Abstract
The data presented in this article relates to the manuscript entitled ‘Engineering of Chinese hamster ovary cell lipid metabolism results in an expanded ER and enhanced recombinant biotherapeutic protein production’, published in the Journal Metabolic Engineering [1]. In the article here, we present data examining the overexpression of the lipid metabolism modifying genes SCD1 and SREBF1 in CHO cells by densitometry of western blots and by using mass spectrometry to investigate the impact on specific lipid species. We also present immunofluorescence data at the protein level upon SCD1 and SREBF1 overexpression. The growth profile data during batch culture of control CHO cells and CHO cells engineered to overexpress SCD1 and SREBF1 during batch culture are also reported. Finally, we report data on the yields of model secretory recombinant proteins produced from control, SCD1 or SREBF1 engineered cells using a transient expression systems.
Keywords: Chinese hamster ovary (CHO) cells, Lipid metabolism engineering, Cell line engineering, Difficult to express protein, Recombinant protein, Endoplasmic reticulum, Transcription factor engineering
Specifications Table
| Subject | Biotechnology |
| Specific subject area | Lipid Metabolism Metabolic Engineering and Recombinant Protein Expression from CHO Cells |
| Type of data | Table Image Chart Graph Figure |
| How data were acquired | Viable cell concentration and culture viability measurements were collected on a ViCell instrument (Beckman Coulter). Secreted recombinant protein concentrations in cell culture supernatants were determined using an Octet® instrument (ForteBio) with IgG calibrators and protein A biosensors. Western blot analysis was undertaken on standard laboratory equipment. Microscopy images were collected on a Zeiss LSM 880/Elyra/Axio Observer Z1 confocal microscope. Mass spectrometry data was collected on a Synapt G2Si (Waters) mass spectrometer. The data was analyzed using the Waters software UNIFI searching a Waters Lipid Maps database. |
| Data format | Raw; this manuscript, reference [3] and http://doi.org/10.5281/zenodo.3610075. doi: 10.5281/zenodo.3610075 Analyzed |
| Parameters for data collection | Control, SCD1 and SREBF1 engineered Chinese hamster ovary cells were cultured under standard batch conditions and transfected to transiently express a range of model secreted biotherapeutic recombinant proteins. |
| Description of data collection | Western blot and densitometry analysis was undertaken to investigate SCD1 and SREBF1 expression. Immunofluorescence analysis was undertaken to analyze expression and cellular localization of SCD1 and SREBF1. Cell counting was undertaking to determine viable cell numbers and culture viability during batch culture. An Octet® instrument and western blotting was used to estimate secreted recombinant protein yields. Mass spectrometry was undertaken to investigate specific lipids in control, SCD1 and SREBF1 engineered cells. |
| Data source location | Institution: University of Kent City/Town/Region: Canterbury Country: UK |
| Data accessibility | With the article, and/or in Refs. [1,3], and in the Zendo data repository; https://doi.org/10.5281/zenodo.3610075. |
| Related research article | Budge JD, Knight TJ, Povey J, Roobol J, Brown IR, Singh G, Dean A, Turner S, Jaques CM, Young RJ, Racher AJ, Smales CM (2020) Engineering of Chinese hamster ovary cell lipid metabolism results in an expanded ER and enhanced recombinant biotherapeutic protein production, Metabolic Engineering, 57:203–216. https://doi.org/10.1016/j.ymben.2019.11.007. |
Value of the Data
|
1. Data description
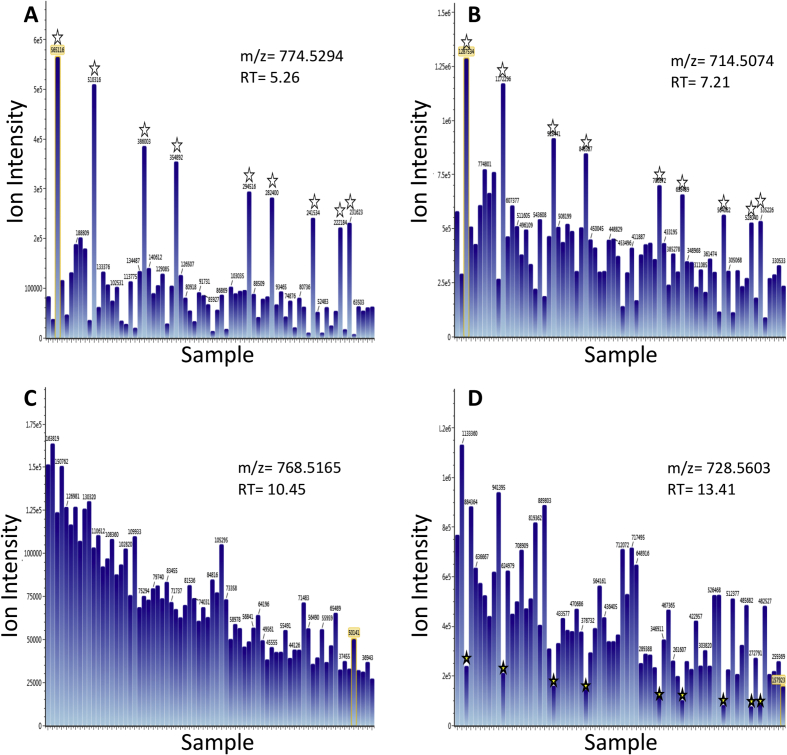
This article reports experimental data on the overexpression of the lipid metabolism modifying genes stearoyl CoA desaturase 1 (SCD1) and sterol regulatory element binding factor 1 (SREBF1) in Chinese hamster ovary (CHO) cells and the yields of model secretory recombinant proteins achieved from these during batch culture in a transient expression mode. A summary of vectors bearing genes for production of model recombinant biotherapeutic molecules used during this study is reported (Table 1). Details of antibodies used for analysis, including the manufacturer and reference, the organism produced in, its applications in the studies reported in this article and in Ref. [1], and the dilution used, are outlined in Table 2. Analysis of exogenous SCD1 and SREBF1 protein expression by densitometry of western blots, and the cellular localization by immunofluorescence, is reported in Fig. 1. The raw Western blot and microscopy image files can be found in the Zendo data repository [3]. The batch-culture growth profiles of control and SCD1 or SREBF1 engineered cells are shown along with the profiles when such cells are used to create stable model secretory recombinant protein expressing cell pools in Fig. 2. Analysis of transient secreted recombinant protein yields achieved from control and SCD1 and SREBF1 engineered CHO cell pools is analyzed in Fig. 3. Analysis of the yield of a protein considered by the authors to be difficult to express when transiently expressed in CHO cells alongside lipid metabolism modifying genes is shown in Fig. 4. Analysis of specific cellular lipids in control and lipid metabolism modifying engineered CHO cells using mass spectrometry is also reported in Fig. 5. The raw mass spectrometry data files can be found in the Zendo data repository ([3] doi: 10.5281/zenodo.3610075).
Table 1.
A summary of vectors bearing genes for production of model recombinant biotherapeutic molecules used and in Budge et al. [1]. The type of recombinant molecule produced by the vector and number of genes required for expression of these molecules are outlined. The figures in which the vectors have been used and a brief description of the method by which they have been utilized are summarized.
| Vector | Recombinant Molecule Type | No. of Genes | Brief Method |
|---|---|---|---|
| cB72.3 | IgG4 | 2 | Vector used to generate cells stably expressing cB72.3 molecule using hosts previously engineered to overexpress LMM molecules. Vector used to transiently produce cB72.3 molecule. |
| Fc-Fusion Protein | Fc fusion protein (FcFP) | 1 | Vector used to generate cells stably expressing an Fc fusion protein using hosts previously engineered to overexpress LMM molecules. Vector used to transiently produce an Fc fusion protein. |
| IL2-F-Control | Interleukin 2 fused bispecific antibody | 3 plus LMM gene | Vectors bearing both IL2-F genes and LMM genes used to transiently co-transfect. |
| IL2-F-SCD1 | |||
| IL2-F-SREBF1 | |||
| IL-2-F | Interleukin 2 fused bispecific antibody | 3 | Vector used to generate cells stably expressing IL2-F molecule using hosts previously engineered to overexpress LMM molecules. |
| DTE-IgG1 | IgG1 | 2 | Vector used to generate cells stably expressing DTE-IgG1 molecule using hosts previously engineered to overexpress LMM molecules. |
Table 2.
Details of antibodies used for analysis including the manufacturer and reference, the organism produced in and its applications reported in this article and Budge et al. [1], and the dilution used.
| Antibody | Company/Catalogue Number | Produced In | Use and Dilution |
|---|---|---|---|
| Primary Antibodies | |||
| Anti-V5 | Sigma/V8012 | Mouse | Immunofluorescence- 1:500 |
| Anti-calnexin | Abcam/ab22595 | Rabbit | Immunofluorescence- 1:200 |
| Anti-γ chain | Sigma/I9764 | Rabbit | Western Blot- 1:2000 |
| Anti-SCD1 | Cell Signalling/2438 | Rabbit | Western Blot- 1:1000 |
| Anti-SREBP1 | Abcam/ab3259 | Mouse | Western Blot- 1:1000 |
| Anti-L7α | Kind gift from Dr. Anne Roobol | Rabbit | Western Blot (used at supplied concentration) |
| Secondary Antibodies | |||
| Anti-Mouse FITC conjugate | Sigma/F0257 | Goat | Immunofluorescence 1:250 |
| Anti-Rabbit FITC conjugate | Sigma/F9887 | Goat | Immunofluorescence 1:250 |
| Anti-Mouse HRP Peroxidase conjugate | Sigma/A4416 | Goat | Western Blot- 1:2000 |
| Anti-Rabbit HRP Peroxidase conjugate | Sigma/A6154 | Goat | Western Blot- 1:2000 |
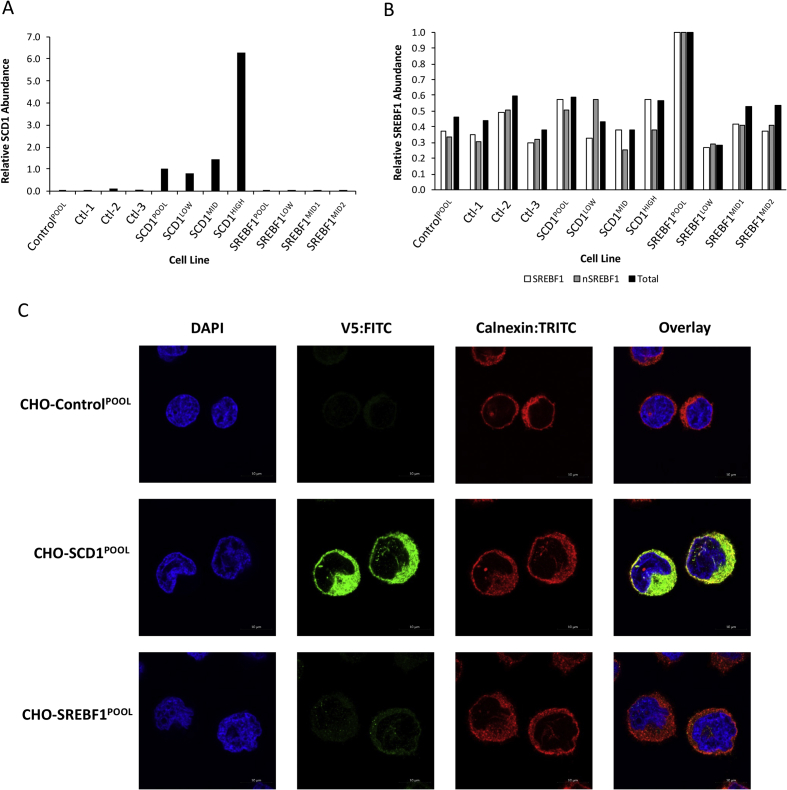
Fig. 1.
Quantitative analysis of relative overexpression levels and cellular localization of SCD1 and SREBF1 in engineered cells. Figure A shows the relative abundance of SCD1 as calculated using densitometry from the western blots reported in Fig. 2A and B of [1], whilst figure B shows the relative abundance of SREBF1 species. The values have been normalized to L7α loading control and subsequently normalized to either CHO-SCD1POOL (A) or CHO-SREBF1POOL (B) values. Cellular localization of overexpressed SCD1 and SREBF1 proteins in CHO-ControlPOOL, CHO-SCD1POOL and CHO-SREBF1POOL cell pools as determined by immunofluorescent detection using an anti-V5 antibody conjugated with a FITC secondary antibody (C). An anti-calnexin antibody conjugated with a TRITC secondary antibody was used to highlight the position of the ER. Images were obtained using confocal microscopy.
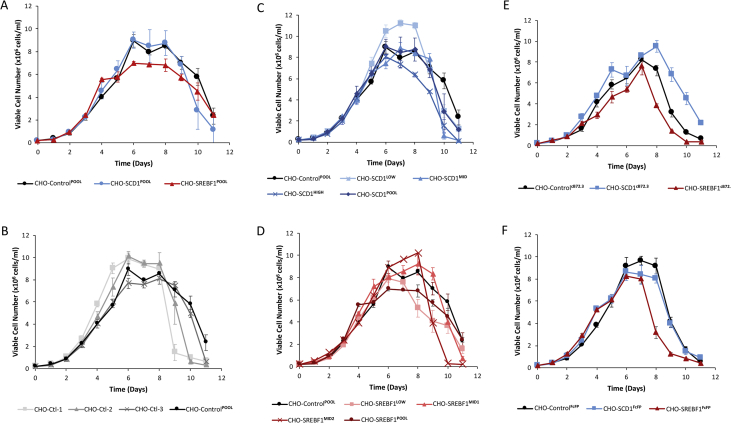
Fig. 2.
Growth profiles of CHOK1SV GS-KO™ cells engineered to overexpress lipid metabolism modifying genes SCD1 and SREBF1. Batch cultures were seeded at 0.2 × 106 viable cells/ml in a total culture volume of 20 ml and cell concentrations measured using a ViCell every 24 h. The CHO-ControlPOOL growth profile is shown in A-D for reference whilst lipid metabolism modified pool data are shown in (A), control monoclonal measurements are shown in (B), SCD1 monoclonal samples are shown in (C) and SREBF1 monoclonal measurements are shown in (D). Cell pools were constructed using lipid modified polyclonal cell pools as hosts such that they stably express either the cB72.3 monoclonal antibody or FcFP molecule. Growth profiles of these cells are shown in Figure (E) for cB72.3 expressing pools and Figure (F) for FcFP expressing pools. n = 3 for each data point and error bars show ± one standard deviation.
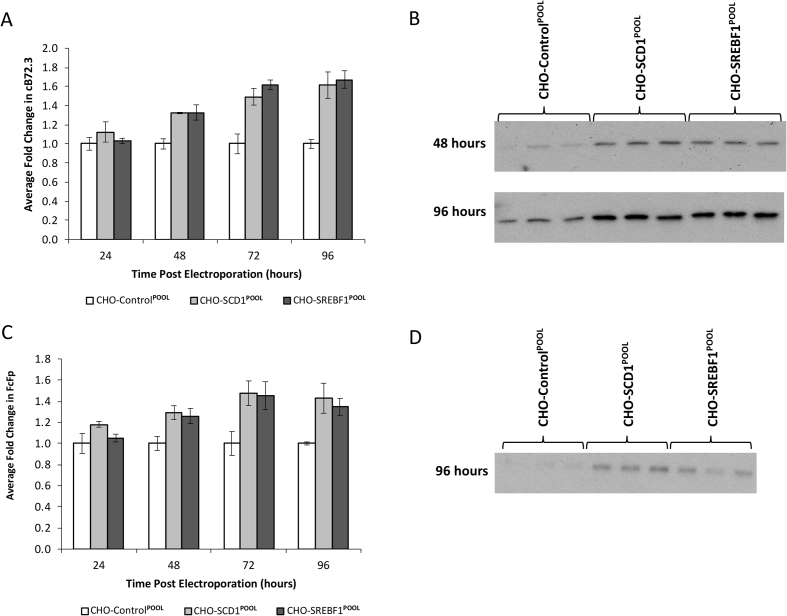
Fig. 3.
Product analysis post transient transfection in lipid modified cells. Previously engineered host cells (CHO-ControlPOOL, CHO-SCD1POOL, CHO-SREBF1POOL) were transiently transfected with vectors containing genes for expression of either cB72.3 (A and B) or FcFP (C and D). Quantitative analysis using an Octet® was carried out on supernatant samples harvested at 24, 48, 72 and 96 h post transfection for cB72.3 (A) and FcFP (C) transfections. Western blot analysis of cB72.3 (B) and FcFP (D) molecules were carried out on supernatant samples harvested at 96 h, and 48 h in the case of cB72.3. n = 3 for each data point and error bars show ± one standard deviation.
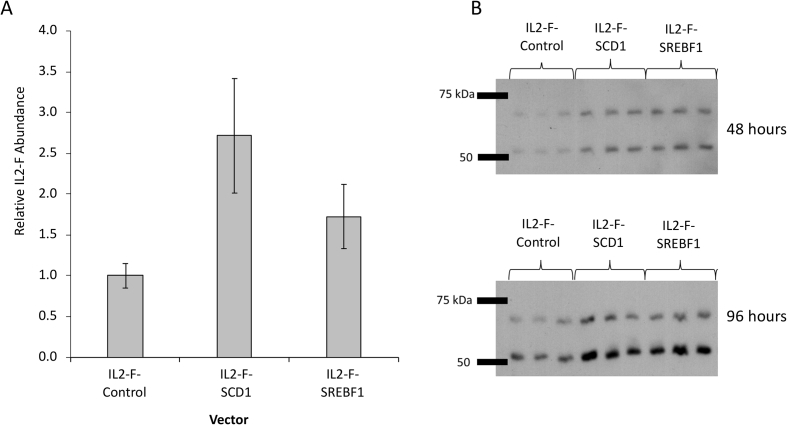
Fig. 4.
Product analysis post transient transfection with vectors bearing genes for a recombinant product and a lipid modifying gene on the same expression vector. Vectors containing genes necessary for expression of an antibody fusion molecule (IL2-F) with either no additional LMM gene (IL2-F-Control), the SCD1 gene (IL2-F-SCD1) or the SREBF1 gene (IL2-F-SREBF1) on the same vector. Quantitative analysis using an Octet® was carried out on supernatant samples harvested at 96 h post transfection to measure relative product concentration (A). Western blot analysis was also carried out on reduced samples. A heavy chain band (approximately 50 kDa) and an antibody heavy chain-IL2 fusion band (approximately 70 kDa) associated with the antibody fusion molecule is observed (B). n = 3 for each data point and error bars show ± one standard deviation.
Fig. 5.
Analysis of specific cellular lipids in control and LMM engineered CHO cells using mass spectrometry. Figures A to D show relative quantities of specific lipid species as indicated by ion intensities at specific retention times. Stars represent data obtained from SCD1 high samples which are either upregulated (A and B), downregulated (D), or unchanged (C) compared to the control and other SCD1 engineered cell pools and clones. Retention times (RT) and m/z ions are indicated within the figure.
2. Experimental design, materials, and methods
2.1. Cloning and construction of vectors used for cell engineering and transient experiments
A summary of the details of vectors generated during this study is described in Table 1. The CHO specific SCD1 gene sequence was amplified via polymerase chain reaction (PCR) using cDNA from Lonza's CHOK1SV™ cell line as the template and the primers 5′-TATGGTACCATGCCGGCC-3’ (forward) and 5′-ATACTCGAGCGGCTACTCTT-3’ (reverse). The SREBF1 mouse gene sequence was isolated from an OriGene (Rockville, MD) derived vector bearing the cDNA clone for the mouse SREBF1 gene (OriGene CAT no. MC205184, NCBI accession no. NM_011480) and amplified via PCR using the primers 5′-TATGCGGCCGCATGGACGAG-3’ (forward) and 5′-ATATCTAGACTGCTGGAAGTGACGGTGGTTC-3’ (reverse). SCD1 or SREBF1 genes were cloned into the pcDNA3.1V5-His/TOPO vector (Thermo Fisher Scientific) using KpnI and XhoI or NotI and XbaI restriction enzymes respectively (as underlined in oligonucleotide sequences) in order to generate vectors with the capacity to overexpress the genes of interest which were in frame with the V5 tag in CHO cells. The SREBF1 CHO cell gene sequence (NCBI accession no. NM_001244003.1) was synthesized by GeneART (ThermoFisher Scientific, USA).
Vectors were also constructed for the expression of model secretory recombinant proteins to assess the impact of cell line engineering on the secreted expression of these model molecules. In particular, we generated vectors from an in-house Lonza vector for a chimeric B72.3 (cB72.3) antibody as a model IgG4 molecule (considered to be easy to express by the authors) and a second Lonza vector was utilized which contained sequences for appropriate expression of a model Fc-fusion protein (FcFP). A vector for expression of a model IgG1 antibody (DTE-IgG1) considered by the authors to be difficult to express was also constructed. The construction of vectors for expression of an interleukin-2 fused bispecific antibody (IL2-F), containing the three genes required for expression of the IL2-F molecule, were built by having the individual genes synthesized by GeneART and then cloning these into a Lonza expression vector where each gene was under the control of a CMV promoter. Variations of the IL2-F vector were also generated such that they also contained sequences for appropriate expression of lipid metabolism modifying (LMM) genes SCD1 (IL2-F-SCD1) or SREBF1 (IL2-F-SREBF1), under the control of a CMV promoter, or a control (IL2-F-Control) lacking any LMM genes but inclusive of an empty expression cassette. All vectors contained a glutamine synthetase (GS) gene under the control of an SV40 promoter for use as a metabolic selection marker where necessary.
2.2. Cell culture and cell line construction
The process by which stably expressing SCD1 and SREBF1 cell pools and clones were established is described elsewhere [1] as is the process for generating stable recombinant protein secreting cell pool and lines. For the purpose of establishing the growth profiles of cell pools and lines under batch culture conditions, cultures were initially seeded with 0.2 × 106 viable cells/ml in 20 ml of CD-CHO medium (ThermoFisher Scientific) and then cultured in a 5% CO2 balanced air environment in a shaking incubator at 140 rpm at 37 °C. Cell concentrations and culture viabilities were routinely determined using a ViCell (Beckman Coulter) instrument and 1 ml of culture sample. Culture viability was calculated as the number of viable cells as a proportion of total cells.
2.3. Transient expression of secretory recombinant biotherapeutic proteins
Transient transfections were performed using a GenePulser Xcell electroporator (Bio-Rad). Vector DNA (20 μg) diluted in 100 μl TE buffer and 700 μl of cells (1 × 107 viable cells) were added to a cuvette. The DNA/cell mix was electroporated at 300 V and 900 μF in a cuvette with a 0.4 mm electrode gap. Medium (1 ml at 37 °C) was then added to the cuvette immediately after electroporation. The electroporated cells were then added to a flask containing 17.2 ml CD-CHO medium and a further 1 ml of medium was used to wash the cuvette and added to the culture such that a final volume of 20 ml was achieved in a 125 ml Erlenmeyer flask (Corning®). Cells were cultured in a 5% CO2 balanced air environment and batch transient cultures were incubated in a shaking incubator at 140 rpm at 37 °C.
2.4. Western blotting
Western blotting was undertaken as described elsewhere [2] and the details of the primary and secondary antibodies used in this study are outlined in Table 2. Quantitative densitometry was analyzed using ImageJ software.
2.5. Lipid extraction from cells and mass spectrometry analysis
The process for extracting lipids for analysis by mass spectrometry from cultured CHO cells and the process for the analysis of the mass spectrometry data is described in Ref. [1].
2.6. Immunofluorescence analysis
Cells were analyzed by immunofluorescence to determine the expression of exogenous ectopic SCD1 and SREBF1, and the localization of these proteins. The procedure for such analysis is described in Ref. [1] and the antibody details described in Table 2. Images were captured using a Zeiss LSM 880/Elyra/Axio Observer Z1 confocal microscope instrument.
2.7. Determination of secreted recombinant protein concentrations
Secreted recombinant protein concentrations in cell culture supernatants were determined using an Octet® instrument (ForteBio) with IgG calibrators and protein A biosensors.
Acknowledgments
We thank Mr Kevin Howland for help with lipid mass spectrometry analysis.
This worked was funded via the Biotechnology and Biological Sciences Research Council via Industrial CASE PhD studentships (JB and TK) and grant BB/N023501/1 (to CMS, Kent) and InnovateUK (to AJR, Lonza Biologics, grant No. 102621).
Conflict of Interest
AD, ST, CMJ, RJY and AJR are employed by Lonza Biologics, who developed and license the GS Gene Expression System®.
Lonza is the assigned owner of, and CMS, JDB, TJK, and RYJ are named inventors on, the filed patent Modulation of lipid metabolism for protein production, patent number WO2017191165A1.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Budge J.D., Knight T.J., Povey J., Roobol J., Brown I.R., Singh G., Dean A., Turner S., Jaques C.M., Young R.J., Racher A.J., Smales C.M. Engineering of Chinese hamster ovary cell lipid metabolism results in an expanded ER and enhanced recombinant biotherapeutic protein production. Metab. Eng. 2020;57:203–216. doi: 10.1016/j.ymben.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roobol A., Roobol J., Carden M.J., Smith M.E., Hershey J.W., Bastide A., Knight J.R., Willis A.E., Smales C.M. The chaperonin CCT interacts with and mediates the correct folding and activity of three subunits of translation initiation factor eIF3: b, i and h. Biochem. J. 2014;458:213–224. doi: 10.1042/BJ20130979. [DOI] [PubMed] [Google Scholar]
- 3.Budge J.D., Knight T.J., Povey J., Roobol J., Brown I.R., Singh G., Dean A., Turner S., Jaques C.M., Young R.J., Racher A.J., Smales C.M. Raw data to accompany the manuscript ‘Data for enginering lipid metabolism of Chinese hamster ovary (CHO) cells for enhanced recombinant protein production’ published in Data in Brief [Data set] Zenodo. 2020 doi: 10.1016/j.dib.2020.105217. [DOI] [PMC free article] [PubMed] [Google Scholar]