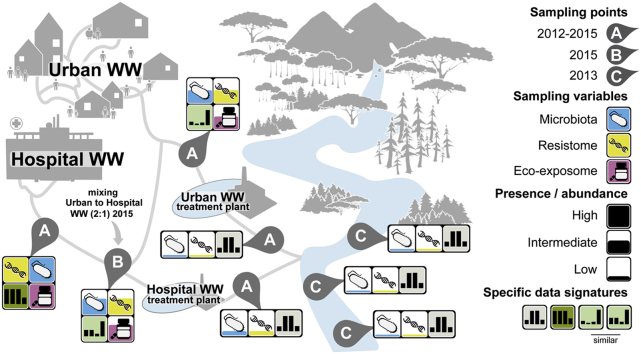
Abstract
Wastewaters (WW) are important sources for the dissemination of antimicrobial resistance (AMR) into the environment. Hospital WW (HWW) contain higher loads of micro-pollutants and AMR markers than urban WW (UWW). Little is known about the long-term dynamics of H and U WW and the impact of their joined treatment on the general burden of AMR. Here, we characterized the resistome, microbiota and eco-exposome signature of 126 H and U WW samples treated separately for three years, and then mixed, over one year. Multi-variate analysis and machine learning revealed a robust signature for each WW with no significant variation over time before mixing, and once mixed, both WW closely resembled Urban signatures. We demonstrated a significant impact of pharmaceuticals and surfactants on the resistome and microbiota of H and U WW. Our results present considerable targets for AMR related risk assessment of WW.
Keywords: Hospital and urban wastewater, Antimicrobial resistance, Resistome (ARGs and MGEs), Microbiota, WW signatures, Hospital and urban wastewater eco-exposome
Abbreviations: AMR, Antimicrobial Resistance; ARG, Antibiotic-Resistant Gene; ARB, Antibiotic-Resistant Bacteria; MGE, Mobile Genetic Element; WW, Wastewater; HWW, Hospital Wastewater; UWW, Urban Wastewater
Graphical abstract
Highlights
-
•
Hospital and urban wastewater resistome and microbiota display specific signatures stable over time.
-
•
Hospital and urban wastewater have a distinct eco-exposome that is associated with their respective resistome and microbiota.
-
•
Mixing hospital with urban wastewater hides the " hospital signature” due to the dilution rate.
1. Introduction
The natural environment and its biodiversity serve as a wide reservoir of genetic determinants implicated in resistance to antimicrobial compounds (Allen et al., 2010; Canton, 2009). Human activity has a significant impact on the terrestrial and aquatic microbial ecosystems through chemical pollutants that are spread via urban, agricultural and industrial waste and which pose an important selective pressure for antimicrobial resistance (AMR) (Flandroy et al., 2018). For instance, urban and hospital wastewaters (UWW and HWW) contain a high diversity of antimicrobial resistance genes (ARGs) and chemicals (Buelow et al., 2018; Jin et al., 2018; Manaia et al., 2018; Wang et al., 2018). It is generally accepted that the implementation of efficient wastewater treatment plants (WWTP) is essential in order to reduce the amounts of chemicals, ARGs and antibiotic resistant bacteria (ARBs) that reach the environment (Buelow et al., 2018; Chonova et al., 2016; Larsson et al., 2018). Despite the global reduction of ARBs, ARGs, and chemical compounds such as antibiotics (ABs), biocides and heavy metals through WW treatment, effluents from urban, hospital and industrial wastewater that are re-introduced into the environment, still contain moderate levels of those ‘contaminants’ (Manaia et al., 2018; Pallares-Vega et al., 2019; Raheem et al., 2018; Zhu et al., 2017). HWWs have been reported to contain particularly high amounts of ARGs, ARBs and ABs (Buelow et al., 2018; Rodriguez-Mozaz et al., 2015; Wang et al., 2018). It has been debated whether HWW contributes significantly to the load of ARGs in the UWW systems, and whether separate treatment for HWWs should be applied (Buelow et al., 2018; Chonova et al., 2016; Lienert et al., 2011). Recent work has shown that HWW has limited impact on the relative levels of ARGs and mobile genetic elements (MGEs) associated with ARGs, like integrons, in hospital receiving urban wastewater (WW) (Buelow et al., 2018; Pallares-Vega et al., 2019; Stalder et al., 2014). However, most studies usually analyzed a limited number of samples and yet, longitudinal studies that monitor WW dynamics are so far lacking, which limits the possibility of assessing the risk for AMR mediated through WW.
We collected 126 WW samples (UWW, HWW and mixed WW) in a French city during a period of approximately four years: 34 months with separate treatments for H and U WW (applying the conventional secondary (activated sludge) treatment process) and 11 months with H and U WW mixed 1:2 (HWW:UWW) into one system. From those we investigated the dynamics and relationship of the respective resistome and microbiota with the measured eco-exposome (pharmaceuticals, mainly antibiotics; surfactants and heavy metals). Multi-variate analysis and machine learning were employed to evaluate signatures of the resistome, microbiota and eco-exposome of HWW compared to UWW.
2. Materials and methods
2.1.1. Sampling and study design
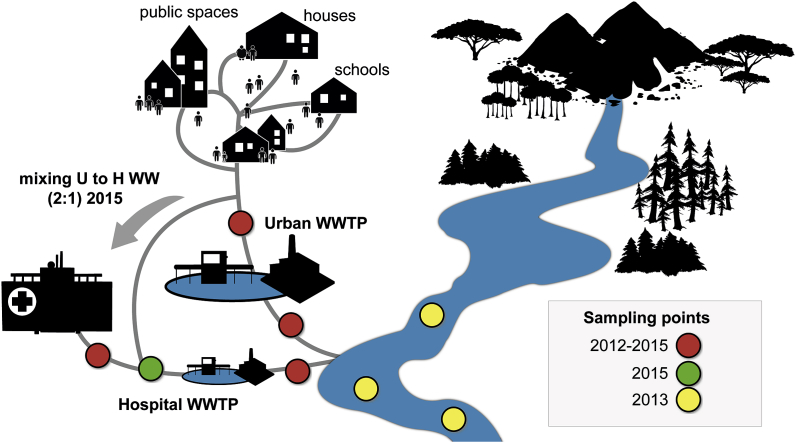
126 Urban and hospital wastewater (UWW and HWW) samples were collected in Scientrier (Bellecombe WWTP), Haute-Savoie, France (Chonova et al., 2018) as part of the multi-disciplinary project SIPIBEL (Chonova et al., 2018). The study site was implemented as an observatory for untreated and treated H and U WW and to evaluate their impact (during separate and subsequently mixed treatment) on the environment (e.g. the effluent receiving river). The CHAL hospital (Centre Hospitalier Alpes Léman) opened in February 2012 and includes 450 beds (140 m3/d), whereas the Bellecombe WWTP was collecting UWW of approximately 21.000 inhabitant equivalents (5200 m3/d). For more details of the SIPIBEL project, study set up, WWT and sample collection refer to Chonova et al., 2018 and Wiest et al. (Chonova et al., 2018; Wiest et al., 2018), and to Fig. 1. Samples were collected in monthly intervals (untreated and treated samples) by flow proportional sampling, from March 2012 through November 2015 (Chonova et al., 2018). From March 2012 to December 2014, UWW and HWW were treated separately applying the same conventional (activated sludge) WWT (Chonova et al., 2018; Wiest et al., 2018). Then, in the period from January 2015 through November 2015, UWW was mixed into the HWW (1:2 ratio HWW:UWW, the ratio was fixed by a local operating constraint) and added to the separate HWW treatment line resulting in a controlled mixed WW (MWW)(Chonova et al., 2018). In addition, 12 water samples of the effluent receiving river up (river upstream) and downstream (river downstream sampling point 1 and 2) of the effluent release pipes have been collected during the winter months of 2013 (January, February, November, and December).
Fig. 1.
Sampling site. Samples were collected in monthly intervals (untreated and treated samples) by flow proportional sampling, from March 2012 through November 2015. From March 2012 to December 2014, UWW and HWW were treated by separate wastewater treatment plants (WWTPs). During the period from January 2015 through November 2015, UWW was mixed into the HWW (1:2 ratio HWW:UWW) and added to the separate HWW treatment line resulting in mixed WW (MWW). In addition, 12 water samples of the effluent receiving river up (river upstream) and downstream (river downstream sampling point 1 and 2) of the effluent release pipes were collected during the winter months of 2013 (January, February, November, and December).
2.2. DNA extraction/sample preparation
Water samples were filtered for microorganisms, using a filtration ramp (Sartorius, Göttingen, Germany), on sterile 47 mm diameter filter with pore size of 0.45 μm (Sartorius, Göttingen, Germany). Microorganisms were recovered from filters and subject to DNA extraction for downstream analysis, using the Power water DNA extraction kit (MoBio Laboratories Inc., Carlsbad, CA, USA). DNA concentration was determined by Qubit Fluoremetric Quantitation (Thermo fisher scientific, Waltham, MA USA) assays according the manufacturer’s instructions. All DNA samples were diluted or concentrated to a final concentration of 10 ng/μl for downstream qPCR and 16S rRNA analysis.
2.3.1. High-throughput qPCR
Nanolitre-scale quantitative PCRs to quantify levels of genes that confer resistance to antimicrobials and heavy metals were performed as described previously (Buelow et al., 2018, 2017), with some modifications in the collection of primers. The primer sequences and their targets are provided in the supplementary data (Supplementary Table 1). In total we targeted 88 individual resistance genes conferring resistance to antibiotics, quaternary ammonium compounds, or heavy metals, grouped into 16 resistance gene classes. The genes targeted include ARGs that are most commonly detected in the gut microbiota of healthy individuals (Forslund et al., 2013; Hu et al., 2013), clinically relevant ARGs (including genes encoding extended spectrum β-lactamases (ESBLs), carbapenemases, and vancomycin resistance), and heavy metal and quaternary ammonium compound resistance genes suggested to favor cross and co – selection for ARGs in the environment (Gnanadhas et al., 2013; Pal et al., 2015). We also targeted genetic elements as important transposase gene families (Zhu et al., 2013) and class 1, 2 and 3 integron integrase genes (by primers described previously (Barraud et al., 2010)), that are important vectors for ARGs in the clinics and often used as proxy for anthropogenic pollution (Gillings et al., 2015). Finally, we targeted the bacterial 16S rRNA gene by universal primers. Gene targets and grouping of genes into gene classes/according to function are detailed in Supplementary Tables 1 and 2 Primer design and validation prior and after Biomark analysis has been done as described earlier (Buelow et al., 2018, 2017). Real-Time PCR analysis was performed using the 96.96 BioMark™ Dynamic Array for Real-Time PCR (Fluidigm Corporation, San Francisco, CA, U.S.A) as described previously (Buelow et al., 2018, 2017). Thermal cycling and real-time imaging was performed at the Plateforme Génomique GeT – INRA Transfert (https://get.genotoul.fr/en/), and Ct values were extracted using the BioMark Real-Time PCR analysis software.
2.3.2. Calculation of normalized abundance and cumulative abundance
Calculations for normalized and cumulative abundance of individual genes and allocated gene classes was done as described previously (Buelow et al., 2018, 2017). Specifically, normalized abundance of resistance genes was calculated relative to the abundance of the 16S rRNA gene (2∧(-(CTARG – CT16S rRNA)). Cumulative abundance of each resistance gene class was calculated based on the sum of the normalized abundance (2∧(-(CTARG – CT16S rRNA)) of all individual genes detected within a class, for each sample. The differences in cumulative abundance over the indicated time periods and sample locations (untreated and treated hospital and urban wastewaters, 2012–2014; up and downstream river samples, 2013; untreated and treated mixed wastewaters, 2015) are shown as an averaged fold-change ± standard deviation. The non-parametric Mann-Whitney test was used to test for significance; p values were corrected for multiple testing by the Benjamin-Hochberg procedure (Benjamini and Hochberg, 1995) with a false discovery rate of 0.05. Averaged normalized abundance data for allocated gene classes is provided in Supplementary Table 3.
2.4. qPCR to determine absolute copy numbers of 16S rRNA genes (bacterial biomass)
The qPCRs for the determination of 16S rRNA gene copy numbers per liters of water as a proxy for the bacterial biomass was performed as described previously by Stalder et. al. (Stalder et al., 2014).
2.5. 16S rRNA gene sequencing and sequence data pre-processing
Extracted DNA samples for 16S rRNA sequencing were prepared following a dual barcoded two-step PCR procedure for amplicon sequencing for Illumina. Primers of the first PCR step included universal CS1 and CS2 tags targeting the V4 region of the hypervariable region of the 16S rRNA gene using the 16SrRNA primer sequences of the earth microbiota project (http://press.igsb.anl.gov/earthmicrobiota/protocols-and-standards/16s/). Samples were sequenced following the Illumina protocol for a 2 × 301 MiSeq run (Illumina, Inc., San Diego, CA). Sequence reads from the Illumina MiSeq were demultiplexed and classified in the following manner: The Python application dbcAmplicons (https://github.com/msettles/dbcAmplicons) was used to identify and assign reads to the appropriate sample by both expected barcode and primer sequences. Barcodes were allowed to have at most 1 mismatch (hamming distance) and primers were allowed to have at most 4 mismatches (Levenshtein distance) as long as the final 4 bases of the primer matched the target sequence perfectly. Reads were then trimmed of their primer sequence and merged into a single amplicon sequence using the application FLASH (Magoc and Salzberg, 2011).
2.5.1. 16S rRNA data analysis
Illumina MiSeq forward and reverse reads were processed using the MASQUE pipeline (https://github.com/aghozlane/masque). Briefly, raw reads are filtered and combined followed by dereplication. Chimera removal and clustering are followed by taxonomic annotation of the resulting OTUs by comparison to the SILVA database. A BIOM file is generated that combines both OTU taxonomic assignment and the number of matching reads for each sample. Relative abundance levels form bacterial taxa (Order level) were obtained and analyzed. The obtained relative abundance OTU tables (Order level) were analyzed with Microsoft excel (Supplementary Table 4), multi-variate analysis package (see below) and by means of a machine learning approach employing a random forest algorithm (see below).
2.6. Chemical analysis
All chemical data measured here and used for analysis where extracted from the SIPIBEL database. Solid-phase extraction (SPE) and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) were used to measure the antibiotics ciprofloxacin, sulfamethoxazole and vancomycin and the pharmaceutical carbamazepine as detailed elsewhere (Wiest et al., 2018). Heavy metals (Zn, Cu, Ni, Pb, Cr, Gd, Hg, As and Cd) were measured with inductively coupled plasma combined with atomic emission spectroscopy (ICP-AES). Concentration of surfactants (anionic, cationic and non-ionic surfactants) were measured following standard methods approved by the French organization of standardization AFNOR as described by Wiest et. al. (Wiest et al., 2018).
2.7. Multivariate analyses
Multivariate statistical techniques were used to test the influence of waste water treatment or sampling time (independent variables) on the microbiota and the resistome (dependent variables) of the different sample groups (urban, hospital, mixed), including all individual genes and genes allocated into gene classes in two independent datasets. Statistically significant influence of the treatment or the sampling time on the microbiota and the resistome were assessed by Redundancy Analysis (RDA) with 499 Monte Carlo permutations. For testing the influence of sampling time, we used sampling year or season as independent variables, and all different sample groups together (i.e., to potentially identify general patterns influencing all groups at the same time), and individually, as dependent variables. Such analysis revealed the percentage of variance that is explained by sampling time or WW treatment in each case, and whether the influence of the independent variables is statistically significant or not. The relationship between the resistome and the microbiota and the measured chemicals in the different raw water samples (eco-exposome) was visualized by means of Principal Component Analysis (PCA) biplots. Moreover, the influence of the measured chemicals (eco-exposome) on the microbiota and resistome was statistically assessed by RDA. A variation partitioning analysis was performed to assess which group of chemicals (Metals, Pharmaceuticals, Surfactants) explains the largest share of the variation of the microbiota and resistome datasets, and to explore whether the interactive effects of the groups of chemicals would have a larger influence on those datasets than the individual groups themselves. All multivariate analyses were performed with the Canoco v5.0 software (Ter Braak and Smilauer, 2012), using a significance level of 0.05.
2.8. Machine learning
A Random Forest Algorithm (RFA) was used in order to predict a response variable (wastewater sample type (urban, hospital or mixed)) of each sample independently, using measurements on individual gene, gene class and microbiota level (predictor variables). To run the RFA the R-package randomForest was used: Breiman and Cutler’s Random Forests for Classification and Regression, a software package for the R-statistical environment (Breiman, 2001). In summary, the RFA follows the pseudo-steps: (I) the response variable and predictor variables are chosen by the user; (II) a predefined number of independent bootstrap samples are drawn from the dataset with replacement, and a classification tree is fit to each sample containing roughly 2/3 of the data, for which predictor variable selection on each node split in the tree is conducted using only a small random subset of predictor variables; (III) the complete set of trees, one for each bootstrap sample, composes the random forest (RF), from which the status (classification) of the response variable is predicted as an average (majority vote) of the predictions of all trees. Compared to single classification trees, RFA increases prediction accuracy, since the ensemble of slight different classification results adjusts for the instability of the individual trees and avoids data overfitting (Hastie et al., 2009). The Mean Decrease Accuracy (MDA), or Breiman-Cutler importance, was employed as a measure of predictor variable importance, for which classification accuracy after data permutation of a predictor variable is subtracted from the accuracy without permutation, and averaged over all trees in the RF to give an importance value [2]. It should be noted that since all predictor variables were of numeric nature, using RFA is equivalent to regression over classification trees. For the results presented here and in supplementary text, only the 2.5% of top RFA scores were considered (as presented by the resulting MDA distribution of all predictor variables), thus selecting the subset of predictor variables which appear statistically more informative than expected in the background of all predictor variables (i.e. we assume that 95% of the RFA scores fall between the 2.5th and 97.5th percentiles, as done elsewhere (Lourenco et al., 2017)).
3. Results
3.1. HWW and UWW resistome and microbiota signatures
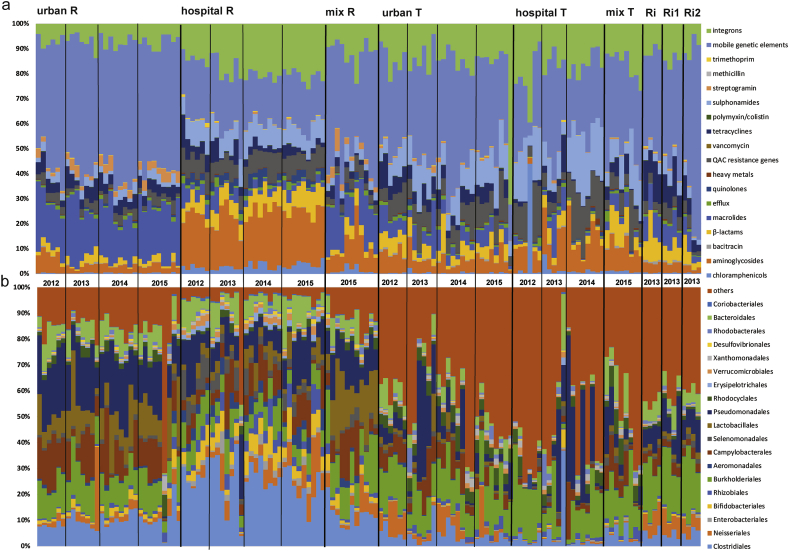
We evaluated the resistome and microbiota dynamics of monthly WW (N = 126) and river (N = 12) samples. H and U WW samples were treated separately through 2012 and 2014, and were mixed at a ratio of 1:2 (HWW:UWW) throughout the year 2015 (Fig. 1). H and U WW samples exhibited a distinct signature with respect to the proportional makeup of their resistome (Fig. 2a) and microbiota (Fig. 2b). Analyzing the data with a Random Forest machine learning approach showed that the distinct H and U WW signatures resulted in a high prediction accuracy. When using the resistome as predictor (on the level of gene classes), 93.5% and 96.7% of untreated HWW and UWW samples respectively, could be correctly classified (Supplementary Fig. 1a). We further analyzed the data on the individual gene level to increase resolution of the machine learning approach. Using individual genes resulted in similarly high predictions (93.5% prediction for untreated HWW and 100% prediction for untreated UWW) (Supplementary Fig. 1b). Similarly, when using the microbiota, 96.8% of untreated HWW and 89% of untreated UWW samples were correctly classified (Supplementary Fig. 1c).
Fig. 2.
Proportional abundance of the resistome and microbiota in untreated (R) and treated (T) HWW, UWW and MWW, as well as river samples up (Ri) and downstream (Ri1 and Ri2) of the waste water release pipes throughout the sampling period (2012–2015). a: proportional abundance of resistome (ARG classes, heavy metals, integron integrase genes and mobile genetic elements) for all samples. b: proportional abundance of the microbiota (displaying the 20 most abundant bacteria at the order level for all samples, where “others” represents the percentage of the remaining taxa).
For the treated H and U WW the machine learning prediction accuracy was lower compared to the untreated WW sources but still considerably high for all predictor levels and in particular for the microbiota (≥80%) (Supplementary Fig. 2).
For the MWW overall classification success was lower (Supplementary Figs. 1 and 2), given a much lower sample size and likely due to the mixing of the two wastewater sources hampering clear signatures.
3.2. Temporal dynamics of H and U WW
Redundancy analysis (RDA) was performed to assess putative significant influences of time on the variability of resistome and microbiota compositions using sampling year or season as independent variables (Monte Carlo (MC) permutations (n = 499)). Analysis was carried out for all different sample groups together (untreated U, H, M WW and treated U, H, M WW) to potentially identify general patterns influencing all groups at the same time. The same analysis was also performed per individual sample group (untreated and treated H, U and M WW alone, respectively).
Over the first 3 years before mixing of untreated HWW and UWW, neither year (p = 0.6 resistome and p = 0.4 microbiota respectively) nor season (p = 0.9 and p = 0.3) had a significant impact on the resistome (resistance gene classes) and microbiota composition of the WW (Supplementary Table 5a). However, by analyzing the sample groups separately, a yearly and seasonal impact on the level of individual resistance genes in HWW was demonstrated (p = 0.004 year, p = 0.032 season; Supplementary Table 5b). Redundancy analysis revealed the relationship between individual genes and gene classes with seasons, pointing towards a correlation of increased normalized abundance of individual genes and gene classes detected during summer season (Supplementary Figs. 3a and 4). The fact that the hospital was only installed in February 2012 (Chonova et al., 2018) may explain why the year 2012 is particular for HWW resistome, with overall lower normalized abundance of the resistome and no obvious trend towards the summer season in 2012. After HWW and UWW mixing (in 2015), no significant variation for the resistome and microbiota composition throughout the seasons could be observed considering all sample groups (untreated MWW, HWW and UWW; p = 0.6 resistome and p = 0.07 microbiota respectively; Supplementary Table 5a).
For treated WW, the analysis exhibited more variation of the resistome and microbiota composition compared to the untreated sources over the years (2012–2014 for H and U WW; Fig. 2, Supplementary Table 5c). Redundancy analysis revealed that in particular the microbiota of treated H and U WW effluents varied between the years (p = 0.016 for all groups, p = 0.024 for treated HWW and p = 0.006 for treated UWW, Supplementary Tables 5c and 5d) while no significant seasonal impact could be observed. The resistome varied between the years for treated HWW on both the gene class and the individual gene levels (p = 0.012 and p = 0.004, Supplementary Table 5d), whereas for the treated UWW this variation was only significant on the individual gene level (p = 0.032). For the treated mixed WW (MWW), significant seasonal variation was observed for the microbiota composition (p = 0.02), while for the resistome no significant variation could be observed (Supplementary Table 5d).
3.3. Human gut bacteria in HWW and UWW
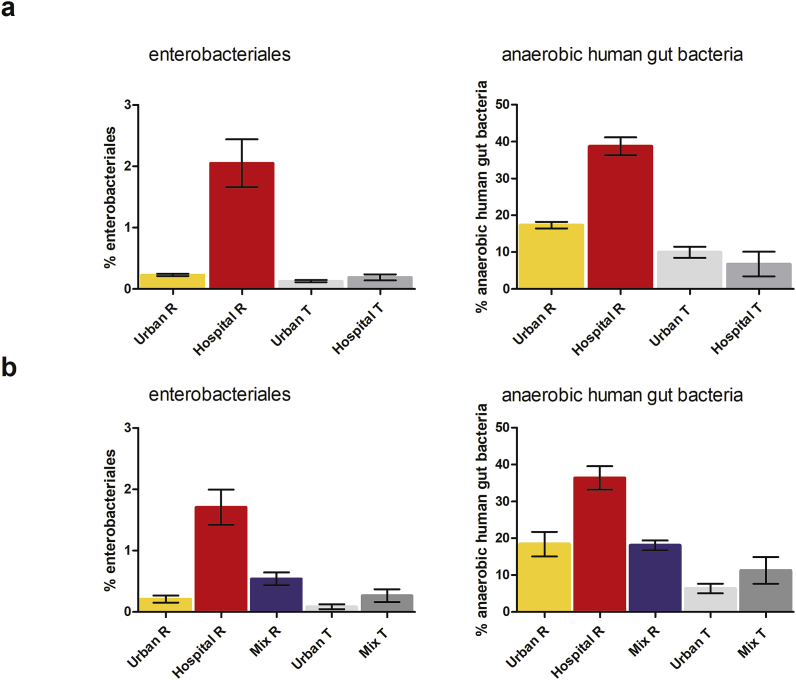
The microbiota of the respective WW was assessed by 16S rRNA sequencing. We calculated the relative abundance of anaerobic human gut bacteria, as well as Enterobacteriales, since many of these Gram-negative bacteria are also pathogens. The orders Clostridiales, Bifidobacteriales and Bacteroidales which represent the most important and abundant anaerobic human gut bacteria (Costea et al., 2018; Rajilic-Stojanovic and de Vos, 2014), were grouped together and are referred to as anaerobic human gut bacteria. Untreated HWW contained significantly higher levels of anaerobic human gut bacteria (38% ± 11 standard deviation) and Enterobacteriales (2% ± 1.5) compared to all other samples (Fig. 3a). Interestingly, anaerobic human gut bacteria and Enterobacteriales are comparable in their relative abundance for untreated UWW and MWW, indicating a significant dilution effect when mixing UWW with HWW (Fig. 3b) on the monitored human gut microbiota markers in WW. The WW treatment allowed a significant (p < 0.05) decrease of the relative abundance of these orders for HWW and UWW (Fig. 3) whereas the reduction for MWW was only marginally significant (p = 0.05) (Fig. 3b).
Fig. 3.
Relative abundance of anaerobic human gut bacteria (Clostridiales, Bifidobacteriales and Bacteroidales) and Enterobactertiales. a: for untreated (n = 21) and treated HWW (n = 19) and untreated (n = 21) and treated (n = 20) UWW, averaged over the numbers of samples collected for each sample group between March 2012 and December 2014 +- standard deviation. b: for untreated (n = 10) HWW, untreated (n = 9) and treated (n = 8) UWW and untreated (n = 10) and treated (n = 8) MWW samples (at the experimental ratio of 1:2 HWW:UWW) averaged over the numbers of samples collected for each sample group between January 2015 and November 2015 +- standard deviation.
3.4. The untreated HWW, UWW and MWW resistome
To verify that the differences we observed for the ARGs/MGEs between effluents were not due to variations in the bacterial biomass, we assessed the absolute copy numbers of 16S rRNA genes per liter of water, and showed that the bacterial biomass was comparable for the untreated WW sources (HWW, UWW and MWW) (Supplementary Fig. 5). Ratios of HWW over UWW and HWW over MWW were calculated based on the averaged cumulative abundance of the resistome in untreated HWW and UWW (Table 1).
Table 1.
Average fold changes for gene classes cumulative abundance of untreated HWW over UWW (2012–2015; significant differences indicated by asterisk *; p values ≤ 0.004) and HWW over MWW (2015, significant differences indicated by asterisk *; p values ≤ 0.03) ± Standard Deviation. Significant differences were calculated by comparing the normalized cumulative abundance values of individual gene classes for all samples belonging to each sample group using the non-parametric Mann-Whitney test. Fold changes were calculated for individually paired samples for each gene class/sample group. NA indicates that gene classes were undetectable in either one or both of the sample groups.
| Gene classes conferring resistance to: | Fold change untreated Hospital WW/Urban WW | Fold change untreated Hospital WW/Mixed WW |
|---|---|---|
| chloramphenicol | 84 (±93)* | 13 (±11)* |
| aminoglycosides | 43 (±31)* | 8 (±6)* |
| bacitracin | 8 (±13)* | 7 (±4)* |
| beta-lactams | 26 (±22)* | 9 (±6)* |
| macrolides | 1 (±1) | 0.6 (±0.3) |
| (multi-drug) Efflux | 9 (±11)* | 4 (±4)* |
| quinolones (qnr) | 161 (±326)* | 10 (±8)* |
| heavy metals | 7 (±9)* | 4 (±3)* |
| quaternary ammonium compounds(QACs) | 18 (±14)* | 7 (±4)* |
| vancomycin | 12 (±35)* | 18 (±20)* |
| tetracycline | 4 (±3)* | 3 (±2)* |
| polymixin | 8 (±9)* | 4 (±4)* |
| sulphonamides | 19 (±12)* | 7 (±4)* |
| methicillin | NA (undetectable in UWW) | NA (undetectable in all but one MWW sample) |
| streptogramin | 0.2 (±0.2)* | 0.2 ((±0.2)* |
| trimethoprim | 8 (±6)* | 5 (±3)* |
| Gene classes grouped according to function: | ||
| transposase genes (MGEs) | 3 (±1)* | 2 (±1) |
| integron integrase genes | 16 (±9)* | 6 (±3)* |
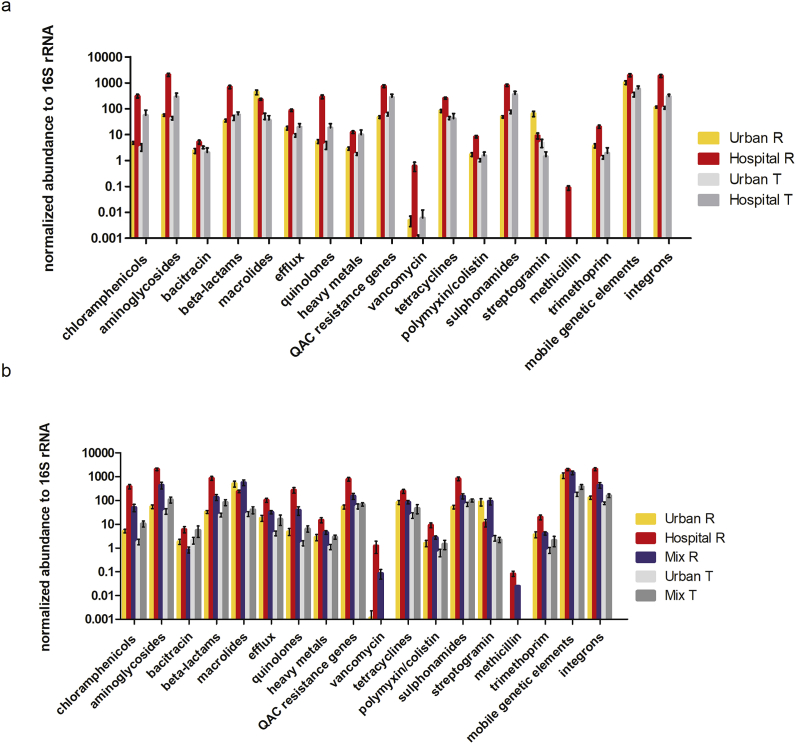
The untreated HWW contained significantly more gene classes compared to the untreated UWW, between 3 (transposase genes) and 161-fold (qnr genes encoding quinolone resistance) higher (p values ≤ 0.004; Table 1). When WW were mixed at the experimental ratio of 1:2 (HWW:UWW), the untreated MWW contained significantly less resistance gene classes compared to untreated HWW, between 3 and 22-fold lower (p values ≤ 0.03). Interestingly, there was no significant difference for the genes encoding resistance to macrolides for both HWW over UWW and HWW over MWW comparisons (Table 1, Fig. 4). The only resistance gene significantly lower in HWW compared to UWW or MWW (p < 0.0001), was the streptogramin resistance gene vatB. The mecA gene encoding resistance to methicillin was undetectable in all UWW and in all but one MWW samples (Table 1).
Fig. 4.
Averaged normalized cumulative abundance of ARG classes, heavy metals, MGEs and integrons over all collected samples per sample type + - standard deviation. a: averaged normalized cumulative abundance of ARG classes, heavy metals, QACs, MGEs and integrase genes in untreated (n21) and treated HWW (n = 19), untreated (n = 21) and treated (n = 20) UWW averaged over the numbers of samples collected for each water type in the given time interval (March 2012–December 2014) +- standard deviation. b: averaged normalized cumulative abundance of ARG classes, heavy metals, QACs, MGEs and integrase genes in untreated (n = 10) HWW, untreated (n = 9) and treated (n = 8) UWW and untreated (n = 10) and treated (n = 8) MWW (at the experimental ratio 1:2 HWW:UWW) samples averaged over the numbers of samples collected for each sample group in the given time interval (January 2015–November 2015) +- standard deviation.
3.5. Resistome before and after WW treatment
The absolute copy numbers of 16S rRNA genes per liter of water for all WW sources decreased by 2–3 log after WW treatment (Supplementary Fig. 5) indicating that secondary WW treatment did reduce total bacterial loads in similar orders of magnitude in the hospital and urban WW treatment plant. To estimate the impact of WW treatment on the resistome, fold changes of normalized cumulative abundance of untreated over treated H, U and M WW were calculated. The normalized cumulative abundance of all gene classes significantly decreased and was between 78 times (for genes conferring resistance to quinolones) and 5 times (for genes conferring resistance to QACs, sulphonamides and genes encoding transposase genes) lower in the treated HWW compared to untreated HWW (p < 0.003) (Table 2; Fig. 4).When comparing untreated UWW to treated UWW, we showed a significant reduction (p < 0.05) in the normalized cumulative abundance for nine resistance gene classes with fold changes between 43 (for the streptogramin resistance gene vatB) and three (for genes encoding resistance to aminoglycosides) times (Table 2, Fig. 4). No significant reduction in normalized abundance was observed after treatment of UWW for gene classes conferring resistance to bacitracin, beta-lactams, quinolones, heavy metals and quaternary ammonium compounds, and for genes encoding integron integrases. Surprisingly, sulphonamide resistance encoding genes were found to be significantly enriched in normalized abundance after UWW treatment (p < 0.05) (Table 2, Fig. 4). For MWW, a similar removal efficacy as for UWW could be observed (Table 2, Fig. 4b), with significant decrease for the normalized cumulative abundance of 10 gene classes and with fold changes between 41 (for the streptogramin resistance gene vatB) and three (for the genes encoding integron integrase genes) times. No significant decrease for gene classes conferring resistance to bacitracin, beta-lactams, tetracycline, heavy metals, QACs and sulphonamides could be detected.
Table 2.
Average fold changes for gene classes of untreated WW over treated WW for H, U and M WW (p values ≤ 0.03) ± Standard Deviation. * = significantly lower; + = significantly higher. Significant differences were calculated by comparing the normalized cumulative abundance values of individual gene classes for all samples belonging to each sample group using the non-parametric Mann-Whitney test. Fold changes were calculated for individually paired samples for each gene class/sample group. Average fold change ± Standard Deviation are depicted in the table for comparison. NA indicates that gene classes were undetectable in either one or both of the sample groups.
| Gene classes conferring resistance to: | Fold change H untreated WW/H treated WW | Fold change U untreated WW/U treated WW | Fold change M untreated WW/M treated WW |
|---|---|---|---|
| chloramphenicol | 34 (±47)* | 5 (±12)* | 16 (±27)* |
| aminoglycosides | 15 (±14)* | 3 (±5) * | 7 (±10)* |
| bacitracin | 12 (±30)* | 1 (±2) | 1 (±1) |
| beta-lactams | 30 (±42) * | 4 (±14) | 10 (±21) |
| erythromycin (macrolides) | 48 (±68)* | 27 (±55)* | 23 (+17)* |
| (multi-drug) efflux | 29 (±43)* | 9 (±27)* | 13 (±17)* |
| quinolones | 78 (±123)* | 3 (±5) | 19 (±29)* |
| heavy metals | 15 (±32) * | 3 (±3) | 2 (±2) |
| quaternary ammonium compounds(QACs) | 5 (±5) * | 1 (±2) | 2 (±2) |
| vancomycin | NA (undetectable in all but one sample for treated HWW) | NA (undetectable in all but one sample for treated UWW) | NA (undetectable in all samples for treated MWW) |
| tetracycline | 51 (±77)* | 13 (±40)* | 6 (±5) |
| polymixin | 29 (±52)* | 5 (±12) * | 7 (±7)* |
| sulphonamides | 5 (±6)* | 1 (±1)+ | 2 (±1) |
| Streptogramin | 17 (±43)* | 43 (±77)* | 41 (±20)* |
| methicillin | NA (undetectable in all treated HWW samples) | NA (undetectable in all treated UWW samples) | NA (undetectable in all but one treated MWW sample) |
| trimethoprim | 66 (±101)* | 9 (±28)* | 6 (±7)* |
| Gene classes grouped according to function: | |||
| transposase genes (MGEs) | 5 (±5)* | 6 (±7)* | 5 (±3)* |
| integron integrase genes | 7 (±6)* | 1 (±1) | 3 (±4)* |
3.6. Impact of treated hospital WW on the receiving river
We also analyzed 12 river samples up and downstream the WWTPs to evaluate putatively associated risks with the release of the treated WW into the effluent receiving river and downstream environment (Supplementary Fig. 6). The resistome for the river samples collected for the sites up (Ri) and downstream (Ri1 and Ri2) of the effluent release pipes during winter season 2013 was not significantly different for either of the three sampling sites (Supplementary Fig. 6). There was no significant difference of the normalized abundance for any of the detected gene classes in the river samples compared to treated UWW. On the contrary, the normalized abundance of nine resistance gene classes including genes encoding MGEs and integron integrase genes was significantly lower (p < 0.04) in all river samples when compared to treated HWW (Supplementary Fig. 6).
3.7. Correlation analysis between the eco-exposome and the microbiome and resistome of hospital and urban WW
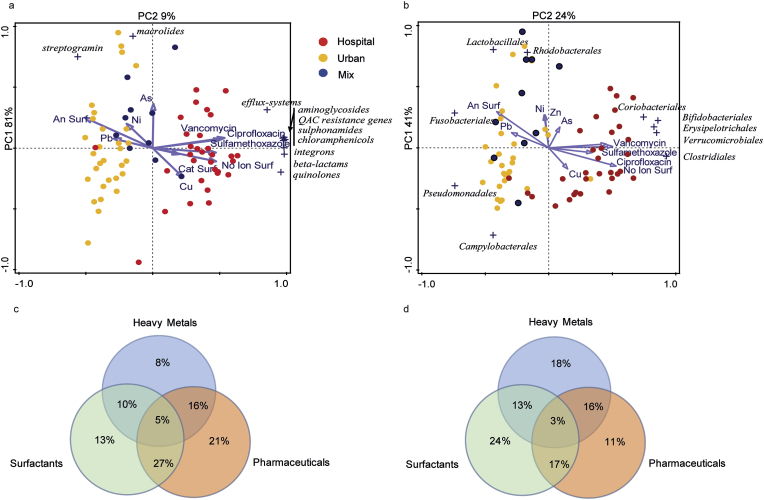
In order to estimate the eco-exposome in the WW, selected pharmaceutical and chemical compounds including antibiotics, surfactants and heavy metals were quantified in untreated WW (Supplementary Table 6). The association between the resistome/microbiota and the eco-exposome was visualized by means of Principal Component Analysis (PCA) biplots. Here, HWW and UWW form two distinct clusters, while the MWW clusters closely to the UWW for both resistome and microbiota (Fig. 5a and b). Antibiotics, non-ionic and cationic surfactants are closely associated with the respective HWW resistome and microbiota (length of arrows), while anionic surfactants and some metals are more associated with the resistome and microbiota of the UWW (Fig. 5a and b). Redundancy analysis (RDA) was performed to statistically asses the association of the monitored eco-exposome with the variation of the resistome and microbiota. The results of the Monte-Carlo permutations indicated that the eco-exposome significantly influences the resistome and microbiota (p-values 0.002).
Fig. 5.
Principal component analysis showing the relationship between the eco-exposome (heavy metals, pharmaceuticals and surfactants) and the resistome (a) and microbiota (b) of untreated HWW, UWW and MWW samples; and Venn diagrams showing the results of the variation partitioning analysis with the different measured chemical classes and the resistome (c) and microbiota (d). In the PCA analysis, dots refer to urban (yellow), hospital (red) and mixed (blue) untreated WW samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Finally, a variation partitioning analysis was performed to study which group of the measured compounds (heavy-metals, pharmaceuticals, surfactants) might have a larger contribution on the resistome and microbiota variation, and to explore whether the interaction between these compounds has a stronger influence than the individually grouped compounds. Pharmaceuticals (the antibiotics ciprofloxacin, sulfamethoxazole and vancomycin, and the neurological drug carbamazepine) are more associated with the variance for the resistome, while the surfactants are associated with the variation for the microbiota (Fig. 5c and d). Finally, we showed that the interaction between pharmaceuticals and surfactants contributes more to the variability in the resistome than the individual compounds alone (Fig. 5c and d). We also collected data on the consumption of three antibiotics, ciprofloxacin, sulfamethoxazole and vancomycin, by the hospital pharmacy over the period of 2012–2014, that were summarized here as gram per season (Supplementary Table 7 and Supplementary Fig. 3c). The studied hospital site has just been opened in the winter month of February 2012 which is probably why antibiotic consumption by the hospital pharmacy was low during winter 2012. No obvious correlation between summer peaks for the measured antibiotics in WW and their respective consumption by the hospital pharmacy was shown. (Supplementary Table 7 and Supplementary Table 8, and Supplementary Figs. 3b and 3c).
4. Discussion
In the context of globally increasing AMR, wastewaters (WW) have been identified as sources for the spread of AMR determinants (ARB, ARGs, MGEs) and chemical pollutants (often pharmaceutical residues) that may favor AMR selection during wastewater treatment and in the receiving environment. In this study we thoroughly monitored the resistome and microbiota dynamics of untreated and treated (applying conventional secondary WWT) hospital and urban WW over four years throughout the seasons in France. We identified distinct and robust resistome and microbiota signatures, in particular for untreated HWW and UWW, indicating that HWW and UWW form distinct and stable ecological niches over time. Performing machine learning (ML) classified each of the WW with high accuracy and revealed top predictive genes, gene classes and taxa for the respective WW sources before and after WW treatment (Supplementary Fig. 8). Interestingly, when collapsing data-sets obtained from untreated and treated samples of the respective WW sources, ML was able to predict HWW and UWW in general with more than 93% certainty on all predictor levels (individual genes, gene classes, taxa) (Supplementary Fig. 9). These top 10 predictors for HWW and UWW provide considerable marker gene classes, individual genes and taxa for the respective WW sources that present targets for the monitoring and management of HWW and UWW. For example, the streptogramin resistance gene vatB, and the transposase gene ISS1N were significantly more abundant in UWW compared to HWW, and seem to be specifically indicative for UWW (Supplementary Figs. 8 and 9).
Based on the ML and multi variate analysis, macrolide resistance genes also contribute to the specific resistome signature for UWW (Supplementary Figs. 8 and 9, Fig. 5a). Considering the fact that macrolide and streptogramin antibiotics are more frequently prescribed in the community compared to the hospital environment in France could explain the high abundance of these gene classes in UWW (Robert et al., 2012). The antibiotics ciprofloxacin, sulfamethoxazole and vancomycin were detected in higher concentrations in HWW compared to UWW, which may point towards a relationship of the measured antibiotics and the detected resistome in HWW (Fig. 5a and c). Interestingly, the qnr genes encoding quinolone resistance in HWW were the ones with the highest fold increase (161-fold) between HWW and UWW. As these genes are located on plasmids, their higher abundance in HWW indirectly reflects the likely abundance of bacteria harboring genetic elements involved in resistance dissemination as plasmids in HWW. Indeed, qnr genes have been described mainly in Enterobacteriales (Carattoli, 2009; Rozwandowicz et al., 2018) and we found that the relative abundance of Enterobacteriales is higher in HWW than in UWW (Fig. 3). The human gut microbiota is an important reservoir for ARGs (Ruppe et al., 2018; Sommer et al., 2010) and recently evidence-based data showed that the occurrence and abundance of (human) fecal pollution correlates with high amounts of ARGs in anthropogenically impacted environments (Karkman et al., 2019; Su et al., 2017). The significant dilution of human gut bacteria in MWW observed here, even with an increased proportional contribution of HWW to UWW (HWW ∼33.4%) compared to other study sites (Buelow et al., 2018; Pallares-Vega et al., 2019; Stalder et al., 2014), is hence likely to explain the significant reduction of the abundance of gene classes after mixing HWW with UWW.
With respect to the reduction of normalized abundance of ARGs and MGEs through secondary UWW treatment, we observed that the normalized cumulative abundance of six gene classes did not significantly decrease while one gene class (sulphonamides) did significantly increase (Table 2, Fig. 4). This may contribute to the dissemination of AMR into the environment as discussed previously (Di Cesare et al., 2016; Karkman et al., 2016; Lee et al., 2017; Pallares-Vega et al., 2019). In addition, the resistome monitored here, exhibits a high mobilization potential due to the high proportion of MGEs, integrons and ARGs that are usually located on plasmids, detected in all WW samples, as well as in the receiving river waters (Fig. 1, MGEs and integrons account for up to 60% of the resistome of treated effluents and river waters). However, specific IS transposase genes are differentially abundant in both WW types (Supplementary Fig. 10). For example, ISS1N, a member of the IS6 family, common in Gram-positive lactic acid bacteria (Haandrikman et al., 1990) is more abundant in UWW (here, identified as a specific UWW marker, Supplementary Figs. 8 and 9), whereas IS26, and Tn3 are more abundant in HWW. These IS are commonly found in Gram-negative bacteria in association with ARGs (Harmer et al., 2014; Nicolas et al., 2015). In order to improve WWT with respect to ARG and ARB removal, advanced WW treatment such as disinfection by UV radiation or ozone treatment, or physical treatment by ultrafiltration of WW, are reported to be more efficient in reducing ARB and ARGs compared to conventionally applied secondary WW treatment (Gouliouris et al., 2019; Jäger et al., 2018).
The exposome, a term originally coined in the context of human health epidemiology and referring to “the totality of human environmental exposures”(Wild, 2005), here specifically refers to the chemical compounds quantified in our longitudinal study that represent partially the environmental or “eco-exposome” (Escher et al., 2017) of the WW microbiota. For our study we focused on surfactants, antibiotics and heavy metals. Anthropogenic pollution through micro pollutants present in WW has previously shown to have a negative impact on the environment (Cairns et al., 2017; Flandroy et al., 2018; Hendry et al., 2017; Palmer and Hatley, 2018; Pereira and Tagkopoulos, 2019). For example, high concentrations of antibiotics and cationic surfactants found in HWW (Rodriguez-Mozaz et al., 2015; Szekeres et al., 2017; Varela et al., 2014), and pharmaceutical production sites (Bengtsson-Palme et al., 2018; Bengtsson-Palme and Larsson, 2016) have been correlated with a higher abundance of ARBs, ARGs, as well as higher abundance of MGEs and integrons (Rodriguez-Mozaz et al., 2015; Rowe et al., 2017; Stalder et al., 2014), whereas anionic surfactants that are generally abundant in urban WW effluents (untreated and treated UWWs; “grey waters”) are associated with toxicity to aquatic and terrestrial environments (Jardak et al., 2016; Shafran et al., 2005; Siggins et al., 2016). Our study reports similar findings. We observed that cationic surfactants and antibiotics are more abundant in HWW (Supplementary Table 6) and significantly correlated to the HWW resistome (Fig. 5), which reflects the frequent use of antibiotics and active-surface agents such as quaternary ammonium compounds in hospitals. Interestingly, we also found that qac genes were significantly higher in HWW compared to UWW. The urban WW eco-exposome on the other hand was found to be enriched with anionic surfactants (Supplementary Table 6) which in turn were associated with the UWW and MWW resistome and microbiota (Fig. 5). Other parameters present in the eco-exposome not measured here might also have an additional influence on the microbiota and resistome of WWs. For example, streptogramin and macrolide antibiotics were not quantified in this study to investigate their putative association with the elevated presence of macrolide and streptogramin resistance genes in UWW. Overall, measures to reduce the load of micro-pollutants in WWs and the environment as a measure to reduce AMR dissemination are currently discussed and warrant further attention (Kraemer et al., 2019; Pereira and Tagkopoulos, 2019; Singer et al., 2016). Finally, the observed peaks and variation during summer season for detected antibiotics, individual resistance genes and gene classes in HWW (Supplementary Fig. 3a and 3b and 4) may be due to dry season and warm temperatures during summer that could result in decreased flow rate of HWW. Interestingly, recently a potential association of rising temperatures with the globally increasing AMR burden has been reported (Kaba et al., 2020; MacFadden et al., 2018).
Our findings give further emphasis to the requirement of implementing and optimizing sanitation systems and operational WWTPs on a global level, particular in countries/continents with poor water sanitation infrastructure and correlated high occurrence of multi-resistant bacteria (Burgmann et al., 2018). The elimination of pollution by means of human feces, and bacterial taxa associated, through advanced or selective WW treatment, may further aid in limiting the release of ARGs associated with human gut bacteria and pathogens (Gouliouris et al., 2019; Karkman et al., 2019; Pehrsson et al., 2016). Finally, the data generated by this study are of important interest to policy makers concerning the risks associated with H and U WW, their putative implication into the dissemination of AMR, and provide further evidence towards the necessity of environmental pollution management in the battle of AMR and other important global health factors such as the preservation of biodiversity and the prevention of climate change (Hendry et al., 2017).
5. Conclusion
-
•
Hospital and urban wastewater resistome and microbiota are remarkably stable over time and display unique signatures.
-
•
The differences between H and U WW signatures are in line with the differences in antibiotic exposure in both settings, and may in addition be shaped by non-antibiotic substances in the monitored eco-exposome.
-
•
We confirm that Hospital WW contains significantly higher loads of ARGs, MGEs and integrons, and demonstrate that it is significantly enriched with a human gut associated microbiota compared to urban WW.
-
•
We suggest that WW mixing of H and U WW (1:2) bares no greater risk than separate treatment in order to lower the emissions of ARGs, MGEs and ARB by HWW.
-
•
Advanced removal of micro-pollutants (such as heavy metals, biocides, surfactants and antibiotics) is crucial to reduce the environmental risk posed by the WW eco-exposome and its putative impact on AMR.
Author contributions
C.D., M-C.P., and E.B. designed the study, C.D. provided access to the SIPIBEL data collection, M.G. and E.B. performed experiments, E.B., A.R., J.L. and S.P.K. performed data analysis, L.W. performed chemical analysis (SIPIBEL project), E.B., M-C.P. and C.D. wrote the manuscript with contribution of all other co-authors.
Data availability
16S rRNA sequence data are available at the European Nucleotide Archive (ENA) under the accession number PRJEB29948. All other important raw data needed to reconstruct the findings of our study are made available in the supplementary material.
Funding
E. Buelow has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement RESOLVE 707999-standard EF.
A. Rico is supported by a postdoctoral grant provided by the Spanish Ministry of Science, Innovation and University (IJCI-2017-33,465).
J. Lourenço is supported by a Lectureship from the Department of Zoology, University of Oxford.
16S rRNA sequence Data collection and analyses performed by the IBEST Genomics Resources Core at the University of Idaho were supported in part by NIH COBRE grant P30GM103324.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the SIPIBEL Consortium and the SIPIBEL field observatory on the hospital’s effluents and urban wastewater treatment plants for displaying data and measurements.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.wroa.2020.100045.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Allen H.K., Donato J., Wang H.H., Cloud-Hansen K.A., Davies J., Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- Barraud O., Baclet M.C., Denis F., Ploy M.C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010;65:1642–1645. doi: 10.1093/jac/dkq167. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J., Kristiansson E., Larsson D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018;42 doi: 10.1093/femsre/fux053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J., Larsson D.G. Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 2016;86:140–149. doi: 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57(1):289–300. [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Buelow E., Bayjanov J.R., Majoor E., Willems R.J.L., Bonten M.J.M., Schmitt H., van Schaik W. Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol. 2018;94(7) doi: 10.1093/femsec/fiy087. Ecol. [DOI] [PubMed] [Google Scholar]
- Buelow E., Bello Gonzalez T.D.J., Fuentes S., de Steenhuijsen Piters W.A.A., Lahti L., Bayjanov J.R., Majoor E.A.M., Braat J.C., van Mourik M.S.M., Oostdijk E.A.N., Willems R.J.L., Bonten M.J.M., van Passel M.W.J., Smidt H., van Schaik W. Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome. 2017;5 doi: 10.1186/s40168-017-0309-z. 88-8017-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgmann H., Frigon D., H Gaze W., M Manaia C., Pruden A., Singer A.C., F Smets B., Zhang T. Water and sanitation: an essential battlefront in the war on antimicrobial resistance. FEMS Microbiol. Ecol. 2018;94 doi: 10.1093/femsec/fiy101. [DOI] [PubMed] [Google Scholar]
- Cairns J., Becks L., Jalasvuori M., Hiltunen T. Sublethal streptomycin concentrations and lytic bacteriophage together promote resistance evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R. Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2009;15(Suppl. 1):20–25. doi: 10.1111/j.1469-0691.2008.02679.x. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Resistance plasmid families in enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonova T., Keck F., Labanowski J., Montuelle B., Rimet F., Bouchez A. Separate treatment of hospital and urban wastewaters: a real scale comparison of effluents and their effect on microbial communities. Sci. Total Environ. 2016;542:965–975. doi: 10.1016/j.scitotenv.2015.10.161. [DOI] [PubMed] [Google Scholar]
- Chonova T., Lecomte V., Bertrand-Krajewski J.L., Bouchez A., Labanowski J., Dagot C., Levi Y., Perrodin Y., Wiest L., Gonzalez-Ospina A., Cournoyer B., Sebastian C. The SIPIBEL project: treatment of hospital and urban wastewater in a conventional urban wastewater treatment plant. Environ. Sci. Pollut. Res. Int. 2018;25:9197–9206. doi: 10.1007/s11356-017-9302-0. [DOI] [PubMed] [Google Scholar]
- Costea P.I., Hildebrand F., Arumugam M., Backhed F., Blaser M.J., Bushman F.D., de Vos W.M., Ehrlich S.D., Fraser C.M., Hattori M., Huttenhower C., Jeffery I.B., Knights D., Lewis J.D., Ley R.E., Ochman H., O’Toole P.W., Quince C., Relman D.A., Shanahan F., Sunagawa S., Wang J., Weinstock G.M., Wu G.D., Zeller G., Zhao L., Raes J., Knight R., Bork P. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2018;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cesare A., Eckert E.M., D’Urso S., Bertoni R., Gillan D.C., Wattiez R., Corno G. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 2016;94:208–214. doi: 10.1016/j.watres.2016.02.049. [DOI] [PubMed] [Google Scholar]
- Escher B.I., Hackermüller J., Polte T., Scholz S., Aigner A., Altenburger R., Böhme A., Bopp S.K., Brack W., Busch W., Chadeau-Hyam M., Covaci A., Eisenträger A., Galligan J.J., Garcia-Reyero N., Hartung T., Hein M., Herberth G., Jahnke A., Kleinjans J., Klüver N., Krauss M., Lamoree M., Lehmann I., Luckenbach T., Miller G.W., Müller A., Phillips D.H., Reemtsma T., Rolle-Kampczyk U., Schüürmann G., Schwikowski B., Tan Y.-M., Trump S., Walter-Rohde S., Wambaugh J.F. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ. Int. 2017;99:97–106. doi: 10.1016/j.envint.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandroy L., Poutahidis T., Berg G., Clarke G., Dao M.C., Decaestecker E., Furman E., Haahtela T., Massart S., Plovier H., Sanz Y., Rook G. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci. Total Environ. 2018;627:1018–1038. doi: 10.1016/j.scitotenv.2018.01.288. [DOI] [PubMed] [Google Scholar]
- Forslund K., Sunagawa S., Kultima J.R., Mende D.R., Arumugam M., Typas A., Bork P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–1169. doi: 10.1101/gr.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M.R., Gaze W.H., Pruden A., Smalla K., Tiedje J.M., Zhu Y.G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015;9:1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadhas D.P., Marathe S.A., Chakravortty D. Biocides--resistance, cross-resistance mechanisms and assessment. Expet Opin. Invest. Drugs. 2013;22:191–206. doi: 10.1517/13543784.2013.748035. [DOI] [PubMed] [Google Scholar]
- Gouliouris T., Raven K.E., Moradigaravand D., Ludden C., Coll F., Blane B., Naydenova P., Horner C., Brown N.M., Corander J., Limmathurotsakul D., Parkhill J., Peacock S.J. Detection of vancomycin-resistant Enterococcus faecium hospital-adapted lineages in municipal wastewater treatment plants indicates widespread distribution and release into the environment. Genome Res. 2019;29:1–9. doi: 10.1101/gr.232629.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandrikman A.J., van Leeuwen C., Kok J., Vos P., de Vos W.M., Venema G. Insertion elements on lactococcal proteinase plasmids. Appl. Environ. Microbiol. 1990;56:1890–1896. doi: 10.1128/aem.56.6.1890-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer C.J., Moran R.A., Hall R.M. 2014. Movement of IS26-Associated Antibiotic Resistance Genes Occurs via a Translocatable Unit that Includes a Single IS26 and Preferentially Inserts Adjacent to Another IS26. mBio 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. Springer Series in Statistics. second ed., second ed. Springer-Verlag; New York: 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- Hendry A.P., Gotanda K.M., Svensson E.I. Human influences on evolution, and the ecological and societal consequences. Philos. Trans. R. Soc. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yang X., Qin J., Lu N., Cheng G., Wu N., Pan Y., Li J., Zhu L., Wang X., Meng Z., Zhao F., Liu D., Ma J., Qin N., Xiang C., Xiao Y., Li L., Yang H., Wang J., Yang R., Gao G.F., Wang J., Zhu B. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013;4:2151. doi: 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- Jäger T., Hembach N., Elpers C., Wieland A., Alexander J., Hiller C., Krauter G., Schwartz T. Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment, and the disseminated loads released to the environment. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardak K., Drogui P., Daghrir R. Surfactants in aquatic and terrestrial environment: occurrence, behavior, and treatment processes. Environ. Sci. Pollut. Res. Int. 2016;23:3195–3216. doi: 10.1007/s11356-015-5803-x. [DOI] [PubMed] [Google Scholar]
- Jin L., Wang R., Wang X., Wang Q., Zhang Y., Yin Y., Wang H. Emergence of mcr-1 and carbapenemase genes in hospital sewage water in Beijing, China. J. Antimicrob. Chemother. 2018;73:84–87. doi: 10.1093/jac/dkx355. [DOI] [PubMed] [Google Scholar]
- Kaba H.E.J., Kuhlmann E., Scheithauer S. Thinking outside the box: association of antimicrobial resistance with climate warming in Europe – a 30 country observational study. Int. J. Hyg Environ. Health. 2020;223:151–158. doi: 10.1016/j.ijheh.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Karkman A., Johnson T.A., Lyra C., Stedtfeld R.D., Tamminen M., Tiedje J.M., Virta M. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2016;92 doi: 10.1093/femsec/fiw014. Epub 2016 Jan 31. [DOI] [PubMed] [Google Scholar]
- Karkman A., Parnanen K., Larsson D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019;10 doi: 10.1038/s41467-018-07992-3. 80-8018-07992–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer S.A., Ramachandran A., Perron G.G. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7 doi: 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson D.G.J., Andremont A., Bengtsson-Palme J., Brandt K.K., de Roda Husman A.M., Fagerstedt P., Fick J., Flach C.F., Gaze W.H., Kuroda M., Kvint K., Laxminarayan R., Manaia C.M., Nielsen K.M., Plant L., Ploy M.C., Segovia C., Simonet P., Smalla K., Snape J., Topp E., van Hengel A.J., Verner-Jeffreys D.W., Virta M.P.J., Wellington E.M., Wernersson A.S. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018;117:132–138. doi: 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- Lee J., Jeon J.H., Shin J., Jang H.M., Kim S., Song M.S., Kim Y.M. Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci. Total Environ. 2017:605–606. doi: 10.1016/j.scitotenv.2017.06.250. 906–914. [DOI] [PubMed] [Google Scholar]
- Lienert J., Koller M., Konrad J., McArdell C.S., Schuwirth N. Multiple-criteria decision analysis reveals high stakeholder preference to remove pharmaceuticals from hospital wastewater. Environ. Sci. Technol. 2011;45:3848–3857. doi: 10.1021/es1031294. [DOI] [PubMed] [Google Scholar]
- Lourenco J., Watkins E.R., Obolski U., Peacock S.J., Morris C., Maiden M.C.J., Gupta S. Lineage structure of Streptococcus pneumoniae may be driven by immune selection on the groEL heat-shock protein. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-08990-z. 9023-017-08990-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadden D.R., McGough S.F., Fisman D., Santillana M., Brownstein J.S. Antibiotic resistance increases with local temperature. Nat. Clim. Change. 2018;8:510–514. doi: 10.1038/s41558-018-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinforma. Oxf. Engl. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaia C.M., Rocha J., Scaccia N., Marano R., Radu E., Biancullo F., Cerqueira F., Fortunato G., Iakovides I.C., Zammit I., Kampouris I., Vaz-Moreira I., Nunes O.C. Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ. Int. 2018;115:312–324. doi: 10.1016/j.envint.2018.03.044. [DOI] [PubMed] [Google Scholar]
- Nicolas E., Lambin M., Dandoy D., Galloy C., Nguyen N., Oger C.A., Hallet B. The tn3-family of replicative transposons. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D.G. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015;16 doi: 10.1186/s12864-015-2153-5. 964-015-2153–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares-Vega R., Blaak H., van der Plaats R., de Roda Husman A.M., Leal L.H., van Loosdrecht M.C.M., Weissbrodt D.G., Schmitt H. Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: a cross-sectional study. Water Res. 2019;161:319–328. doi: 10.1016/j.watres.2019.05.100. [DOI] [PubMed] [Google Scholar]
- Palmer M., Hatley H. The role of surfactants in wastewater treatment: impact, removal and future techniques: a critical review. Water Res. 2018;147:60–72. doi: 10.1016/j.watres.2018.09.039. [DOI] [PubMed] [Google Scholar]
- Pehrsson E.C., Tsukayama P., Patel S., Mejia-Bautista M., Sosa-Soto G., Navarrete K.M., Calderon M., Cabrera L., Hoyos-Arango W., Bertoli M.T., Berg D.E., Gilman R.H., Dantas G. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B.M.P., Tagkopoulos I. Benzalkonium chlorides: uses, regulatory status, and microbial resistance. Appl. Environ. Microbiol. AEM. 2019;85(13) doi: 10.1128/AEM.00377-19. 00377-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raheem A., Sikarwar V.S., He J., Dastyar W., Dionysiou D.D., Wang W., Zhao M. Opportunities and challenges in sustainable treatment and resource reuse of sewage sludge: a review. Chem. Eng. J. 2018;337:616–641. [Google Scholar]
- Rajilic-Stojanovic M., de Vos W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J., Pean Y., Varon E., Bru J.P., Bedos J.P., Bertrand X., Lepape A., Stahl J.P., Gauzit R. Societe de pathologie infectieuse de langue francaise (SPILF), Observatoire national de l’epidemiologie de la resistance bacterienne aux antibiotiques (ONERBA), Surveillance de la prescription des antibiotiques (SPA) Group. Point prevalence survey of antibiotic use in French hospitals in 2009. J. Antimicrob. Chemother. 2012;67:1020–1026. doi: 10.1093/jac/dkr571. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S., Chamorro S., Marti E., Huerta B., Gros M., Sanchez-Melsio A., Borrego C.M., Barcelo D., Balcazar J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015;69:234–242. doi: 10.1016/j.watres.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Rowe W.P.M., Baker-Austin C., Verner-Jeffreys D.W., Ryan J.J., Micallef C., Maskell D.J., Pearce G.P. Overexpression of antibiotic resistance genes in hospital effluents over time. J. Antimicrob. Chemother. 2017;72:1617–1623. doi: 10.1093/jac/dkx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- Ruppe E., Ghozlane A., Tap J., Pons N., Alvarez A.S., Maziers N., Cuesta T., Hernando-Amado S., Clares I., Martinez J.L., Coque T.M., Baquero F., Lanza V.F., Maiz L., Goulenok T., de Lastours V., Amor N., Fantin B., Wieder I., Andremont A., van Schaik W., Rogers M., Zhang X., Willems R.J.L., de Brevern A.G., Batto J.M., Blottiere H.M., Leonard P., Lejard V., Letur A., Levenez F., Weiszer K., Haimet F., Dore J., Kennedy S.P., Ehrlich S.D. Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat. Microbiol. 2018;4:112–123. doi: 10.1038/s41564-018-0292-6. [DOI] [PubMed] [Google Scholar]
- Shafran A.W., Gross A., Ronen Z., Weisbrod N., Adar E. Effects of surfactants originating from reuse of greywater on capillary rise in the soil. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2005;52:157–166. [PubMed] [Google Scholar]
- Siggins A., Burton V., Ross C., Lowe H., Horswell J. Effects of long-term greywater disposal on soil: a case study. Sci. Total Environ. 2016;557–558:627–635. doi: 10.1016/j.scitotenv.2016.03.084. [DOI] [PubMed] [Google Scholar]
- Singer A.C., Shaw H., Rhodes V., Hart A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M.O., Church G.M., Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence. 2010;1:299–303. doi: 10.4161/viru.1.4.12010. [DOI] [PubMed] [Google Scholar]
- Stalder T., Barraud O., Jove T., Casellas M., Gaschet M., Dagot C., Ploy M.C. Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J. 2014;8:768–777. doi: 10.1038/ismej.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J.Q., An X.L., Li B., Chen Q.L., Gillings M.R., Chen H., Zhang T., Zhu Y.G. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome. 2017;5 doi: 10.1186/s40168-017-0298-y. 84-8017-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres E., Baricz A., Chiriac C.M., Farkas A., Opris O., Soran M.L., Andrei A.S., Rudi K., Balcazar J.L., Dragos N., Coman C. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. Barking Essex 1987. 2017;225:304–315. doi: 10.1016/j.envpol.2017.01.054. [DOI] [PubMed] [Google Scholar]
- Ter Braak C.J.F., Smilauer P. Microcomputer Power; Ithaca USA: 2012. Canoco Reference Manual and User’s Guide: Software for Ordination. [Google Scholar]
- Varela A.R., Andre S., Nunes O.C., Manaia C.M. Insights into the relationship between antimicrobial residues and bacterial populations in a hospital-urban wastewater treatment plant system. Water Res. 2014;54:327–336. doi: 10.1016/j.watres.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang P., Yang Q. Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci. Total Environ. 2018;621:990–999. doi: 10.1016/j.scitotenv.2017.10.128. [DOI] [PubMed] [Google Scholar]
- Wiest L., Chonova T., Berge A., Baudot R., Bessueille-Barbier F., Ayouni-Derouiche L., Vulliet E. Two-year survey of specific hospital wastewater treatment and its impact on pharmaceutical discharges. Environ. Sci. Pollut. Res. Int. 2018;25:9207–9218. doi: 10.1007/s11356-017-9662-5. [DOI] [PubMed] [Google Scholar]
- Wild C.P. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Prev. Biomark. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Zhu Y.G., Johnson T.A., Su J.Q., Qiao M., Guo G.X., Stedtfeld R.D., Hashsham S.A., Tiedje J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.G., Zhao Y., Li B., Huang C.L., Zhang S.Y., Yu S., Chen Y.S., Zhang T., Gillings M.R., Su J.Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017;2:16270. doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA sequence data are available at the European Nucleotide Archive (ENA) under the accession number PRJEB29948. All other important raw data needed to reconstruct the findings of our study are made available in the supplementary material.