Sox9) and a control virus encoding for only eGFP (Ad-eGFP) were used in this study. Both adenoviruses were validated previously 20. Chondrocytes were infected with 75 MOI of adenovirus in suspension on a nutator at 37˚C for one hour. During infection, cells were at a concentration of 16.5 million cells/mL. The medium that was used during infection consisted of Dulbecco’s Modified Eagle Medium (DMEM) (Fisher Scientific, # 11–965-118) with 10% non-heat-inactivated fetal bovine serum (Fisher Scientific, # 10–437-028), 1% non-essential amino acids (Fisher Scientific, # 11–140-050), 1% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (Fisher Scientific, # 15–630-080), 1% penicillin/streptomycin (Fisher Scientific, # 15–140-122), 1% L-glutamine (Fisher Scientific, # 25–030-081), 0.28 mM L-ascorbic acid (Sigma-Aldrich, # A5960–25G), 0.4 mM L-proline (Sigma-Aldrich, # H54409–25G), and 0.5 µg/mL Fungizone (Fisher Scientific, # 15–290-026); this medium will be referred to as “3D cell culture medium” from this point forward. Depending on which tissues were being cultivated, the 3D cell culture medium also contained either 10 µM of SB431542 (Tocris, catalog # 1614), which is an inhibitor of the TGF-β type I receptor (ALK5), or an equivalent volume of the vehicle in which the inhibitor was suspended, DMSO.
Scaffoldless 3D cell culture
Immediately after adenovirus infection, chondrocytes were plated in 24-mm-diameter Transwell supports (Sigma-Aldrich, catalog # CLS3412) in 6-well plates. Cells were plated at a density of 7 million cells/cm2 or 33 million cells per Transwell support (i.e., 2 mL of the cell suspension described in the previous paragraph were added to each Transwell support). In the bottom of the wells underneath the Transwell supports, 2.5 mL of 3D cell culture medium containing either SB431542 or DMSO were added. Cultures were placed in an incubator at 37˚C and allowed to sit for 24 hours. After the 24 hours, medium was changed. Specifically, 1.5 mL of 3D cell culture medium containing either SB431542 or DMSO were added to the top of the Transwell supports, and 2.5 mL of the same medium were added to the bottom of the wells underneath the Transwell supports. After this initial medium change, medium was changed every 12 hours. Also, 72 hours after plating cells, the cultures were placed on a rotator (Rotomix, Type 50800) inside the 37˚C incubator at approximately 150 rpm to circulate the culture medium and facilitate nutrient diffusion. The cultures were left on the rotator for the remainder of the cultivation period. At the end of the experiment, the Transwell membranes with their attached 3D tissues were cut out of the Transwell supports using a sharp blade. Each 3D tissue was then cut into six pieces, and the pieces were used for either flow cytometry, Western blot, histology, or thickness testing, all of which are described below.
Flow cytometry
Tissues were harvested after 5 days of cultivation for flow cytometry. Cells were extracted from the tissues. Specifically, a piece of each harvested tissue was placed in a 6-well plate with 3 mL of 0.25% trypsin-EDTA (Fisher Scientific, catalog # 25–200-072) for 5 minutes at room temperature. To help dissociate the tissue, the tissue was agitated with a pipette tip, and the trypsin-EDTA solution was pipetted up and down repeatedly. Next, 3D cell culture medium was added to the wells, and the cell suspension was spun down for 5 minutes at 2000 rpm. Cells were resuspended in 1 mL of PBS. An Attune NxT Flow Cytometer was used to identify the percentage of cells expressing eGFP.
Western blot
Tissues were harvested after 5 and 21 days of cultivation for FLAG-Sox9 and SMAD3 western blots respectively. Fifteen μg of protein lysate per sample were separated by reducing electrophoresis on 4–20% polyacrylamide gels (Bio-Rad Laboratories, # 456–8096). Protein was transferred from gels to polyvinylidene fluoride membranes (Bio-Rad Laboratories, # 162–0177) using a Trans-Blot Turbo Transfer System (Bio-Rad Laboratories, # 1704150). Membranes were blocked with either 3% bovine serum albumin (Fisher Scientific, # BP1605–100) (for pSMAD3 blots) or 3% milk (Santa Cruz Biotechnology, # sc-2325) (for Smad2/3 and FLAG-Sox9 blots). Membranes were then incubated overnight with either anti-pSMAD3 primary antibody (1:1000, Cell Signaling Technology, # 9520S, raised in rabbit, monoclonal), anti-Smad2/3 primary antibody (1:1000, Cell Signaling Technology, # 8685S, raised in rabbit, monoclonal), or anti-FLAG primary antibody (1:1000, Sigma-Aldrich, # F1804, raised in mouse, monoclonal). To assess whether equivalent amounts of protein were loaded in all wells of the FLAG-Sox9 western blots, an anti-cyclophilin B primary antibody (1:1000, Abcam, # ab16045, raised in rabbit, polyclonal) was used. Membranes were washed with Tris-buffered saline containing 0.1% Tween 20 (TBST) and incubated with HRP-conjugated anti-mouse (1:1000, Santa Cruz Biotechnology, # sc-2055, raised in goat) or anti-rabbit (1:2000, Cell Signaling Technology, # 7074S, raised in goat) secondary antibodies. Images of Western blots were acquired on a ChemiDoc MP system (Bio-Rad Laboratories). Western blot quantification was performed with ImageJ (version 1.50i).
Histology
Tissues were harvested after 21 days of cultivation for histology. Tissues were fixed in 4% paraformaldehyde at 4˚C overnight. Samples were processed then embedded in paraffin. Sections were cut at a thickness of 6 µm and mounted on UltraClear slides (Denville, # M1021). Sections were stained with either hematoxylin and eosin (H&E), toluidine blue, or picrosirius red. H&E and picrosirius red protocols were described previously 22. The toluidine blue protocol is described as follows. Sections were deparaffinized and rehydrated in 2 washes of xylene for 5 minutes each, 2 washes of 100% ethanol for 2 minutes each, and 1 wash of 90% ethanol for 1 minute. After a rinse with distilled water for 2 minutes, slides were immersed in 0.1% toluidine blue for 15 seconds. This was followed by 5 quick rinses in cold tap water. Sections were then dehydrated in 90% ethanol for 5 seconds, followed by 2 washes of 100% ethanol for 5 seconds each and 2 washes of xylene for 1 minute each.
Compression test for tissue thickness
Tissues were harvested after 21 days of cultivation for thickness testing. A previous protocol for thickness testing was used with modifications 4. The protocol is described as follows. A computer-controlled electromechanical test system (Bose LM1 ElectroForce TestBench) was fitted with a 250-g load cell (Bose). A custom-made stainless steel compression platen was secured onto the load cell. Tissue was cut to a 0.5-cm square and placed in the center of a 35-mm dish. The dish was placed directly under the center of the compression platen. The compression platen was lowered until it touched the surface of the tissue as indicated by a change in force measured by the load cell. The position of the compression platen was recorded at this point. The tissue was removed from the dish, and the compression platen was lowered further until touching the bottom of the dish as indicated by a change in force measured by the load cell. The position of the compression platen was recorded again at this point. The difference in the two position recordings was calculated to determine tissue thickness.
Statistical methods
Microsoft Excel (version 14.4.0) was used for all statistical tests unless otherwise noted. An Anderson–Darling test was performed to assess the data for normality. All data was normally distributed. An f-test was performed to assess whether variances between groups were equal. All groups in the flow cytometry data set had equal variance with each other. All groups in the thickness data set had equal variance with each other. GraphPad Prism (version 6.0f) was used to perform ANOVAs. For the flow cytometry data set, the ANOVA p-value was greater than 0.05. For the thickness data set, the ANOVA p-value was less than 0.05. For the thickness data set, pairwise comparisons were performed using the Student’s t-test (two-tailed, unpaired) for equal variance. For all statistical analyses, a p-value of less than 0.05 was considered statistically significant. All graphs were made with GraphPad Prism.
Results
Generation of cartilage-like tissues in vitro.
To generate 3D cartilage-like tissues in vitro, chondrocytes were plated in Transwell supports, and the cultures were placed on a rotator (Figure 1). TGF-β signaling was attenuated in a subset of cultures using an antagonist of the ALK5/ TGF-β type I receptor, SB431542. Treatment with the antagonist resulted in reduction of phosphorylated SMAD3 (pSMAD3) protein levels (Figure 2A) indicating that TGF-β signaling had been inhibited. FLAG-tagged Sox9 was overexpressed via adenovirus (Ad-Sox9) in a subset of cultures that were treated with SB431542, to test the hypothesis that Sox9 could compensate for loss of TGF-β to promote formation of cartilage and verify the utility of the culture system in identifying anabolic agents 19. Ad-Sox9 expressed GFP through an IRES (Internal Ribosomal Entry Site) located 3’ to the Sox9 coding sequence. All other cultures were infected with a control adenovirus (Ad-eGFP) to account for effects of the adenovirus itself. On average, approximately 65% of the cells in all tissues fluoresced green, as determined by fluorescence microscopy (Figure 2B, C) and FACS (Figure 2D) confirming that the majority of cells were infected with adenovirus and that infection was comparable between different tissues. FLAG-Sox9 protein expression in Ad-Sox9-infected cells was also confirmed by western blot (Figure 2E).
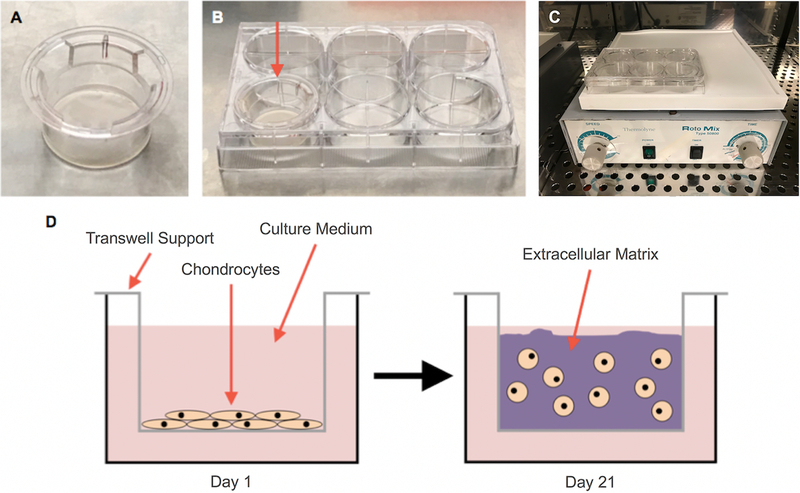
Figure 1: Set-up for generating scaffoldless tissue-engineered cartilage.
Transwell supports (A) were placed in 6-well plates (B), and chondrocytes were plated in the Transwell supports. To facilitate nutrient diffusion during cultivation, cultures were placed on a rotator (C). On day 1 of the cultivation period, cells were seeded at high density onto the top surface of the Transwell membranes, and by day 21, chondrocytes produced enough extracellular matrix to form cartilage-like tissues (D).
Figure 2: TGF-β signaling and Sox9 expression.
Three different tissue types were generated: “Control,” “SB431542,” and “SB431542 + Ad-Sox9.” Control cultures were treated with DMSO vehicle and infected with Ad-eGFP. SB431542 cultures were treated with SB431542 and infected with Ad-eGFP. SB431542 + Ad-Sox9 cultures were treated with SB431542 and infected with Ad-Sox9. (n = 3 biological replicates for each group) (A). Cells that were infected with adenovirus fluoresced green, giving tissues an overall green fluorescent hue (B and C). Flow cytometry was used to determine that approximately 65% of cells in each tissue type were infected with adenovirus (n = 3 for each group); the middle, top, and bottom horizontal bars for each group represent the average, one standard deviation above the average, and one standard deviation below the average, respectively (D). SB431542 + Ad-Sox9 cultures exhibited expression of FLAG-Sox9 protein (n = 3 for each group) (E).
Antagonism of TGF-β/Alk5 disrupts cartilage formation in vitro and Sox9 promotes cartilage formation in Alk5 inhibited cultures.
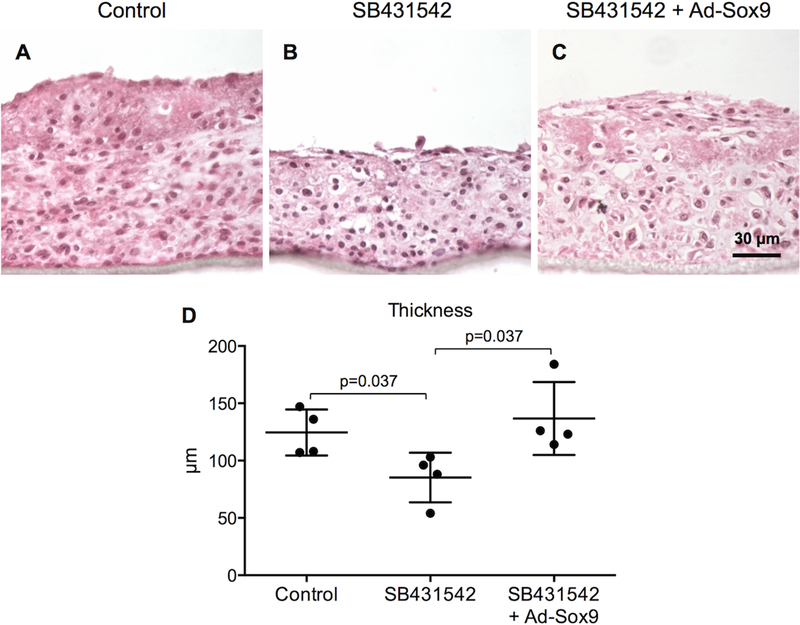
To assess general histological characteristics of the in vitro-generated tissues, H&E staining was performed. SB431542-treated tissues exhibited reduced cartilage formation when compared to control tissues (Figures 3A, B). Expression of Ad-Sox9 in SB431542-treated tissues resulted in cultures with a similar amount of cartilage as seen in controls indicating Sox9 could promote cartilage formation even in the absence of TGF-β signaling (Figures 3A–3C). Next, an electromechanical test system was used to measure the tissue thicknesses. Based on these measurements, samples in which TGF-β signaling was antagonized with SB431542 were statistically thinner than controls, whereas expression of Sox9 in SB431542 treated cultures resulted in tissue thickness similar to controls and greater than that of tissues treated with SB431542 alone (Figure 3D). Overall, H&E staining and electromechanical testing showed that antagonism of TGF-β signaling resulted in reduced cartilage formation and Sox9 acted to compensate and promote cartilage formation in samples with low TGF-β signaling.
Figure 3: Sox9 compensates for reduced cartilage formation in SB431542 treated cultures.
H&E staining was used to visualize general histological characteristics of the in vitro tissues. (n = 3 biological replicates for each group) (A–C). An electromechanical test system showed that the thicknesses of control, SB431542, and SB431542 + Ad-Sox9 tissues were 124 ± 20 µm, 85 ± 22 µm, and 137 ± 32 µm respectively (reported as mean ± standard deviation). There were statistically significant differences between control and SB431542 tissues (p = 0.037), SB431542 and SB431542 + Ad-Sox9 tissues (p = 0.037), but not control and SB431542 + Ad-Sox9 tissues (p = 0.535) (n = 4 for each group); the middle, top, and bottom horizontal bars for each group represent the average, one standard deviation above the average, and one standard deviation below the average, respectively (D).
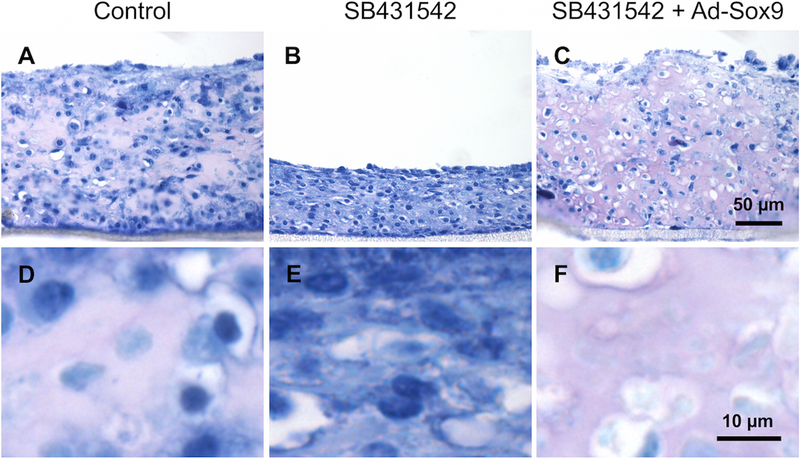
To assess proteoglycan expression in the in vitro-generated tissues, toluidine blue staining was performed. Toluidine blue is a metachromatic stain—larger amounts of glycosaminoglycans on proteoglycans in the extracellular matrix result in the matrix staining purple, while smaller amounts of glycosaminoglycans result in the matrix staining blue 23. The matrix in control tissues exhibited purple staining as expected for cartilage (Figure 4A, D). The matrix in SB431542-treated tissues was blue, which is indicative of low proteoglycan content (Figure 4B, E). The matrix in SB431542 + Ad-Sox9 tissues exhibited purple staining similar to controls (Figure 4C, F). The results suggest that TGF-β regulates proteoglycan expression in the in vitro generated cartilage samples and that expression of Sox9 can compensate for loss of TGF-β signaling in the tissue.
Figure 4: Sox9 prevents reduction in proteoglycan content in SB431542 treated cultures.
Toluidine blue staining was performed to assess proteoglycan content in the extracellular matrix of the in vitro tissues. D–F show zoomed-in regions of the matrices in A–C. The matrix of control tissues exhibited purple staining (A and D), the matrix of SB431542 tissues exhibited blue staining (B and E), and the matrix of SB431542 + Ad-Sox9 tissues exhibited purple staining (C and F) (n = 3–4 for each group).
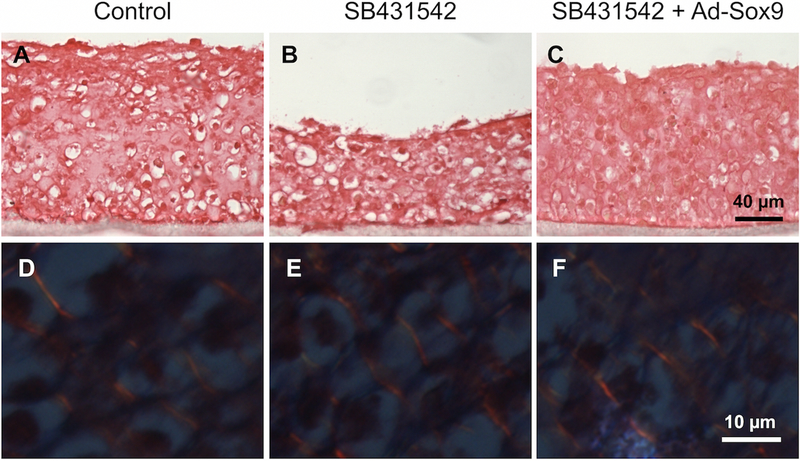
To assess overall collagen expression, collagen fiber thickness, and collagen fiber orientation in the engineered tissues, picrosirius red staining followed by polarized light microscopy was performed. First, picrosirius red staining was observed under white light. There were no noticeable differences in staining between any of the tissues (Figures 5A–5C). Next, picrosirius red staining was observed under polarized light so that individual collagen fibers could be identified. All tissues exhibited bright orange collagen fibers that were approximately 1–2 µm in diameter. The fibers were oriented diagonally throughout the tissues and appeared evenly spaced and organized (Figures 5D–5F). There were no noticeable differences in the thickness, the orientation, or the organization of these fibers between any of the tissues. Overall, the results suggest that changes in tissue thickness and proteoglycan expression described previously were not associated with changes in collagen fiber thickness, orientation, or organization.
Figure 5: Collagen matrices formed without noticeable differences between engineered tissues.
Picrosirius red staining was performed to assess overall collagen expression, collagen fiber orientation, and collagen fiber thickness. When observed under white light, there were no noticeable differences in overall collagen expression between tissues (A–C). When observed under polarized light, the tissues exhibited diagonal collagen fibers (D–F, bright orange lines). However, no noticeable differences were observed between tissues in terms of the orientation or thickness of the collagen fibers (n = 3 for each group).
Discussion
Previously, we and others showed that TGF-β signaling through Alk5 and Smad3 promote cartilage formation and prevent OA 6, 7. Mice with reduced TGF-β signaling exhibit thinner cartilage, reduced proteoglycan staining, and increased hypertrophy compared to control mice 3, 4. Accordingly, in the present study, when TGF-β signaling was antagonized during the formation of cartilage tissue in vitro, the resulting cartilage was thinner and contained less proteoglycan than the control tissue. Collagen organization did not appear to be affected by reduced TGF-β signaling. Overall, attenuation of TGF-β signaling in the in vitro-generated cartilage recapitulated multiple phenotypes observed in mice with alterations in TGF-β signaling.
To validate the use of the model for identification of agents that can promote cartilage formation and compensate for attenuation of TGF-β signaling, we tested whether Sox9, a downstream target of TGF-β 20, 21, could promote cartilage formation in the context of reduced TGF-β signaling. Previous studies showed that increased SOX9 expression may be associated with increased cartilage thickness and increased proteoglycan expression in vivo 18, 24, 25. In the present study, Ad-Sox9 compensated for the disrupted cartilage formation that was associated with reduced TGF-β signaling, resulting in tissue similar to the control. To more fully validate the model, additional molecules already known to be capable of preventing or treating OA would need to be tested. The present study serves as an initial validation study.
We and others previously showed TGF-β regulates Sox9 activity and that this regulation is dependent on Smad3 21, 26. Smad3 is a well-characterized mediator of TGF-β signaling, so we assessed pSMAD3 protein levels to confirm that the drug antagonist was working. pSMAD3 levels were reduced in SB431542-treated tissues and were similarly reduced in SB431542 tissues infected with Ad-Sox9 as expected since Sox9 is downstream of the receptor and SMAD3. The results also suggest that Sox9 functions downstream of TGF-β to promote cartilage formation and can thus compensate for attenuation of TGF-β signaling. Previous studies showed that Sox9 regulates various proteoglycan-related genes, such as aggrecan (ACAN), decorin (DCN), and 3’-phosphoadenosine 5’-phosphosulfate synthase 2 (PAPSS2) 20, 27. One hypothesis for how Sox9 acted in the present study is that it regulated proteoglycan-related genes, such as ACAN, DCN, and PAPSS2, resulting in an overall increase in proteoglycan expression and tissue thickness. However, additional work needs to be performed to test this hypothesis. Also, previous work showed that treatment of OA with TGF-β ligand increased proteoglycan expression in cartilage but caused osteophyte formation and synovial hyperplasia 28, 29. To avoid the negative effects of TGF-β ligand treatment while taking advantage of its positive effects, one approach could involve targeting anabolic-specific components of the TGF-β signaling pathway. We previously showed that TGF-β regulates the half-life and phosphorylation of Sox9 protein 20, 21. This fact, combined with the findings of the present study and previous studies on Sox9 18, 19, suggest that further investigation into targeting Sox9 for prevention of OA or repair and regeneration of cartilage could be useful.
Previous studies showed that in vitro tissue-engineered cartilage models can be used to study a range of cartilage signaling pathways, including mechanical trauma signaling pathways 11 and cytokine signaling pathways that involve tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1) 9, 10. Tissue-engineered cartilage models that are focused on mechanical trauma and cytokines are very different from the model described here with alterations in TGF-β signaling. For example, an mRNA microarray study compared mice with destabilization of the medial meniscus (DMM) to control mice 30. The DMM mice did not exhibit altered expression of Tgfbr2, parathyroid hormone-like hormone (Pthlh), and two genes encoding for matrix-modifying enzymes, Papss2 and procollagen lysine 2-oxoglutarate 5-dioxygenase 2 (Plod2). However, mice with reduced TGF-β signaling due to expression of a dominant-negative receptor exhibited altered expression of Pthlh, Papss2, and Plod2 3, 4. Also, an mRNA microarray study assessed TNF-α-related 3D cartilage models 10. Multiple genes, including matrix metalloproteinase 13 (MMP-13), were regulated differently in the TNF-α-related model when compared to the DNIIR mouse model with reduced TGF-β signaling. Overall, using a range of tissue-engineered cartilage models, including mechanical trauma models, TNF-α/IL-1-related models, and TGF-β-related models, may help assess new prophylactic and therapeutic molecules for their efficacy against cartilage tissues that exhibit multiple kinds of altered signaling pathways.
The cartilage-like tissues in the present study were approximately 14 times larger in area compared to the cartilage-like tissues in previous studies that used Transwell and Millicell supports 13, 14, 16. The present study used Transwell supports with diameters of 2.4 cm in 6-well plates, whereas previous studies used Transwell and Millicell supports that were small enough to fit into 24-well plates 13, 14, 16. The present study showed that relatively large pieces of cartilage-like tissue can be easily produced in vitro using off-the-shelf components instead of custom-made components described in previous studies 31, 32. While the present study did not focus on generating tissues for implantation, relatively large Transwell supports, such as those used in the present study, could be further investigated for easily generating clinical-scale tissues for implantation.
While the in vitro tissues in the present study exhibited collagen matrices, there were no clear differences in the collagen matrices between tissues. A potential reason for this is that the in vitro collagen matrices may not be mature like in vivo collagen matrices. For example, in mice, perpendicular collagen fibers begin to appear at 8 days of age, and by 12 days of age, the deep zone of articular cartilage exhibits thickened perpendicular collagen fibers with diameters of approximately 5–15 µm 33. In the present study, the collagen fibers were approximately 1–2 µm in diameter, and there were no regions exhibiting perpendicular fibers. While attenuation of TGF-β signaling and expression of Sox9 did not affect the collagen matrices formed in vitro for the present study, in vivo models with more mature collagen matrices may reveal different results. However, this hypothesis needs to be tested. Also, to create more mature collagen matrices in vitro, one approach could involve applying compressive load to the in vitro cultures since perpendicular collagen fibers first appear in load-bearing regions in vivo 33. Overall, in terms of the collagen matrix, the present study showed that organized collagen matrices can be generated in relatively large-sized cartilage-like tissues in vitro.
There are advantages and limitations to the cartilage model in the present study. First, SB431542 was added to cultures at the same time that cells were producing extracellular matrix. This approach mimics our previous TGF-β-related OA mouse model where TGF-β signaling was attenuated during postnatal cartilage maturation and resulted in OA 3–5. A direct comparison of the in vitro model in the present study to our previous DNIIR mouse model emphasizes the efficiency of the in vitro model. Specifically, tissue thinning and reduced proteoglycan expression appeared after only 3 weeks in the in vitro model, while the same characteristics appeared after 2–5 months in the mouse model 4, 5. The in vitro model has the potential to accelerate studies compared to the mouse model. However, in human OA, TGF-β signaling is hypothesized to decrease with age. Therefore, a more clinically relevant in vitro model might involve attenuation of TGF-β signaling after the tissue has already developed. Specifically, future experiments could focus on administering SB431542 to cultures after cells have already produced matrix. Second, the present study focuses on attenuation of TGF-β’s anabolic activity and does not focus on catabolic events known to contribute to human OA. Reduction of TGF-β signaling and reduction of associated anabolic effects during aging are thought to reduce the ability of cartilage to counteract catabolic signaling such as IL-1 signaling, contributing to OA 7. Future work could assess whether loss of TGF-β signaling results in the inability to counteract catabolic signaling pathways. Specifically, IL-1 signaling, various degradative enzymes, and loss of matrix components into the culture medium could be investigated.
Conclusions
TGF-β signaling was attenuated in an easy-to-use in vitro tissue-engineered cartilage model, resulting in alterations in cartilage formation. Expression of FLAG-Sox9 in cartilage tissues with attenuated TGF-β signaling compensated for the effects of reduced TGF-β signaling, allowing cartilage to form. The results suggest that Sox9 functions downstream of TGF-β and serves as preliminary validation of the model’s usefulness in screening for agents that promote cartilage formation.
Acknowledgements
We would like to thank the UAB Experimental Biomechanics Core for providing the electromechanical test system. The study was funded by HHMI 56005705 (PI Patel) to RDC, T32 AR069516 (PI Bridges) to RDC, and R01 AR062507 to RS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of Interest Disclosure
The authors have no conflicts of interest to declare.
References
- 1.Valdes AM, Spector TD, Tamm A, Kisand K, Doherty SA, Dennison EM, Mangino M, Tamm A, Kerna I, Hart DJ, Wheeler M, Cooper C, Lories RJ, Arden NK, Doherty M. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis and rheumatism. 2010;62(8):2347–52. [DOI] [PubMed] [Google Scholar]
- 2.Verdier MP, Seite S, Guntzer K, Pujol JP, Boumediene K. Immunohistochemical analysis of transforming growth factor beta isoforms and their receptors in human cartilage from normal and osteoarthritic femoral heads. Rheumatology international. 2005;25(2):118–24. [DOI] [PubMed] [Google Scholar]
- 3.Chavez RD, Sohn P, Serra R. Prg4 prevents osteoarthritis induced by dominant-negative interference of TGF-ss signaling in mice. PloS one. 2019;14(1):e0210601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramaswamy G, Sohn P, Eberhardt A, Serra R. Altered responsiveness to TGF-beta results in reduced Papss2 expression and alterations in the biomechanical properties of mouse articular cartilage. Arthritis research & therapy. 2012;14(2):R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serra R, Johnson M, Filvaroff EH, LaBorde J, Sheehan DM, Derynck R, Moses HL. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. The Journal of cell biology. 1997;139(2):541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. The Journal of cell biology. 2001;153(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaney Davidson EN, Scharstuhl A, Vitters EL, van der Kraan PM, van den Berg WB. Reduced transforming growth factor-beta signaling in cartilage of old mice: role in impaired repair capacity. Arthritis research & therapy. 2005;7(6):R1338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scharstuhl A, van Beuningen HM, Vitters EL, van der Kraan PM, van den Berg WB. Loss of transforming growth factor counteraction on interleukin 1 mediated effects in cartilage of old mice. Annals of the rheumatic diseases. 2002;61(12):1095–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass KA, Link JM, Brunger JM, Moutos FT, Gersbach CA, Guilak F. Tissue-engineered cartilage with inducible and tunable immunomodulatory properties. Biomaterials. 2014;35(22):5921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlichting N, Dehne T, Mans K, Endres M, Stuhlmuller B, Sittinger M, Kaps C, Ringe J. Suitability of porcine chondrocyte micromass culture to model osteoarthritis in vitro. Molecular pharmaceutics. 2014;11(7):2092–105. [DOI] [PubMed] [Google Scholar]
- 11.Mohanraj B, Meloni GR, Mauck RL, Dodge GR. A high-throughput model of post-traumatic osteoarthritis using engineered cartilage tissue analogs. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2014;22(9):1282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Athanasiou KA, Eswaramoorthy R, Hadidi P, Hu JC. Self-organization and the self-assembling process in tissue engineering. Annual review of biomedical engineering. 2013;15:115–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchi VJ, Weber JF, Waldman SD, Backstein D, Kandel RA. Formation of Hyaline Cartilage Tissue by Passaged Human Osteoarthritic Chondrocytes. Tissue Eng Part A. 2017;23(3–4):156–65. [DOI] [PubMed] [Google Scholar]
- 14.Hayes AJ, Hall A, Brown L, Tubo R, Caterson B. Macromolecular organization and in vitro growth characteristics of scaffold-free neocartilage grafts. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2007;55(8):853–66. [DOI] [PubMed] [Google Scholar]
- 15.Hu JC, Athanasiou KA. A self-assembling process in articular cartilage tissue engineering. Tissue Eng. 2006;12(4):969–79. [DOI] [PubMed] [Google Scholar]
- 16.Murdoch AD, Grady LM, Ablett MP, Katopodi T, Meadows RS, Hardingham TE. Chondrogenic differentiation of human bone marrow stem cells in transwell cultures: generation of scaffold-free cartilage. Stem cells. 2007;25(11):2786–96. [DOI] [PubMed] [Google Scholar]
- 17.Natoli RM, Responte DJ, Lu BY, Athanasiou KA. Effects of multiple chondroitinase ABC applications on tissue engineered articular cartilage. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2009;27(7):949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cucchiarini M, Orth P, Madry H. Direct rAAV SOX9 administration for durable articular cartilage repair with delayed terminal differentiation and hypertrophy in vivo. Journal of molecular medicine. 2013;91(5):625–36. [DOI] [PubMed] [Google Scholar]
- 19.Cucchiarini M, Thurn T, Weimer A, Kohn D, Terwilliger EF, Madry H. Restoration of the extracellular matrix in human osteoarthritic articular cartilage by overexpression of the transcription factor SOX9. Arthritis and rheumatism. 2007;56(1):158–67. [DOI] [PubMed] [Google Scholar]
- 20.Chavez RD, Coricor G, Perez J, Seo HS, Serra R. SOX9 protein is stabilized by TGF-beta and regulates PAPSS2 mRNA expression in chondrocytes. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2017;25(2):332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coricor G, Serra R. TGF-beta regulates phosphorylation and stabilization of Sox9 protein in chondrocytes through p38 and Smad dependent mechanisms. Scientific reports. 2016;6:38616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkhatib B, Liu C, Serra R. Tgfbr2 is required in Acan-expressing cells for maintenance of the intervertebral and sternocostal joints. JOR spine. 2018;1(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekiya I, Vuoristo JT, Larson BL, Prockop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao L, Yang F, Liu G, Yu D, Li H, Fan Q, Gan Y, Tang T, Dai K. The promotion of cartilage defect repair using adenovirus mediated Sox9 gene transfer of rabbit bone marrow mesenchymal stem cells. Biomaterials. 2011;32(16):3910–20. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Yang F, Cornelia R, Tang W, Swisher S, Kim H. Hypoxia-inducible factor-1 is a positive regulator of Sox9 activity in femoral head osteonecrosis. Bone. 2011;48(3):507–13. [DOI] [PubMed] [Google Scholar]
- 26.Furumatsu T, Tsuda M, Taniguchi N, Tajima Y, Asahara H. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J Biol Chem. 2005;280(9):8343–50. [DOI] [PubMed] [Google Scholar]
- 27.Tew SR, Li Y, Pothacharoen P, Tweats LM, Hawkins RE, Hardingham TE. Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2005;13(1):80–9. [DOI] [PubMed] [Google Scholar]
- 28.Bakker AC, van de Loo FA, van Beuningen HM, Sime P, van Lent PL, van der Kraan PM, Richards CD, van den Berg WB. Overexpression of active TGF-beta-1 in the murine knee joint: evidence for synovial-layer-dependent chondro-osteophyte formation. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2001;9(2):128–36. [DOI] [PubMed] [Google Scholar]
- 29.van Beuningen HM, Glansbeek HL, van der Kraan PM, van den Berg WB. Osteoarthritis-like changes in the murine knee joint resulting from intra-articular transforming growth factor-beta injections. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2000;8(1):25–33. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner MD, Vincent TL, Driscoll C, Burleigh A, Bou-Gharios G, Saklatvala J, Nagase H, Chanalaris A. Transcriptional analysis of micro-dissected articular cartilage in post-traumatic murine osteoarthritis. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2015;23(4):616–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ford AC, Chui WF, Zeng AY, Nandy A, Liebenberg E, Carraro C, Kazakia G, Alliston T, O’Connell GD. A modular approach to creating large engineered cartilage surfaces. J Biomech. 2018;67:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitney GA, Mera H, Weidenbecher M, Awadallah A, Mansour JM, Dennis JE. Methods for producing scaffold-free engineered cartilage sheets from auricular and articular chondrocyte cell sources and attachment to porous tantalum. BioResearch open access. 2012;1(4):157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes LC, Archer CW, ap Gwynn I. The ultrastructure of mouse articular cartilage: collagen orientation and implications for tissue functionality. A polarised light and scanning electron microscope study and review. European cells & materials. 2005;9:68–84. [DOI] [PubMed] [Google Scholar]