Summary
Human induced pluripotent stem cell (hiPSC) culture has become routine, yet the cost of pluripotent cell media, frequent medium changes, and the reproducibility of differentiation have remained restrictive. Here, we describe the formulation of a hiPSC culture medium (B8) as a result of the exhaustive optimization of medium constituents and concentrations, establishing the necessity and relative contributions of each component to the pluripotent state and cell proliferation. The reagents in B8 represent only 3% of the costs of commercial media, made possible primarily by the in-lab generation of three E. coli-expressed, codon-optimized recombinant proteins: fibroblast growth factor 2, transforming growth factor β3, and neuregulin 1. We demonstrate the derivation and culture of 34 hiPSC lines in B8 as well as the maintenance of pluripotency long term (over 100 passages). This formula also allows a weekend-free feeding schedule without sacrificing capacity for differentiation.
Keywords: human induced pluripotent stem cell, pluripotent state, culture media, weekend-free, differentiation, chemically defined, FGF2
Highlights
-
•
The B8 hiPSC culture medium formula is a result of exhaustive optimization
-
•
The reagents in B8 represent only 3% of the costs of commercial media
-
•
B8 is suitable for the derivation and culture of hiPSC lines long term (>100 passages)
-
•
This formula allows weekend-free feeding without sacrificing differentiation capacity
In this article, Burridge and colleagues describe the formulation of a hiPSC culture medium called B8 as a result of exhaustive optimization. The reagents in B8 represent only 3% of the costs of commercial media. B8 is suitable for both derivation and long-term culture of hiPSCs and allows a weekend-free feeding schedule without sacrificing capacity for differentiation.
Introduction
Human induced pluripotent stem cells (hiPSCs) are functionally immortal and can proliferate without limit while maintaining the potential to differentiate to, hypothetically, all ∼220 cell lineages within the human body. hiPSC generation has become routine due to the simplicity of amplification of CD71+ blood proerythroblasts (Chou et al., 2011, Chou et al., 2015, Tan et al., 2014) or myeloid cells (Eminli et al., 2009, Staerk et al., 2010) and commercial Sendai virus-based reprogramming factor expression (Fujie et al., 2014, Fusaki et al., 2009). This simplicity has resulted in increased enthusiasm for the applications of hiPSC-derived cells across many fields, including regenerative medicine, disease modeling, drug discovery, and pharmacogenomics.
However, these applications require the culture of either large quantities of hiPSCs or hiPSC lines derived from large numbers of patients, and three major restrictions have become evident: (1) the cost of large-scale pluripotent cell culture, which is prohibitive for high-patient-number projects; (2) the time-consuming requirement for daily medium changes, including on the weekend, which is particularly problematic for laboratories in industry; (3) interline variability in differentiation efficacy, which is highly dependent on pluripotent culture consistency and methodology.
Our understanding of the medium conditions required to culture human pluripotent stem cells (hPSCs) has progressed steadily over the last 15 years, with significant breakthroughs coming from the discovery of the necessity for high concentrations of fibroblast growth factor 2 (FGF2) (Xu et al., 2005); the use of transforming growth factor β1 (TGF-β1) (Amit et al., 2004); and the elimination of knockout serum replacement with the TeSR formula, which contains 19 components (Ludwig et al., 2006a, Ludwig et al., 2006b, Ludwig and Thomson, 2007), followed by the first robust chemically defined formula, E8 (Beers et al., 2012, Chen et al., 2011), which consists of just eight components. A number of alternative non-chemically defined pluripotent formulations have been described, including CDM-BSA (Hannan et al., 2013, Vallier et al., 2005, Vallier et al., 2009), DC-HAIF (Singh et al., 2012, Wang et al., 2007), hESF9T (Furue et al., 2008, Yamasaki et al., 2014), FTDA (Breckwoldt et al., 2017, Frank et al., 2012, Piccini et al., 2015), and iDEAL (Marinho et al., 2015) (Figure S1A).
Each of the available formulations consists of a core of three major signaling components: (1) insulin or insulin growth factor (IGF1), which bind INSR and IGF1R to signal the PI3K/AKT pathway promoting survival and growth; (2) FGF2 and/or neuregulin 1 (NRG1), which bind FGFR1/FGFR4 or ERBB3/ERBB4, respectively, activating the PI3K/AKT/mTOR and MAPK/ERK pathways; and (3) TGF-β1, NODAL, or Activin A, which bind TGFBR1/2 and/or ACVR2A/2B/1B/1C to activate the TGF-β signaling pathway. NODAL is used less commonly in pluripotent medium formulations due to the expression of the NODAL antagonists LEFTY1/2 in hPSCs (Besser, 2004, Sato et al., 2003), resulting in a requirement for high concentrations in vitro (Chen et al., 2011). In addition, numerous growth-factor-free formulae utilizing small molecules to replace some or all growth factors have been described (Burton et al., 2010, Desbordes et al., 2008, Kumagai et al., 2013, Tsutsui et al., 2011); however, these have not successfully translated to common usage. Recently, a growth-factor-free formula, AKIT, was demonstrated (Yasuda et al., 2018), combining inhibitors of GSK3B (1-azakenpaullone), DYRK1 (ID-8), and calcineurin/NFAT (tacrolimus/FK506), albeit with much reduced proliferation and colony growth, as well as increased interline variability in growth. Finally, more than 15 commercial pluripotent media are also available of which the formulae are proprietary and not disclosed to researchers. These media represent the major cost for most hiPSC labs and considerably restrict research efforts. Some of these media formulae are suggested to support hiPSC growth without daily medium changes, or “weekend-free,” likely by using improved buffering and methods to stabilize FGF2 (such as including heparin), which otherwise degrades quickly at 37°C (Chen et al., 2012, Furue et al., 2008), and including bovine serum albumin (BSA), which acts as a multifaceted antioxidant.
Here, we demonstrate a novel medium formula (B8), thoroughly optimized to support a high growth rate under low seeding density conditions, that requires minimal medium exchanges, is low cost, and maintains differentiation reproducibility. This formula is capable of supporting both hiPSC generation and long-term culture for >100 passages. Production of B8 supplement aliquots suitable for making 100 L of B8 medium is simple for any research lab with basic equipment, with the reagents required for complete bottles of medium costing ∼US$16 per liter. A full protocol is provided, including detailed instructions for recombinant protein production in three simple steps. All plasmids for protein production are available through Addgene. With the commoditization of these protocols, we believe it is possible to substantially increase what is achievable with hiPSCs due to the near elimination of pluripotent cell culture costs and minimization of labor associated with cell culture.
Results
Optimization of Medium Constituents with a Short-Term Assay
We began with simple cost reductions to our existing hiPSC culture medium formula based on E8 that we have used extensively (Figure 1A). First, we optimized suitable concentrations of matrices on which the hiPSCs are grown. Although laminin-511 (Rodin et al., 2010), laminin-521 (Rodin et al., 2014), vitronectin (Braam et al., 2008), and Synthemax II (Melkoumian et al., 2010) are suitable for hiPSC culture (Burridge et al., 2014), none are appropriately cost effective, and in the case of vitronectin or Synthemax II, also not suitable for subsequent cardiomyocyte differentiation (Burridge et al., 2014). Matrigel, although an undefined product (Hughes et al., 2010), is a cost-effective and commonly used matrix with substantial data for using it at 50 μg cm−2 (Ludwig et al., 2006a). Comparing two similar commercial products, Matrigel (Corning) and Cultrex/Geltrex (Trevigen/Gibco), we found that both can be used at concentrations as low as approximately 2 μg cm−2 (a 1:1,000 dilution) (Figure S1A) and were subsequently used at a conservative approximately 2.5 μg cm−2 (a 1:800 dilution) for all future experiments.
Figure 1.
Optimization of Basic Human Pluripotent Stem Cell Medium Constituents with a Short-Term Growth Assay
Results are normalized to initial medium component concentrations shown with a dark gray bar. Optimized component concentrations are shown with a diagonal hash. Optimizations were completed using hiPSC line 19c3 and a short-term 6-day growth assay.
(A) Concentrations of initial medium formula components.
(B) Schematic of assay growth schedule showing seeding of cells at 10,000 cells per well of a 12-well plate, the day on which medium was changed (E8Y/E8), and the use of a PrestoBlue cell viability assay on day 6 to assess final cell number.
(C) Comparison of the effect on relative growth of concentrations of recombinant human insulin (n = 19).
(D) Recombinant human IGF1 LR3 (n = 3).
(E) L-ascorbic acid 2-phosphate (n = 19).
(F) Transferrin (n = 22).
(G) Sodium selenite (n = 18).
(H) FGF2-K128N (n = 6).
(I) TGF-β1 (n = 22). n = full independent experimental replicates, Mann-Whitney test, ∗p ≤ 0.05, ∗∗∗p ≤ 0.005, ∗∗∗∗p ≤ 0.0001, n.s., not significant. The significance bar refers to the significance between the conditions at the beginning and the end of the bar.
We next established a suitable pluripotent growth assay modeled on that used by Ludwig et al. (2006b) and Chen et al. (2011) after establishing that neither automated cell counting of dissociated cells (6-well) nor small-format plate reader-based cell viability assays (i.e., 96-well) were suitably robust (Figure 1B). This protocol involved dissociating ∼75% confluent hiPSCs to single cells with TrypLE and seeding 10,000 cells per well in a 12-well plate in the medium to be tested in the presence of a Rho kinase inhibitor for 24 h. This medium was then changed every day for 5 days, followed by a PrestoBlue viability assay using a plate reader to assess the number of viable cells present. We utilized this 6-day protocol rather than our typical 4-day protocol to enhance the detection of variance between formulae during sub-optimal low-density culture. All initial experiments were performed using a version of FGF2 we generated in house with a mutation (K128N) previously demonstrated to enhance thermostability (Chen et al., 2012). In a first round of optimizations we assessed a range of concentrations for each of the six major E8 medium components (Figures 1C–1I). During this stage we established that insulin was essential, and the effect of insulin was dose dependent up to 15 μg mL−1 (Figure 1C). Insulin could be replaced only by very high levels of IGF1 LR3 (≥1 μg mL−1), although this was not cost effective (Figure 1D). Ascorbic acid 2-phosphate was not essential, as previously demonstrated (Prowse et al., 2010), but higher levels (≥150 μg mL−1) enhanced growth (Figure 1E). Of note, this level of ascorbic acid 2-phosphate is similar to the level optimized in the cardiac differentiation medium CDM3 (Burridge et al., 2014). Transferrin was also not essential to the medium formula, but improved growth in a dose-dependent manner, with 20 μg mL−1 exhibiting sufficient growth while maintaining cost efficiency (Figure 1G). A source of selenium was shown to be essential, although concentrations of sodium selenite between 20 and 100 ng mL−1 did not significantly affect growth, and sodium selenite became toxic at ≥200 ng mL−1 (Figure 1F). FGF2-K128N was optimal at ≥60 ng mL−1 (Figure 1H), and TGF-β1 was sufficient at ≥1 ng mL−1 in this simple one-passage growth assay (Figure 1I).
Optimization of Additional Medium Components
In previous work, Chen et al. (2011) showed that transferrin improves single-cell clonality in the absence of the Rho-associated kinase (ROCK1/2) inhibitor Y27632. We could not replicate this observation in our assay (Figure 2A), suggesting that the inclusion of a ROCK1/2 inhibitor for at least the first 24 h after passage is sufficient. We also confirmed that the addition of 2 μM thiazovivin for the first 24 h significantly improved growth over 10 μM Y27632 and was an ∼5× more cost-effective choice (Figures 2B and 2C). Some recent hiPSC growth formulae have suggested that the addition of high levels (2×) of non-essential amino acids (NEAA) and/or low levels (0.1×) of chemically defined lipids enhances growth (Figure S1B). In our assay NEAA did not augment growth and the addition of lipids was inhibitory at all but the lowest levels (Figure 2D). Supplementation with BSA, a common hPSC medium component, did not have positive or negative effects on growth and was excluded to maintain a chemically defined formula (Figure 2E). The DMEM/F12 basal medium we use from Corning contains higher levels of sodium bicarbonate (∼29 mM or 2,438 mg L−1) compared with DMEM/F12 from other manufacturers (Figure S1C). Supplementation of Gibco DMEM/F12, which contains 14 mM sodium bicarbonate, with 20 mM additional sodium bicarbonate has recently been demonstrated to be advantageous to hiPSC growth rate by suppressing acidosis of the medium (Liu et al., 2018). In our hands, the standard 29 mM sodium bicarbonate was sufficient (Figure 2F). We also assayed the effect of pH and osmolarity on growth. We found that pH 7.0–7.1, measured at room temperature and atmospheric CO2 and modified with HCl or NaOH (Figure 2G), and a lower osmolarity of 290–310 mOsm L−1, modified with NaCl or water (Figure 2H), promoted the highest growth rate, in contrast to pH 7.2 and 350 mOsm L−1 demonstrated by Ludwig et al. (Ludwig et al., 2006b) or pH 7.4 and 340 mOsm L−1 used by Chen et al. (Chen et al., 2011). Of interest here, low-osmolarity KnockOut DMEM (270 mOsm L−1) has historically been used for mouse embryonic stem cell culture based on the approximation of mouse embryonic tissue (Lawitts and Biggers, 1992). All of our experiments were optimized for growth at 5% O2 and 5% CO2, a common compromise for optimal hypoxia, which enhances reprogramming efficiency (Chen et al., 2011), pluripotency (Forristal et al., 2010), and genome stability (Narva et al., 2013), without the significant cost increase from high N2 usage to obtain lower O2 levels or from 10% CO2, suggested by some (Ludwig et al., 2006b), that necessitates higher levels of sodium bicarbonate.
Figure 2.
Optimization of Additional Human Pluripotent Stem Cell Medium Constituents with a Short-Term Growth Assay
Results are normalized to initial medium component concentrations shown with a dark gray bar. Optimized component concentrations are shown with a diagonal hash. Optimizations were completed using hiPSC line 19c3 and a short-term 6-day growth assay.
(A) Comparison of the suitability of recombinant transferrin (10 μg mL−1) to support clonal growth with and without ROCK1/2 inhibition using Y27632 (10 μM) during the first 24 h after passage (n = 3).
(B) Comparison of the effect on relative growth of concentrations of ROCK1/2 inhibitor Y27632 only during first 24 h after passage (n = 5).
(C) Comparison of the effect on relative growth of concentrations of ROCK1/2 inhibitor thiazovivin during first 24 h after passage (n = 16).
(D) Comparison of the effect on relative growth of the addition of non-essential amino acids (NEAA) and chemically defined lipids (n = 5).
(E) Fatty acid-free albumin (n = 4).
(F) Sodium bicarbonate (n = 5).
(G) pH (n = 9).
(H) Osmolarity (n = 8).
(I) FGF2-G3 (n = 3). n = full independent experimental replicates, Mann-Whitney test, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.005, n.s., not significant. The significance bar refers to the significance between the conditions at the beginning and the end of the bar.
In our original hiPSC culture medium formula, we used commercial recombinant human FGF2, and this accounted for >60% of the medium reagent cost. This led us to investigate the production of FGF2 in house, for which we generated an FGF2 sequence with E. coli-optimized codon usage to enhance yield and a K128N point mutation to improve thermostability (Chen et al., 2012). This sequence was inserted into a recombinant protein production plasmid (pET-28a) that utilizes an upstream 6×His tag for purification that was not cleaved during processing (Figure S2). We subsequently generated two additional thermostable FGF2 variants: FGF1-4X (Chen et al., 2012) and FGF2-G3 (Dvorak et al., 2018). We found that FGF2-G3, with nine point mutations, was more potent than FGF2-K128N, showing a similar effect on growth rate at 5 ng mL−1 compared with FGF2-K128N at 100 ng mL−1 (Figure 2I) when tested using this short-term assay.
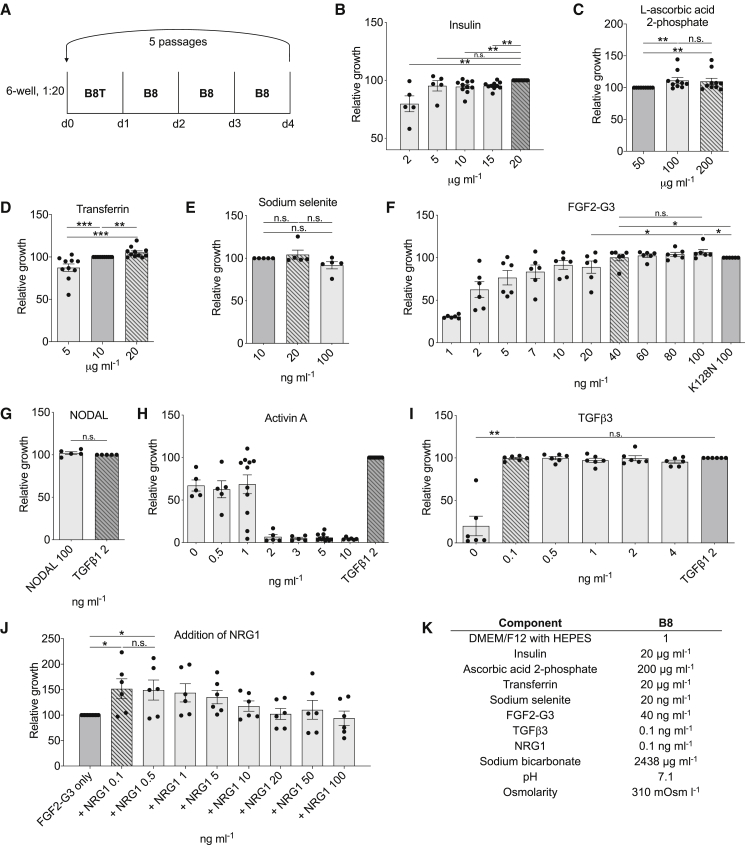
Optimization of Medium Components Using a Five-Passage Assay
Our initial single-passage experiments were useful for preliminary optimizations, yet we were aware from previous experiments that some variables, such as the elimination of TGF-β1, had minimal effects in one passage and would have detectable negative effects only in longer-term experiments. We began a second round of optimizations using a five-passage version of our growth assay (Figure 3A). Each experiment was independently repeated at least five times. These experiments again confirmed optimal concentrations of insulin (20 μg mL−1; Figure 3B), ascorbic acid 2-phosphate (200 μg mL−1; Figure 3C), transferrin (20 μg mL−1; Figure 3D), sodium selenite (20 ng mL−1; Figure 3E), and FGF2-G3 (40 ng mL−1; Figure 3F).
Figure 3.
Optimization of B8 Medium Constituents with a Long-Term Growth Assay
Results are normalized to initial medium component concentrations shown with a dark gray bar. Optimized component concentrations are shown with a diagonal hash. Optimizations were completed in hiPSC line 19c3 using a long-term five-passage, 4-day growth assay. Optimized component concentrations shown with a diagonal hash.
(A) Schematic of passaging schedule.
(B) Comparison of the effect on relative growth of recombinant human insulin concentrations (n = 10).
(C) L-ascorbic acid 2-phosphate (n = 13).
(D) Recombinant transferrin (n = 10).
(E) Sodium selenite (n = 5).
(F) In-house made FGF2-G3 (n = 6).
(G) NODAL (n = 5).
(H) Activin A (n = 5).
(I) In-house made TGF-β3 after 9 passages compared with commercial TGF-β1 (n = 9).
(J) Addition of NRG1 to 40 ng mL−1 FGF2-G3 (n = 6).
(K) Final B8 formula. n = full independent experimental replicates, Mann-Whitney test, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.005, n.s., not significant. The significance bar refers to the significance between the conditions at the beginning and the end of the bar.
We next looked for alternatives to TGF-β1, as this was now the costliest component (∼40% of total reagent cost) and found that this could not be used at lower concentrations than previously suggested (2 ng mL−1) when using our five-passage assay (Figure S3A). We found that other Activin/NODAL/TGF-β1 signaling sources such as NODAL were able to maintain growth to the same level as TGF-β1 (Figure 3G), whereas Activin A was not suitable (Figure 3H). Activin A in combination with TGF-β1 also has had a negative effect on growth (Figure S3B). TGF-β1 is a homodimer of two TGFB1 gene products, and therefore the recombinant protein is commonly produced in mammalian cells, making it complex to produce for basic research labs. These mammalian cells provide post-translational modifications crucial for biological activity, such as glycosylation, phosphorylation, proteolytic processing, and formation of disulfide bonds, which are not present in E. coli. To overcome this issue, we generated a version of TGF-β1 (TGF-β1m) that is unable to form dimers due to a point mutation. This monomeric protein is predicted to be ∼20-fold less potent than TGF-β1 but can be easily produced in large quantities in E. coli (Kim et al., 2015). Our initial experiments demonstrated that the TGFB1 sequence with E. coli-optimized codon usage was expressed in inclusion bodies. To overcome this, we inserted the sequence into a protein production plasmid (pET-32a) designed to produce a thioredoxin fusion protein allowing expression in the cytoplasm. It has also been demonstrated that TGF-β3 is more potent than TGF-β1 (Huang et al., 2014). Comparing TGF-β1, TGF-β1m, TGF-β3, and TGF-β3m, we found that TGF-β3 offered the best combination of being able to be produced in E. coli and suitable for use at 0.1 ng mL−1 (Figures 3I, S3C, and S3D) and was therefore selected for the final formula.
Finally, two human embryonic stem cell (hESC) medium formulations that we studied, DC-HAIF and iDEAL, contain both FGF2 and NRG1 (Figure S1B). We found that supplementation with all tested levels of NRG1 enhanced growth by >15% over FGF2-G3 alone (Figures 3J and S3E), although NRG1 is not able to support growth in the absence of FGF2 (Figure S3F). We similarly generated recombinant NRG1 protein using pET-32a, which prevented production as inclusion bodies, with subsequent cleavage using thrombin to form an active protein (Figures S3G and S4). Our final B8 medium formulation was derived from these five-passage assay optimizations and is shown in Figure 3K.
Demonstration of the Suitability of B8 for hiPSC Generation and Weekend-Free Culture
The gold-standard demonstration of the suitability of a hiPSC medium is the capacity to generate hiPSC lines and maintain them in long-term culture. Our standard hiPSC line 19c3 has been cultured for >100 passages (p) in B8 (Figure 4A) and maintained expression of markers of undifferentiated status (SSEA4 and TRA-1-60) by flow cytometry (p131 at time of assay). We also generated hiPSC lines from 37 patients using established protocols but using B8. These hiPSC lines maintained SSEA4 and TRA-1-60 expression in culture (up to p65 at time of assay) (Figure 4A). In B8 hiPSCs maintained an hESC-like morphology (Figure 4B); positive immunofluorescent staining for SSEA4, POU5F1, SOX2, and TRA-1-60 (Figure 4C); and normal karyotype (Figure 4D). We then compared this B8 formula (Figure 3K) with our traditional E8 formula and found growth similar to that in B8 across commonly used split ratios (Figure 5A). It has been demonstrated that, unlike commercial FGF2, thermostable variants of FGF2 such as FGF2-G3 are capable of inducing pERK in FGF-starved cells, even after the medium had previously been stored for extended periods at 37°C (Chen et al., 2012, Dvorak et al., 2018). To confirm that our FGF2-G3 performed similarly, we performed a comparable assay and corroborated that FGF2-G3 was stable after 7 days at 37°C, whereas commercial FGF2 was not capable of stimulating pERK after 2 days at 37°C. As common weekend-free medium formulae such as StemFlex have reverted to using albumin and likely contain heparin, both of which are not chemically defined, we also assessed the function of these components in this assay and found that both improved the stability of FGF2-G3 (Figure 5B).
Figure 4.
Qualification of B8 as Suitable for hiPSC Generation and Culture
(A) Demonstration of the expression of markers of undifferentiated status in hiPSC line 19c3 cultured in B8 from passage (p) 21 to p131 (left) and 37 individual hiPSC lines derived in B8 from p12 to p65 (right), assessed by flow cytometry.
(B) Phase-contrast images (original magnification 10×) of hiPSC line 19c3 p44 cultured in B8. Scale bar, 100 μm.
(C) Expression of markers of undifferentiated status in a variety of B8-derived hiPSC lines. Scale bar, 100 μm.
(D) Example G-banding karyotype analysis of four hiPSC lines derived in B8.
Figure 5.
Qualification of B8 as Suitable for hiPSC Generation and Culture
(A) 19c3 hiPSC growth at low seeding densities in B8 compared with E8 (n = 8). n = full experimental replicates, Mann-Whitney test, ∗p ≤ 0.05, n.s., not significant.
(B) Assessment of stimulation of phospho-ERK after media had been stored at 37°C for 2 or 7 days, comparing in-house generated FGF2-G3 (with or without 0.5 mg mL−1 fatty acid-free albumin or 100 ng mL−1 heparin) with a commercial FGF2 (Peprotech). hiPSCs were starved of FGF2 for 24 h and then treated with the indicated medium for 1 h before collection for western blot, except for “No starve” cells, which were maintained in B8 with FGF2-G3. Total ERK was used as a loading control.
With the understanding that B8 supports significantly enhanced growth of hiPSCs across a variety of low seeding densities, which are sub-optimal for growth (Figure 5A), we established the suitability of this formula to skip days of medium change. We established a matrix of medium change days both with and without thiazovivin (Figure 6A) and showed that daily B8 medium changes (top row) were, surprisingly, one the least suitable for growth rate. B8T treatment without medium changes was the least effective of the thiazovivin-containing protocols (bottom row), whereas B8T followed by B8 (dark gray bar) was a suitable compromise between growth rate and extended exposure to thiazovivin. With the knowledge of the influence of various medium change-skipping timelines, we devised two 7-day schedules consisting of passaging cells every 3.5 days that would allow for the skipping of medium changes on the weekends, a major caveat of current hiPSC culture protocols (Figures 6B and 6C). Our data demonstrated that a weekend-free 3.5-day schedule with medium exchange after the first 24 h (WF) was suitable over a 25-passage timeline, resulting in 90 population doublings compared with 102 with daily medium changes (B8 daily). The addition of 0.5 mg mL−1 albumin enhanced this WF protocol to 98 population doublings. In contrast, no medium exchange (i.e., passage only, WF no Δ), either with or without albumin, was not as suitable (Figure 6D). Further experiments with various doses of albumin or heparin further confirmed that these do not have an additive effect on weekend-free growth in B8 (Figure 6E). One of the major caveats of skipping medium changes is that differentiation efficiency and reproducibility are diminished. We used our existing cardiac (Burridge et al., 2014), endothelial (Patsch et al., 2015), and epithelial differentiation (Li et al., 2015, Qu et al., 2016) protocols to demonstrate that B8 supported differentiation to similarly high levels with either daily medium changes or our weekend-free protocol (Figures 6F–6H).
Figure 6.
Optimization of Weekend-Free Passaging Schedule that Is Still Compatible with Monolayer Differentiation
(A) Establishment of an optimal 3.5-day medium change schedule. Experiments were completed with hiPSC line 19c3. Gray box represents B8T treatment, white box represents B8 treatment, length of box represents number of days of culture in that medium (n = 8).
(B) Weekend-free (WF) passage and medium change schedule. Gray shaded represent days of medium change. 1:20 refers to passage split ratio.
(C) Weekend-free schedule without medium change (WF no Δ).
(D) Comparison of growth when using the WF and WF no Δ schedules, with or without addition of 0.5 mg mL−1 fatty acid-free albumin over 25 passages.
(E) Comparison with the addition of varying levels of fatty acid-free albumin (mg mL−1) to a WF schedule (n = 4).
(F) Cardiac differentiation efficiency when using WF schedule (n = 5).
(G) Endothelial differentiation efficiency when using WF schedule (n = 6).
(H) Epithelial differentiation efficiency when using WF schedule with or without addition of 0.5 mg mL−1 albumin (A) (n = 7). n = full independent experimental replicates.
Discussion
The hiPSC field predominantly utilizes two hiPSC culture media, the commercial mTeSR1 and Essential 8. These two media, along with six other published hiPSC culture medium formulae, have much in common due to their shared history (Figure S1B). Despite this, there are many medium components that have not had their concentration optimized or necessity demonstrated. In many cases, especially the formulation of TeSR, optimization experiments were discussed yet no data were provided. In the experiments here, we found that only five components were truly essential for hiPSC culture: insulin, sodium selenite, FGF2, DMEM/F12 (Figure 1), and TGF-β1. The importance of the fifth component, TGF-β1, was evident only in the five-passage assay (Figures 1I, 3I, S3A, S3C, and S3D). The other three components, ascorbic acid 2-phosphate, transferrin, and NRG1, are dispensable for hiPSC growth, although their removal results in a reduced growth rate.
During the development of this assay platform, we found that precise measurement of relative growth was a suitable surrogate for metrics of the pluripotent state (such as NANOG expression). When cells began to spontaneously differentiate, such as when TGF-β1 is omitted from the formula, the resulting slowing of growth was easily detectable. We noted a number of surprising results during this optimization. For example, we could not find a positive or negative effect of the addition of albumin (Figures 2D and 6D), despite it being a common constituent of many academic and commercial medium formulae. We propose that high levels of ascorbic acid in our formula may have eliminated some of the need for the antioxidant role of albumin. We found that Activin A was not suitable, either with or without TGF-β1 (Figures 3H and S3B), despite its inclusion in a variety of other formulae. We do show that at very low doses Activin A could support growth, albeit to a lesser extent than TGF-β1. In some medium formulae, such as hESF9T, there is a positive correlation between higher levels of TGF-β1 (up to 10 ng mL−1) and expression of NANOG (Yamasaki et al., 2014), while others have found only minimal differences above 0.1 ng mL−1 (Frank et al., 2012), which is in line with our results. Similarly, it was shown that higher levels of human serum albumin correlated with higher levels of POU5F1, SOX2, and NANOG (Frank et al., 2012), yet albumin-free formulae have been used extensively without issue.
One of our major efforts during the development of B8 was the minimization of medium costs. Typical list prices of 1 L of commercial medium such as Essential 8 or mTeSR1 are US$450–US$550 (Figure S5A) plus shipping. In our previous E8-based formula, the growth factors FGF2 and TGF-β1 represented more than 80% of the total medium reagent costs, and we found that this could be much higher if laboratories could not negotiate discounts from commercial recombinant protein suppliers, resulting in a reagent cost of ∼$115 per liter (Figure S5A). The generation of recombinant proteins in house, shown here to be major factor in reducing medium costs, although daunting, was highly simplified by using three commercial products: MagicMedia for E. coli growth, B-PER II for lysis, and Ni-NTA spin columns for purification. Combined, these eliminate much of the complexity in recombinant protein production, including the need for high-performance liquid chromatography or fast protein liquid chromatography. Each of these components could easily be replaced by more cost-effective procedures, such as inducing protein expression using isopropyl β-D-1-thiogalactopyranoside, making lysis buffers, or using higher-throughput columns, albeit with greater complexity. Our optimization of the plasmids and generation of thioredoxin fusion proteins where necessary eliminate much of the complexity associated with inclusion bodies and the resulting refolding processes otherwise required. A typical 1-L E. coli culture, which requires 2 days and basic laboratory skills, will usually provide 15 mg of FGF2-G3, enough for ∼375 L of B8. Similarly, a 1-L culture of TGF-β3 or NRG1 will commonly provide enough protein for years of work (1000 L of B8 medium). The recombinant protein production can be outsourced to a core facility, such as http://rppc.mccormick.northwestern.edu, to remove some of the major burden of complexity, with only minimal costs and high yield of ∼80 mg L−1 of E. coli. This optimization results in a medium reagent cost of ∼$16 per liter and leaves insulin and transferrin as the major remaining reagent costs, although the concentrations of these components can be reduced to 5 μg mL−1 with only minor impact on growth rate (Figures 3B and 3D), reducing reagent costs to ∼$11 per liter (Figure S5C). The difference between growth rate of hiPSCs in either formula is likely undetectable in normal culture.
Our costs described here include only the generation or purchase of reagents at list price, and numerous other costs associated with compounding media in house will influence cost effectiveness. Savings will be less efficient for labs that use very small quantities of B8; this issue may be overcome by multiple laboratories pooling resources or an institute's core facility taking responsibility for supplement aliquot generation.
The final major reagent cost is the basal medium, DMEM/F12, now representing ∼75% of the cost of the reagents in the B8 formula (Figure S5A). It is simple to generate DMEM/F12 from powder to reduce costs, although the requirement for suitable water quality and maintaining these systems, and the use of filter sterilization, adds cost and complexity to the medium-making process and therefore may not be appropriate for all but the highest usage labs. DMEM/F12 is a combination of 52 amino acids, vitamins, inorganic salts, and other components derived from DMEM, a high-nutrient (amino acid and vitamin)-concentration medium, originally optimized for fibroblast growth, and F12, a rich and complex fatty acid-containing medium optimized for CHO cells to generate a complex “catch-all” formula. Few experiments have been completed to compare alternatives to DMEM/F12 for hiPSC culture; indeed, Chen et al. (2011) showed comparable results between DMEM/F12 and the comparatively simple MEMα, suggesting that there is much to learn in this respect.
hiPSC culture without daily medium changes introduces a number of potential caveats; for example, we know that the L-glutamine in the medium is unstable at 37°C and that the concentration is reduced by about a third over 4 days. We also know that cells are producing lactate and ammonia, reducing the pH of the medium, although this is buffered partially by the HEPES and sodium bicarbonate. Finally, hiPSCs release autocrine or paracrine factors into the medium that may induce differentiation, and the increase in these factors over time has not been decoupled from the use of nutrients and production of metabolic waste. We chose not to maintain exposure to ROCK1/2 inhibitors as there exist data suggesting that this induces epithelial-to-mesenchymal transition (Maldonado et al., 2016). Furthermore, ROCK inhibitors have a wide range of off-target effects (Andrews et al., 2010). These issues may result in undesired effects, although this remains to be validated experimentally, and formulations do exist that maintain constant ROCK1/2 inhibitor exposure (Tsutsui et al., 2011).
As hiPSC projects become larger and move beyond proof-of-principle experiments, the labor required to maintain cells becomes burdensome, especially the need for daily medium changes 7 days a week. Here we show that our B8 formula is well suited to weekend-free schedules. A major issue with some commercial media is that although a weekend-free schedule is feasible, growth of hiPSCs is considerably slower, and it is recommended to grow cells as low-density colonies. These low-density colonies are not compatible with subsequent monolayer differentiation protocols, as have become commonplace with the majority of lineages. The optimization of B8 specifically for fast monolayer growth, along with the incorporation of thermostable FGF2-G3, overcomes many of these issues while maintaining compatibility with common differentiation protocols and has become standard practice within our laboratory.
Experimental Procedures
Detailed methods are provided in the Supplemental Information.
Human Induced Pluripotent Cell Culture
All pluripotent and reprogramming cell cultures were maintained at 37°C with 5% CO2 and 5% O2. Differentiation cultures were maintained at 5% CO2 and atmospheric (∼21%) O2. E8 medium was made in house as previously described (Burridge et al., 2015, Chen et al., 2011). Other medium components tested were human Long R3 IGF1 (Sigma, 91590C), thiazovivin (LC Labs, T-9753), recombinant human TGF-β3 (Cell Guidance Systems, GFH109), sodium bicarbonate (Sigma, S5761), NEAA (Gibco, 11140050), Chemically Defined Lipid Concentrate (Gibco, 11905031), fatty acid-free BSA (GenDEPOT, A0100), and heparin sodium salt (Sigma H3149-250KU). The pH was adjusted with 1 N HCl (Sigma, H9892) or 1 N NaOH (Sigma, S2770) and measured at room temperature and atmospheric CO2 using a SevenCompact pH meter (Mettler Toledo). Osmolarity was adjusted with sodium chloride (Sigma, S5886) or cell culture water (Corning, 25-055-CV) and measured with a Vapro 5600 vapor pressure osmometer (Wescor).
Medium Variable Optimization Protocol
The hiPSC line 19c3 (p20–p60), derived from a healthy male, was used for medium variable optimization. Prior to assay, cells were grown to 75% confluence after 4 days of culture as above. Cells were dissociated with TrypLE (Gibco, 12604013) for 3 min at 37°C and resuspended in DMEM/F12, transferred to a 15-mL conical tube (Falcon), and centrifuged at 200 × g for 3 min (Sorvall ST40). The pellet was resuspended in DMEM/F12 and diluted to 1 × 105 cells mL−1, and 10,000 cells were plated per well in Matrigel (1:800)-coated 12-well plates (Greiner) in the medium to be tested along with 2 μM thiazovivin for the first 24 h. Media were changed daily and cells were grown for 6 days. This lower than normal seeding density was used to allow the discovery of factors detectable only under more extreme conditions and therefore provide data on the robustness of the formulation. Cell growth was then assessed using PrestoBlue (Invitrogen, A13262), by adding 100 μL of PrestoBlue to the 1 mL of existing medium in each well and incubating for 2 h at 37°C. Fluorescence (560 nm excitation, 590 nm emission) was then measured using a Varioskan LUX (Thermo Scientific) plate reader with “top read” function.
Population Doubling Level Assessment
Population doubling level (PDL) was calculated according to the following formula:
| PDL = 3.32[log10(n/n0)], |
where n = cell number and n0 = number of cells seeded.
Cardiac Differentiation
Differentiation into cardiomyocytes was performed according to a previously described protocol with slight modifications (Burridge et al., 2014, Burridge et al., 2015). Briefly, hiPSCs were split at a 1:20 ratio using 0.5 mM EDTA as above and grown in B8 medium for 4 days, reaching ∼75% confluence. At the start of differentiation (day 0), B8 medium was changed to CDM3 (chemically defined medium, three components) (Burridge et al., 2014), consisting of RPMI 1640 (Corning, 10-040-CM), 500 μg mL−1 fatty acid-free BSA (GenDEPOT, A0100), and 200 μg mL−1 l-ascorbic acid 2-phosphate (Wako, 321-44823) supplemented with 6 μM of glycogen synthase kinase 3-β inhibitor CHIR99021 (LC Labs, C-6556). On day 1, the medium was changed to CDM3, and on day 2 the medium was changed to CDM3 supplemented with 2 μM Wnt inhibitor Wnt-C59 (Biorbyt, orb181132). The medium was then changed on day 4 and then every other day with CDM3. Contracting cells were noted from day 7. On day 14 of differentiation, cardiomyocytes were dissociated using DPBS for 20 min at 37°C followed by 1:200 Liberase TH (Roche, 5401151001) diluted in DPBS for 20 min at 37°C, centrifuged at 300 × g for 5 min, and filtered through a 100-μm cell strainer (Falcon) and analyzed.
Endothelial Differentiation
hiPSCs were grown to ∼60% confluence and differentiated according to an adapted version of a protocol previously described (Patsch et al., 2015). On day 5 of differentiation, endothelial cells were dissociated with Accutase (Corning, 25058Cl) for 5 min at 37°C, centrifuged at 300 × g for 5 min, and analyzed.
Epithelial Differentiation
hiPSCs were split at a 1:20 ratio using 0.5 mM EDTA as above and grown in B8T medium for 1 day, reaching ∼15% confluence at the start of differentiation. Surface ectoderm differentiation was performed according to an adapted version of previously described protocols (Li et al., 2015, Qu et al., 2016). On day 4 of differentiation, epithelial cells were dissociated with Accutase for 5 min at 37°C, centrifuged at 300 × g for 5 min, and analyzed.
Statistical Methods
Data were analyzed in Excel or R and graphed in GraphPad Prism 8. Detailed statistical information is included in the corresponding figure legends. Data are presented as means ± SEM. Comparisons were conducted via a Mann-Whitney test, with significant differences defined as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, and ∗∗∗∗p < 0.0001. No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Author Contributions
P.W.B. conceived and supervised the project. H.-H.K. completed most experiments, along with K.A.F., E.A.P., C.J.W., H.F., M.V.O., M.J., M.R.-T., M.B., and T.M. Recombinant protein production was completed by X.G. and plasmid generation by J.-M.D. C.E. provided technical support, and A.L.G. provided recombinant DNA construction. P.W.B. wrote the paper. All authors approved the manuscript.
Acknowledgments
This work was supported by NIH NCI grant R01 CA220002, American Heart Association Transformational Project Award 18TPA34230105, a Dixon Foundation Translational Research Grants Innovation Award (P.W.B.), and the Fondation Leducq (P.W.B., A.L.G.).
Published: January 9, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.12.007.
Supplemental Information
References
- Amit M., Shariki C., Margulets V., Itskovitz-Eldor J. Feeder layer- and serum-free culture of human embryonic stem cells. Biol. Reprod. 2004;70:837–845. doi: 10.1095/biolreprod.103.021147. [DOI] [PubMed] [Google Scholar]
- Andrews P.D., Becroft M., Aspegren A., Gilmour J., James M.J., McRae S., Kime R., Allcock R.W., Abraham A., Jiang Z. High-content screening of feeder-free human embryonic stem cells to identify pro-survival small molecules. Biochem. J. 2010;432:21–33. doi: 10.1042/BJ20101022. [DOI] [PubMed] [Google Scholar]
- Beers J., Gulbranson D.R., George N., Siniscalchi L.I., Jones J., Thomson J.A., Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J. Biol. Chem. 2004;279:45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- Braam S.R., Zeinstra L., Litjens S., Ward-van Oostwaard D., van den Brink S., van Laake L., Lebrin F., Kats P., Hochstenbach R., Passier R. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008;26:2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- Breckwoldt K., Letuffe-Breniere D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Holmstrom A., Wu J.C. Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Curr. Protoc. Hum. Genet. 2015;87:21.3.1–21.3.15. doi: 10.1002/0471142905.hg2103s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton P., Adams D.R., Abraham A., Allcock R.W., Jiang Z., McCahill A., Gilmour J., McAbney J., Kane N.M., Baillie G.S. Identification and characterization of small-molecule ligands that maintain pluripotency of human embryonic stem cells. Biochem. Soc. Trans. 2010;38:1058–1061. doi: 10.1042/BST0381058. [DOI] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Yu P., Hou Z., Thomson J.A. Thermal stability of fibroblast growth factor protein is a determinant factor in regulating self-renewal, differentiation, and reprogramming in human pluripotent stem cells. Stem Cells. 2012;30:623–630. doi: 10.1002/stem.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou B.K., Mali P., Huang X., Ye Z., Dowey S.N., Resar L.M., Zou C., Zhang Y.A., Tong J., Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou B.K., Gu H., Gao Y., Dowey S.N., Wang Y., Shi J., Li Y., Ye Z., Cheng T., Cheng L. A facile method to establish human induced pluripotent stem cells from adult blood cells under feeder-free and xeno-free culture conditions: a clinically compliant approach. Stem Cells Transl. Med. 2015;4:320–332. doi: 10.5966/sctm.2014-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbordes S.C., Placantonakis D.G., Ciro A., Socci N.D., Lee G., Djaballah H., Studer L. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak P., Bednar D., Vanacek P., Balek L., Eiselleova L., Stepankova V., Sebestova E., Kunova Bosakova M., Konecna Z., Mazurenko S. Computer-assisted engineering of hyperstable fibroblast growth factor 2. Biotechnol. Bioeng. 2018;115:850–862. doi: 10.1002/bit.26531. [DOI] [PubMed] [Google Scholar]
- Eminli S., Foudi A., Stadtfeld M., Maherali N., Ahfeldt T., Mostoslavsky G., Hock H., Hochedlinger K. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forristal C.E., Wright K.L., Hanley N.A., Oreffo R.O., Houghton F.D. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2010;139:85–97. doi: 10.1530/REP-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Zhang M., Scholer H.R., Greber B. Small molecule-assisted, line-independent maintenance of human pluripotent stem cells in defined conditions. PLoS One. 2012;7:e41958. doi: 10.1371/journal.pone.0041958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie Y., Fusaki N., Katayama T., Hamasaki M., Soejima Y., Soga M., Ban H., Hasegawa M., Yamashita S., Kimura S. New type of Sendai virus vector provides transgene-free iPS cells derived from chimpanzee blood. PLoS One. 2014;9:e113052. doi: 10.1371/journal.pone.0113052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furue M.K., Na J., Jackson J.P., Okamoto T., Jones M., Baker D., Hata R., Moore H.D., Sato J.D., Andrews P.W. Heparin promotes the growth of human embryonic stem cells in a defined serum-free medium. Proc. Natl. Acad. Sci. U S A. 2008;105:13409–13414. doi: 10.1073/pnas.0806136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan N.R., Segeritz C.P., Touboul T., Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Schor S.L., Hinck A.P. Biological activity differences between TGF-beta1 and TGF-beta3 correlate with differences in the rigidity and arrangement of their component monomers. Biochemistry. 2014;53:5737–5749. doi: 10.1021/bi500647d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.S., Postovit L.M., Lajoie G.A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- Kim Y.V., Gasparian M.E., Bocharov E.V., Chertkova R.V., Tkach E.N., Dolgikh D.A., Kirpichnikov M.P. New strategy for high-level expression and purification of biologically active monomeric TGF-beta1/C77S in Escherichia coli. Mol. Biotechnol. 2015;57:160–171. doi: 10.1007/s12033-014-9812-7. [DOI] [PubMed] [Google Scholar]
- Kumagai H., Suemori H., Uesugi M., Nakatsuji N., Kawase E. Identification of small molecules that promote human embryonic stem cell self-renewal. Biochem. Biophys. Res. Commun. 2013;434:710–716. doi: 10.1016/j.bbrc.2013.03.061. [DOI] [PubMed] [Google Scholar]
- Lawitts J.A., Biggers J.D. Joint effects of sodium chloride, glutamine, and glucose in mouse preimplantation embryo culture media. Mol. Reprod. Dev. 1992;31:189–194. doi: 10.1002/mrd.1080310305. [DOI] [PubMed] [Google Scholar]
- Li L., Song L., Liu C., Chen J., Peng G., Wang R., Liu P., Tang K., Rossant J., Jing N. Ectodermal progenitors derived from epiblast stem cells by inhibition of Nodal signaling. J. Mol. Cell Biol. 2015;7:455–465. doi: 10.1093/jmcb/mjv030. [DOI] [PubMed] [Google Scholar]
- Liu W., Ren Z., Lu K., Song C., Cheung E.C.W., Zhou Z., Chen G. The suppression of medium acidosis improves the maintenance and differentiation of human pluripotent stem cells at high density in defined cell culture medium. Int. J. Biol. Sci. 2018;14:485–496. doi: 10.7150/ijbs.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T., Thomson J.A. Defined, feeder-independent medium for human embryonic stem cell culture. Curr. Protoc. Stem Cell Biol. 2007 doi: 10.1002/9780470151808.sc01c02s2. Chapter 1, Unit 1C.2. [DOI] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., Thomson J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Ludwig T.E., Levenstein M.E., Jones J.M., Berggren W.T., Mitchen E.R., Frane J.L., Crandall L.J., Daigh C.A., Conard K.R., Piekarczyk M.S. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- Maldonado M., Luu R.J., Ramos M.E., Nam J. ROCK inhibitor primes human induced pluripotent stem cells to selectively differentiate towards mesendodermal lineage via epithelial-mesenchymal transition-like modulation. Stem Cell Res. 2016;17:222–227. doi: 10.1016/j.scr.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Marinho P.A., Chailangkarn T., Muotri A.R. Systematic optimization of human pluripotent stem cells media using design of experiments. Sci. Rep. 2015;5:9834. doi: 10.1038/srep09834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkoumian Z., Weber J.L., Weber D.M., Fadeev A.G., Zhou Y., Dolley-Sonneville P., Yang J., Qiu L., Priest C.A., Shogbon C. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat. Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- Narva E., Pursiheimo J.P., Laiho A., Rahkonen N., Emani M.R., Viitala M., Laurila K., Sahla R., Lund R., Lahdesmaki H. Continuous hypoxic culturing of human embryonic stem cells enhances SSEA-3 and MYC levels. PLoS One. 2013;8:e78847. doi: 10.1371/journal.pone.0078847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsch C., Challet-Meylan L., Thoma E.C., Urich E., Heckel T., O'Sullivan J.F., Grainger S.J., Kapp F.G., Sun L., Christensen K. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini I., Rao J., Seebohm G., Greber B. Human pluripotent stem cell-derived cardiomyocytes: genome-wide expression profiling of long-term in vitro maturation in comparison to human heart tissue. Genome Data. 2015;4:69–72. doi: 10.1016/j.gdata.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowse A.B., Doran M.R., Cooper-White J.J., Chong F., Munro T.P., Fitzpatrick J., Chung T.L., Haylock D.N., Gray P.P., Wolvetang E.J. Long term culture of human embryonic stem cells on recombinant vitronectin in ascorbate free media. Biomaterials. 2010;31:8281–8288. doi: 10.1016/j.biomaterials.2010.07.037. [DOI] [PubMed] [Google Scholar]
- Qu Y., Zhou B., Yang W., Han B., Yu-Rice Y., Gao B., Johnson J., Svendsen C.N., Freeman M.R., Giuliano A.E. Transcriptome and proteome characterization of surface ectoderm cells differentiated from human iPSCs. Sci. Rep. 2016;6:32007. doi: 10.1038/srep32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S., Domogatskaya A., Strom S., Hansson E.M., Chien K.R., Inzunza J., Hovatta O., Tryggvason K. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat. Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Niaudet C., Simonson O.E., Salmela E., Hansson E.M., Domogatskaya A., Xiao Z., Damdimopoulou P., Sheikhi M. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 2014;5:3195. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
- Sato N., Sanjuan I.M., Heke M., Uchida M., Naef F., Brivanlou A.H. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A.L., Sun Y., Menendez L., Kulik M., Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J., Dawlaty M.M., Gao Q., Maetzel D., Hanna J., Sommer C.A., Mostoslavsky G., Jaenisch R. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.K., Toh C.X., Ma D., Yang B., Liu T.M., Lu J., Wong C.W., Tan T.K., Li H., Syn C. Human finger-prick induced pluripotent stem cells facilitate the development of stem cell banking. Stem Cells Transl. Med. 2014;3:586–598. doi: 10.5966/sctm.2013-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Valamehr B., Hindoyan A., Qiao R., Ding X., Guo S., Witte O.N., Liu X., Ho C.M., Wu H. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat. Commun. 2011;2:167. doi: 10.1038/ncomms1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Vallier L., Touboul T., Brown S., Cho C., Bilican B., Alexander M., Cedervall J., Chandran S., Ahrlund-Richter L., Weber A. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells. 2009;27:2655–2666. doi: 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- Wang L., Schulz T.C., Sherrer E.S., Dauphin D.S., Shin S., Nelson A.M., Ware C.B., Zhan M., Song C.Z., Chen X. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Rosler E., Jiang J., Lebkowski J.S., Gold J.D., O'Sullivan C., Delavan-Boorsma K., Mok M., Bronstein A., Carpenter M.K. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells. 2005;23:315–323. doi: 10.1634/stemcells.2004-0211. [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Taguchi Y., Shimamoto A., Mukasa H., Tahara H., Okamoto T. Generation of human induced pluripotent stem (Ips) cells in serum- and feeder-free defined culture and TGF-Beta1 regulation of pluripotency. PLoS One. 2014;9:e87151. doi: 10.1371/journal.pone.0087151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S.Y., Ikeda T., Shahsavarani H., Yoshida N., Nayer B., Hino M., Vartak-Sharma N., Suemori H., Hasegawa K. Chemically defined and growth-factor-free culture system for the expansion and derivation of human pluripotent stem cells. Nat. Biomed. Eng. 2018;2:173–182. doi: 10.1038/s41551-018-0200-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.