Abstract
Circular RNAs (circRNAs) are broad and diverse endogenous non-coding RNAs. Emerging evidence has revealed that circRNAs play pivotal roles in cancers, regulating the gene expression by acting as a microRNA (miRNA) sponge. However, the biological functions of circRNAs in bladder cancer (BCa) remain largely unknown. In this study, we identified an altered circRNA, termed circFUT8, by screening RNA sequencing data generated from three BCa tissues and matched adjacent normal bladder tissues. Quantitative real-time PCR analysis demonstrated that circFUT8 was downregulated in BCa tissues and correlated with patients’ prognosis, histological grade, and lymph node (LN) metastasis. Functionally, gain- and loss-of-function assays indicated that circFUT8 inhibited the migration and invasion of BCa cell lines in vitro and LN metastasis in vivo. Mechanistically, circFUT8 directly bound to miR-570-3p and partially abrogated its oncogenic role, and miR-570-3p could suppress the expression of tumor suppressor gene Krüpple-like factor 10 (KLF10) by binding its 3′ untranslated region (3′ UTR). Moreover, we found that circFUT8 promoted the expression of KLF10 by competitively sponging miR-570-3p. In conclusion, circFUT8 functions as a tumor suppressor in BCa cells by targeting the miR-570-3p/KLF10 axis and may serve as a potential biomarker and therapeutic target for the management of BCa patients with LN metastasis.
Keywords: circFUT8, bladder cancer, lymph node metastasis, miR-570-3p, KLF10, Slug
Introduction
Bladder cancer (BCa) is one of the most frequently diagnosed cancers worldwide and the most lethal malignancy in the urinary system,1,2 with 437,442 newly diagnosed cases in 2016.3 Approximately 20% to 30% newly diagnosed BCa cases are classified into a muscle-invasive subtype or tend to be metastatic,4 and the 5-year survival rate is only 5% with metastatic BCa.5 Unfortunately, no effective therapies are applicable in clinic for BCa patients with metastasis. Lymph node (LN) metastasis is a crucial route of metastasis and a significantly prognostic factor for BCa.6,7 Thus, a better understanding of molecular mechanisms about LN metastasis in BCa is critical for developing some efficient treatment strategies.
Circular RNAs (circRNAs) are identified as a new class of non-coding RNAs, which are abundant and involved in multiple biological functions in eukaryotic organisms.8 circRNAs possess a covalently closed-loop structure generated by alternative splicing and back-splicing of pre-mRNA transcripts,9 which renders a stronger resistance to exonucleases and consequent stability. Initially, circRNAs were considered as non-functional junk RNAs produced by abnormal RNA splicing.10 With the development of next-generation sequencing, increasingly more circRNAs have been discovered in eukaryotic cells, which indicates that circRNAs have regulatory potency rather than by-products of RNA splicing.11
MicroRNAs (miRNAs) play an important role in post-transcriptional regulation of gene expression by direct base pairing to target mRNA in untranslated regions (UTRs). The miRNA sponging has been discovered as a classic function of circRNAs, which indicates that circRNAs serve as competitive endogenous RNAs (ceRNAs) to regulate mRNA translation by binding to miRNAs.12 Recent studies have uncovered that circRNAs are abnormally expressed in human tumors and correlated with tumor progression, diagnosis, prognosis, and metastasis assessment.13, 14, 15 For instance, the famous circRNA ciRS-7 induces colorectal cancer cells into a more aggressive phenotype by the inhibition of miR-7 and subsequent activation of EGFR and RAF1, providing a novel biomarker in prognosis assessment.13 Moreover, it has been certified that circYAP1 is a sponge of miR-367-5p to regulate the proliferation and invasion of gastric cancer cells14 and that circMTO1 serves as a sponge of miRNA-9 to suppress hepatocellular carcinoma progression.15 Despite several studies revealing that circRNAs could regulate the progression of BCa,16, 17, 18 current findings about tumor-metastasis-related circRNAs in BCa remain largely unknown.
In this study, we identified a circRNA, circFUT8 (circBase: hsa_circ_0003028), from published RNA sequencing (GenBank: GEO97239) of BCa tissues and matched adjacent normal bladder tissues and verified the results using our established BCa cell line metastasis model.17 Furthermore, we confirmed that circFUT8 was originated from exon 3 of the FUT8 gene and dramatically decreased in BCa tissues and cell lines. Low expression of circFUT8 in BCa patients was associated with poor prognosis, high histological grade, and LN metastasis. Further mechanism studies demonstrated that circFUT8 inhibited the migration and invasion of BCa cells via sponging miR-570-3p to promote Krüpple-like factor 10 (KLF10) expression.
Results
Validation and Characterization of circFUT8 in BCa Cells
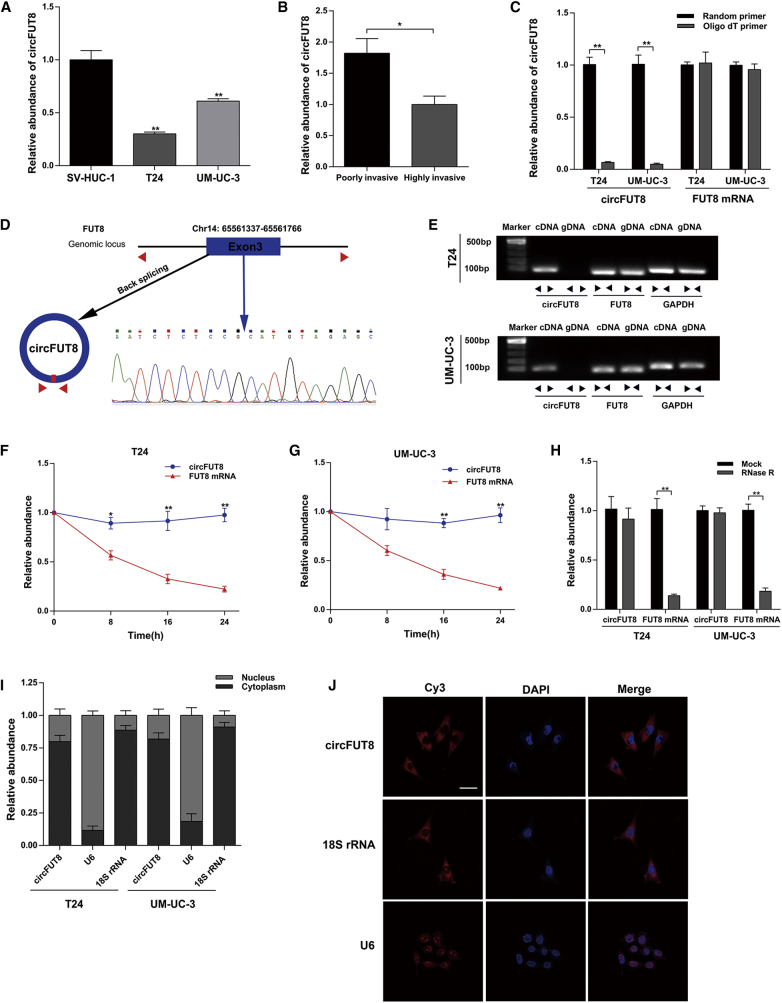
RNA sequencing results of three matched normal and BCa tissues can be downloaded from the GenBank database (GenBank: GEO97239). Based on these data, we screened a significantly downregulated circRNA, circFUT8 (chr14: 65561337-65561766) in BCa. circFUT8 was chosen for further study for the following three reasons: (1) consistent with RNA sequencing results, the abundance of circFUT8 was lower in T24 and UM-UC-3 cell lines compared with that in the normal human urothelial cell line SV-HUC-1 (Figure 1A); (2) circFUT8 was significantly decreased in a highly invasive T24 cell subline, compared with a poorly invasive cell subline (Figure 1B), by using our previously established BCa cell line metastasis model;17 and (3) the further study of circFUT8 is necessary, because its biological functions are rare to our knowledge. The detailed screening processes of circFUT8 are supplied in Figures S1A and S1B.
Figure 1.
The Identification and Abundance of circFUT8 in BCa and the Subcellular Location of circFUT8
(A) The relative abundance of circFUT8 in SV-HUC-1, T24, and UM-UC-3 was detected by quantitative real-time PCR. (B) The relative abundance of circFUT8 in our established poorly and highly invasive T24 cell sublines. (C) The abundance of circFUT8 and FUT8 mRNA was detected by quantitative real-time PCR with reverse transcription products using random primer or oligo(dT) primer. (D) Schematic diagram demonstrating that exon 3 derived from FUT8 formed the circFUT8. The back-splicing junction site of circFUT8 was validated by Sanger sequencing. The blue arrow indicates the special back-splicing junction of circFUT8. (E) The existence of circFUT8 was validated in the T24 and UM-UC-3 cell lines by gel electrophoresis using the products of quantitative real-time PCR. (F and G) The relative abundance of circFUT8 and FUT8 mRNA in the (F) T24 and (G) UM-UC-3 cell lines was detected by quantitative real-time PCR with actinomycin D treatment at the indicated time points. The relative expression of circFUT8 was normalized to the expression of 0 h. (H) The relative abundance of circFUT8 and FUT8 mRNA in T24 and UM-UC-3 cell lines was detected by quantitative real-time PCR with or without RNase R treatment. (I) Nuclear and cytoplasmic RNA fractionation experiment showed that circFUT8 was mainly localized in the cytoplasm, and the abundance of circFUT8 was normalized to the value measured in the cytoplasm. (J) FISH assay of the T24 cell line indicated that circFUT8 was mainly distributed in the cytoplasm. Nuclei were stained blue by DAPI. circFUT8, U6, and 18S rRNA were labeled with Cy3 and stained red. Scale bar, 50 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05, **p < 0.01, Student’s t-test.
In browsing the human reference genome (GRCh37/hg19), the result indicated that circFUT8 was derived from exon 3 of the FUT8 gene. Due to the deficiency of 3′ polyadenylated tail, circFUT8 was almost undetectable by quantitative real-time PCR when reverse-transcription products using oligo(dT) primers compared with random primers, while FUT8 mRNA was not (Figure 1C). Sanger sequencing was conducted, and the result certified the existence of the back-splicing junction site (Figure 1D). We also designed the convergent primers and divergent primers to amplify the linear and circRNA of FUT8 by quantitative real-time PCR, and cDNA and genomic DNA (gDNA) were used as the template. The nucleic acid products of quantitative real-time PCR were validated by 1% agarose gel electrophoresis. As previously expected, circFUT8 was only amplified by divergent primers in cDNA but not in gDNA (Figure 1E). Furthermore, an actinomycin D assay showed that the half-life of the circFUT8 transcript exceeded 24 h, suggesting that the circular form of FUT8 was more stable than the linear form in BCa cell lines (Figures 1F and 1G). In addition, RNA extracts from BCa cells were pretreated with RNase R. Compared with linear FUT8 mRNA, quantitative real-time PCR results showed that the circular form of FUT8 was resistant to RNase R (Figure 1H).
Nuclear and cytoplasmic extraction assays in T24 and UM-UC-3 cell lines indicated that the abundance of circFUT8 was obviously higher in cytoplasm than in nucleus (Figure 1I). The images of fluorescence in situ hybridization (FISH) also showed that the majority of circFUT8 was localized in the cytoplasm of the T24 cell line (Figure 1J). Taken together, the stable circFUT8 was relatively low expressed in BCa cell lines and mainly distributed in cytoplasm.
circFUT8 Is Downregulated in BCa Tissues and Associated with Prognosis, Histological Grade, and LN Metastasis
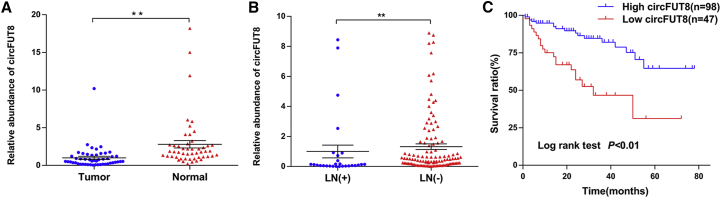
To explore the expression of circFUT8 in BCa, RNAs extracted from paired BCa tissues were used for quantitative real-time PCR. The result indicated that circFUT8 was significantly downregulated in BCa tissues compared with the matched adjacent normal tissues (Figure 2A).
Figure 2.
The Abundance and Clinical Significance of circFUT8 in BCa Patients
(A) Quantitative real-time PCR analysis indicated that the circFUT8 was significantly downregulated in 50 BCa tissues compared with their matched adjacent normal tissues. **p < 0.01, Wilcoxon matched-pairs signed-rank test. (B) The relative abundance of circFUT8 in patients with different LN status. **p < 0.01, Mann-Whitney test. (C) Kaplan-Meier survival curve indicates that patients with low circFUT8 expression had a poor overall survival rate.
Then, 145 BCa tissues with follow-up information were adopted to assess the association of circFUT8 with patients’ prognosis and clinical parameters. First, the expression of circFUT8 was analyzed in patients, and the result indicated that LN metastasis was associated with low circFUT8 (Figure 2B). Besides, we differentiated this cohort into a high-circFUT8 group and a low-circFUT8 group. A chi-square test was carried out, and results shown in Table 1 indicated that the expression of circFUT8 was correlated with LN metastasis and histological grade, but not other clinical parameters. Additionally, overall survival analysis conducted by Kaplan-Meier method and log-rank test demonstrated low-circFUT8 expression exhibited poor survival rate (Figure 2C).
Table 1.
Correlation between circFUT8 Expression and Clinicopathological Characteristics of BCa Patients
| Variable | No. of Cases (and %) | circFUT8 |
p | |
|---|---|---|---|---|
| Low | High | |||
| Age (Years) | ||||
| ≤65 | 74 (51.0) | 21 | 53 | 0.289 |
| >65 | 71 (49.0) | 26 | 45 | |
| Gender | ||||
| Male | 123 (84.8) | 37 | 86 | 0.156 |
| Female | 22 (15.2) | 10 | 12 | |
| Tumor Stage | ||||
| Ta–T1 | 59 (40.7) | 14 | 45 | 0.064 |
| T2–T4 | 86 (59.3) | 33 | 53 | |
| LN Status | ||||
| LN− | 117 (80.7) | 27 | 90 | 0.000a |
| LN+ | 28 (19.3) | 20 | 8 | |
| Multifocality | ||||
| Unifocal | 135 (93.1) | 43 | 92 | 0.728 |
| Multifocal | 10 (6.9) | 4 | 6 | |
| Histological Grade | ||||
| Low | 22 (15.2) | 3 | 19 | 0.048a |
| High | 123 (84.8) | 44 | 79 | |
| Tumor Size | ||||
| <3cm | 120 (82.8) | 41 | 79 | 0.323 |
| ≥3cm | 25 (17.2) | 6 | 19 | |
| Total | 145 | 47 | 98 | |
p < 0.05 was considered to be statistically significant (chi-square test).
circFUT8 Inhibits the Migration and Invasion of BCa Cell Lines and Can Be Regulated by DHX9
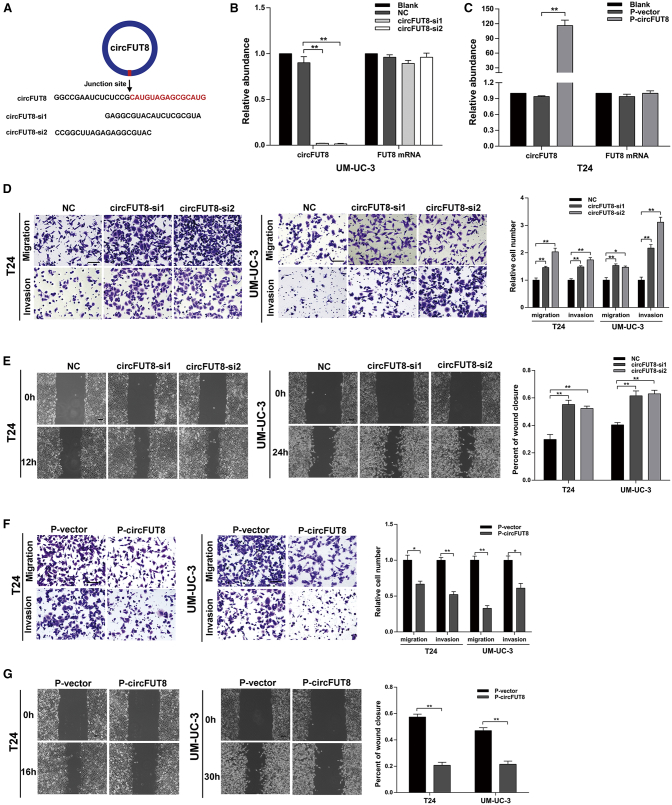
To evaluate the biological role of circFUT8 in BCa cells, gain- and loss-of-circFUT8 assays were applied in our study. Two small interfering RNAs (siRNAs) targeting the back-splicing junction site of circFUT8 were designed (Figure 3A), and the data indicated a significantly decreased level of circFUT8 after siRNA transfection but no effect on the mRNA level of FUT8 (Figure 3B; Figure S2A). Similarly, the quantitative real-time PCR data also showed the significant upregulation of circFUT8 but no obvious change in FUT8 mRNA level in stably overexpressed circFUT8 BCa cell lines (Figure 3C; Figure S2B). Compared with the negative-control cells, the circFUT8-knockdown cells exhibited the enhanced ability of migration and invasion in wound-healing and Transwell assays (Figures 3D and 3E). Moreover, the stable overexpression of circFUT8 cells showed the reverse ability in the same assays (Figures 3F and 3G). DExH-box helicase 9 (DHX9) is a well-known nuclear RNA helicase that can inhibit the production of circRNAs by binding to their flanking inverted complementary sequences.19 In our study, we found an upregulation of circFUT8 after silencing DHX9 (Figure S2C), suggesting that DHX9 may be a potential regulator.
Figure 3.
circFUT8 Acts as a Tumor Suppressor in BCa Cells
(A) Schematic diagram showing two targeted siRNAs. siRNAs targeted the back-splicing junction site of circFUT8. (B and C) Quantitative real-time PCR analysis of circFUT8 and FUT8 mRNA in UM-UC-3 cells treated with two siRNAs (B) and T24 cells with stable overexpression of circFUT8 (C). (D and E) Wound-healing and Transwell assays indicated that the migration and invasion abilities of BCa cell lines were enhanced after silencing circFUT8. (F and G) Stable overexpression of circFUT8 inhibited the migration and invasion abilities of BCa cell lines evaluated by Transwell and wound-healing assays, and the time for Transwell assays: 10 h (T24) and 25 h (UM-UC-3) for migration and 15 h (T24) and 34 h (UM-UC-3) for invasion. Scale bars, 100 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.
circFUT8 Acts as a Molecular Sponge for miR-570-3p in BCa Cells
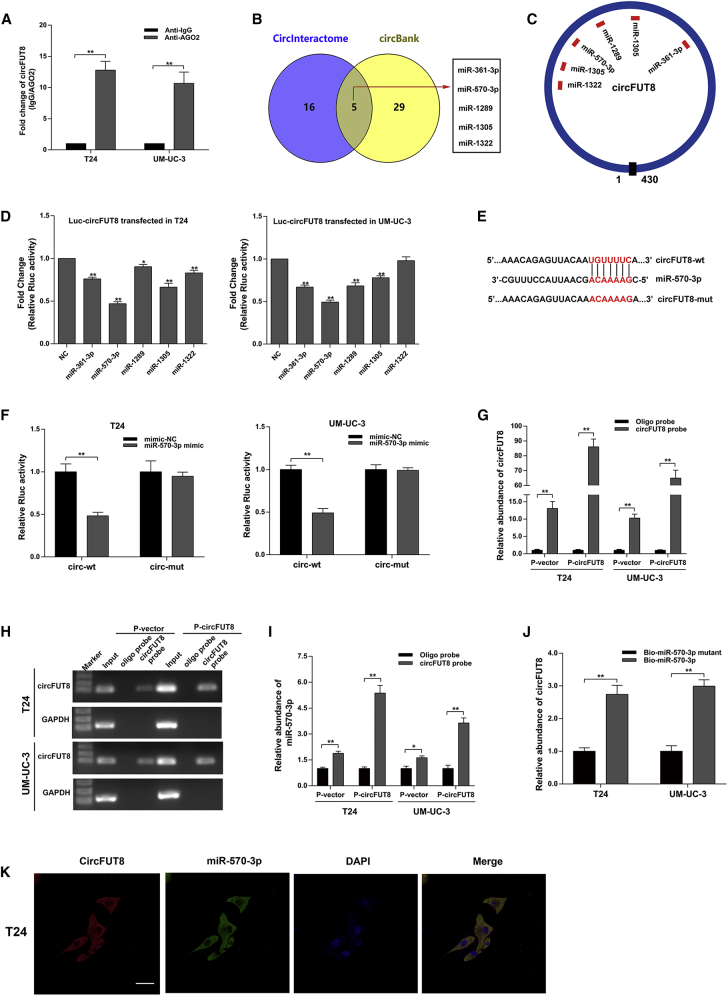
circRNAs have been reported to act as the sponge to miRNAs in cancers.14,15 circFUT8 was abundant and stale in the cytoplasm of BCa cells, and we wondered whether circFUT8 regulated the migration and invasion of BCa cells by binding certain miRNAs. It is well known that miRNAs modulate mRNA translation in an AGO2-dependent manner. By RNA immunoprecipitation (RIP) assay for AGO2 in T24 and UM-UC-3 cells, circFUT8 was specially enriched by quantitative real-time PCR (Figure 4A), indicating that circFUT8 is involved in the miRNA-mediated mRNA translation.
Figure 4.
circFUT8 Serves as a Sponge for miR-570-3p in BCa Cells
(A) The enrichment of circFUT8 was detected by quantitative real-time PCR after RIP assay for AGO2. (B) Potential target miRNAs of circFUT8 were predicted by CircInteractome and circBank. (C) Schematic diagram demonstrated the predicted binding sites of the five miRNAs in circFUT8. (D) Luciferase reporter assay: BCa cells transfected with psiCHECK-2-wild-type circFUT8 were co-transfected with the five miRNA mimics, respectively. (E and F) The characteristics of psiCHECK-2-wild-type circFUT8 (circ-wt) and psiCHECK-2 mutant-type circFUT8 (circ-mut) (E). Luciferase reporter assay indicated that miR-570-3p mimic significantly reduced the Rluc activity of circ-wt but not circ-mut (F). (G and H) circFUT8 was assessed by quantitative real-time PCR (G) after RNA pull-down assay, and the quantitative real-time PCR products from input and the RNA pull-down assay were used for gel electrophoresis (H). Oligo probe was used as negative control for RNA pull-down assay, and GAPDH was considered the internal control of gel electrophoresis. (I) miR-570-3p was analyzed by quantitative real-time PCR after RNA pull-down assay using circFUT8 probe. (J) circFUT8 captured by biotin-coupled miR-570-3p or miR-570-3p mutant mimics in T24 and UM-UC-3 cells was quantified by quantitative real-time PCR. (K) FISH assay demonstrates the co-localization of circFUT8 and miR-570-3p in the T24 cell line. Nuclei were stained with DAPI. The circFUT8 probe was labeled with Cy3, and the miR-570-3p probe was labeled with Cy5. Scale bar, 50 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.
To screen certain miRNAs binding to circFUT8, we acquired 5 overlapped miRNAs predicted by CircInteractome (https://circinteractome.nia.nih.gov/index.html) and circBank (http://www.circbank.cn/) (Figures 4B and 4C).20 The predicted miRNAs by CircInteractome and circBank are also listed in Table S1.
To identify whether the 5 candidate miRNAs could bind to circFUT8, we constructed a luciferase reporter plasmid by inserting circFUT8 sequence into the psiCHECK-2 vector. Subsequently, 5 miRNA mimics were co-transfected with circFUT8 luciferase reporter plasmid in BCa cells. The results showed that the miR-570-3p mimic could reduce Renilla luciferase (Rluc) activity more effectively (Figure 4D). We further mutated the binding site of miR-570-3p in the luciferase reporter plasmid and confirmed that the miR-570-3p had no effect on Rluc activity (Figures 4E and 4F).
To further verify the interaction between miR-570-3p and circFUT8 in BCa cells, we designed the biotin-coupled circFUT8 probe hybridizing with the back-splicing junction site and performed a pull-down assay. circFUT8 was detected by quantitative real-time PCR and agarose gel electrophoresis after pull-down assay to assess the efficiency of circFUT8 probe (Figures 4G and 4H). After purifying the RNAs bound to circFUT8, the results showed that the enrichment of miR-570-3p was significantly increased compared with oligo probe (Figure 4I). Furthermore, we transfected the biotin-coupled miR-570-3p mimic or miR-570-3p mutant mimic into BCa cells with stable overexpression of circFUT8. The RNAs captured by biotin-coupled miR-570-3p mimic or mutant were collected. As expected, the biotin-coupled miR-570-3p mimic captured circFUT8 effectively (Figure 4J). Moreover, the co-localization between circFUT8 and miR-570-3p in BCa cells was certified by RNA-FISH assay (Figure 4K). Taken together, these results supported that circFUT8 could directly bind to miR-570-3p, and miR-570-3p was the ideal target of circFUT8 for the following study.
miR-570-3p Promotes Migration and Invasion of BCa Cell Lines by Targeting KLF10
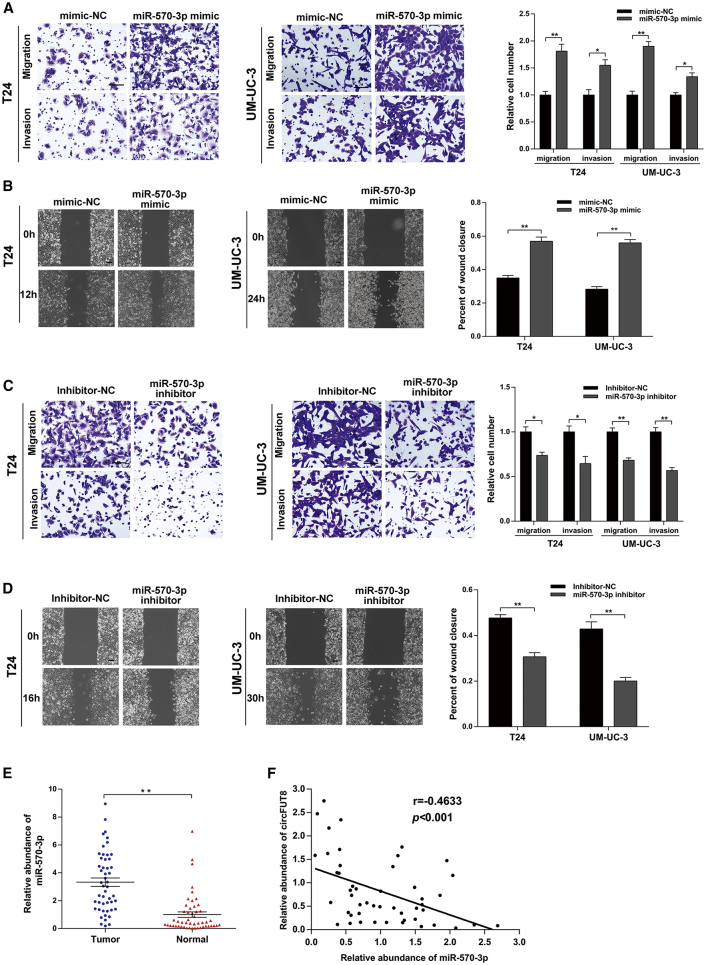
To elucidate the function of miR-570-3p in BCa cells, the mimic and inhibitor of miR-570-3p were used in our study. The miR-570-3p mimic could significantly promote the migration and invasion of BCa cells in Transwell and wound-healing assays (Figures 5A and 5B), and miR-570-3p inhibitor led to the opposite effect (Figures 5C and 5D). Additionally, the expression of miR-570-3p in BCa tissues and matched normal bladder tissues was also detected by quantitative real-time PCR. Compared with matched normal tissues, the result showed that miR-570-3p was significantly upregulated in BCa tissues (Figure 5E), suggesting an oncogenic role of miR-570-3p. Additionally, we also found a negative correlation between circFUT8 and miR-570-3p expression in BCa tissues (Figure 5F).
Figure 5.
miR-570-3p Promotes the Migration and Invasion of BCa Cells
(A and B) The migration and invasion abilities of T24 and UM-UC-3 cells transfected with miR-570-3p mimic were measured by Transwell (A) and wound-healing (B) assays. (C and D) The migration and invasion abilities of T24 and UM-UC-3 cells transfected with miR-570-3p inhibitor were evaluated by Transwell (C) and wound-healing (D) assays, and the times for Transwell assays were as follows: 10 h (T24) and 25 h (UM-UC-3) for migration; 15 h (T24) and 34h (UM-UC-3) for invasion. (E) The abundance of miR-570-3p in BCa tissues and matched normal bladder tissues detected by quantitative real-time PCR (n = 50). (F) The correlation between circFUT8 and miR-570-3p in BCa tissues (n = 49, and one extreme value was excluded). Scale bars, 100 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.
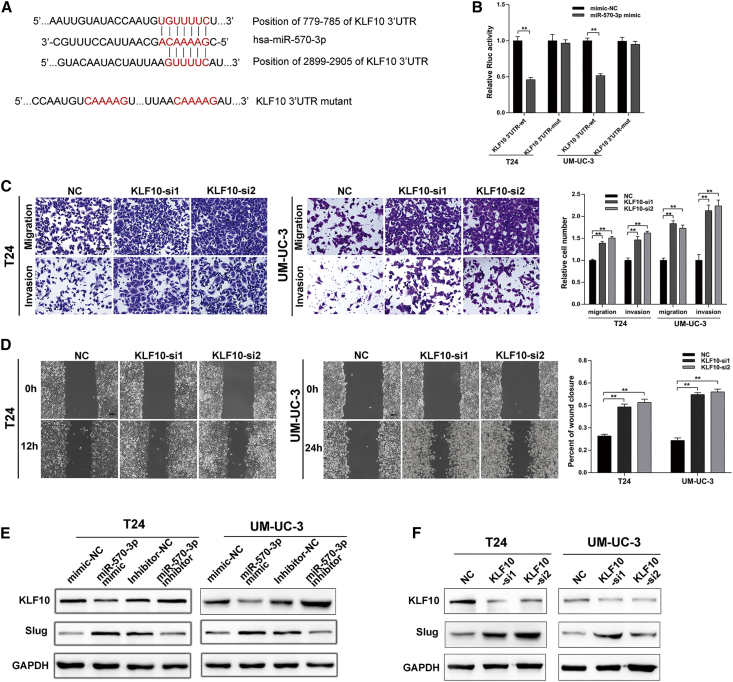
Subsequently, we used miRbase and miRTarBase to predict a tumor-associated gene, KLF10, as the potential target gene of miR-570-3p (Figure 6A). To confirm whether miR-570-3p directly targeted KLF10, the 3′ UTR of KLF10 mRNA containing a binding site with miR-570-3p was cloned into a luciferase reporter plasmid. Then, the reporter plasmid and miR-570-3p were co-transfected into BCa cells, and relative Rluc activity was significantly reduced (Figure 6B). Furthermore, we constructed a mutated 3′ UTR of KLF10-mRNA-associated luciferase reporter plasmid and abolished the binding site of miR-570-3p, and the result showed that miR-570-3p abrogated its ability to decrease Rluc activity (Figure 6B).
Figure 6.
KLF10 Inhibits the Migration and Invasion of BCa Cells and Is Regulated by miR-570-3p
(A) Schematic diagram showing two potential binding sites between miR-570-3p and the 3′ UTR of KLF10 and also showing the mutated bases in the 3′ UTR of KLF10 for the following luciferase reporter assay. (B) A luciferase reporter assay in BCa cells was applied to detect the interaction between miR570-3p mimic and psiCHECK-2-wild-type KLF10 (KLF10 3′ UTR-wt) or psiCHECk-2 mutant type KLF10 (KLF10 3′ UTR-mut) plasmids. (C and D) The migration and invasion abilities of T24 and UM-UC-3 cells were enhanced after silencing KLF10 assessed by Transwell and wound-healing assays. (E and F) Western blot assay indicated that miR-570-3p inhibited KLF10 expression and promoted the expression of Slug (E) and that knockdown of KLF10 by siRNAs could upregulate the expression of Slug (F). Scale bars, 100 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.
Transcript factor KLF10 was characterized as a tumor-suppressor gene in cancers.21,22 Metastatic brain tumors from lung adenocarcinoma indicated that KLF10 was at an abnormally low level in brain tumors.21 KLF10 was also identified as an anti-metastasis gene by the inhibition of Slug gene transcription.22 In our study, functional assays revealed that KLF10 inhibited the migration and invasion of BCa cell lines (Figures 6C and 6D).
Additionally, transfection of miR-570-3p mimic reduced the expression of KLF10 in protein level, and the opposite effect was conducted by the miR-570-3p inhibitor (Figure 6E). We next certified that KLF10 negatively regulated the expression of Slug (Figure 6F). Collectively, these findings indicated that miR-570-3p regulated the expression of KLF10 by targeting its 3′ UTR and that KLF10 could inhibit the migration and invasion of BCa cell lines by reducing the expression of Slug.
circFUT8 Promotes the Expression of KLF10 by Sponging the miR-570-3p
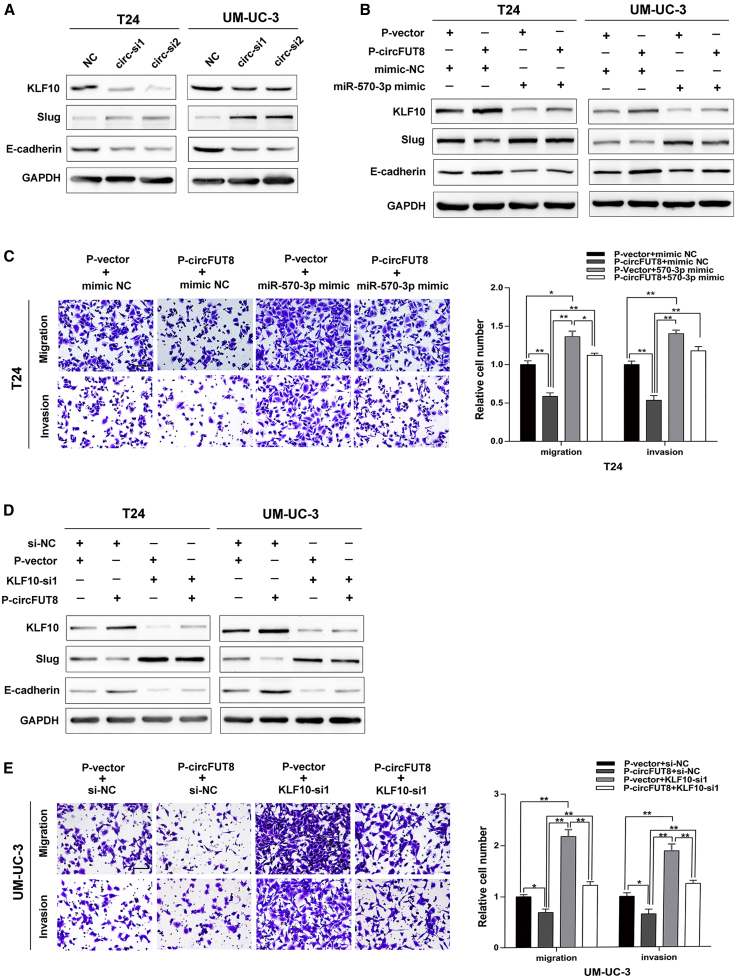
Given that circFUT8 and KLF10 shared the same binding site in miR-570-3p (Figures 4E and 6A), we hypothesized that circFUT8 might exert its biological effect in BCa cells, depending on KLF10, by competitively binding to miR-570-3p. By altering the circFUT8 in BCa cells, western blot analysis demonstrated that the expression of KLF10 was positively correlated with the level of circFUT8, and this biological effect could be partially abrogated by ectopic expression of miR-570-3p (Figures 7A and 7B). Subsequently, we verified that miR-570-3p could impair the effect of circFUT8 on inhibiting migration and invasion of BCa cells by co-transfection (Figure 7C; Figure S3A).
Figure 7.
Validation of circFUT8-Related Downstream Genes and the Existence of the circFUT8/miR-570-3p/KLF10 Axis
(A) Western blot analysis of KLF10, Slug, and E-cadherin after knockdown of circFUT8 in T24 and UM-UC-3 cell lines. (B) The upregulation of Slug and the downregulation of KLF10 and E-cadherin in T24 and UM-UC-3 cells transfected with miR-570-3p mimics were partially reversed by the overexpression of circFUT8. (C) The enhanced migration and invasion abilities of T24 cells by miR-570-3p mimics were partially reversed by the overexpression of circFUT8 using Transwell assay. (D) The upregulation of Slug and the downregulation of KLF10 and E-cadherin in T24 and UM-UC-3 cells transfected with KLF10 siRNA were partially reversed by the overexpression of circFUT8, as detected by western blot. (E) The enhanced migration and invasion abilities of UM-UC-3 cells by KLF10 siRNA were partially reversed by the overexpression of circFUT8 using Transwell assay. The times for Transwell assays were as follows: 10 h (T24) and 25 h (UM-UC-3) for migration and 15 h (T24) and 34 h (UM-UC-3) for invasion. Scale bars, 100 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.
As the downstream of KLF10, Slug and E-cadherin could be regulated by circFUT8, and this effect was partially abrogated by modulating the expression of KLF10 (Figure 7D). To confirm the role of KLF10 in biological function mediated by circFUT8, we attempted to detect the migration and invasion of BCa cells by simultaneously modulating the expression of circFUT8 and KLF10. Overexpression of circFUT8 decreased the ability of BCa cells to migrate and invade, but co-transfection of KLF10 siRNA partially abolished this effect (Figure 7E; Figure S3B). Taken together, these results uncovered that the circFUT8/miR-570-3p/KLF10/Slug axis played a vital role in the migration and invasion of BCa cells.
The Role of circFUT8 and miR-570-3p in LN Metastasis
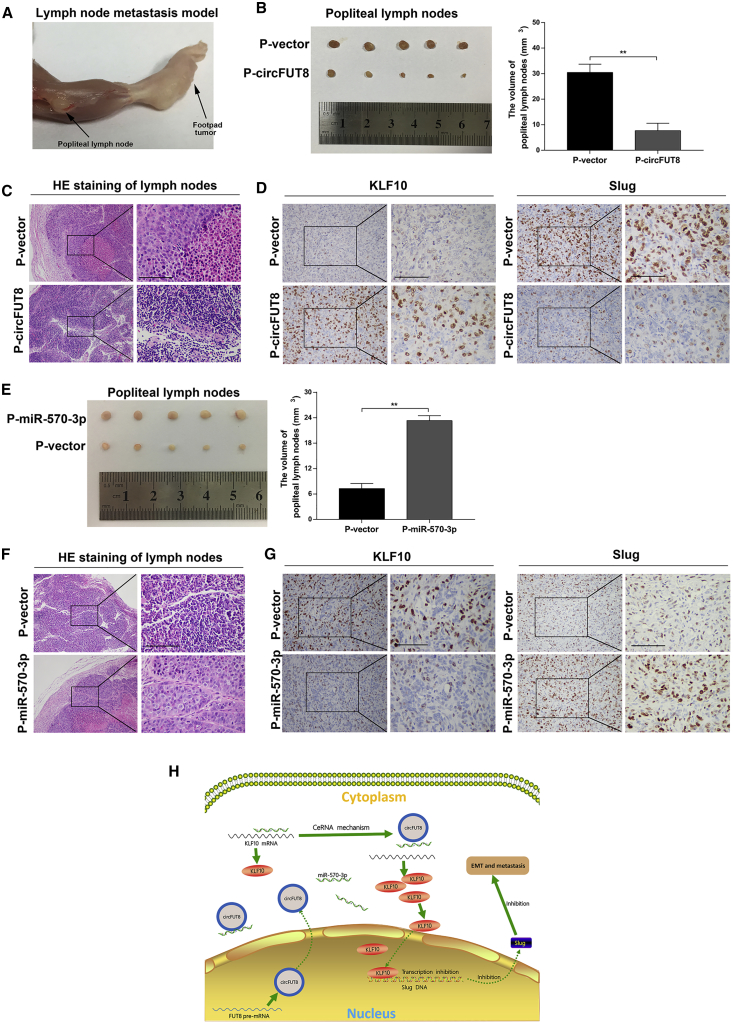
To elucidate the roles of circFUT8 and miR-570-3p in LN metastasis of BCa, a popliteal LN metastasis model was used in BALB/c nude mice. Lymph drainage of footpads was directional to popliteal LN, which supplied a sensitive measurement to learn the lymphatic metastasis of BCa (Figure 8A). UM-UC-3 cells with stable overexpression of target RNA or vector were injected into the footpads of nude mice.
Figure 8.
The Effects of circFUT8 and miR-570-3p on LN Metastasis of BCa Cells In Vivo
(A) Representative image of the nude mouse model of popliteal LN metastasis. (B) The image of enucleated popliteal LNs. The volume of LNs was significantly decreased after circFUT8 treatment for 4 weeks. (C) Representative images of H&E staining of the popliteal LNs indicate that tumor-associated LN metastasis existed in the vector group but not the circFUT8 group. (D) IHC staining of KLF10 and Slug in tumor from footpads with vector and circFUT8 treatment. (E) The volume of LNs was significantly increased after miR-570-3p treatment for 3 weeks. (F) H&E staining of the popliteal LNs indicates that miR-570-3p promoted the LN metastasis of UM-UC-3 cells. (G) IHC staining of KLF10 and Slug in tumor from footpads with vector and miR-570-3p treatment. (H) Schematic diagram indicates that circFUT8 inhibited the metastasis of BCa cells through the miR-570-3p/KLF10/Slug axis. Scale bars, 100 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.
Strikingly, the volume of LNs was smaller in the overexpression of the circFUT8 group than in that of the vector group 4 weeks post-injection, and hematoxylin and eosin (H&E) staining of LNs indicated that BCa-associated LN metastasis existed in the vector group, suggesting an inhibiting effect of circFUT8 on lymphatic metastasis (Figures 8B and 8C). Immunohistochemistry (IHC) of footpad tumors indicated that the expression level of KLF10 was significantly increased in the circFUT8 overexpression group compared with that in the vector group, but the effect was opposite on Slug (Figure 8D).
Whereas the volume of the popliteal LNs was significantly smaller in the control group than in the miR-570-3p group 3 weeks post-injection (Figure 8E), the LN metastasis was mainly distributed in miR-570-3p group detected by H&E staining (Figure 8F). By IHC staining of primary tissues of footpad, results certified that miR-570-3p reduced the expression of KLF10 and promoted Slug expression (Figure 8G).
Collectively, our results in vivo revealed that circFUT8 acted with an inhibitory role in the LN metastasis of BCa, whereas miR-570-3p displayed the opposite role.
Discussion
LN metastasis leads a poor prognosis for BCa patients, and, currently, treatment strategies for LN metastasis are limited in clinic.23,24 Thus, exploring the molecular mechanisms underlying LN metastasis and screening promising targets may facilitate clinical prevention and therapy. Several long non-coding RNAs involved in LN metastasis of BCa have been reported, including Dancer and Blacat2.25,26 Emerging evidence suggests that circRNAs play a critical role in the initiation and progression of several types of cancer, serving as biomarkers to facilitate diagnosis and prognosis.27, 28, 29 Here, we reported a circRNA termed circFUT8 and systematically elucidated its role in the metastasis of BCa. The level of circFUT8 was downregulated in BCa tissues and correlated with LN metastasis, histological grade, and prognosis. Overexpression of circFUT8 inhibited the migration and invasion of BCa in both cultured BCa cell lines and mouse models. Mechanistically, we demonstrated that circFUT8 modulated the aggressiveness of BCa by regulation of the miR-570-3p/KLF10/Slug axis.
Accumulating investigations have implied that circRNAs have abundant biological functions and act as miRNA sponges and RNA-binding protein sponges and can even be translated into peptides.29, 30, 31, 32 Several investigations have revealed that circRNAs are differentially expressed in several human tumors and exert pivotal roles in proliferation and metastasis;27,28 and miRNA sponge is the most frequently studied function of circRNA in cancers, briefly, diminishing the miRNA’s regulation of the target gene by competitively binding the base-paired part of miRNA. circHIPK3 has been reported to have a regulatory role in human cancer cells by sponging multiple miRNAs.33 It has been confirmed that circPVT1 is involved in a cell cycle of gastric cancer by indirectly regulating the expression of E2F2 as a sponge of miR-125.34 In this study, we identified that circFUT8 was downregulated in BCa. A previous study illustrated that the invert complementary sequences in genome promoted the formation of circRNAs and that DHX9 could bind to these special sequences and inhibit the formation of circRNAs.19 In our study, we revealed that the expression of circFUT8 was elevated by silencing DHX9, suggesting that DHX9 was a potential upstream regulator of circFUT8.
AGO2 is involved in the RNA-induced silencing complex, which facilitates the repressing or degrading of target mRNAs by miRNAs.35 The subcellular location revealed that circFUT8 was mainly distributed in cytoplasm, and circFUT8 was specially enriched by RIP assay for AGO2, which strongly supports the hypothesis that circFUT8 may function as a miRNA sponge. The prediction of targeted miRNAs and several assays, such as pull-down assay, dual-luciferase reporter assay, and biotin-coupled miRNA capture assay, indicated that circFUT8 harbored the binding site with miR-570-3p. The RNA-FISH assay also demonstrated that circFUT8 and miR-570-3p were co-located in the cytoplasm of BCa cells. To our knowledge, the function of miRNA-570-3p has not been studied in many cancers, including BCa. The quantitative real-time PCR data demonstrated that miR-570-3p was upregulated in BCa tissues compared with the matched normal bladder tissue, and the function assay showed that miR-570-3p promoted the migration and invasion of BCa cell lines in vitro and LN metastasis in vivo, implying that miR-570-3p exerted an oncogenic role in BCa. Subsequent biological analyses confirmed that circFUT8 could reverse the oncogenic role of miR-570-3p in BCa. All together, our experiments strongly suggested that circFUT8 might regulate gene expression to modulate the biological characteristics of BCa by sponging miR-570-3p.
KLF10, originally called TGF-β (transforming growth factor-β)-inducible early gene-1 (TIEG1), was a zinc-finger-containing transcription factor and identified as an early response gene after TGF-β treatment in human osteoblasts.36 Here, we identified that KLF10 was the direct downstream of miR-570-3p based on the following evidence: miR-570-3p possessed the complementary sequence with the 3′ UTR of KLF10 mRNA predicted by miRbase and miRTarBase, and luciferase report assays also indicated that the binding reaction existed between miR-570-3p and the 3′ UTR of KLF10 mRNA. Next, we found that miR-570-3p reversely regulated the expression of KLF10. The previous evidence revealed that KLF10 was downregulated in a series of cancers and inhibited the radiosensitivity, growth, and metastasis of cancer cells.37, 38, 39, 40 An important investigation discovered that KLF10 significantly inhibited tumor metastasis by inhibiting the transcription of Slug.22 In our study, western blot analysis was performed, and the results supported the finding that KLF10 negatively regulated the expression of Slug. Here, we proposed that KLF10 played a central role in determining the effect of epithelial-to-mesenchymal transition (EMT) by the regulation of Slug and its downstream E-cadherin.
To summarize, we identified that circFUT8 was downregulated in BCa tissues and that low circFUT8 was associated with poor prognosis, high histological grade, and LN metastasis. Also, circFUT8 inhibited the migration and invasion of BCa cells and harbored the binding site with miR-570-3p. Furthermore, our results demonstrated that miR-570-3p promoted tumor metastasis by inhibiting KLF10-mediated Slug signaling in BCa cells. Additionally, circFUT8 promoted KLF10 expression by competitively sponging miR-570-3p. Thus, we speculated that the regulatory signal of circFUT8/miR-570-3p/ KLF10/Slug was involved in LN metastasis of BCa, which may have considerable potential as a prognostic predictor and therapeutic target for BCa with LN metastasis.
Materials and Methods
Ethics Statement and Patient Tissue Specimens
All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health and approved by the Ethics Committee of Sun Yat-sen University. 145 BCa tissues with follow-up information and 50 matched adjacent normal bladder tissues were obtained from patients who underwent surgery in the Sun Yat-sen Memorial Hospital, Sun Yat-sen University, between 2010 and 2017. The use of human BCa tissues was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. Written informed consent was obtained from all patients involved in this study. We defined the BCa stage according to the criteria of the 2009 seventh edition of the TNM Classification of Malignant Tumours from the International Union Against Cancer.
Cell Culture and Isolation of T24 Sublines
Normal human urothelial cell line SV-HUC-1 and the human BCa cell lines T24 and UM-UC-3 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). T24 was cultured in RPMI-1640 medium (GIBCO, Gaithersburg, MD, USA), UM-UC-3 was cultured in DMEM (GIBCO, Gaithersburg, MD, USA), and SV-HUC-1 was cultured in F-12K medium (GIBCO, Gaithersburg, MD, USA). The cells were cultured in the humid atmosphere at 37°C with 5% CO2 and all aforementioned different culture mediums were supplemented with 10% fetal bovine serum (FBS; Biological Industries, Beit Haemek, Israel) and 1% penicillin/streptomycin (GIBCO, Gaithersburg, MD, USA), respectively.
The repeated Transwell assay was used to isolate highly and poorly invasive T24 sublines, and the detailed process was described in our previous study.17 After eight rounds of selection, T24 cells that totally failed to invade through the chamber were defined as poorly invasive T24 subline cells. On the other hand, the highly invasive T24 subline cells successfully invaded through the chamber during all selection rounds. The characteristics of the T24 sublines were confirmed by Transwell and wound-healing assays before use in our study.
RNA Extraction, RNase Treatment, Actinomycin D Assay, and PCR Assay
Total RNA was isolated from cells and tissues by RNAiso Plus (TaKaRa Bio, Shiga, Japan). gDNA was isolated using the MiniBEST Universal Genomic DNA Extraction Kit v.5.0 (TaKaRa Bio, Shiga, Japan). For actinomycin D assay, T24 and UM-UC-3 cells were treated with 2 μg/mL actinomycin D (Sigma, St. Louis, MO, USA) to block transcription for 8, 16, and 24 h. RNase R treatment was processed in RNAs extracted from T24 and UM-UC-3 at 37°C with RNase R (Epicentre Technologies, Chicago, IL, USA) for 30 min under the protocol of a previous study.17 The Nuclear and Cytoplasmic Extraction Kit (Life Technologies, Camarillo, CA, USA) was applied to isolate RNA from nuclear and cytoplasm extracts according to the manufacturer’s instructions. Complementary DNA was synthesized using random primers and the reverse transcription kit PrimeScript RT Master Mix (TaKaRa Bio, Shiga, Japan). For miRNA, the target miRNA reverse transcription primer and PrimeScript RT Master Mix (TaKaRa Bio, Shiga, Japan) were used to synthesize the complementary DNA. Quantitative real-time PCR was carried out with the TB Green Premix Ex TaqII (TaKaRa Bio, Shiga, Japan) in a Quantstudio Dx system (Applied Biosystems, Singapore, Singapore). For quantitative real-time PCR, GAPDH was used as an internal standard control for circRNA and mRNA, and the abundance of miRNA was normalized to small nuclear U6. The data of relative expression were calculated by the 2−ΔΔCT method.
Plasmid Construction and Transfection
The circFUT8 sequence was cloned into the plenti-ciR-GFP-T2A vector (IGE Biotechnology, Guangzhou, China) to construct its overexpression plasmid. The luciferase reporter plasmids of circFUT8 and KLF10 3′ UTR were synthesized using the psiCHECK-2 vector (Synbio Technologies, Suzhou, China). siRNA and miRNA mimic or inhibitor were transfected using Lipofectamine RNAiMax (Invitrogen, Waltham, MA, USA). X-tremeGENE (Sigma, St. Louis, MO, USA) was applied for plasmid transfection. The transfection procedure was under the guidance of the manufacturer’s protocol. Lentiviruses containing miR-570-3p plasmid (PCDH-CMV-MCS-EF1a-GFP-puro vector) for overexpression were constructed by iGene (Guangzhou, China). Lentivirus packaging for circFUT8 and stable transfection cell screening were carried out as described in a previous study.17
Cell Migration and Invasion
For wound-healing assays, the transfected cells were cultured to nearly 100% confluence in six-well plates, and the wound was created by scratching the cell layer using sterile 200-μL plastic pipette tips. The cells were further cultured in medium containing 1% FBS. The relative migration of cells was calculated as diminishing wound distance normalized to the 0-h control.
For Transwell invasion and migration assays, 7 × 104 transfected cells in 200 μL 1% FBS medium were seeded in the top chamber of a 24-well Transwell chamber (8 μm, Corning, Costar, Corning, NY, USA) with or without pre-coated Matrigel (BD Biosciences, USA). 600 μL medium containing 10% FBS was placed into the lower chamber as a chemoattractant. If not specially indicated, the time of Transwell assay was 8 h (T24) and 22 h (UM-UC-3) for migration or 12h (T24) and 30h (UM-UC-3) for invasion. Then, the chambers were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. The cells on the upper membrane surface were removed by a cotton swab, and the migrated or invaded cells on the lower membrane surface were photographed by microscopy (Nikon, Tokyo, Japan) at 200× magnification. The aforementioned experiments were performed three times.
FISH
BCa cells were cultured to 50% confluence in the confocal dish before this assay. The Cy3-labeled probe to circFUT8 hybridizing with the back-splicing junction site and the Cy5-labeled probe to miRNA-570-3p were designed and synthesized at GenePharma (Shanghai, China). The signals of the probes were processed by using a FISH kit (GenePharma, China) according to the manufacturer’s instructions. Briefly, the cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min. After adding the mixture of probe, the confocal dish was incubated at 73°C for 5 min and at 37°C for 15 h in the dark. The nuclei were stained by 4’, 6-diamidino-2-phenylindole (DAPI). The images were acquired on a Zeiss LSM 800 confocal microscope (Carl Zeiss, Jena, Germany).
RIP
The RIP assay was carried out using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA, USA) according to the manufacturer’s protocol. Briefly, anti-AGO2 and control IgG (Millipore, Burlington, MA, USA) were incubated with magnetic beads at room temperature for 30 min. Next, cells (1 × 107) were lysed in lysis buffer, and the cell lysate was conjugated to coated magnetic beads on a rotator at 4°C overnight. The beads were then washed, and the co-precipitated RNA was isolated using RNAiso Plus. The extracted RNA was examined by quantitative real-time PCR to investigate the enrichment of circFUT8.
Luciferase Reporter Assay
Cells were seeded in 24-well plates at a density of 4 × 104 cells per well 24 h before transfection. The cells were co-transfected with luciferase reporter plasmids and miRNA mimics. After 48 h, the firefly and Rluc activities were measured by a dual-luciferase reporter assay system (Promega, Madison, WI, USA). The relative Rluc activity was normalized to firefly luciferase, and each assay was repeated in three independent experiments.
Biotin-Coupled Probe Pull-Down Assay
The biotin-coupled probe targeted circFUT8 hybridizing with the back-splicing junction site was designed, and a random oligo probe was used as a negative control. The details of pull-down assay were carried out as previously described.17,18 Briefly, the circFUT8 probe and oligo probe were incubated with streptavidin magnetic beads (Life Technologies, USA) at room temperature for 2 h. About 1 × 107 cells were fixed with 1% formaldehyde, lysed in lysis buffer, and incubated with the probe-coated magnetic beads at 4°C overnight. Finally, the magnetic beads were washed five times with the lysis buffer, and the RNA complexes bound to the beads were eluted and extracted with RNAiso Plus for quantitative real-time PCR.
Biotin-Coupled miRNA Capture
The stable overexpression of circFUT8 BCa cells were transfected with biotin-coupled miRNA mimic (GenePharma, Shanghai, China). After 48 h, the cells were harvested and lysed in lysis buffer. To prepare the blocked beads, streptavidin magnetic beads were washed with lysis buffer three times and blocked with yeast tRNA for 2 h on a rotator at 4°C. Then, the cell lysis buffer was incubated with the blocked beads at 4°C overnight. The beads were washed with lysis buffer five times, and the RNAs specifically binding to miRNA were purified with the RNeasy Mini Kit (QIAGEN, Hilden, Germany). The abundance of circFUT8 was detected by quantitative real-time PCR in three independent experiments.
Xenografts in Mice
BALB/c nude mice (4 weeks old) were purchased from the Experimental Animal Center, Sun Yat-sen University, and the animal experiment procedures were approved by the Animal Ethic Committee of Sun Yat-sen University. To evaluate the roles of circFUT8 and miR-570-3p in LN metastasis of BCa, the popliteal lymphatic metastasis model was applied in our study. The stably expressed circFUT8 or miR-570-3p UM-UC-3 cells were constructed by the lentivirus-transduced method. The footpads of nude mice were injected with 5 × 106 constructed UM-UC-3 cells suspended in 50 μL PBS. After 3 or 4 weeks, the primary tumors in footpad and popliteal LNs were paraffin embedded for further analysis.
Western Blot
Cells were collected and lysed with RIPA lysis buffer (Beyotime, Nanjing, China) with protease and phosphatase inhibitors added (CWBio, Beijing, China). Protein samples were separated by SDS-polyacrylamide-gel electrophoresis and transferred to polyvinylidene fluoride membranes. Then the membranes were incubated with the following primary antibodies at 4°C for 12 h: KLF10 (Santa Cruz Biotechnology, 1:1,000), Slug (Cell Signaling Technology, 1:1,000), E-cadherin (Cell Signaling Technology, 1:1,000), and GAPDH (Abcam, 1:5,000). Then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at room temperature for 1 h. The protein-band signal was detected with the ECL Detection Kit (Millipore, Darmstadt, Germany) and visualized using an Optimax X-ray Film Processor (Protec, Oberstenfeld, Germany).
H&E Staining and IHC Assay
This experiment was performed as previously described.41 The paraffin-embedded tissues from nude mice were cut into 4-μm slides for H&E and IHC. The primary antibodies used in this study were KLF10 (Santa Cruz Biotechnology, 1:200) and Slug (Cell Signaling Technology, 1:200). The images were obtained by using a Nikon Eclipse 80i system (Nikon, Tokyo, Japan).
Sequence Information Used in This Study
The sequences of the primers, oligonucleotides, and probes used in this study are provided in Table S2.
Statistical Analysis
Statistical analysis was performed with SPSS v.19.0 (IBM, SPSS, Chicago, IL, USA). The chi-square test was carried out to assess the association of the circFUT8 level with the patients’ clinicopathological parameters. Overall survival analysis was assessed by Kaplan-Meier method and log-rank test. Comparison between two groups was performed by Student’s t test, Wilcoxon rank-sum test, or Mann-Whitney test. The data were indicated as the mean ± standard error of the mean (SEM), and a p value less than 0.05 was considered statistically significant.
Author Contributions
T.L. and H.Q. conceived of the study and carried out its design. Q.H., D.Y., W.D., and J.B. performed the experiments. Q.H., L.H., M.Y., and J.H. conducted the statistical analyses. Q.H. wrote the paper, and W.D. revised the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 81772719, 81472384, and 81773026), the Guangdong Science and Technology Development Fund (2017B020227007), and the Project Supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.12.014.
Contributor Information
Haide Qin, Email: qinhaide@163.com.
Tianxin Lin, Email: lintx@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Antoni S., Ferlay J., Soerjomataram I., Znaor A., Jemal A., Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur. Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Ebrahimi H., Amini E., Pishgar F., Moghaddam S.S., Nabavizadeh B., Rostamabadi Y., Aminorroaya A., Fitzmaurice C., Farzadfar F., Nowroozi M.R. Global, regional and national burden of bladder cancer, 1990 to 2016: results from the GBD Study 2016. J. Urol. 2019;201:893–901. doi: 10.1097/JU.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 4.Burger M., Catto J.W., Dalbagni G., Grossman H.B., Herr H., Karakiewicz P., Kassouf W., Kiemeney L.A., La Vecchia C., Shariat S., Lotan Y. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 5.Kamat A.M., Hahn N.M., Efstathiou J.A., Lerner S.P., Malmström P.U., Choi W., Guo C.C., Lotan Y., Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 6.Zargar-Shoshtari K., Zargar H., Lotan Y., Shah J.B., van Rhijn B.W., Daneshmand S., Spiess P.E., Black P.C. A multi-institutional analysis of outcomes of patients with clinically node positive urothelial bladder cancer treated with induction chemotherapy and radical cystectomy. J. Urol. 2016;195:53–59. doi: 10.1016/j.juro.2015.07.085. [DOI] [PubMed] [Google Scholar]
- 7.Raza S.J., Al-Daghmin A., Zhuo S., Mehboob Z., Wang K., Wilding G., Kauffman E., Guru K.A. Oncologic outcomes following robot-assisted radical cystectomy with minimum 5-year follow-up: the Roswell Park cancer institute experience. Eur. Urol. 2014;66:920–928. doi: 10.1016/j.eururo.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 11.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 12.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H., Liu Y., Bian Z., Zhang J., Zhang R., Chen X., Huang Y., Wang Y., Zhu J. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 Kip1 axis. Mol. Cancer. 2018;17:151. doi: 10.1186/s12943-018-0902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 16.Xie F., Li Y., Wang M., Huang C., Tao D., Zheng F., Zhang H., Zeng F., Xiao X., Jiang G. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol. Cancer. 2018;17:144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H., Bi J., Dong W., Yang M., Shi J., Jiang N., Lin T., Huang J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aktaş T., Avşar Ilık İ., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 20.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zohrabian V.M., Nandu H., Gulati N., Khitrov G., Zhao C., Mohan A., Demattia J., Braun A., Das K., Murali R., Jhanwar-Uniyal M. Gene expression profiling of metastatic brain cancer. Oncol. Rep. 2007;18:321–328. [PubMed] [Google Scholar]
- 22.Mishra V.K., Subramaniam M., Kari V., Pitel K.S., Baumgart S.J., Naylor R.M., Nagarajan S., Wegwitz F., Ellenrieder V., Hawse J.R., Johnsen S.A. Krüppel-like transcription factor KLF10 suppresses TGFβ-induced epithelial-to-mesenchymal transition via a negative feedback mechanism. Cancer Res. 2017;77:2387–2400. doi: 10.1158/0008-5472.CAN-16-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May M., Herrmann E., Bolenz C., Tiemann A., Brookman-May S., Fritsche H.M., Burger M., Buchner A., Gratzke C., Wülfing C. Lymph node density affects cancer-specific survival in patients with lymph node-positive urothelial bladder cancer following radical cystectomy. Eur. Urol. 2011;59:712–718. doi: 10.1016/j.eururo.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Shariat S.F., Ehdaie B., Rink M., Cha E.K., Svatek R.S., Chromecki T.F., Fajkovic H., Novara G., David S.G., Daneshmand S. Clinical nodal staging scores for bladder cancer: a proposal for preoperative risk assessment. Eur. Urol. 2012;61:237–242. doi: 10.1016/j.eururo.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z., Chen X., Xie R., Huang M., Dong W., Han J., Zhang J., Zhou Q., Li H., Huang J., Lin T. DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing IL-11-STAT3 signaling and CCND1 expression. Mol. Ther. 2019;27:326–341. doi: 10.1016/j.ymthe.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He W., Zhong G., Jiang N., Wang B., Fan X., Chen C., Chen X., Huang J., Lin T. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J. Clin. Invest. 2018;128:861–875. doi: 10.1172/JCI96218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y., Guo Z., Fang N., Jiang W., Fan Y., He Y., Ma Z., Chen Y. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed. Pharmacother. 2019;117:109151. doi: 10.1016/j.biopha.2019.109151. [DOI] [PubMed] [Google Scholar]
- 29.Hang D., Zhou J., Qin N., Zhou W., Ma H., Jin G., Hu Z., Dai J., Shen H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783–2791. doi: 10.1002/cam4.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:1147. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Sheu-Gruttadauria J., Xiao Y., Gebert L.F., MacRae I.J. Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J. 2019;38:e101153. doi: 10.15252/embj.2018101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramaniam M., Harris S.A., Oursler M.J., Rasmussen K., Riggs B.L., Spelsberg T.C. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang V.H., Tsai Y.C., Tsai Y.L., Peng S.L., Chen S.L., Chang T.M., Yu W.C., Ch’ang H.J. Krüpple-like factor 10 regulates radio-sensitivity of pancreatic cancer via UV radiation resistance-associated gene. Radiother. Oncol. 2017;122:476–484. doi: 10.1016/j.radonc.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Song K.D., Kim D.J., Lee J.E., Yun C.H., Lee W.K. KLF10, transforming growth factor-β-inducible early gene 1, acts as a tumor suppressor. Biochem. Biophys. Res. Commun. 2012;419:388–394. doi: 10.1016/j.bbrc.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 39.Yang N., Chen J., Zhang H., Wang X., Yao H., Peng Y., Zhang W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017;8:e2975. doi: 10.1038/cddis.2017.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin W., Chen B.B., Li J.Y., Zhu H., Huang M., Gu S.M., Wang Q.Q., Chen J.Y., Yu S., Wu J., Shao Z.M. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol. Cell. Biol. 2012;32:50–63. doi: 10.1128/MCB.06152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Gu P., Xie R., Han J., Liu H., Wang B., Xie W., Xie W., Zhong G., Chen C. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J. Cell. Mol. Med. 2017;21:1266–1279. doi: 10.1111/jcmm.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.