Abstract
This study aim at investigating the function of microRNA (miR)-34b-5p in breast cancer prognosis and development. qRT-PCR was used for miR-34b-5p expression examination in breast cancer samples. CCK8, immunohistochemistry, scratch wound healing, transwell assays were performed for cell experiments. Subcutaneously implanted tumor model was carried out for animal experiment. Western blot was conducted for protein expression detection. Bioinformatics analysis was performed for exploring the underlying mechanism. MiR-34b-5p expression was down-regulated in breast cancer samples and cells, and miR-34d-5p could inhibit cell viability, migration and invasion also delay tumor growth in vivo. Low miR-34b-5p expression showed a bad prognosis of breast cancer patients. MiR-34b-5p functioned as a tumor suppressor by targeting ARHGAP1, and ARHGAP1 knockdown could reverse the effect of miR-34b-5p inhibitor on breast cancer cells. MiR-34b-5p exerts an anti-tumorigenesis role in breast cancer cells by targeting ARHGAP1, which is profitable for the breast cancer diagnosis and molecular treatment.
Keywords: microRNA-34b-5p, breast cancer, tumor growth, invasion, ARHGAP1
Introduction
Breast cancer is a malignant tumor that severely impacts female inhabitants’ health and life. Global cancer statistics for 2018 showed that lung cancer and breast cancer are the two most common types of cancer in terms of the number of new cases [1]. In 2018, there were 2.1 million new breast cancer patients, accounting for 11.6% of the global burden of cancer incidence [2]. Although the clinical therapeutic effect of breast cancer has been improving in recent years, the incidence of breast cancer still ranks first in the incidence of female malignant tumors in China, and shows a younger trend in the affected population [3,4]. The occurrence and development of breast cancer is a very complicated pathological process, and the pathogenesis of breast cancer is still ignorant. Current studies suggest that the occurrence of breast cancer may be related to abnormal gene expression caused by epigenetic modification [5,6].
MicroRNAs (miRNAs) are a class of small non-coding RNAs composed of 18-22 nucleotides, which play an indispensable part in plenty of cancers. In breast cancer, a number of miRNAs are reported down-regulated in tumor tissues and exert anti-oncogenesis effect on cell proliferation and invasion, such as miR-224-5p [6], miR-30 family [7], miR-26-5p [8]. Preliminary studies showed that miR-34b-5p expression was decreased in thyroid carcinoma [9], and it associated with lung injury [10] and pulmonary fibrosis [11]. Kato et al. reported that miR-34 was is required for the DNA damage response human breast cancer cells [12]. Zhou et al. investigated that miR-34b/c-5p was associated with breast cancer cell proliferation and apoptosis [13]. The role of miR-34b-5p in breast cancer prognosis, cell migration and invasion, and underlying mechanism involved are still unclear.
In this work, we studied the expression of miR-34b-5p in breast cancer samples and examined the effect of miR-34b-5p expression on cell proliferation, colony formation, migration and invasion of breast cancer cells. The downstream target gene of miR-34b-5p was also investigated. Our data will be profitable for overcoming the difficulties of breast cancer diagnosis and therapy.
Materials and methods
Patients’ samples collection
Seventy-nine paired breast cancer tissues and adjacent non-tumor tissues were collected from May, 2011 to October, 2017 in Women’s Hospital, School of Medicine, Zhejiang University (Hangzhou, China). The experiments were performed with accordance to Declaration of Helsinki, and were approved by the Ethics Committee of Women’s Hospital, School of Medicine, Zhejiang University. The written informed consent was obtained from every patient with breast cancer involved in our study. The clinicopathological parameters of breast cancer patients were shown in Table 1.
Table 1.
The correlation of miR-34b-5p expression and clinicopathologic feature of breast cancer patients
| Features | MiR-34b-5p expression | P value | |
|---|---|---|---|
|
| |||
| High (n=32) | Low (n=47) | ||
| Age | |||
| >60 | 19 | 26 | P>0.05 |
| ≤60 | 13 | 21 | |
| Tumor size | |||
| <3 cm | 17 | 16 | P<0.05 |
| ≥3 cm | 15 | 31 | |
| TNM stage | |||
| I-II | 30 | 20 | P<0.05 |
| III-IV | 2 | 27 | |
| Lymphatic metastasis | |||
| Metastasis | 14 | 29 | P>0.05 |
| No metastasis | 18 | 22 | |
Immunohistochemistry (IHC) analysis
Paraffin-embedded tissues were deparaffinized, hydrated in ethyl alcohol and incubated with 0.3% H2O2 for elimination of endogenous peroxidase activity. After rupture of nuclear membrane with 0.1% Triton X-100 for 30 mins and blockage with 5% normal donkey serum, slides were incubated with primary antibody at 4°C overnight and secondary antibody at room temperature for 1 h. Immunohistochemistry results were captured using Nikon Eclipse 80i microscope.
Cell lines and culture
Brest cancer cell lines including T47D, SK-BR-3, HCC197, MDA-MB-231, and MCF-7, and normal mammary cells (MCF10A) were purchased from ATCC Company (Manassas, VA, USA). Cells were cultured in DMEM medium (Beyotime, Shanghai, China) supplemented with 10% (Beyotime) at 37°C in a humidified atmosphere containing 5% CO2.
Cell transfection
MiR-34b-5p mimic, miR-34b-5p inhibitor and corresponding controls (miR-NC), siARHGAP1 and negative controls (siNC), and oeARHGAP1 and negative control (NC) were synthesis from Genechem (Shanghai, China), and were transfected into breast cancer cells by a Lipofectamine 2000 (Invitrogen, NewYork, USA) according to the manufacturer’s specification. After transfection for 48 hours, transfection results were examined by qRT-PCR and further experiments were also carried out.
qRT-PCR
Total RNAs from cells and tissues were obtained by using a Total RNA extraction kit (Solarbio). The reverse transcription reaction was carried out using TaqMan® MicroRNA Reverse Transcription (RT) Kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) (bio-rad CFX96) was used for SYBR Premix Ex Taq RR420A (Takara). β-actin and U6 were used for endogenous reference of ARHGAP1 and miR-34b-5p using 2-ΔΔCT method, respectively. The primers (Genechem) used are as following: miR-34b-5p: Forward, GGGTAGGCAGTGTCATTAGC; Reverse, AACAACCAACACAACCCAAC. ARHGAP1: Forward, CGTTTCCTGACTGCTTTCC; Reverse, TGGCCTTGAGGGTGATG.
CCK8 assay
Cells were digested and inoculated in 96-well plate. The total volume of culture medium was 100 ml (3000 cells) per well. The cells were placed in 37°C and CO2 incubator. The cells were adhered to the cell wall and replaced with culture medium. The culture medium mixed with 10% CCK-8 reagent was added into each hole. The cells were incubated at 37°C for 1 h. The absorbance was measured at 450 nm after room temperature equilibrium was taken out from incubator. Cell viability was measured at 24, 48 and 72 h and cell growth curve was drawn.
Clone formation analysis
After transfection, breast cancer cells were collected by trypsin working fluid and then washed in conditioned medium twice. Cells were suspended in conditioned medium and the concentration of cell suspension was adjusted to 1 × 103/ml. 4 ml culture medium was added to 6-well culture plate, and 0.4 ml cell suspension was added to each hole after balancing in a CO2 incubator. After 15 days, abandon the culture medium, wash the cells carefully with PBS twice, add 1 ml/hole of polyformaldehyde, fix for 10 minutes, discard the fixative solution, wash it slowly with running water, add LML crystal violet to each hole for 30 minutes, wash the dye solution slowly with running water, and dry it in the fume cupboard. Using gel imaging system to count the number of clones, scan and save the image.
Wound healing assay
We performed wound healing assay for detecting cell migration. Breast cancer cells were incubated in a 6-well plate with a density of 1.0 × 105. When the cells were fully confluent, the cells were scratched at the tip of the sterile pipette, and then washed twice by PBS to remove the separated cells. Microscopic images of wounds and migrating cells were taken at 0 and 24 hours.
Transwell assay
To investigate cell invasion of MDA-MB-231 cells and MCF-7 cells, transwell assay was performed. The transwell chamber was treated with Matrigel (Solarbio, Beijing, China), MDA-MB-231 cells and MCF-7 cells were cultured in the upper chamber (Solarbio) with non-serum medium and maintained at 37°C in 5% CO2 overnight. Invasive breast cancer cells were fixed with methanol for 10 minutes, then stained with crystal violet (Yuanye, Shanghai, China) and then calculated by microscopy.
Bioinformatics analysis
The expressions of miR-34b-5p and ARHGAP1 in breast cancer tissues and normal tissues were analyzed from TCGA database. The binding sites of miR-34b-5p to ARHGAP1 were predicted by the bioinformatics website miRBase (http://mirbase/org) and TargetScan (http://www.targetscan.org).
Dual luciferase reporter assay
In the luciferase reporter assay, HEK293 cells were transfected with using 10 ng Renilla luciferase vector with Lipofectamine® 2000 and 200 ng of ARHGAP1 3’-UTR or ARHGAP1-mut and 20 mM miR-34b-5p mimic/control (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were collected 48 h after transfection and were analyzed using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA). The luciferase activity was detected using the GloMax fluorescence reader (Promega Corporation), and the authors detected luciferase activity. The pRL-CMV Renilla luciferase reporter was used for contrast correction in transfection efficiency. Each experiment was performed in triplicate.
Subcutaneously implanted tumor model
The animal experiments were performed complied with the International Association for the Study of Pain (IASP), and approved by the Animal Ethics Committee of Women’s Hospital, School of Medicine, Zhejiang University. MCF-7 and MDA-MB-231 cells in logarithmic growth phase were digested and counted with 0.25% trypsin when the fusion degree reached 70%-80%. MCF-7 and MDA-MB-231 cells were suspended in a mixture of PBS and Matrigel (ratio 3:1) to form a single cell suspension. The cell concentration was adjusted to 8 × 106/200 μl. Twelve BALB/C female nude mice (4-6 weeks old; 18-20 g) were fed in a pathogen-free laboratory. They were divided into two groups with 6 mice in each group. The right dorsal skin of nude mice was disinfected with 75% alcohol. Then 200 ml of homogeneous suspension of MCF-7 and MDA-MB-231 cells cells (containing 8 × 106 cells) was extracted by sterile syringe (1 ml) and injected subcutaneously into the right dorsal skin of nude mice. The size of subcutaneous tumors was measured with vernier caliper every 7 days. The length (a) and short diameter (b) of the transplanted tumors were recorded. The volume of the transplanted tumors was calculated and the growth curve was drawn according to the volume formula: V (mm3) = (a × b2)/2. On the 35st day after inoculation, nude mice were killed under anesthesia, and the tumors were removed and weighed.
Western blotting
Protein lysates were extracted using radio immunoprecipitation assay (RIPA) buffer with 1 mM phenylmethane sulfonyl fluoride. Samples were subsequently sonicated for 2 min and centrifuged. The supernatants were collected and used for protein analysis. Lysates were separated on 8% polyacrylamide gels and transferred onto PVDF membrane. The membranes were blocked with phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBST) and 5% nonfat milk (w/v) for 1 hour at room temperature. After they were washed with PBST, the membranes were probed with antibodies overnight at 4°C. Anti-ARHGAP1 (ab224231, 1:1000) and anti-GAPDH (ab181602, 1:3000) were obtained from Abcam (Cambridge, MA, USA). The membranes were washed again with PBST, then horseradish peroxidase (HRP) labeled IgG at 1:5000 dilution was added at room temperature for 1 h, and the blots were developed using ECL Western blotting reagents.
Statistical analysis
All the experiments were repeated at least three times. The results were exhibited as mean ± SD. Student’s t-test or one-way ANOVA was used to analyze data from two or multiple groups, respectively. Pearson’s χ2 test was used to analyze the correlation between clinicopathological features and miR-34b-5p expression in breast cancer patients. Spearman’s correlation analysis was used to determine the correlations between the levels of miR-34b-5p and ARHGAP1 in breast cancer tissues. P<0.05 was considered as statistical significant.
Results
The expression of miR-34b-5p was declined in breast cancer
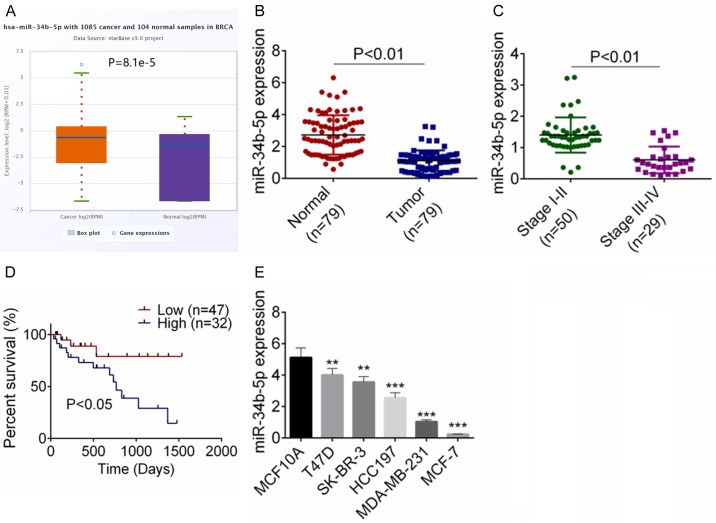
Preliminary data from TCGA database showed that miR-34b-5p was markedly down-regulated in breast cancer tissues as compared with non-tumor tissues (P<0.01, Figure 1A). We subsequently evaluated the miR-34b-5p expressions by qRT-PCR in 79 breast cancer samples and the relevant non-tumor tissues. Figure 1B showed an obvious decrease of miR-34b-5p expression in breast cancer tissues compared with the normal tissues (P<0.01). Moreover, breast cancer patients in stage III-IV exhibited a lower miR-34b-5p expression than the patients in stage I-II (P<0.01, Figure 1C). Also, lower expression of miR-34b-5p indicated an unsatisfied prognosis (P<0.05, Figure 1D). The correlation of clinicopathological characteristics of breast cancer patients and miR-34b-5p expression was analyzed and shown in Table 1. Low miR-34b-5p expression was correlated with TNM stage and tumor size.
Figure 1.
The expression of miR-34b-5p was decreased in breast cancer. A. From TCGA database, miR-34b-5p expression was decreased in breast cancer tissues. B. MiR-34b-5p expression was decreased in 79 breast cancer tissues by qRT-PCR assay. C. The expression of miR-34b-5p in different stages of breast cancer patients was examined by qRT-PCR assay. D. Low expression of miR-34b-5p showed a bad prognosis. E. The expression of miR-34b-5p in breast cancer cell lines (T47D, SK-BR-3, HCC197, MDA-MB-231, MCF-7) and MCF10A cells was detected by qRT-PCR assay. Data are shown as mean ± SD. **P<0.01, ***P<0.001.
The miR-34b-5p expressions in five breast cancer cell lines (T47D, SK-BR-3, HCC197, MDA-MB-231, MCF-7) and a normal cell line (MCF10A) were examined, and Figure 1E revealed that miR-34b-5p expression was descended in tumor cells (P<0.05).
Over-expression of miR-34b-5p showed inhibiting effect on cell growth in vivo and in vitro
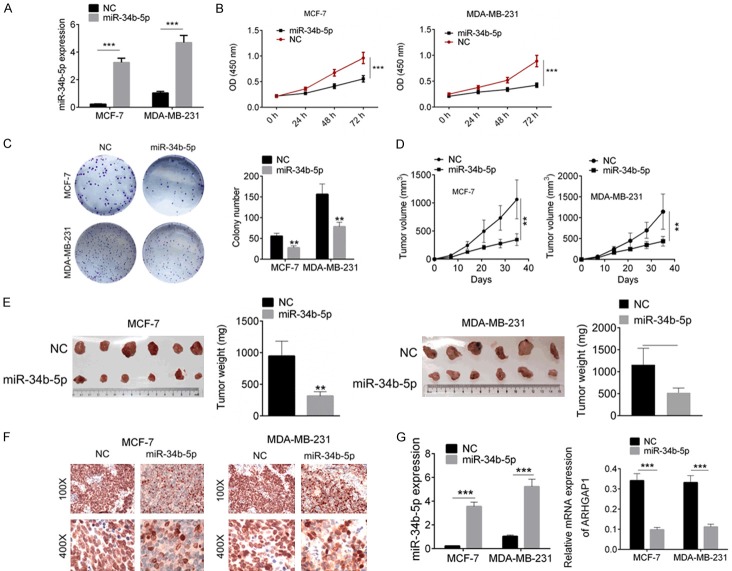
Compared with other breast cancer cell lines, MDA-MB-231 cells and MCF-7 cells displayed the lowest expression of miR-34b-5p and were therefore treated with miR-34b-5p mimic transfection (Figure 2A). Cellular functional experiments were afterwards carried out. Cell proliferation of MDA-MB-231 and MCF-7 cells with miR-34b-5p mimic transfection was evidently decreased as compared to the miR-NC cells by CCK8 assay (P<0.01, Figure 2B). Cell colony-formation ability of breast cancer cells with miR-34b-5p mimic transfection was also inhibited detected by crystal violet staining assay (P<0.01, Figure 2C). We then studied the effect of miR-34b-5p on tumor growth based on a subcutaneously implanted tumor model in nude mouse. Tumor volume was recorded every seven days, and the mice were finally sacrificed after 35 days. We observed in Figure 2D, 2E, miR-34b-5p effectively slowed the implanted tumor growth and reduced tumor volume (P<0.01). The Ki67 expression in miR-NC and miR-34b-5p mimic tumor tissues was identified by immunohistochemical analysis, and miR-34b-5p suppressed Ki67 expression (P<0.01, Figure 2F). Furthermore, the expressions of miR-34b-5p and ARHGAP1 in tumor tissues were examined by RT-PCR analysis, and the results showed that miR-34b-5p expression was increased in miR-34b-5p overexpression group, and ARHGAP1 expression was decreased in miR-34b-5p overexpression group (P<0.001, Figure 2G).
Figure 2.
Over-expression of miR-34b-5p inhibited cell proliferation in vivo and in vitro. A. MDA-MB-231 and MCF-7 cells were transfected with miR-34b-5p mimics and miR-NC, and miR-34b-5p expression was identified by qRT-PCR. B. Cell proliferation was examined by CCK8 assay. C. Cell colony formation was tested by crystal violet staining. D, E. Subcutaneously implanted tumor model in nude mouse was used for evaluating the effect of miR-34b-5p mimic on tumor growth. F. Ki67 expression in tumor tissues was detected by immunohistochemical analysis. G. The expressions of miR-34b-5p and ARHGAP1 were qualified by RT-PCR analysis. Data are shown as mean ± SD. **P<0.01, ***P<0.001.
MiR-34b-5p inhibited tumor cell migration and invasion of breast cancer
We then performed scratch assay and Transwell assay to explore the biological function of miR-34b-5p on cell migration and invasion. As shown in Figure 3A, MDA-MB-231 and MCF-7 cells with miR-34b-5p mimic transfection exhibited a lower migration ability than the cells with miR-NC transfection (P<0.01). In addition, cell migration rates of miR-34b-5p mimictransfected MDA-MB-231 cells (64.7 ± 8.72%) and MCF-7 cells (56.4 ± 6.31%) were also crippled as compared with the miR-NC cells (P<0.01, Figure 3B).
Figure 3.
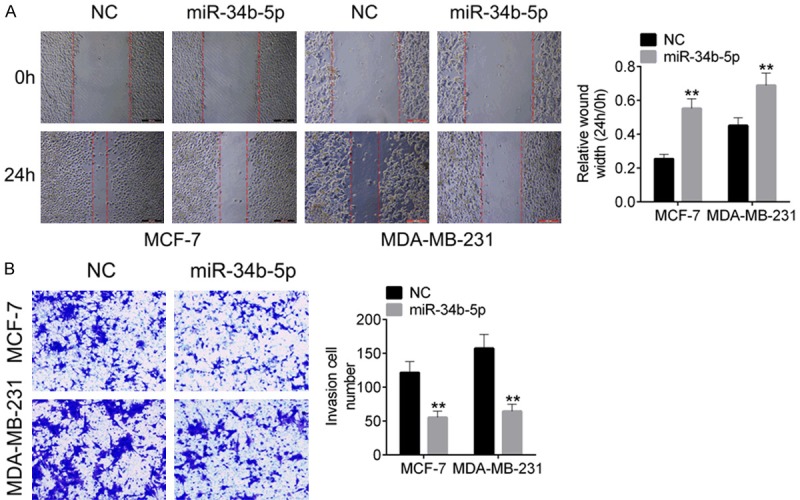
MiR-34b-5p mimic suppressed cell migration and invasion of MDA-MB-231 and MCF-7 cells. A. Cell migration of MDA-MB-231 and MCF-7 cells transfected with miR-34b-5p mimic or miR-NC was identified by wound healing. B. Cell invasion of breast cancer cells was tested by transwell assay. Data are shown as mean ± SD. **P<0.01, ***P<0.001.
MiR-34b-5p targeted ARHGAP1
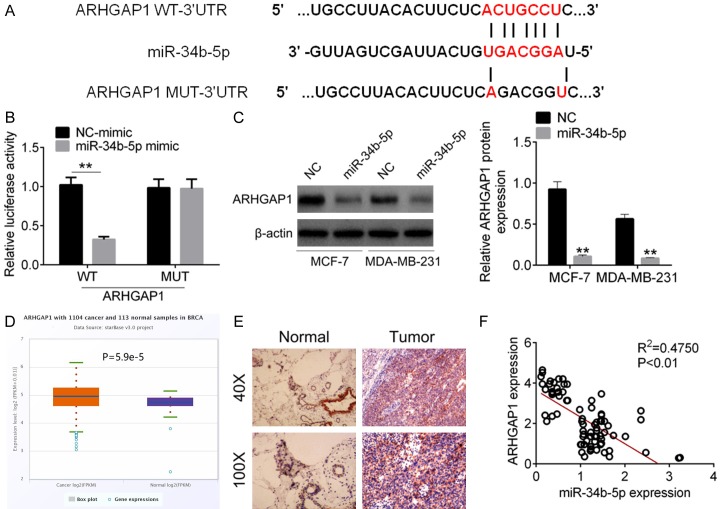
We further investigate the underlying molecular mechanism by which miR-34b-5p regulated cellular physiology. The target genes of miR-34b-5p were analyzed by Targetscan, miRmap and microT, ARHGAP1 was selected as the target of miR-34b-5p (Figure 4A). Dual-luciferase reporter assay proved that ARHGAP1 3’UTR could combine with miR-34b-5p, and therefore decreased the relative fluorescence intensity (P<0.01, Figure 4B). In breast cancer cells, miR-34b-5p inhibited the protein expression of ARHGAP1 (P<0.01, Figure 4C). We also found that ARHGAP1 was over-expression in breast cancer samples from the TCGA data (P=5.9e-5, Figure 4D). The immunohistochemical analysis of tumor tissues in breast cancer patients demonstrated the protein level of ARHGAP1 in was increased (Figure 4E). Furthermore, correlation analysis revealed that the expression of ARHGAP1 was increased with the decreased expression of miR-34b-5p (Figure 4F).
Figure 4.
The target gene of miR-34b-5p was ARHGAP1. A. The binding sites of miR-34b-5p and ARHGAP1 3’UTR. B. Luciferase reporter gene was used for verifying the binding sites. C. Protein expression of ARHGAP1 in MDA-MB-231 and MCF-7 cells transfected with miR-NC and miR-34b-5p mimic was tested by western blot assay. D. ARHGAP1 was over-expressed in breast cancer from TCGA database. E. The protein expression of ARHGAP1 was increased in breast cancer tissues by immunohistochemical analysis. F. The ARHGAP1 expression was negatively correlated with miR-34b-5p expression in breast cancer samples. Data are shown as mean ± SD. **P<0.01, ***P<0.001.
Knockdown of ARHGAP1 restored the effect of miR-34b-5p inhibitor
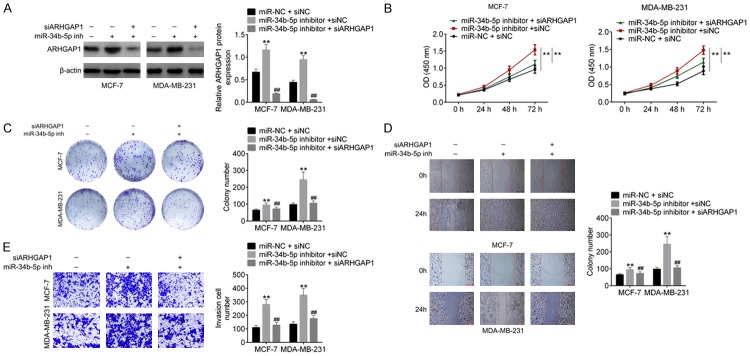
We finally performed the rescue experiments to verify the fact that miR-34b-5p regulated cellular physiology by targeting ARHGAP1. MDA-MB-231 cells and MCF-7 cells were transfected miR-34b-5p inhibitor and then co-transfected with siARHGAP1. After transfection, ARHGAP1 protein levels in miR-NC + siNC, miR-34b-5p inhibitor + siNC, miR-34b-5p inhibitor + siARHGAP1 cells were detected by western blot. The results showed that ARHGAP1 expression was increased in miR-34b-5p inhibitor + siNC cells and descended in miR-34b-5p inhibitor + siARHGAP1 cells (P<0.01, Figure 5A). Additionally, Figure 5B-E showed that miR-34b-5p inhibitor promoted cell proliferation, colony formation ability, cell migration and invasion of MDA-MB-231 cells and MCF-7 cells, while ARHGAP1 knockdown evidently abolished the effect of miR-34b-5p inhibitor.
Figure 5.
ARHGAP1 knockdown abolished the effect of miR-34b-5p inhibitor on breast cancer cells. A. MDA-MB-231 and MCF-7 cells were transfeceted with miR-NC + siNC, miR-34b-5p inhibitor + siNC, miR-34b-5p inhibitor + siARHGAP1, then the protein expression of ARHGAP1 was examined by western blot analysis. B-E. Cell proliferation, colony formation, migration and invasion were examined by CCK8, crystal violet staining, wound healing and transwell, respectively. Data are shown as mean ± SD. **P<0.01, ***P<0.001.
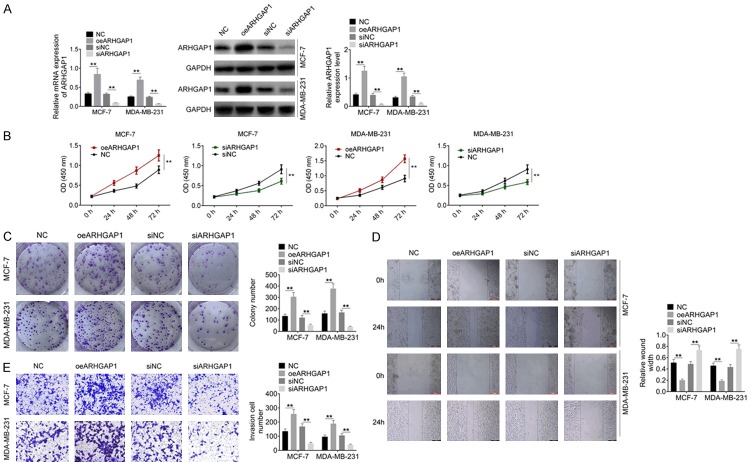
ARHGAP1 regulated cell proliferation, migration and invasion of breast cancer cells
To further explore the biological function of ARHGAP1 in breast cancer cells, MDA-MB-231 cells and MCF-7 cells were transfected with oeARHGAP1 or siARHGAP1 (Figure 6A). Cell proliferation, migration and invasion were subsequently evaluated. As shown in Figure 6B-E, over-expression of ARHGAP1 significantly promoted cell proliferation, cell clony formation, migration and invasion compared with the control group (P<0.05). Moreover, down-regulation of ARHGAP1 showed the opposite effect on MDA-MB-231 cells and MCF-7 cells (P<0.05).
Figure 6.
ARHGAP1 regulated cell activities of breast cancer cells. A. ARHGAP9 overexpression and knockdown were performed in breast cancer cells, and transfection efficiency was examined by RT-PCR and western blot anaylsis. B-E. Cell proliferation, colony formation, migration and invasion were examined by CCK8, crystal violet staining, wound healing and transwell, respectively. Data are shown as mean ± SD. **P<0.01.
Discussion
MiRNAs are important non-coding single-stranded small molecules that regulate many physiological and pathological processes. They can inhibit or degrade the expression of the target genes at the transcriptional level by binding with 3’UTR of the downstream targets. Some miRNAs with abnormal expression were demonstrated as potential indicators for breast cancer diagnosis and treatment. We first found that miR-34b-5p expression was down-regulated evidently in breast cancer samples compared with the normal tissues from the RNA sequencing data in TCGA. We collected breast cancer tumor samples and the non-tumor controls, and verified that miR-34b-5p expression was descended in breast cancer. As previous studies investigated, abnormal expression of miRNAs also indicated poor/favorable prognosis in some maladies. For example, high expressions of miR-155 and miR-206 were associated with poor prognosis in breast cancer [14,15], and low expression of miR-145 showed a well prognosis [15]. In our study, high miR-34b-5p expression exerted a better prognosis than that of low miR-34b-5p expression. Besides, from the analysis of clinical features, low miR-34b-5p expression was associated with TNM stage and lymphatic metastasis.
Functional experiments were consequently carried out for evaluating the effect of miR-34b-5p on breast cancer cells. Excessive cell proliferation is an essential element for tumor growth and development. A lot of studies indicated low expression of miRNA gave rise to cell proliferation, such as miR-135a [16], miR-26b [17], and miR-107 [18]. In our study, we investigated that miR-34b-5p over-expression showed inhibiting effect on cell proliferation and cell clonality. A subcutaneously implanted tumor model in nude mouse was then used to explore the effect of miR-34b-5p on tumor growth in vivo. The results displayed that miR-34b-5p mimic remarkably slowed tumor growth and reduced the size of tumors. It is well-known that Ki67 is a representative factor of tumor genesis [19,20], and therefore the expression of Ki67 in tumor tissues was examined. In miR-34b-5p mimic group, Ki67 expression was obviously lower in miR-34b-5p overexpression tumors than that in the negative control group. Intensive cell migration and invasion in carcinomas are always as the necessary activities for the tumor metastasis. Previous reports illuminated that miR-34b over-expression could repress migration and invasion of non-small-cell lung cancer cells [21], metastatic prostate cancer cells [22] and melanoma cells [23]. In the present study, we firstly found that miR-34b-5p mimic could weaken the migration and invasion capabilities of breast cancer cells. MiRNAs play indispensable roles in cancer progression via inhibiting or degrading the expression of the target genes at the transcriptional level by binding with 3’UTR of the downstream targets. Preliminary reports penetrated that miR-34b-5p could binding with 3’UTR of IGFBP2 [24], tissue inhibitor of metalloproteinase 3 (TIMP3) [11] and progranulin [10]. In our work, we proved that miR-34b-5p could target ARHGAP1 to regulate cell phenotype of breast cancer cells. Based on the TCGA data, ARHGAP1 was over-expressed in breast cancer samples. Furthermore, ARHGAP1 expression was associated with the migration and invasion of cervical carcinoma [25], bone metastatic microenvironment [26], and ewing sarcroma [27]. The rescue experiments showed that siARHGAP1 abolished the effect of miR-34b-5p inhibitor on breast cancer cells. Moreover, we investigated that ARHGAP1 promoted cell proliferation, migration and invasion of breast cancer cells, and knockdown of ARHGAP1 showed anti-tumor effect on breast cancer cells.
In conclusion, our study illustrated that miR-34b-5p expression was associated better prognosis of breast cancer, and evidently affected cell proliferation, migration and invasion. ARHGAP1 was found to be the target gene of miR-34b-5p, which was involved in the regulation of cell proliferation, migration and invasion by miR-34b-5p. Our data provided new insights for the breast cancer diagnosis and molecular treatment.
Acknowledgements
This study was supported by the Fund of Department of Education of Zhejiang Province (Y201737788 and Y201636178).
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Ma J, Goding SA, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis CE, Fedewa SA, Goding SA, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 4.Agbaria S, Haim A, Fares F, Zubidat AE. Epigenetic modification in 4T1 mouse breast cancer model by artificial light at night and melatonin-the role of DNA-methyltransferase. Chronobiol Int. 2019;36:629–643. doi: 10.1080/07420528.2019.1574265. [DOI] [PubMed] [Google Scholar]
- 5.Al-Nakhle H, Smith L, Bell SM, Burns PA, Cummings M, Hanby AM, Lane S, Parker MD, Hughes TA, Speirs V. Regulation of estrogen receptor beta1 expression in breast cancer by epigenetic modification of the 5’ regulatory region. Int J Oncol. 2013;43:2039–2045. doi: 10.3892/ijo.2013.2112. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y, Li Z, Xie J, Wang P, Zhu J, Li Y, Wang Y. MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochem Biophys Res Commun. 2018;506:793–798. doi: 10.1016/j.bbrc.2018.10.150. [DOI] [PubMed] [Google Scholar]
- 7.Croset M, Pantano F, Kan C, Bonnelye E, Descotes F, Alix-Panabieres C, Lecellier CH, Bachelier R, Allioli N, Hong SS, Bartkowiak K, Pantel K, Clezardin P. miRNA-30 family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis-associated genes. Cancer Res. 2018;78:5259–5273. doi: 10.1158/0008-5472.CAN-17-3058. [DOI] [PubMed] [Google Scholar]
- 8.Wilke CM, Hess J, Klymenko SV, Chumak VV, Zakhartseva LM, Bakhanova EV, Feuchtinger A, Walch AK, Selmansberger M, Braselmann H, Schneider L, Pitea A, Steinhilber J, Fend F, Bosmuller HC, Zitzelsberger H, Unger K. Expression of miRNA-26b-5p and its target TRPS1 is associated with radiation exposure in post-Chernobyl breast cancer. Int J Cancer. 2018;142:573–583. doi: 10.1002/ijc.31072. [DOI] [PubMed] [Google Scholar]
- 9.Maroof H, Islam F, Dong L, Ajjikuttira P, Gopalan V, McMillan N, Lam AK. Liposomal delivery of miR-34b-5p induced cancer cell death in thyroid carcinoma. Cells. 2018;7 doi: 10.3390/cells7120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie W, Lu Q, Wang K, Lu J, Gu X, Zhu D, Liu F, Guo Z. miR-34b-5p inhibition attenuates lung inflammation and apoptosis in an LPS-induced acute lung injury mouse model by targeting progranulin. J Cell Physiol. 2018;233:6615–6631. doi: 10.1002/jcp.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu RP, Lu YY, Zhang XJ. MiR-34b-5p knockdown attenuates bleomycin-induced pulmonary fibrosis by targeting tissue inhibitor of metalloproteinase 3 (TIMP3) Eur Rev Med Pharmacol Sci. 2019;23:2273–2279. doi: 10.26355/eurrev_201903_17276. [DOI] [PubMed] [Google Scholar]
- 12.Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Wang L, Dong D, Wang Z, Ji W, Yu M, Zhang F, Niu R, Zhou Y. MiR-34b/c-5p and the neurokinin-1 receptor regulate breast cancer cell proliferation and apoptosis. Cell Prolif. 2019;52:e12527. doi: 10.1111/cpr.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan Y, Huang X, Quan X. Expression of miRNA-206 and miRNA-145 in breast cancer and correlation with prognosis. Oncol Lett. 2018;16:6638–6642. doi: 10.3892/ol.2018.9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad A, Zhang W, Wu M, Tan S, Zhu T. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genomics. 2018;40:243–251. doi: 10.1007/s13258-017-0624-6. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013;13:7. doi: 10.1186/1475-2867-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XY, Luo QF, Wei CK, Li DF, Li J, Fang L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int J Clin Exp Med. 2014;7:32–40. [PMC free article] [PubMed] [Google Scholar]
- 19.Sahin S, Isik GI, Cakir A, Seckin S, Uluoglu O. Clinicopathological significance of the proliferation markers Ki67, RacGAP1, and topoisomerase 2 alpha in breast cancer. Int J Surg Pathol. 2016;24:607–613. doi: 10.1177/1066896916653211. [DOI] [PubMed] [Google Scholar]
- 20.Jurikova M, Danihel L, Polak S, Varga I. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang XF, Zhao LX, Guo SP, Wei S, Zhai JF, Zhou QH. miR-34b inhibits the migration/invasion and promotes apoptosis of non-small-cell lung cancer cells by YAF2. Eur Rev Med Pharmacol Sci. 2019;23:2038–2046. doi: 10.26355/eurrev_201903_17244. [DOI] [PubMed] [Google Scholar]
- 22.Fang LL, Sun BF, Huang LR, Yuan HB, Zhang S, Chen J, Yu ZJ, Luo H. Potent inhibition of miR-34b on migration and invasion in metastatic prostate cancer cells by regulating the TGF-beta pathway. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18122762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazar J, Khaitan D, DeBlasio D, Zhong C, Govindarajan SS, Kopanathi S, Zhang S, Ray A, Perera RJ. Epigenetic regulation of microRNA genes and the role of miR-34b in cell invasion and motility in human melanoma. PLoS One. 2011;6:e24922. doi: 10.1371/journal.pone.0024922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zhang X, Li Z, Abdalla BA, Chen Y, Nie Q. MiR-34b-5p mediates the proliferation and differentiation of myoblasts by targeting IGFBP2. Cells. 2019;8 doi: 10.3390/cells8040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JP, Liu Y, Yin YH. ARHGAP1 overexpression inhibits proliferation, migration and invasion of C-33A and SiHa cell lines. Onco Targets Ther. 2017;10:691–701. doi: 10.2147/OTT.S112223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto K, Ochi H, Sunamura S, Kosaka N, Mabuchi Y, Fukuda T, Yao K, Kanda H, Ae K, Okawa A, Akazawa C, Ochiya T, Futakuchi M, Takeda S, Sato S. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc Natl Acad Sci U S A. 2018;115:2204–2209. doi: 10.1073/pnas.1717363115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satterfield L, Shuck R, Kurenbekova L, Allen-Rhoades W, Edwards D, Huang S, Rajapakshe K, Coarfa C, Donehower LA, Yustein JT. miR-130b directly targets ARHGAP1 to drive activation of a metastatic CDC42-PAK1-AP1 positive feedback loop in Ewing sarcoma. Int J Cancer. 2017;141:2062–2075. doi: 10.1002/ijc.30909. [DOI] [PMC free article] [PubMed] [Google Scholar]