Abstract
Strains Marseille-P4001 and Marseille-P3668 are new species from the order Bacteroidales isolated from healthy French volunteers. They are anaerobic Gram-negative rod-shaped bacteria. They exhibited 92.68% and 96.68% 16S rRNA sequence identities with Parabacteroides gordonii strain MS-1 and Parabacteroides chinchillae JCM 17104, respectively, the phylogenetically closest species. Their respective draft genomes measured 5.23 Mb and 3.73 Mb with 39.2 mol% and 40.8 mol% of G + C content. Using a taxonogenomics method, we propose here a brief description of Parabacteroides pacaensis sp. nov., strain Marseille-P4001T and Parabacteroides provencensis sp. nov., strain Marseille-P3668T as new bacterial species.
Keywords: Culturomics, human gut microbiota, new bacteria, Parabacteroides pacaensis sp. nov., Parabacteroides provencensis sp. nov
Introduction
It is important to understand the implications of bacterial diversity in normal physiological functions and in the disease [1]. Culturomics is a concept developing different culture conditions in order to enlarge our knowledge of the human microbiota through the discovery of previously uncultured bacteria [[2], [3], [4], [5]]. Once a bacterium was isolated, we used a taxono-genomics approach, including matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), phylogenetic analysis, main phenotypic description and genome sequencing, to describe it [6,7].
Here we describe Parabacteroides pacaensis sp. nov., strain Marseille-P4001T (= CSUR P4001), and Parabacteroides provencensis sp. nov., strain Marseille-P3668T (= CSUR P3668), according this taxono-genomics concept.
Isolation and growth conditions
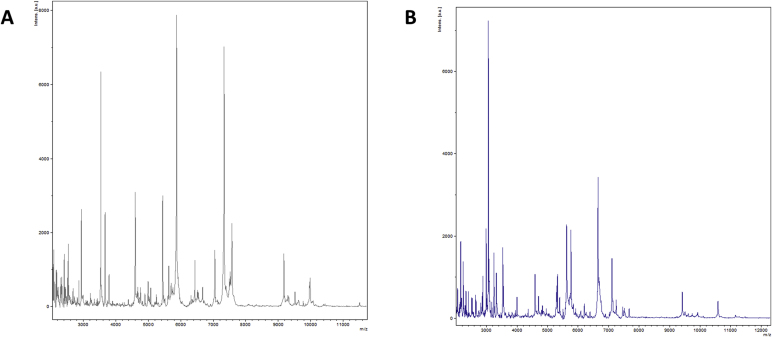
We isolated two unidentified bacterial strains from the fresh stools of two volunteers living in France. A screening was made by MALDI-TOF MS on a Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany) as previously described [8]. The obtained spectra (Fig. 1) were imported into MALDI Biotyper 3.0 software (Bruker Daltonics) and analysed against the main spectra of the bacteria included in two databases (Bruker and constantly updated URMS databases). The study was validated by the ethics committee of Institut Fédératif de Recherche IFR48 under number 2016-010. Strains Marseille-P4001T and Marseille-P3668Twere first isolated after 7 days of pre-incubation in an anaerobic blood culture bottle (Becton-Dickinson Diagnostics, Le Pont-de-Claix, France) supplemented with 5% sheep blood at 37°C.
Fig. 1.
MALDI-TOF MS Reference mass spectrum of Parabacteroides pacaensis sp. nov. strain Marseille-P4001T (a) and Parabacteroides provencensis sp. nov., strain Marseille-P3668T (b). The reference spectrum was generated by comparison of spectra from 12 individual colonies.
Phenotypic characteristics
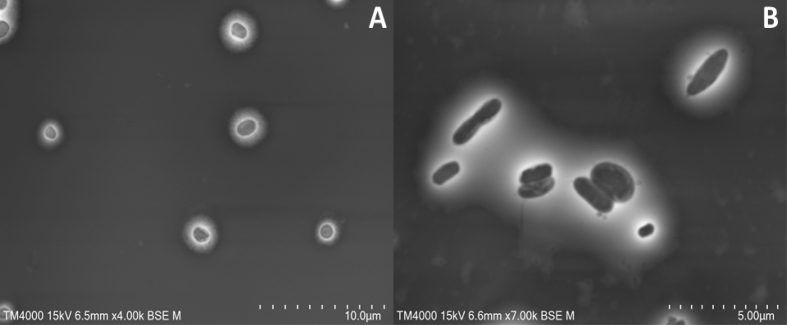
After the isolation step, the strain Marseille-P4001T and strain Marseille-P3668T were cultured with the aim to get pure and isolated colonies on blood agar. The colonies of Marseille-P4001 and Marseille-P3668 had almost the same morphological aspect, namely beige, small and smooth. Bacterial cells were Gram-negative for both strains. The sporulation test (10 min at 80°C) was negative. Different growth temperatures (20, 28, 32, 37, 45 and 56°C), pH (5, 6, 7, 7.5, 8 and 8.5), and atmospheres (aerobic, anaerobic and microaerophilic (CampyGEN, Oxoid, Basingstoke, UK)) were tested on 5% sheep-blood-enriched Columbia agar. Strain Marseille-P4001T grows at 28 and 37°C in anaerobic conditions at pH 7. Strain Marseille-P3668T grows from 28 to 45°C (optimally at 37°C) at pH ranging from 6 to 8.5 (optimally at pH 7) in anaerobic conditions. API ZYM (bioMérieux, Marcy l’Étoile, France) was performed to determine specific enzymatic properties for both strains. The results are tabulated in Table 1. Using API 50 CH strips (bioMérieux) the carbohydrate metabolism of both strains was evaluated according to the manufacturer’s instructions (Table 2). For strain Marseille-P4001T the following positive reactions were noted: esterase (C4), leucine arylamidase, α-galactosidase, β-galactosidase, N-acetyl-β-glycosaminidase, alkaline phosphatase, esculin ferric citrate, d-melezitose, d-saccharose, d-mannitol, methyl-αd-glucopyranoside and glycogen. All the other reactions tested were negative. Strain Marseille-P3668T had positive reactions for alkaline phosphatase, leucine arylamidase, α-galactosidase, β-galactosidase, naphthol-AS-BI-phosphohydrolase, phosphatase acid, N-acetyl-β-glycosaminidase, α-fucosidase, esculin ferric citrate and dulcitol. All the other reactions tested were negative. Strain Marseille-P4001T and strain Marseille-P3668T showed catalase-positive and oxidase-negative activities. A comparative study of the biochemical characteristics of those strains with other closely related Parabacteroides species is presented in Table 3. For scanning electron microscopy, a colony was collected from agar and immersed in a 2.5% glutaraldehyde fixative solution for each strain. The slide was gently washed in water, air-dried and examined with a TM4000 microscope. The cells of strain Marseille-P4001 appear to be rod-shaped with a mean length of 1.5 μm and a mean diameter of 0.5 μm. The cells of strain Marseille-P3668 are rod-shaped with a mean length of 2 μm and a mean diameter of 0.7 μm (Fig. 2).
Table 1.
Phenotypic characterization of Parabacteroides pacaensis strain Marseille-P4001T sp. nov. and Parabacteroides provencensis sp. nov. strain Marseille-P3668T, based on analytical profile index (API) ZYM tests
| Tests | Characteristics | P4001T | P3668T |
|---|---|---|---|
| API ZYM | Alkaline phosphatase | + | + |
| Esterase (C4) | + | – | |
| Esterase lipase (C8) | – | – | |
| Lipase (C14) | – | – | |
| Leucine arylamidase | + | + | |
| Valine arylamidase | – | – | |
| Cystine arylamidase | – | – | |
| Trypsin | – | – | |
| α-chymotrypsin | – | – | |
| Acid phosphatase | – | + | |
| Naphthol-AS-BI-phosphohydrolase | – | + | |
| α-galactosidase | + | + | |
| β-galactosidase | + | + | |
| β-glucuronidase | – | – | |
| α-glucosidase | – | – | |
| β-glucosidase | – | – | |
| N-acetyl-β-glucosaminidase | + | + | |
| α-mannosidase | – | – | |
| α-fucosidase | – | + | |
| Glycerol | – | – |
Table 2.
Phenotypic characterization of Parabacteroides pacaensis strain Marseille-P4001T sp. nov. and Parabacteroides provencensis sp. nov. strain Marseille-P3668T, based on API 50 CH test
| Tests | Characteristics | P4001T | P3668T |
|---|---|---|---|
| 50 CH | Erythritol | – | – |
| d-arabinose | – | – | |
| l-arabinose | – | – | |
| d-ribose | – | – | |
| d-xylose | – | – | |
| l-xylose | – | – | |
| d-Adonitol | – | – | |
| Methyl βd-xylopyranoside | – | – | |
| d-galactose | – | – | |
| d-glucose | – | – | |
| d-fructose | – | – | |
| d-mannose | – | – | |
| l-sorbose | – | – | |
| l-rhamnose | – | – | |
| Dulcitol | – | + | |
| Inositol | – | ||
| d-mannitol | + | – | |
| d-sorbitol | – | – | |
| Methyl αd-mannopyranoside | – | – | |
| Methyl αd-glucopyranoside | + | – | |
| N-acetyl-glucosamine | – | – | |
| Amygdalin | – | – | |
| Arbutin | – | – | |
| Esculin ferric citrate | + | + | |
| Salicin | – | – | |
| d-cellobiose | – | – | |
| d-maltose | – | – | |
| d-lactose | – | – | |
| d-melibiose | – | – | |
| d-saccharose | + | – | |
| d-trehalose | – | – | |
| Inulin | – | – | |
| d-melezitose | + | – | |
| d-raffinose | – | – | |
| Amidon | + | – | |
| Glycogen | – | – | |
| Xylitol | – | – | |
| Gentiobiose | – | – | |
| d-turanose | – | – | |
| d-xylose | – | – | |
| d-tagalose | – | – | |
| d-fucose | – | – | |
| l-fucose | – | – | |
| d-arabitol | – | – | |
| l-arabitol | – | – | |
| Potassium gluconate | – | – | |
| Potassium 2-ketogluconate | – | – | |
| Potassium 5-ketogluconate | – | – |
Table 3.
Comparison of differential characteristics of Parabacteroides pacaensis sp. nov., Parabacteroides provencensis sp. nov., Parabacteroides timonensis and Parabacteroides chartae
| Property | P. pacaensis | P. provencensis | P. timonensis | P. chartae |
|---|---|---|---|---|
| Cell diameter (μm) | 0.5 | 0.7 | 0.5 | 0.7–1 |
| Oxygen requirement | – | – | – | – |
| Gram stain | – | – | – | – |
| Salt requirement | – | – | – | – |
| Motility | – | – | – | – |
| Endospore formation | – | – | – | – |
| Alkaline phosphatase | + | + | + | + |
| Catalase | + | + | + | – |
| Oxidase | – | – | – | NA |
| Urease | – | – | – | – |
| β-Galactosidase | + | + | + | + |
| N-acetyl-glucosamine | – | + | + | + |
| Arabinose | – | – | + | + |
| Lipase (C8) | + | – | + | + |
| Mannose | – | – | + | + |
| Mannitol | + | – | + | – |
| Sucrose | + | – | + | + |
| d-Glucose | – | – | + | + |
| d-Fructose | – | – | + | – |
| d-Maltose | – | – | + | + |
| Source | Human | Human | Human | Environment |
Fig. 2.
Scanning electron microscopy of stained Parabacteroides pacaensis sp. nov., strain Marseille-P4001T (a) and Parabacteroides provencensis sp. nov., strain Marseille-P3668T (b) (Hitachi TM4000). Scales and acquisition settings are shown on the figure.
Strain identification
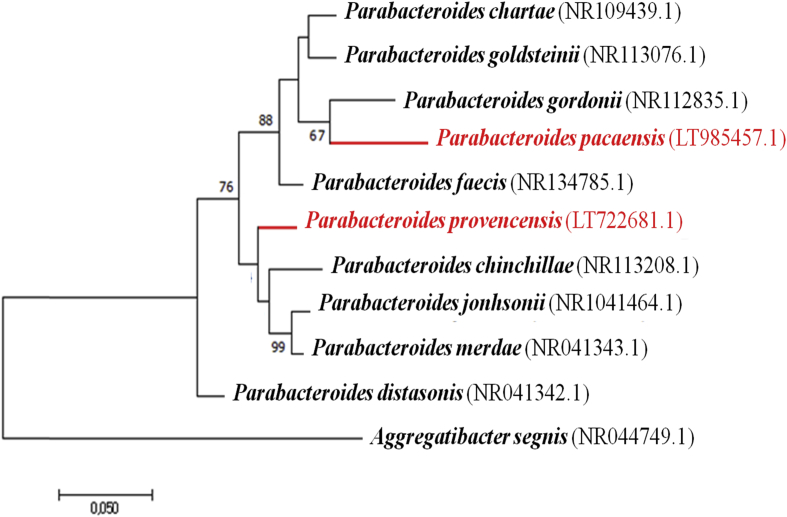
The 16S rRNA gene was sequenced to classify the bacteria. Amplification was performed by using the primer pair fD1 and rP2 (Eurogentec, Angers, France) and sequencing used the Big Dye® Terminator v1.1 Cycle Sequencing Kit and 3500xL Genetic Analyzer capillary3500xL sequencer (Thermofisher, Saint-Aubin, France), as previously described [9]. The 16S rRNA nucleotide sequences were assembled and corrected using CodonCode Aligner software (http://www.codoncode.com). Strain Marseille-P4001T exhibited a 92.68% sequence identity with Parabacteroides gordonii strain MS-1 (GenBank accession number NR112835.1) and strain Marseille-P3668T exhibited a 96.68% sequence identity with Parabacteroides chinchillae JCM 17104 (GenBank accession number NR113208.1), the phylogenetically closest species with standing in nomenclature (Fig. 3). Considering these phylogenetic values lower than the thresholds fixed to delineate new bacterial taxa [10,11], we consequently classify these strains as members within the genus Parabacteroides belonging to family Tannerellaceae.
Fig. 3.
Phylogenetic trees highlighting the position of Parabacteroides pacaensis sp. nov. and Parabacteroides provencensis sp. nov. based on the 16S rRNA gene sequences relative to the most closely related type strains within the genus Parabacteroides GenBank accession numbers are indicated in parentheses. Sequences were aligned using MUSCLE with default parameters, phylogenetic inference was obtained using the Maximum likelihood method and the MEGA 7 software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree. The scale bar indicates a 5% nucleotide sequence divergence.
Genome sequencing
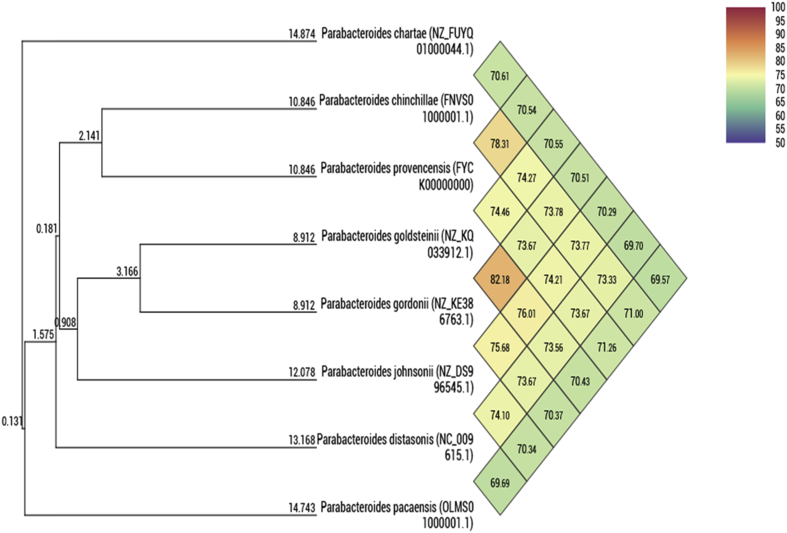
Genomic DNA was extracted using the EZ1 biorobot (Qiagen, Courtaboeuf, France) with the EZ1 DNA tissue Kit and then sequenced on the MiSeq technology (Illumina, San Diego, CA, USA) with the Nextera Mate Pair sample prep kit and Nextera XT Paired end (Illumina), as previously described [12]. The assembly was performed with a pipeline incorporating different software (Velvet [13], Spades [14] and Soap Denovo [15]) and trimmed data (MiSeq and Trimmomatic [16] softwares) or untrimmed data (only MiSeq software). GapCloser software [17] was used to reduce assembly gaps. Scaffolds <800 bp and scaffolds with a depth value <25% of the mean depth were removed. The best assembly was selected by using different criteria (number of scaffolds, N50, number of N). The genome of Parabacteroides pacaensis strain Marseille-P4001T is 5 238 628 bp long with a 39.21 mol% G + C content. Hence, the genome of Parabacteroides provencensis strain Marseille-P3668T is 3 732 078 bp long with a 40.8 mol% G + C content. The degree of genomic similarity of strain Marseille-P4001T and Marseille-P3668T with closest species was estimated using the OrthoANI software [18]. Values among closely related species (Fig. 4) ranged from 78.31% between Parabacteroides chinchillae and Parabacteroides provencensis to 82.18% between Parabacteroides goldsteinii and Parabacteroides gordonii; 71.26% of similarity is shared between P. provencensis and P. pacaensis.
Fig. 4.
Heatmap generated with OrthoANI values calculated using the OAT software between Parabacteroides pacaensis sp. nov. and Parabacteroides provencensis sp. nov., and other closely related species with standing in nomenclature.
Conclusion
Based on the results from unique phenotypic characteristics, including API gallery tests, MALDI-TOF spectrum, and phylogenetic and genomic analysis such as 16S rRNA sequence similarity <95% and OrthoANI value <95% with the phylogenetically closest species with standing in nomenclature, we formally propose strain Marseille-P4001T and strain Marseille-P3668T as type strains of Parabacteroides pacaensis sp. nov and Parabacteroides provencensis sp. nov., respectively.
Description of Parabacteroides pacaensis sp. nov.
Parabacteroides pacaensis (pa.ca'en.sis N.L. masc. adj. pacaensis, derived from the abbreviation PACA, for the region of Provence Alpes Côte d’Azur, where the strain was first isolated). The strain grows in varied conditions. Optimum growth of colonies was obtained at 37°C on 5% sheep-blood-enriched Columbia agar after 3 days in an anaerobic atmosphere. They appear smooth and small. Parabacteroides pacaensis is a Gram-negative rod-shaped bacterium with a mean length of 1.4 μm and a mean diameter of 0.5 μm. Strain Marseille-P4001T produced esterase (C4), leucine arylamidase, α- and β-galactosidase, N-acetyl-β-glycosaminidase and alkaline phosphatase, and metabolized esculin ferric citrate, d-melezitose, d-saccharose, d-mannitol, methyl-αd-glucopyranoside and glycogen. No activities were observed with trypsin, α-glucosidase, glycerol, d-arabinose, d-ribose, d-xylose, d-glucose, d-fructose, d-mannose, l-rhamnose, d-lactose, d-fucose and d-arabitol. Strain Marseille-P4001T is catalase-positive and oxidase-negative. The genome size of Parabacteroides pacaensis sp. nov., strain Marseille-P4001T is about 5.24 Mb long with 39.2 mol% G + C content. The GenBank accession number for the 16S rRNA gene sequence of strain Marseille-P4001T is LT985457 and for the whole genome shotgun project is OLMS01000001-OLMS01000014. This strain was isolated from the fresh stool of a healthy French volunteer.
Description of Parabacteroides provencensis sp. nov.
Parabacteroides pacaensis (pro.ven.cen'cis, N.L. fem. adj. provencensis, pertaining to Provence, the region of France where the type strain was isolated). The strain grows in varied conditions. Optimum growth of colonies was obtained at 37°C on 5% sheep-blood-enriched Columbia Agar after 3 days in anaerobic conditions. They appear smooth and small. Parabacteroides pacaensis is a Gram-negative rod-shaped bacterium with a mean length of 2 μm and a mean diameter of 0.7 μm. Strain Marseille-P3668T produced alkaline phosphatase, leucine arylamidase, α- and β-galactosidase, naphthol-AS-BI-phosphohydrolase, acid phosphatase, N-acetyl-β-glycosaminidase, and α-fucosidase and metabolize only esculin ferric citrate and Dulcitol. But any activities were observed with trypsin, α-glucosidase, glycerol, d-arabinose, d-ribose, d-xylose, d-glucose, d-fructose, d-mannose, l-rhamnose, d-lactose, d-fucose and d-arabitol. Strain Marseille-P3668T is catalase-positive and oxidase-negative. The genome size of P. provencensis strain Marseille-P3668T is about 3.73 Mb long with 40.8 mol% G + C content. The GenBank accession number for the 16S rRNA gene sequence of strain Marseille-P3668T is LT722681 and for the whole genome shotgun project is FYCK01000001-FYCK01000021. This strain was isolated from the fresh stool of a healthy French volunteer.
Nucleotide sequence accession number
The 16S rRNA gene and genome sequences were deposited in GenBank under accession numbers LT985457 and OLMS01000001-OLMS01000014, respectively, for Strain Marseille-P4001T and under accession numbers LT722681 and FYCK01000001-FYCK01000021, respectively, for Strain Marseille-P3668T.
Deposit in culture collections
Strain Marseille-P4001T was deposited in our strain collections under number (= CSUR P4001) and Strain Marseille-P3668T under number (= CSUR P3668).
Conflict of interest
None to declare.
Funding sources
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the programme Investissements d’avenir, reference ANR-10-IAHU-03, the Region Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
Acknowledgements
The authors thank Catherine Robert for sequencing the genome and Aurelia Caputo for submitting the genomic sequence to GenBank.
References
- 1.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. https://www.nature.com/articles/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 3.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 5.Lagier J.-C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 8.Lo C.I., Fall B., Sambe-Ba B., Diawara S., Gueye M.W., Mediannikov O. MALDI-TOF mass spectrometry: a powerful tool for clinical microbiology at Hôpital Principal de Dakar, Senegal (West Africa) PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morel A.-S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 10.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 11.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 12.Lo C.I., Sankar S.A., Fall B., Ba B.S., Diawara S., Gueye M.W. High-quality draft genome sequence and description of Haemophilus massiliensis sp. nov. Stand Genomic Sci. 2016;11:31. doi: 10.1186/s40793-016-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G.C., Xu T.J., Zhu R., Zhang Y., Li S.Q., Wang H.W. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience. 2019;8(1) doi: 10.1093/gigascience/giy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]