Abstract
We present an extensive, large-scale, long-term and multitaxon database on phenological and climatic variation, involving 506,186 observation dates acquired in 471 localities in Russian Federation, Ukraine, Uzbekistan, Belarus and Kyrgyzstan. The data cover the period 1890–2018, with 96% of the data being from 1960 onwards. The database is rich in plants, birds and climatic events, but also includes insects, amphibians, reptiles and fungi. The database includes multiple events per species, such as the onset days of leaf unfolding and leaf fall for plants, and the days for first spring and last autumn occurrences for birds. The data were acquired using standardized methods by permanent staff of national parks and nature reserves (87% of the data) and members of a phenological observation network (13% of the data). The database is valuable for exploring how species respond in their phenology to climate change. Large-scale analyses of spatial variation in phenological response can help to better predict the consequences of species and community responses to climate change.
Subject terms: Biodiversity, Phenology
| Measurement(s) | phenological event • climate |
| Technology Type(s) | digital curation |
| Factor Type(s) | geographic location • year |
| Sample Characteristic - Location | Russia • Ukraine • Uzbekistan • Belarus • Kyrgyzstan |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.11687169
Background & Summary
Phenological dynamics have been recognised as one of the most reliable bio-indicators of species responses to ongoing warming conditions1. Together with other adaptive mechanisms (e.g. changes in the spatial distribution and physiological adaptations), phenological change is a key mechanism by which plants and animals adapt to a changing world2,3. Many studies have documented that in the northern hemisphere, spring events have become earlier whereas autumn events are occurring later than before, mostly due to rising temperatures4–6. Despite this broadly shared response, there are systematic differences in phenological responses to climate change among individual species7–9, different taxonomic groups and trophic levels10–12. Further, while some studies have reported that different species are likely to have evolved distinct phenological responses to environmental cues13,14, others suggest that many species are synchronised because phenotypic plasticity in phenological response to climate may maintain local adaptation15,16.
Comprehensive understanding of phenological responses to climate change requires community-wide data that are both long-term and spatially extensive11,17,18. Such data are still not common and, with few exceptions11,17,18, the assessments of broad-scale taxonomic and geographic variations in phenological changes have generally involved meta-analyses5,19, or analyses of large observational databases that either represent mid-latitude systems4,5,20 or are characterized by low species richness13. Therefore, the spatial variation in phenological dynamics of species communities at large scale is still not well known13,17. Yet, this information is essential for understanding how species and communities respond to climate change16. A further common problem with many previously published data sets is publication bias. Few scientific journals are keen to publish papers reporting no detectable signal in species response to climate change – which can result in strongly biased conclusions in meta-analyses (but see12,13). Assembling monitoring data which has been consistently collected over long time and a large spatial extent addresses these problems directly12.
We present a large-scale and long-term dataset that can be used to examine community-level spatial variation in phenological dynamics and its climatic drivers. The database consists of 506,186 observation dates collected in 471 localities in the Russian Federation, Ukraine, Uzbekistan, Belarus and Kyrgyzstan (Fig. 1) over a 129-year period (from 1890 to 2018). During this period, researchers intensively conducted regular observations to record dates at which a predefined list of phenological and climatic events (Fig. 2) occurred. Although 96% of the observations were acquired from 1960 onwards, a few time series are very long. Events measured for plants include e.g. the onset days of leaf unfolding, first flowering time, and leaf fall; for birds they include e.g. days for first spring and last autumn occurrences; for insects, amphibians, reptiles and fungi they include e.g. day of first occurrence in the spring. The plant data were acquired in fixed plots, and the bird data along established routes. Climatic events were recorded as calendar dates when those events took place. Of all phenological dates, 87% were collected by research personnel of nature protected areas and national parks, who followed a systematic protocol. Thus, sampling effort remained nearly constant over time. The remaining 13% of the observations came from a well-established volunteer phenological network of volunteers, who followed a similar systematic protocol.
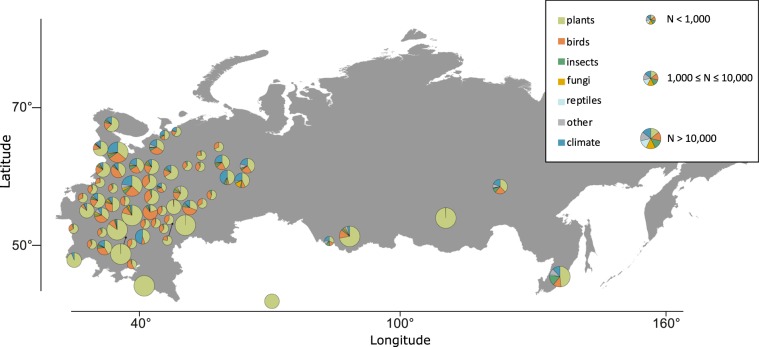
Fig. 1.
Spatial and taxonomic distribution of data. The size of each circle shows the total number of phenological observations, and the coloured sectors the proportions of observations belonging to each taxonomical group. The number of distinct localities in the database is 471, but in the figure data from nearby locations have been pooled into 63 locations which are situated at least 100 km apart.
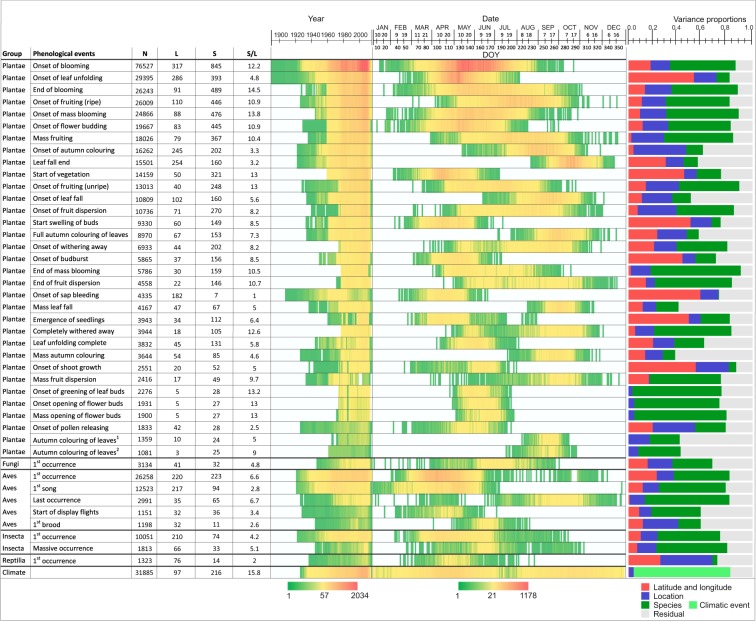
Fig. 2.
Illustration of the structure of the data for phenological events with highest coverage. Each row corresponds to a type of phenological event. For each event, shown are the total number of records (N), the number of locations from which the records originate (L), the number of species that the data involve (S), and the mean number of species per location (S/L). The two heat maps show the temporal coverage of data in terms of years included (reflecting data availability), and in terms of the phenological dates (reflecting the timing of the included events). Further shown is a variance partitioning, with the colours corresponding to the fixed effects of latitude, longitude and their interaction (red), the random effect of the site (blue), the random effect of the taxon or climatic parameter (green), and the residual (grey). The event types are ordered within each taxonomic group according to the total amount of data.
The recording scheme implemented at nature reserves offers unique opportunities for addressing community-level change across replicate local communities21. These data have been systematically collected not as independent monitoring efforts, but using a shared and carefully standardized protocol adapted for each local community. Thus, variability in observation effort is of much less concern than in most other distributed cross-taxon phenological monitoring schemes. To enable analyses of higher-level taxonomical groups, we have included taxonomic classifications for the species in the database.
Methods
Data acquisition
The data were collected by two research programs: the Chronicles of Nature (Letopisi Prirody) monitoring program, and a volunteer network of phenological observers (Fenologicheskii Klub). The Chronicles of Nature monitoring program22 is based on the network of strictly protected areas (zapovedniks) and national parks. The program gradually evolved during early 1900s23 and was formally established in 1940 with the aim of streamlining scientific work in protected areas with standardized methodology among the organizations. The program involves the permanent personnel of each participating organization. The results of the monitoring programs are published annually as Chronicle of Nature books. One printed copy of the books was kept in the office of the participating organization and another copy was sent to the Governmental Environmental Conservation Service (or a corresponding entity depending on the specific point in time).
In the Chronicles of Nature monitoring program, bird phenological events are extracted from route-based observations conducted regularly by ornithologists or professional rangers of the protected areas. Plant phenological events are reported either by botanists who visit permanent monitoring plots or transect, or by rangers who conduct regular walk-throughs within the strictly protected area or national park. The insect phenological data are extracted from standardized trapping data collected by entomologists on permanent plots or transects. The amphibian and reptile data are extracted from standardized trapping data collected by herpetologists. The fungal phenological data are collected by mycologists on permanent plots or transects. The weather event data are collected following a list of pre-defined events (e.g. first day of snowfall) by dedicated personnel or sourced from observations made on a local meteorological station. The types of data collected by each organization depends on the expertise of different taxonomic groups in the scientific personnel. For more details on how the data were collected, see22,24–28.
The network of phenological volunteer observers was established by the Russian Geographic Society in 1848 with questionnaires sent out to selected contacts among scientific community, including teachers and general public29. The participants of the volunteer observation network make observations throughout the year to collect data on a pre-defined limited set of phenological events related to plants, animals, and weather. The species included in the pre-defined lists were selected so that they could be identified reliably without specific taxonomical training.
Data digitalization and unification
The compilation of the data in a common database was initiated in the context of the project “Linking environmental change to biodiversity change: long-term and large-scale data on European boreal forest biodiversity” (EBFB), funded for 2011–2015 by the Academy of Finland, and continued with the help of other funding to OO since 2016. We organized a series of project meetings that were essential for data acquisition, digitalization and unification. These meetings were organized in Ekaterinburg (Russia) by the Institute of Plant and Animal Ecology, Ural Branch of RAS (Russian Academy of Sciences) in 2011; in Petrozavodsk (Russia) by the Forest Research Institute, at the Karelian Research Center, RAS in 2013; in Miass (Russia) by the Ilmen Nature Reserve in 2014; in Krasnoyarsk (Russia) by the Stolby Nature Reserve in 2014; in Artybash (Russia) by the Altaisky Nature Reserve in 2015; in Listvyanka, Lake Baikal (Russia) by the Zapovednoe Pribajkalje Nature Reserve in 2016; in Roztochja (Ukraine) by the Ministry of Natural Resources of Ukraine in 2016; in Puschino (Russia) by the Prioksko-Terrasniy Nature Reserve in 2017, in Vyshinino (Russia) by the Kenozero National Park in 2018, and in St Petersburg (Russia) by the Komarov Botanical Institute of the Russian Academy of Sciences in 2019.
The compilation of the data into a common database was conducted by the database coordinators (EM and CL) in Helsinki (Finland). Those participants that already held the data in digital format submitted it in the original format, and those that had the data only in paper format digitized it using Excel-based templates developed in the project meetings. Submitted data were processed by the database coordinators according to the following steps:
The data were formatted so that each observation (the phenological date of a particular event in a particular locality and year) formed one row in the data table (e.g. un-pivoting tables that involved several years as the columns). The phenological event names were split into event type (e.g. “first occurrence“) and species name.
The event type names (provided originally typically in Russian) were translated into English and the species names (usually provided in Russian) were identified to scientific names, using dictionaries that were partly developed and verified in the project meetings. All scientific names were periodically verified by mapping them to the Global Biodiversity Information Facility (GBIF) backbone taxonomy30.
We associated each data record with the following set of information fields: (1) project name, i.e. the source organization, (2) dataset name, (3) locality name, (4) unique taxon identifier, (5) scientific taxon name, and (6) event type.
We imported the data records in the main database (maintained as an EarthCape database at https://ecn.ecdb.io). During the import, the taxonomic names, locality names, and dataset names were matched against already existing records.
The database was published in Zenodo31.
Updates and limitations
There are at least 150 National Parks and Nature Reserves that collect Chronicles of Nature Book data (in Armenia, Azerbaijan, Belarus, Georgia, Kazakhstan, Kyrgyzstan, Moldova, Russian Federation, Tajikistan, Turkmenistan, Ukraine and Uzbekistan). Out of these, the current database covers data from 62 organizations, with the highest coverage in European Russia (Fig. 2). The collection of new data continues in most parks. Thus, the database is not complete, and we aim to support the database with updates, depending on the interest of new partners to join, as well as resources and funding. The technical validation procedures described below will also be applied to any new information included in the database. The resulting new versions of the database will be released through the Zenodo repository to ensure the long-term availability of the database.
The Chronicles of Nature programme involves several kinds of systematically collected data beyond phenology data: e.g. trapping data on small mammals, count data on birds, and yield data on berries and mushrooms22. We aim to publish these data as separate data papers.
Data Records
The database is organized in six datasets: (1) a classification of taxa included, (2) a list of phenological events included, (3) a list of climatic events included, (4) information on the study site, (5) the phenology data, and (6) an information sources table for phenology data31. All tables are in csv format (columns separated by commas), and their fields are described in Tables 1–6. The tables can be linked to each other using the unique study site names and the unique identifiers for species and climatic evens.
Table 2.
The fields of the phenological events table (phenologicalevents.csv).
| Field name | Description |
|---|---|
| kingdom | The kingdom for which the phenological event is relevant |
| eventtype | The name of the phenological event |
| description | The description of the phenological event |
| bbch | BBCH-scale used to identify the phenological development stages of plants32 |
Table 3.
The fields of the climatic events table (climaticevents.csv).
| Field name | Description |
|---|---|
| group | The type of the climatic event (e.g. related to temperature, snow or ice) |
| eventtype | The name of the climatic event |
| description | The description of the climatic event |
Table 4.
The fields of the study sites table (studysites.csv).
| Field name | Description |
|---|---|
| studysite | The name of the study site |
| latitude | Latitude (typically of center of protected area) |
| longitude | Longitude (typically of center of protected area) |
Table 5.
The fields of the phenology table (phenology.csv).
| Field name | Description |
|---|---|
| project | The name of the project in which the data were collected |
| dataset | The name of the dataset to which the data belongs to |
| studysite | The name of the study site in which the data were collected |
| taxonidentifier | The unique identifier of the taxon (“Climate” for climatic events) |
| taxon | The scientific name of taxon (climatic group for climatic events) |
| eventtype | The type of the event |
| year | The year |
| dayofyear | The date of the observation as the number of days since January 1st in the focal year |
| quality | An indicator variable of any quality issues identified with the data |
Table 1.
The fields of the taxonomy table (taxonomy.csv).
| Field name | Description |
|---|---|
| taxonidentifier | The unique identifier of the taxon |
| taxon | The scientific name of the taxon |
| taxonomiclevel | The highest taxonomical level to which the taxon is described |
| kingdom | Kingdom |
| phylum | Phylum |
| class | Class |
| order | Order |
| family | Family |
| genus | Genus |
| species | Species |
| gbifkey | The GBIF key for the taxon |
| gbifstatus | Whether the GBIF status of the taxon is accepted |
Table 6.
The fields of the information sources table (informationsources.csv).
| Field name | Description |
|---|---|
| project | The name of the project |
| source | The type of information sources (mostly Chronicles of Nature Books of the participating organizations) |
| reference | The references used to extract the phenology data |
Technical Validation
We asked the contributors to carefully check the validity of the phenological dates prior to submission. While uploading the submitted data to the database, we did manual validation checks to pinpoint data records that were suspicious (e.g. summer events recorded in winter), and sent the suspicious data records back for the contributors for correction or validation. However, given the extensive size of the database, it is likely that the database contains a number of erroneous records. Thus, we performed a series of checks to identify spurious data points and to examine for the strength of biological signal in the data.
First, we fitted for each (site – climatic/species name – event type) triplet a von Mises distribution, i.e. the circular normal distribution, where the circularity was used to connect the last day of the previous year to the first year of the next year. We identified as potentially spurious those records that were beyond the 0.9999367 central confidence interval of the fitted distribution (i.e. points located at least four standard deviations away from the mean, assuming a Gaussian distribution). This filtering revealed 322 severe outliers that were returned to the data owners for validation. If the data owner could neither verify nor correct the exceptional date, we marked this data record as suspicious.
Second, for each (site – climatic/species name – event type) triplet we fitted (i) a single von Mises distribution and (ii) a mixture of two von Mises distributions, and compared the fits of the models (i) and (ii) using the Bayesian Information Criteria (BIC). We identified the data as potentially spurious if the mixture model fitted better to the data with BIC difference of 5 or greater, and if the distance between the estimated means of the distributions in the mixture was greater than 30 days. For 214 such cases, we performed a manual examination of the data. This revealed e.g. the use of identical event names with different actual meaning (e.g. first arrival of the Willow Tit Parus montanus, recorded in spring and autumn seasons, and thus related to spring and autumn migration). Next, we repeated exactly the same filtering procedure, but for (climatic/species name – event type) pairs – to ensure that similarly named event types had consistent meaning across all sites.
Major sources of variation in the data
To quantify the main sources of variation and thus to illustrate the types of ecological signals present in the data, we performed a variance partitioning analysis separately for each group of species and phenological events. As predictors, we used species and the location, the latter of which we further explained by the linear effects of latitude, longitude, and their interaction. These analyses were preformed using the LinearModelFit and Variance functions in Mathematica 11.1; Wolfram Research 2018. As an example, let us consider the event type with highest amount of data, which is the onset of blooming for plants. These data consist of 76,527 phenological dates, originating from 317 sites and representing 845 taxa (Fig. 2). We first computed for each site an average day over the species and years, resulting in 317 site-specific dates. These dates describe when plants on average (over years and plant species) have their onset of blooming on each location. While the collection of species included in the study varies from site to site, we still consider these dates meaningful proxies for the overall phenology of the onset of plant blooming. The amount of variation explained by the site-level averages was 33% of the original variance. Out of the variation explained by the site, 54% was further explained by the linear effects of latitude, longitude, and their interaction. We then partitioned the remaining variation (after the effect of site was accounted for) to components that could be attributed to the species (53% of the original variance) and to the residual (14% of the original variance). This analysis provided rather strong support for a strong ecological signal being present in the data, as 86% of the variation among the 76,527 data points could be attributed to the main effects of the location and species, and as ca. half of the variation among the locations could be attributed to a simple geographic trend. We note that the residual variation in this analyses should not be interpreted as erroneous noise, as it contains e.g. variation over time, and thus reflects e.g. the impact of climate change on phenology.
We repeated the above described analysis for all groups of phenological events for which there were at least 1000 data records, as well as climatic events related to temperature, snow, and ice. The results are illustrated in Fig. 2. The amount of explained variance is generally relatively high in all cases, suggesting that much of the variation in the data are explained by location and species.
Acknowledgements
The field work was conducted as part of the monitoring program of nature reserves, Chronicles of Nature. The work was funded by Academy of Finland, grants 250243, 284601, 309581 (OO); the European Research Council, ERC Starting Grant 205905 (OO); Nordic Environment Finance Corporation Grant (OO); Jane and Aatos Erkko Foundation Grant (OO and TR); University of Helsinki HiLIFE Fellow Grant 2017–2020 (OO); the Kone Foundation 44-6977 (MD); Spanish Ramon y Cajal grant RYC-2014-16263 (MD); the Federal Budget for the Forest Research Institute of Karelian Research Centre Russian Academy of Sciences 220-2017-0003, 0220-2017-0005 (LV, SS and JK); the Russian Foundation for Basic Research Grant 16-08-00510 (LK), and the Ministry of Education and Science of the Russian Federation 0017-2019-0009 (Keldysh Institute of Applied Mathematics, Russian Academy of Sciences) (NI, MSh). Special thanks to other colleagues who helped with data collection, especially A. Beshkarev, G. Bushmakova, T. Butorina, A. Esipov, N. Gordienko, E. Kireeva, V. Koltsova, I. Kurakina, V. Likhvar, I. Likhvar, D. Mirsaitov, M. Nanynets, L. Ovcharenko, L. Rassohina, E. Romanova, A. Shelekhov, N. Shirshova, D. Sizhko, I. Sorokin, H. Subota, V. Syzhko, G. Talanova, P. Valizer and A. Zakusov.
Author contributions
O. Ovaskainen acquired the funding, led the project, organized the project meetings, performed the analyses and contributed to the first draft of the paper. E. Meyke organized the project meetings, organized the data into the database, and contributed to the first draft of the paper. C. Lo participated in the project meetings and organized the data into the database. G. Tikhonov participated in the project meetings, performed the technical validation of the data and contributed to the first draft of the paper. M. Delgado organized the project meetings and contributed to the first draft of the paper. T. Roslin participated in the project meetings and contributed to the first draft of the paper. E. Gurarie participated in the project meetings. M. Abadonova collected the original data and participated in the project meetings. O. Abduraimov collected the original data. O. Adrianova collected the original data. T. Akimova collected the original data. M. Akkiev collected the original data. A. Ananin collected the original data, contributed to organizing the data and participated in the project meetings. E. Andreeva collected the original data and participated in the project meetings. N. Andriychuk collected the original data. M. Antipin collected the original data and participated in the project meetings. K. Arzamascev collected the original data. S. Babina organized the project meetings and collected the original data. M. Babushkin collected the original data and participated in the project meetings. O. Bakin collected the original data and participated in the project meetings. A. Barabancova collected the original data. I. Basilskaja collected the original data. N. Belova collected the original data. N. Belyaeva collected the original data and participated in the project meetings. T. Bespalova collected the original data, contributed to organizing the data and participated in the project meetings. E. Bisikalova collected the original data and participated in the project meetings. A. Bobretsov collected the original data and participated in the project meetings. V. Bobrov organized the project meetings. V. Bobrovskyi collected the original data and participated in the project meetings. E. Bochkareva collected the original data. G. Bogdanov collected the original data and participated in the project meetings. V. Bolshakov organized the project meetings. S. Bondarchuk collected the original data and participated in the project meetings. E. Bukharova collected the original data and participated in the project meetings. A. Butunina collected the original data. Y. Buyvolov organized the project meetings, contributed to organizing the data and collected the original data. A. Buyvolova contributed to organizing the data and participated in the project meetings. Y. Bykov collected the original data. E. Chakhireva collected the original data. O. Chashchina organized the project meetings and collected the original data. N. Cherenkova collected the original data and participated in the project meetings. S. Chistjakov collected the original data and participated in the project meetings. S. Chuhontseva organized the project meetings and collected the original data. E. Davydov collected the original data and participated in the project meetings. V. Demchenko collected the original data. E. Diadicheva collected the original data. A. Dobrolyubov collected the original data and participated in the project meetings. L. Dostoyevskaya collected the original data. S. Drovnina collected the original data and participated in the project meetings. Z. Drozdova collected the original data and participated in the project meetings. A. Dubanaev collected the original data. Y. Dubrovsky collected the original data. S. Elsukov collected the original data. L. Epova collected the original data and participated in the project meetings. O. Ermakova collected the original data. O. Ermakova collected the original data and participated in the project meetings. A. Esengeldenova collected the original data. O. Evstigneev collected the original data. I. Fedchenko collected the original data and participated in the project meetings. V. Fedotova collected the original data, contributed to organizing the data and participated in the project meetings. T. Filatova collected the original data and participated in the project meetings. S. Gashev collected the original data and participated in the project meetings. A. Gavrilov collected the original data and participated in the project meetings. I. Gaydysh collected the original data. D. Golovcov collected the original data and participated in the project meetings. N. Goncharova collected the original data and participated in the project meetings. E. Gorbunova collected the original data and participated in the project meetings. T. Gordeeva collected the original data. V. Grishchenko collected the original data. L. Gromyko collected the original data. V. Hohryakov collected the original data, contributed to organizing the data and participated in the project meetings. A. Hritankov collected the original data. E. Ignatenko collected the original data and participated in the project meetings. S. Igosheva collected the original data and participated in the project meetings. U. Ivanova collected the original data. N. Ivanova organized training in data digitalization and participated in the project meetings. Y. Kalinkin collected the original data. E. Kaygorodova collected the original data and participated in the project meetings. F. Kazansky collected the original data and participated in the project meetings. D. Kiseleva collected the original data. A. Knorre organized the project meetings and collected the original data. L. Kolpashikov collected the original data and participated in the project meetings. E. Korobov collected the original data. H. Korolyova collected the original data. N. Korotkikh collected the original data and contributed to organizing the data. G. Kosenkov collected the original data. S. Kossenko collected the original data. E. Kotlugalyamova collected the original data. E. Kozlovsky collected the original data and participated in the project meetings. V. Kozsheechkin collected the original data and participated in the project meetings. A. Kozurak collected the original data. I. Kozyr collected the original data and participated in the project meetings. A. Krasnopevtseva collected the original data and participated in the project meetings. S. Kruglikov collected the original data. O. Kuberskaya collected the original data and participated in the project meetings. A. Kudryavtsev collected the original data and participated in the project meetings. E. Kulebyakina collected the original data, contributed to organizing the data and participated in the project meetings. Y. Kulsha collected the original data. M. Kupriyanova collected the original data and participated in the project meetings. M. Kurbanbagamaev collected the original data. A. Kutenkov organized the project meetings and collected the original data. N. Kutenkova organized the project meetings and collected the original data. N. Kuyantseva organized the project meetings and collected the original data. A. Kuznetsov collected the original data. E. Larin collected the original data, contributed to organizing the data and participated in the project meetings. P. Lebedev organized the project meetings, collected the original data and contributed to organizing the data. K. Litvinov collected the original data and participated in the project meetings. N. Luzhkova collected the original data, contributed to organizing the data and participated in the project meetings. A. Mahmudov collected the original data. L. Makovkina collected the original data. V. Mamontov collected the original data and participated in the project meetings. S. Mayorova collected the original data. I. Megalinskaja collected the original data and participated in the project meetings. A. Meydus collected the original data and participated in the project meetings. A. Minin collected the original data, contributed to organizing the data and participated in the project meetings. O. Mitrofanov collected the original data. M. Motruk collected the original data. A. Myslenkov collected the original data. N. Nasonova collected the original data. N. Nemtseva collected the original data. I. Nesterova collected the original data. T. Nezdoliy collected the original data and participated in the project meetings. T. Niroda collected the original data. T. Novikova collected the original data. D. Panicheva collected the original data and participated in the project meetings. A. Pavlov collected the original data and participated in the project meetings. K. Pavlova collected the original data and participated in the project meetings. P. Petrenko collected the original data and participated in the project meetings. S. Podolski collected the original data. N. Polikarpova contributed to organizing the data and participated in the project meetings. T. Polyanskaya collected the original data. I. Pospelov collected the original data. E. Pospelova collected the original data. I. Prokhorov organized the project meetings. I. Prokosheva collected the original data, contributed to organizing the data and participated in the project meetings. L. Puchnina collected the original data and participated in the project meetings. I. Putrashyk collected the original data. J. Raiskaya collected the original data. Y. Rozhkov collected the original data and participated in the project meetings. O. Rozhkova collected the original data and participated in the project meetings. M. Rudenko collected the original data and participated in the project meetings. I. Rybnikova collected the original data. S. Rykova collected the original data. M. Sahnevich organized the project meetings and collected the original data. A. Samoylov collected the original data. V. Sanko collected the original data. I. Sapelnikova collected the original data, contributed to organizing the data and participated in the project meetings. S. Sazonov collected the original data. Z. Selyunina collected the original data and participated in the project meetings. K. Shalaeva collected the original data. M. Shashkov organized training in data digitalization and participated in the project meetings. A. Shcherbakov collected the original data. V. Shevchyk collected the original data. S. Shubin collected the original data. E. Shujskaja contributed to organizing the data and participated in the project meetings. R. Sibgatullin collected the original data. N. Sikkila collected the original data and participated in the project meetings. E. Sitnikova collected the original data and participated in the project meetings. A. Sivkov collected the original data. N. Skok collected the original data. S. Skorokhodova organized the project meetings and collected the original data. E. Smirnova collected the original data. G. Sokolova collected the original data. V. Sopin collected the original data. Y. Spasovski collected the original data and participated in the project meetings. S. Stepanov collected the original data. V. Stratiy collected the original data. V. Strekalovskaya collected the original data. A. Sukhov collected the original data. G. Suleymanova collected the original data and participated in the project meetings. L. Sultangareeva collected the original data and participated in the project meetings. V. Teleganova collected the original data. V. Teplov collected the original data. V. Teplova collected the original data. T. Tertitsa collected the original data and participated in the project meetings. V. Timoshkin collected the original data. D. Tirski collected the original data. A. Tolmachev collected the original data. A. Tomilin contributed to data management and participated in the project meetings. L. Tselishcheva collected the original data and participated in the project meetings. M. Turgunov collected the original data. Y. Tyukh collected the original data. V. Van collected the original data. E. Vargot collected the original data and participated in the project meetings. A. Vasin collected the original data. A. Vasina collected the original data and participated in the project meetings. A. Vekliuk collected the original data. L. Vetchinnikova collected the original data and participated in the project meetings. V. Vinogradov collected the original data. N. Volodchenkov collected the original data. I. Voloshina collected the original data. T. Xoliqov collected the original data and participated in the project meetings. E. Yablonovska-Grishchenko collected the original data. V. Yakovlev collected the original data. M. Yakovleva organized the project meetings and collected the original data. O. Yantser collected the original data and contributed to organizing the data. Y. Yarema collected the original data. A. Zahvatov collected the original data. V. Zakharov collected the original data and participated in the project meetings. N. Zelenetskiy collected the original data. A. Zheltukhin collected the original data and participated in the project meetings. T. Zubina collected the original data. J. Kurhinen initiated the establishment of the co-operative network, acted as the network coordinator and organized the project meetings.
Code availability
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Tatyana Gordeeva, Sergei Sazonov, Andrei Sivkov, Viktor Teplov, Vladimir Yakovlev
References
- 1.Post E, Forchhammer MC, Stenseth NC, Callaghan TV. The timing of life-history events in a changing climate. Proc. R. Soc. B. 2001;268:15–23. doi: 10.1098/rspb.2000.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koh LP, et al. Species Coextinctions and the Biodiversity Crisis. Science. 2004;305:1632–1634. doi: 10.1126/science.1101101. [DOI] [PubMed] [Google Scholar]
- 3.Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 2012;15:365–377. doi: 10.1111/j.1461-0248.2011.01736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 5.Root T, Price J, Hall K, Schneider S. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- 6.Ovchinnikova T, Fomina VA, Dolzhkovaja NP, Andreeva EB, Sukhovolskii VG. Analysis of changes in the timing of seasonal events in woody plants of the Stolby Reserve in connection with climatic factors. Con. Bor. Zo. 2011;28:54–59. [Google Scholar]
- 7.Parmesan C. Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Glob. Chang. Biol. 2007;13:1860–1872. doi: 10.1111/j.1365-2486.2007.01404.x. [DOI] [Google Scholar]
- 8.Both C, Van Asch M, Bijlsma RG, Van Den Burg AB, Visser ME. Climate change and unequal phenological changes across four trophic levels: Constraints or adaptations? J. Anim. Ecol. 2009;78:73–83. doi: 10.1111/j.1365-2656.2008.01458.x. [DOI] [PubMed] [Google Scholar]
- 9.Cook BI, et al. Sensitivity of spring phenology to warming across temporal and spatial climate gradients in two independent databases. Ecosystems. 2012;15:1283–1294. doi: 10.1007/s10021-012-9584-5. [DOI] [Google Scholar]
- 10.Voigt W, et al. Trophic levels are differentially sensitive to climate. Ecology. 2003;84:2444–2453. doi: 10.1890/02-0266. [DOI] [Google Scholar]
- 11.Thackeray SJ, et al. Trophic level asynchrony in rates of phenological change for marine, freshwater and terrestrial environments. Glob. Chang. Biol. 2010;16:3304–3313. doi: 10.1111/j.1365-2486.2010.02165.x. [DOI] [Google Scholar]
- 12.Thackeray SJ, et al. Phenological sensitivity to climate across taxa and trophic levels. Nature. 2016;535:241–245. doi: 10.1038/nature18608. [DOI] [PubMed] [Google Scholar]
- 13.Menzel A, Sparks TH, Estrella N, Roy DB. Altered geographic and temporal variability in phenology in response to climate change. Glob. Ecol. Biogeogr. 2006;15:498–504. doi: 10.1111/j.1466-822X.2006.00247.x. [DOI] [Google Scholar]
- 14.Hong BC, Shurin JB. Latitudinal variation in the response of tidepool copepods to mean and daily range in temperature. Ecology. 2015;96:2348–2359. doi: 10.1890/14-1695.1. [DOI] [PubMed] [Google Scholar]
- 15.Phillimore AB, Stålhandske S, Smithers RJ, Bernard R. Dissecting the contributions of plasticity and local adaptation to the phenology of a butterfly and its host plants. Am. Nat. 2012;180:655–670. doi: 10.1086/667893. [DOI] [PubMed] [Google Scholar]
- 16.Roy DB, et al. Similarities in butterfly emergence dates among populations suggest local adaptation to climate. Glob. Chang. Biol. 2015;21:3313–3322. doi: 10.1111/gcb.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi H, Takahashi M. Latitudinal patterns in the phenological responses of leaf colouring and leaf fall to climate change in Japan. Glob. Ecol. Biogeogr. 2008;17:556–561. doi: 10.1111/j.1466-8238.2008.00398.x. [DOI] [Google Scholar]
- 18.Primack RB, et al. Spatial and interspecific variability in phenological responses to warming temperatures. Biol. Conserv. 2009;142:2569–2577. doi: 10.1016/j.biocon.2009.06.003. [DOI] [Google Scholar]
- 19.Ge Q, Wang H, Rutishauser T, Dai J. Phenological response to climate change in China: A meta-analysis. Glob. Chang. Biol. 2015;21:265–274. doi: 10.1111/gcb.12648. [DOI] [PubMed] [Google Scholar]
- 20.Walther GR, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- 21.Ovaskainen O, et al. Community-level phenological response to climate change. Proc. Natl. Acad. Sci. USA. 2013;110:13434–9. doi: 10.1073/pnas.1305533110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filonov, K. P. & Nukhimovskaya, Y. D. The Chronicles of Nature in Zapovedniks of the USSR. Methodological Notes. (In Russian, Nauka Press, Moscow, 1990).
- 23.Spetich MA, Kvashnina AE, Nukhimovskya YD, Olin E, Rhodes J. History, administration, goals, value, and long-term data of Russia’s strictly protected scientific nature reserves. Nat. Areas J. 2009;29:71–78. doi: 10.3375/043.029.0109. [DOI] [Google Scholar]
- 24.Dobrovolsky, B. V. Phenology of Insects. (In Russian, Vysshaya Shkola Publishing House, 1969).
- 25.Beideman, I. N. The Study of Plant Phenology in Field Geobotany. (In Russian, Russian Academy of Sciences, 1960).
- 26.Preobrazhenskiy, S. M. & Galahov, N. N. Phenological Monitoring. (In Russian, Detskoe Gosudarstvennoe Izdatelstvo, Moscow, 1948).
- 27.Zharkov, I. V. Basic Nature Observations. (In Russian, USSR Ministry of Agriculture publishing House, Moscow, 1954).
- 28.Beideman, I. N. Methods for Phenology Observations of Plants and Plant Communities. (In Russian, Nauka Press, Novosibirsk, 1972).
- 29.Rural Chronicle, Compiled from Observations Which May Serve to Determine Climate in Russia in 1851, Vol 1. (in Russian, SPb, 1854).
- 30.2017. GBIF Secretariat. GBIF Backbone Taxonomy. [DOI]
- 31.Ovaskainen O, 2020. Chronicles of Nature Calendar, a long-term and large-scale multitaxon database on phenology. Zenodo. [DOI] [PMC free article] [PubMed]
- 32. Meier, U. Growth Stages of Mono- and Dicotyledonous Plants. (Blackwell, 1997).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2017. GBIF Secretariat. GBIF Backbone Taxonomy. [DOI]
- Ovaskainen O, 2020. Chronicles of Nature Calendar, a long-term and large-scale multitaxon database on phenology. Zenodo. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Not applicable.