Abstract
The 31- and 32-nt 5′-fragments of Y4-RNA (Y4RNAfr) exist abundantly in human plasma. The Y4RNAfr can function as 5′-half-tRNA-type sgRNA for tRNase ZL, although we do not know yet what its physiological roles are and what cellular RNAs are its genuine targets. In this paper, we analyzed the effects of the Y4RNAfr on cell viability and transcriptomes using HL60, RPMI-8226, and HEK293 cells, and Y4RNAfr-binding RNAs in A549 cells. Although the Y4RNAfr hardly affected the viability of HL60, RPMI-8226, and HEK293 cells, it significantly affected their transcriptome. The DAVID analysis for > 2-fold upregulated and downregulated genes suggested that the Y4RNAfr may affect various KEGG pathways. We obtained 108 Y4RNAfr-binding RNAs in A549 cells, searched potential secondary structures of complexes between theY4RNAfr and its binding RNAs for the pre-tRNA-like structure, and found many such structures. One of the five best fitted structures was for the MKI67 mRNA, suggesting that the Y4RNAfr can decrease the cellular MKI67 level through guiding the cleavage of the MKI67 mRNA by tRNase ZL. This may be one of the underlying mechanisms for the reported observation that the Y4RNAfr suppresses the proliferation of A549 cells.
Keywords: Y4-RNA, Transcriptome, Next-generation sequencing, KEGG pathway, tRNase ZL, Pre-tRNA
1. Introduction
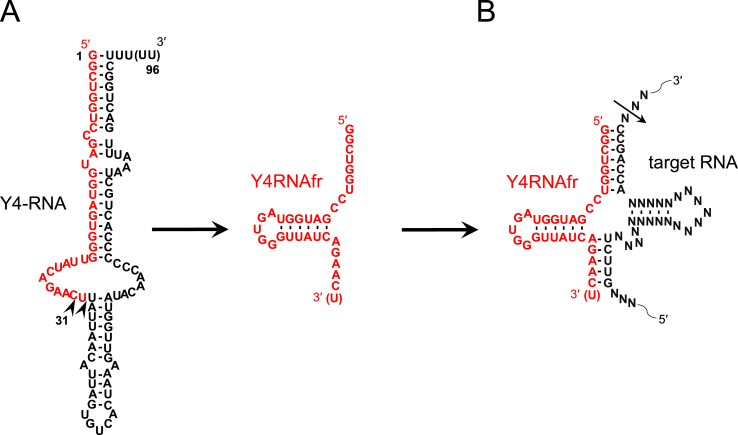
Human Y4-RNA is an ~95-nt non-coding RNA, the gene of which is located on chromosome 7 [1], and appears to be involved in the initiation of DNA replication and RNA quality control [2,3]. We have found that the 31-nt 5′-fragment of Y4-RNA exists abundantly in plasma [4], and have also shown that its 31- and 32-nt 5′-fragments exist in saliva samples after cell removal [5] (Fig. 1A). Repetto et al. [6] have demonstrated that the ~31-nt Y4-RNA fragment exists in sera of coronary artery disease patients more abundantly than in those of controls. Furthermore, Kaudewitz et al. [7] have shown that the ~31-nt Y4-RNA fragment level in plasma correlates with platelet function in patients with acute coronary syndrome and platelet activation markers in a general population. These observations suggest that the ~31-nt Y4-RNA fragment may become a diagnostic/prognostic plasma marker for acute coronary syndrome. In this paper, we collectively call the 3′-heterogenious 31- and 32-nt Y4-RNA fragments Y4RNAfr.
Fig. 1.
Human Y4-RNA and its fragments. (A) Human 94–96-nt full-length Y4-RNA and its 31/32-nt 5′-fragments are shown. Arrowheads on the full-length Y4-RNA indicate the cleavage sites to generate the 31- and 32-nt fragments collectively termed Y4RNAfr. (B) A secondary structure of the complex between the Y4RNAfr and a model target RNA. The expected cleavage site by tRNase ZL is denoted by an arrow.
We have discovered Y4RNAfr by analyzing small plasma RNAs from healthy persons and multiple myeloma patients in the course of investigation for the possibility that the intracellular gene regulatory network via tRNase ZL and small guide RNA (sgRNA) is extended intercellularly [4,8]. sgRNA is defined as ~7–30-nt RNA that can guide tRNase ZL to recognize and cleave target RNA by forming a pre-tRNA-like or micro-pre-tRNA-like complex between the target RNA and the sgRNA [9]. miRNAs and degraded RNAs such as 5′-half-tRNAs and rRNA fragments can function as sgRNA [8,10]. It has been shown that the PPM1F and DYNC1H1 mRNAs can be cleaved and downregulated by tRNase ZL together with the cellular 5′-half-tRNAGlu and the 28S rRNA 3′-terminal fragment, respectively. An sgRNA-mediated gene silencing technology termed tRNase ZL-utilizing efficacious gene silencing (TRUE gene silencing) is based on this gene regulatory network [[11], [12], [13], [14], [15], [16], [17]].
We have demonstrated that Y4RNAfr can also function as 5′-half-tRNA-type sgRNA for tRNase ZL, although we do not know yet what its physiological roles are and what cellular RNAs are its genuine targets [4] (Fig. 1B). In any case, the existence in human plasma of Y4RNAfr and the various other potential sgRNAs suggests that the gene regulatory network via tRNase ZL and sgRNA may be constructed intercellularly as well as intracellularly. In this paper, we investigated what the physiological roles of plasma Y4RNAfr are by analyzing its effects on cell viability and transcriptomes and its binding cellular RNAs using various human cell lines.
2. Materials and methods
2.1. RNA preparation
The 31-nt Y4RNAfr (5′-pGGCUGGUCCGAUGGUAGUGGGUUAUCAGAAC-3′) and the five control RNAs CR1 (5′-pUUUUUCUp-3′), CR2 (5′-pGGCCAGGp-3′), CR3 (5′-pCACCAAUp-3′), CR4 (5′-pCACCAAUAGGAUCCp-3′), and CR5 (5′-pCCUGGCCp-3′), which were fully 2′-O-methylated and 5′/3′-phosphorylated, were synthesized with a DNA/RNA synthesizer and subsequently purified through high-performance liquid chromatography by Nippon Bioservice (Asaka, Saitama, Japan). CR1–CR5 were chosen randomly from our sgRNA library. Likewise, the 31-nt Y4RNAfr (5′-pGGCUGGUCCGAUGGUAGUGGGUUAUCAGAAC-biotin-3′) and the control RNA CR6 (5′-pUCCCUGGUGGUCUAGUGGUUAGGAUUCGGCGCUCU-biotin-3′), which were 5′-phosphorylated and 3′-biotinylated, were synthesized.
2.2. Cell culture
HL60 and RPMI-8226 cells were cultured in RPMI-1640 (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (MP Biomedicals Japan, Tokyo, Japan) and 1% penicillin-streptomycin (Invitrogen Japan, Tokyo, Japan) at 37 °C in 5% CO2 humidified incubator. HEK293 and A549 cells were cultured in DME media (Wako, Osaka, Japan) likewise. HL60, RPMI-8226, HEK293, and A549 cells were from human promyelocytic leukemia, human myeloma, human embryonic kidney, and human non-small cell lung cancer, respectively.
2.3. Cell viability assay
The human cells were plated at 500 or 1000 cells/100 μl/well on a 96-well dish in media containing 1 μM of the naked Y4RNAfr or CR1. After 3 days, the cell viability was quantitated with Cell Counting Kit-8 (DOJINDO, Tokyo, Japan).
2.4. Transcriptome analysis
HL60 and RPMI-8226 cells (at 80,000 cells/500 μl/well on a 24-well dish) and HEK293 cells (at 20,000 cells/500 μl/well on a 24-well dish) were cultured in media without and with 1 μM of the naked Y4RNAfr. As controls, the cells were also cultured in the presence of 1 μM of naked CR2 or CR3 for HL60 cells, 1 μM of naked CR4 for RPMI-8226 cells, and 1 μM of naked CR5 for HEK293 cells. After one day, each total RNA was extracted with RNAiso Plus (Takara Bio, Shiga, Japan). The RNA samples were subjected to DNA microarray analysis with a SurePrint G3 Human GE v2 8 × 60K Microarray (Agilent Technologies, Tokyo, Japan). This microarray analysis was carried out by Takara Bio (Shiga, Japan).
2.5. RNA pull-down
One day after A549 cells were plated at 150,000 cells/2 ml/well on a 6-well dish, the cells were transfected with 200 pmol of the biotinylated Y4RNAfr or CR6 using Lipofectamine 2000 (Thermo Fisher Scientific, Massachusetts, USA) according to manufacturer's protocol. After 18-h incubation, the cells on 6 wells for each RNA treatment were removed with trypsin/EDTA, collected, and washed with phosphate-buffered saline (PBS).
The collected cells were incubated in the presence of 1% formaldehyde at 37 °C for 10 min, and then further incubated in the presence of 0.5 M glycine at 37 °C for 5 min. After washing the formaldehyde-treated cells with PBS three times, the cells were suspended in the cell lysis buffer (10 mM HEPES pH7.5, 20 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT), incubated on ice for 10 min, and homogenized with a Dounce tissue grinder. The homogenate was centrifuged at 3300×g for 7 min, and the supernatant was regarded as a cytoplasmic fraction. The pellet was re-suspended in the nucleus lysis buffer (20 mM HEPES pH7.5, 50 mM KCl, 1.5 mM MnCl2, 1% NP-40, 0.4% sodium deoxycholate, 0.1% N-lauroylsarcosine, 1 mM DTT), sonicated on ice for 7 min, and centrifuged at 16,000×g for 10 min. The supernatant was regarded as a nuclear fraction.
The biotinylated Y4RNAfr or CR6 together with its bound nuclear or cytoplasmic RNAs was purified and collected with Dynabeads M − 270 Streptavidin beads (Thermo Fisher Scientific, Massachusetts, USA) according to manufacturer's protocol. The purified RNA samples were extracted with phenol/chloroform, precipitated with ethanol, and re-dissolved in RNase-free water.
The obtained RNA samples were fragmented to ~60–200-nt RNA with Fragmentation Reagents (Thermo Fisher Scientific, Massachusetts, USA) according to manufacturer's protocol, precipitated with ethanol, and re-dissolved in RNase-free water. The fragmented RNA samples were treated with alkaline phosphatase (Takara Bio, Shiga, Japan) at 50 °C for 30 min, extracted with phenol/chloroform twice, precipitated with ethanol, and re-dissolved in RNase-free water. The dephosphorylated RNA samples were treated with T4 polynucleotide kinase (Takara Bio, Shiga, Japan) and ATP at 37 °C for 30 min, extracted with phenol/chloroform, precipitated with ethanol, and re-dissolved in RNase-free water.
2.6. Next-generation sequencing
A cDNA library was constructed from each of the four RNA samples (~2 μg) using a TruSeq Small RNA Sample Prep Kit (Illumina, California, USA), T4 RNA Ligase 2, truncated (New England Biolabs, Massachusetts, USA), and SuperScript II Reverse Transcriptase (Invitrogen, Massachusetts, USA) by 15 cycles of PCR according to manufacturers’ protocols. The cDNAs from the Y4RNAfr-bound nuclear RNA, the CR6-bound nuclear RNA, the Y4RNAfr-bound cytoplasmic RNA, and the CR6-bound cytoplasmic RNA were indexed with the sequences 5′-ATCACG-3′, 5′-TTAGGC-3′, 5′-ACAGTG-3′, and 5′-CAGATC-3′, respectively. A mixture of the four cDNA libraries was separated by polyacrylamide gel electrophoresis, and ~150–300-base-pair cDNAs, which correspond to ~40–190-nt original RNAs, were recovered and purified. The purified cDNA sample was subjected to the next-generation sequencer MiSeq (Illumina). This sequence analysis was carried out by Takara Bio (Shiga, Japan).
2.7. Bioinformatic analysis
Raw signal data obtained from the analysis with the SurePrint G3 Human GE v2 8 × 60K Microarray were imported to a data analytics platform, Subio Platform version 1.22. The microarray contains multiple probes for one gene in a subset of the genes. The raw data of each cell group were normalized globally to adjust means of the raw signal values, and probes that contained at least one abnormal peak signal (gIsFeatNonUnifOL = 1) were removed to obtain filtered data sets. And probes whose signals were <50 in all samples were also excluded to obtain further filtered data sets. Then we transformed the raw signal values to log2 ratio values by setting the raw signal values in the transcriptome of each Y4RNAfr-treated sample to zero. We extracted genes whose log2 ratio values were < −1.0 and >1.0 in common in comparison to the transcriptomes of the cell samples that were untreated or treated with the control RNAs.
The RNA sequences obtained by MiSeq were mapped on the human reference genome hg19 in Ensembl (http://asia.ensembl.org/Homo_sapiens/Info/Index), the human reference genome hg38 in National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/grc/human), and the human reference genome hg38 in University of California, Santa Cruz (http://genome.ucsc.edu) by Subio (Kagoshima, Japan). The mapped data were analyzed on Subio Platform version 1.22. The Y4RNAfr-pulled-down cellular RNAs, of which raw signal values are more than 10 and more than 5 fold compared with those of RNAs that bound to the control RNA CR6 in at least one of the three mapped data sets, were regarded as Y4RNAfr-bound RNAs.
The genes selected in the DNA microarray analysis and the MiSeq analysis were located on chromosomes using Subio Platform. Data sets of differentially expressed genes and differentially Y4RNAfr-bound RNA genes were further analyzed with a functional annotation tool on the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/summary.jsp) to obtain KEGG pathways implicated in the differences [[18], [19], [20]].
2.8. Search for pre-tRNA-like secondary structures
Each sequence of the genes encoding the Y4RNAfr-bound RNAs was obtained from the database of National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov). Sequences in each gene complementary to the 5′-terminal sequence 5′-GGCUGGU-3′ and the 3′-terminal sequence 5′-AGAAC-3′ of the Y4RNAfr were searched for using Microsoft Word 2016. One or two mismatches and G-U pairs were allowed. Secondary structures between the Y4RNAfr and each gene RNA sequence were constructed manually to find pre-tRNA-like secondary structures.
2.9. Accession number
DNA microarray data and RNA sequence data were deposited in the DNA Data Bank of Japan (www.ddbj.nig.ac.jp) with the accession numbers E-GEAD-332 and DRR198079–DRR198082, respectively.
3. Results
3.1. The Y4RNAfr hardly affects cell viability
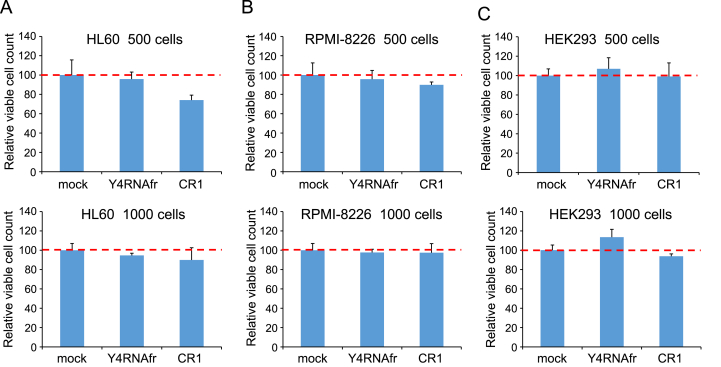
First, we investigated how the Y4RNAfr affects viability of human cultured cells. We cultured HL60 leukemia cells, RPMI-8226 multiple myeloma cells, and HEK293 cells in the presence of the chemically synthesized Y4RNAfr without any transfection reagent for 3 days, and measured their viability. Since the Y4RNAfr was supposed to be added nakedly to the culture media, it was fully 2′-O-methylated and 5′/3′-phosphorylated to protect it from nuclease attack [14]. Under two different cell density conditions, the Y4RNAfr hardly affected the viability of these cells, while the control RNA CR1 tended to decrease their viability (Fig. 2).
Fig. 2.
Cell viability assays. Relative viable cell counts were measured 72 h after HL60 (A), RPMI-8226 (B), or HEK293 (C) cells were cultured in the absence and the presence of 1 μM of the naked Y4RNAfr or CR1. The relative viable cell counts in the absence of the RNAs (mock) are adjusted to 100. Error bars denote standard deviations (n = 3).
3.2. The Y4RNAfr affects transcriptomes
Next, we investigated how the Y4RNAfr affects the transcriptomes of human cells. We cultured HL60, RPMI-8226, and HEK293 cells in the presence of the Y4RNAfr for one day, and performed transcriptome analysis for their total RNA using DNA microarrays. As for the HL60 transcriptome, we extracted genes whose expression level increased and decreased by more than 2 fold in common compared with the expression levels in HL60 cells that were untreated or treated with the control RNA CR2 or CR3, and these genes are listed in Supplementary Table S1. The most increased and decreased 10 genes in comparison with each control transcriptome are also shown in Table 1. Among the top 10 most increased genes, five genes were shared by three comparisons, and two genes were by two comparisons. And among the top 10 most decreased genes, one gene was shared by three comparisons, and four genes were by two comparisons.
Table 1.
The top 10 most increased and most decreased genes.
| HL60 |
RPMI-8226 |
HEK293 |
||||||
|---|---|---|---|---|---|---|---|---|
| CR2 | CR3 | mock | CR4 | mock | CR5 | mock | ||
| increased | 1 | lnc-ARFGEF2-3 | lnc-ARFGEF2-3 | lnc-ARFGEF2-3 | KCNJ8 | KCNJ8 | LOC102723646 | LOC102723646 |
| 2 | DKFZp667F0711 | SPAG17 | DKFZp667F0711 | PABPC1P2 | DISP1 | LINC00948 | LINC00948 | |
| 3 | LINC00472 | lnc-FZD1-1 | LINC00472 | XLOC_l2_000080 | ENPP1 | MAMDC2 | lnc-DPYD-2 | |
| 4 | OR2T2 | DKFZp667F0711 | OR2T2 | ENPP1 | XLOC_l2_000080 | lnc-ZNF280D-1 | LRRTM3 | |
| 5 | LOC100129781 | SPATA31D3 | ABCC13 | lnc-C8orf59-1 | LOC101928796 | LOC100996844 | lnc-ZNF280D-1 | |
| 6 | ABCC13 | LOC102723456 | ZNF793 | LOC101928796 | LRTM2 | SNORD115-5 | lnc-C17orf97-7 | |
| 7 | SKOR2 | LINC00472 | PRR27 | LRTM2 | lnc-FAM49A-1 | lnc-IGF1R-1 | SNORD115-5 | |
| 8 | ZNF793 | OR2T2 | LINC01085 | DISP1 | lnc-COPS4-1 | CTNNA3 | LOC100996844 | |
| 9 | lnc-PIGP-1 | lnc-SLC4A3-8 | lnc-C10orf90-2 | lnc-COPS4-1 | SPINK2 | lnc-C3orf38-2 | lnc-TPBG-3 | |
| 10 |
lnc-FZD1-1 |
ABCC13 |
CCK |
SPINK2 |
LOC101929911 |
MYADML |
LOC340340 |
|
| decreased | 1 | SPATA3-AS1 | lnc-C5orf64-1 | SPATA3-AS1 | MAP7D2 | ADAMTS4 | MCHR2 | LOC102467226 |
| 2 | KRTAP5-4 | lnc-FAM110B-1 | C7orf76 | LRRC63 | lnc-C15orf2-6 | F13A1 | SYNPR | |
| 3 | UNC80 | OR2T34 | GRAP | lnc-USPL1-2 | lnc-TCF4-4 | OR2F1 | LOC101060038 | |
| 4 | lnc-CCDC37-3 | ZNF528 | LOC101927204 | lnc-CD99L2-2 | lnc-PFDN1-1 | LOC102467226 | LOC101927291 | |
| 5 | SIGLEC9 | lnc-RP3-368B9.1.1–4 | lnc-PDGFD-5 | LINC01013 | ZNF365 | LOC102723597 | LOC440446 | |
| 6 | lnc-PDGFD-5 | lnc-PDGFD-5 | lnc-SERPINI1-9 | GALNT13 | lnc-BATF3-2 | APELA | GSTA3 | |
| 7 | lnc-CMPK2-1 | lnc-CMPK2-1 | TTN | LOC102723778 | OR9Q2 | NCRNA00250 | lnc-CMPK2-13 | |
| 8 | lnc-METTL15-4 | XLOC_l2_004640 | XLOC_l2_010969 | lnc-FAM135B-2 | SVILP1 | ITGAL | NCRNA00250 | |
| 9 | LOC101927204 | lnc-SERPINI1-9 | LINC00935 | IFNA7 | HRASLS | ACTA2-AS1 | ACTA2-AS1 | |
| 10 | LOC101928725 | lnc-CLASP2-1 | LOC101927136 | SLCO2A1 | DEFB109P1 | LOC101929756 | LOC100131023 | |
Likewise, genes with more than 2-fold increased and decreased expressions were extracted with respect to RPMI-8226 and HEK293 cells (Supplementary Table S1), and the top 10 most increased and decreased genes in comparison with each control transcriptome are listed in Table 1. As for the RPMI-8226 transcriptomes, eight genes among the top 10 most increased genes were common to two comparisons, and no gene was common among the top 10 most decreased genes. As for the HEK293 transcriptomes, five genes among the top 10 most increased genes were common to two comparisons, and three genes among the top 10 most decreased genes were common.
Further analysis of the transcriptome data found that no gene was commonly upregulated or downregulated among HL60, RPMI-8226, and HEK293 cells but that there were some genes commonly upregulated between two cell types. One, six, and three genes were commonly upregulated between HL60 and RPMI-8226 cells, between HL60 and HEK293 cells, and between RPMI-8226 and HEK293 cells, respectively (Table 2). And there was no gene commonly downregulated between any two cell types.
Table 2.
Commonly upregulated genes between two cell types.
| HL60/RPMI-8226 | HL60/HEK293 | RPMI-8226/HEK293 |
|---|---|---|
| EXOC6B | LRRTM3 | OR5K2 |
| DKFZp667F0711 | ANKRD62 | |
| Q7Z2X8 | lnc-ODZ4-2 | |
| CD200R1 | ||
| PLA2G4E | ||
| lnc-NMT2-1 |
To see if the genes upregulated and downregulated by the Y4RNAfr treatment are spatially connected, we located the genes extracted for each cell type on the chromosomes (Supplementary Fig. S1). In each case, overall, the genes appear to be located dispersedly, and their notable localization was not detected. However, interestingly, three genes, LRRTM3, DKFZp667F0711, and lnc-NMT2-1, out of the six genes commonly upregulated between HL60 and HEK293 cells were located on chromosome 10 (Supplementary Figs. S1A and C).
Furthermore, we performed the DAVID analysis for the extracted genes of each cell's transcriptome, and found various KEGG pathways involved in the genes whose expression levels were changed by the Y4RNAfr (Table 3). The Y4RNAfr appears to affect the olfactory transduction pathway commonly in HL60 and RPMI-8226 cells and the Staphylococcus aureus infection pathway commonly in RPMI-8226 and HEK293 cells (Table 3).
Table 3.
KEGG pathways involved in genes whose expression levels were changed by Y4RNAfr.
| Cell | Term | Count | % | P-Value |
|---|---|---|---|---|
| HL60 | Tight junction | 4 | 3.1 | 5.10E-03 |
| Olfactory transduction | 5 | 3.8 | 7.60E-02 | |
| Histidine metabolism |
2 |
1.5 |
8.60E-02 |
|
| RPMI-8226 | Antigen processing and presentation | 7 | 1.8 | 5.00E-04 |
| Type I diabetes mellitus | 5 | 1.3 | 2.30E-03 | |
| Autoimmune thyroid disease | 5 | 1.3 | 5.00E-03 | |
| Staphylococcus aureus infection | 5 | 1.3 | 5.70E-03 | |
| Asthma | 4 | 1 | 7.20E-03 | |
| Graft-versus-host disease | 4 | 1 | 9.50E-03 | |
| Allograft rejection | 4 | 1 | 1.30E-02 | |
| Toxoplasmosis | 6 | 1.5 | 1.60E-02 | |
| Intestinal immune network for IgA production | 4 | 1 | 2.50E-02 | |
| Tuberculosis | 7 | 1.8 | 3.10E-02 | |
| Neuroactive ligand-receptor interaction | 9 | 2.3 | 3.10E-02 | |
| Viral myocarditis | 4 | 1 | 4.00E-02 | |
| PI3K-Akt signaling pathway | 10 | 2.6 | 4.00E-02 | |
| Phagosome | 6 | 1.5 | 5.00E-02 | |
| Inflammatory bowel disease (IBD) | 4 | 1 | 5.40E-02 | |
| Regulation of actin cytoskeleton | 7 | 1.8 | 6.20E-02 | |
| Leishmaniasis | 4 | 1 | 6.90E-02 | |
| Pathways in cancer | 10 | 2.6 | 7.90E-02 | |
| Influenza A | 6 | 1.5 | 8.30E-02 | |
| Olfactory transduction |
10 |
2.6 |
8.50E-02 |
|
| HEK293 | Complement and coagulation cascades | 4 | 2.5 | 4.60E-03 |
| Staphylococcus aureus infection | 3 | 1.9 | 2.90E-02 | |
| Arachidonic acid metabolism | 3 | 1.9 | 3.60E-02 | |
| Salmonella infection | 3 | 1.9 | 6.30E-02 | |
| Rheumatoid arthritis | 3 | 1.9 | 7.00E-02 | |
| Toll-like receptor signaling pathway | 3 | 1.9 | 9.60E-02 |
3.3. Cellular RNAs that bind to the Y4RNAfr
Next, we investigated what RNAs the Y4RNAfr binds to in cells. We transfected human A549 cells with the 3′-biotinylated Y4RNAfr, and sequenced nuclear and cytoplasmic RNAs that bound to the Y4RNAfr. The RNAs, of which raw signal values are more than 10 and more than 5 fold compared with those of RNAs that bound to the control RNA CR6, were regarded as Y4RNAfr-bound RNAs, and further analyzed. 40 and 68 RNA species were selected as nuclear and cytoplasmic RNAs, respectively, that bind to the Y4RNAfr (Supplementary Table S2). Then we located the genes encoding these RNAs on the chromosomes, and found that, on the whole, these genes are located dispersedly without any notable localization (Supplementary Fig. S2).
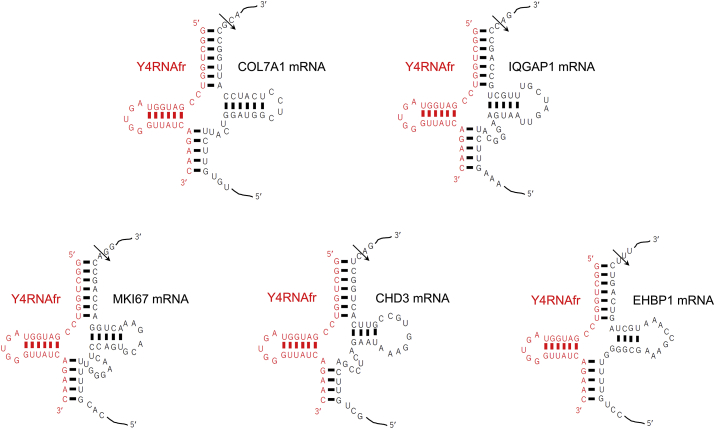
We searched potential secondary structures of cellular RNA/Y4RNAfr complexes for the pre-tRNA-like structure, and found many such structures. The five best fitted structures were obtained for COL7A1, IQGAP1, MKI67, CHD3, and EHBP1 mRNAs (Fig. 3).
Fig. 3.
Potential secondary structures of the complexes between the 31-nt Y4RNAfr and the COL7A1, IQGAP1, MKI67, CHD3, or EHBP1 mRNA. The arrow indicates the expected cleavage site by tRNase ZL.
We also carried out the DAVID analysis for the genes of the 108 Y4RNAfr-bound RNAs, and found 10 KEGG pathways including ascorbate and aldarate metabolism, pentose and glucuronate interconversions, and porphyrin and chlorophyll metabolism, which may be affected by the Y4RNAfr (Table 4). Lastly, it should be noted that the gene encoding one of the cytoplasmic Y4RNAfr-bound RNAs is ELAC2 that encodes tRNase ZL (Supplementary Table S2).
Table 4.
KEGG pathways involved in Y4RNAfr-bound RNA genes.
| Term | Count | % | P-Value |
|---|---|---|---|
| Ascorbate and aldarate metabolism | 10 | 8.3 | 2.00E-13 |
| Pentose and glucuronate interconversions | 9 | 7.4 | 8.60E-11 |
| Porphyrin and chlorophyll metabolism | 9 | 7.4 | 6.90E-10 |
| Retinol metabolism | 10 | 8.3 | 9.40E-10 |
| Drug metabolism - other enzymes | 9 | 7.4 | 1.50E-09 |
| Steroid hormone biosynthesis | 9 | 7.4 | 1.00E-08 |
| Drug metabolism - cytochrome P450 | 9 | 7.4 | 3.70E-08 |
| Metabolism of xenobiotics by cytochrome P450 | 9 | 7.4 | 7.30E-08 |
| Chemical carcinogenesis | 9 | 7.4 | 1.40E-07 |
| Metabolic pathways | 15 | 12.4 | 7.40E-02 |
4. Discussion
In this paper, to investigate what the physiological roles of the plasma Y4RNAfr are, we analyzed the effects of the Y4RNAfr on cell viability and transcriptomes using HL60, RPMI-8226, and HEK293 cells, and Y4RNAfr-binding RNAs in A549 cells. Although the Y4RNAfr hardly affected the viability of HL60, RPMI-8226, and HEK293 cells, it significantly affected their transcriptome. As for the Y4RNAfr-treated HL60 cells, 163 and 33 genes were upregulated and downregulated, respectively, by more than 2 fold in common compared with the untreated cells and the CR2-and CR3-treated cells (Supplementary Table S1). And the DAVID analysis for these genes suggested that the Y4RNAfr may be involved in tight junctions, olfactory transduction, and histidine metabolism (Table 3).
As for the Y4RNAfr-treated RPMI-8226 cells, 509 and 74 genes were upregulated and downregulated, respectively, in common compared with the untreated cells and the CR4-treated cells (Supplementary Table S1). Twenty KEGG pathways including antigen processing and presentation, type I diabetes mellitus, and autoimmune thyroid disease were suggested to be affected by the Y4RNAfr (Table 3).
With respect to HEK293 cells, 211 and 24 genes were upregulated and downregulated, respectively, by the Y4RNAfr in common compared with the untreated cells and the CR5-treated cells (Supplementary Table S1). Six KEGG pathways including complement and coagulation cascades, Staphylococcus aureus infection, and arachidonic acid metabolism appeared to be affected by the Y4RNAfr treatment (Table 3).
With respect to human monocytes/macrophages, it has been shown that the Y4RNAfr regulates cell death and inflammation via TLR7 [21]. In addition, the Y4RNAfr has been demonstrated to modulate cytokine release and PD-L1 expression via TLR7 in monocytes [22].
Concerning human non-small cell lung cancer A549 cells, their proliferation has been shown to be suppressed by the Y4RNAfr [23]. The present analysis suggested that the Y4RNAfr affects the 10 KEGG pathways in A549 cells (Table 4). In addition, among potential secondary structures of cellular RNA/Y4RNAfr complexes, one of the five best structures fitted to the pre-tRNA-like structure was with the MKI67 mRNA (Fig. 3). This suggests that the Y4RNAfr may suppress the proliferation of A549 cells by decreasing the MKI67 level through guiding the cleavage of the MKI67 mRNA by tRNase ZL, since it is known that MKI67 is associated with ribosomal RNA transcription and that the decrease in the MKI67 level suppresses cell proliferation [24].
Our present observations together with other groups’ observations suggest that the Y4RNAfr in plasma has various physiological roles in various types of cells. And finally, the possibilities of modulation of the tRNase ZL level through Y4RNAfr binding to tRNase ZL (ELAC2) mRNA and modulation of the MKI67 level through the cleavage of the MKI67 mRNA by tRNase ZL under the direction of the Y4RNAfr may support the existence of the intercellular gene regulatory network via tRNase ZL and sgRNA.
Funding information
This work was supported by a research grant from Niigata University of Pharmacy and Applied Life Sciences.
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2019.11.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Maraia R.J., Sasaki-Tozawa N., Driscoll C.T. The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucleic Acids Res. 1994;22:3045–3052. doi: 10.1093/nar/22.15.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang A.T., Langley A.R., Christov C.P. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sim S., Wolin S.L. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscipl. Rev. RNA. 2011;2:686–699. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ninomiya S., Kawano M., Abe T. Potential small guide RNAs for tRNase ZL from human plasma, peripheral blood mononuclear cells, and cultured cell lines. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa T., Haino A., Seki M. The Y4-RNA fragment, a potential diagnostic marker, exists in saliva. Non Coding RNA Res. 2017;2:122–128. doi: 10.1016/j.ncrna.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Repetto E., Lichtenstein L., Hizir Z. RNY-derived small RNAs as a signature of coronary artery disease. BMC Med. 2015;13:259. doi: 10.1186/s12916-015-0489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaudewitz D., Skroblin P., Bender L.H. Association of microRNAs and YRNAs with platelet function. Circ. Res. 2016;118:420–432. doi: 10.1161/CIRCRESAHA.114.305663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbarbary R.A., Takaku H., Uchiumi N. Modulation of gene expression by human cytosolic tRNase ZL through 5′-half-tRNA. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata H.S., Takaku H., Takagi M. The T loop structure is dispensable for substrate recognition by tRNase ZL. J. Biol. Chem. 2005;280:22326–22334. doi: 10.1074/jbc.M502048200. [DOI] [PubMed] [Google Scholar]
- 10.Elbarbary R.A., Takaku H., Uchiumi N. Human cytosolic tRNase ZL can downregulate gene expression through miRNA. FEBS Lett. 2009;583:3241–3246. doi: 10.1016/j.febslet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Habu Y., Miyano-Kurosaki N., Kitano M. Inhibition of HIV-1 gene expression by retroviral vector-mediated small-guide RNAs that direct specific RNA cleavage by tRNase ZL. Nucleic Acids Res. 2005;33:235–243. doi: 10.1093/nar/gki164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashima A., Takaku H., Shibata H.S. Gene-silencing by the tRNA maturase tRNase ZL under the direction of small guide RNA. Gene Ther. 2007;14:78–85. doi: 10.1038/sj.gt.3302841. [DOI] [PubMed] [Google Scholar]
- 13.Elbarbary R.A., Takaku H., Tamura M. Inhibition of vascular endothelial growth factor expression by TRUE gene silencing. Biochem. Biophys. Res. Commun. 2009;379:924–927. doi: 10.1016/j.bbrc.2008.12.173. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi M., Elbarbary R.A., Nakashima A. A naked RNA heptamer targeting the human Bcl-2 mRNA induces apoptosis of HL60 leukemia cells. Cancer Lett. 2013;328:362–368. doi: 10.1016/j.canlet.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe N., Narita M., Yamahira A. Induction of apoptosis of leukemic cells by TRUE gene silencing using small guide RNAs targeting the WT1 mRNA. Leuk. Res. 2013;37:580–585. doi: 10.1016/j.leukres.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Iizuka S., Oridate N., Nashimoto M. Growth inhibition of head and neck squamous cell carcinoma cells by sgRNA targeting the cyclin D1 mRNA based on TRUE gene silencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haino A., Ishikawa T., Seki M. TRUE gene silencing: screening of a heptamer-type small guide RNA library for potential cancer therapeutic agents. J. Vis. Exp. 2016 doi: 10.3791/53879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogata H., Goto S., Sato K. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hizir Z., Bottini S., Grandjean V. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2016.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haderk F., Schulz R., Iskar M. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aah5509. pii: eaah5509. [DOI] [PubMed] [Google Scholar]
- 23.Li C., Qin F., Hu F. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8:2. doi: 10.1186/s13578-018-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullwinkel J., Baron-Lühr B., Lüdemann A. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J. Cell. Physiol. 2006;206:624–635. doi: 10.1002/jcp.20494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.