Abstract
Circular RNAs (circRNAs) represent a class of endogenous non-coding RNAs. Recently, it has been reported that circRNAs might serve as novel potential biomarkers for the diagnosis of cancer, including non-small cell lung cancer (NSCLC). Mutations in the ATP binding cassette subfamily A member 3 (ABCA3) have been increasingly associated with lung disease; however, roles for circRNAs at the ABCA3 locus have not been identified. To characterize novel biomarkers in NSCLC, a bioinformatics platform was used to select circRNAs within the ABCA3 gene. Divergent primers were designed for hsa_circ_0037515 and hsa_circ_0037516, and the PCR products were sequenced to verify their existence in lung tissue. To evaluate diagnostic potential, expression levels of hsa_circ_0037515, hsa_circ_0037516, and ABCA3 were measured by quantitative reverse transcription-polymerase chain reaction; differences in expression levels were analyzed using the paired t-test, and receiver operating characteristic (ROC) curves were established. Our results demonstrate that ABCA3 mRNA, hsa_circ_0037515, and hsa_circ_0037516 are significantly downregulated in 61 paired samples of NSCLC compared to adjacent lung tissues (P < 0.01), and that the areas under the ROC curves for the two circRNAs (0.81 and 0.82, respectively) are indicative of diagnostic value. Furthermore, the significance was improved by considering the two circRNAs in combination (area under the ROC curve, 0.90). Our results suggest that hsa_circ_0037515 and hsa_circ_0037516 serve as novel potential biomarkers for the diagnosis of NSCLC.
Keywords: Non-small cell lung cancer (NSCLC), circular RNAs, biomarker, diagnosis, clinical significance
Introduction
Lung cancer is a major worldwide cause of cancer-related mortality, accounting for 1.6 million deaths per year [1]. Most cases (~85%) are caused by non-small cell lung cancer (NSCLC), which consists of adenocarcinoma (AD) and squamous cell carcinoma (SSC) [2]. Despite a variety of treatment approaches, diagnosis is often delayed, and the five-year survival rate after treatment is only 30% [3]. Therefore, research to determine effective biomarkers for early NSCLC is critical.
Circular RNAs (circRNAs) are an abundant class of endogenous non-coding RNAs in human cells, formed by joining 3’- and 5’-ends together through RNA circularization [4]. Intracellular circRNAs are more stable than linear RNAs because their creation involves non-canonical splicing without free 3’ and 5’ ends, which enables them to resist RNA exonucleases [5]. Many studies have shown that circRNAs can function as microRNA (miRNA) sponges that regulate target gene transcription and splicing, which ultimately influences gene expression [6-8]. Recent investigations have demonstrated that specific circRNAs play significant roles in cancers, such as colorectal cancer, bladder carcinoma, and gastric cancer [9-11]. Furthermore, several circRNAs have recently been identified as potential diagnostic or prognostic markers in NSCLC [12,13].
ATP binding cassette subfamily A member 3 (ABCA3) is a highly conserved transporter protein that plays a critical role in regulating pulmonary surfactant homeostasis [14]. Mutations in the ABCA3 gene have been increasingly associated with lung disease [15]. Recent studies indicate that ABCA3 is differentially expressed in a variety of cancers, including breast cancer [16] acute lymphoblastic leukemia [17], and NSCLC [18]. However, roles of circRNAs at the ABCA3 locus have not been identified.
In this study, using 61 paired NSCLC/adjacent non-cancerous tissues, we predicted the location of two circRNAs within exons 8-10 of the ABCA3 gene: hsa_circ_0037515, located at chr16:2367283-2367765 (hg19); and hsa_circ_0037516, located at chr16:2369581-2369841 (hg19) via circBase datebase (http://www.circbase.org/cgi-bin/simplesearch.cgi), subsequently evaluated the expression of these two circRNAs. Our data show that the expression of ABCA3 and both hsa_circ_0037515 and hsa_circ_0037516 was significantly downregulated in NSCLC, with significant diagnostic value for the two circRNAs in combination. On the basis of our findings, these circRNAs may provide novel biomarkers for NSCLC.
Materials and methods
Specimens and clinical information
A total of 61 pairs of fresh lung tissue specimens were collected from NSCLC patients between May 2011 and June 2014 in the Tangdu Hospital. The tissues were maintained at -80°C until use. The clinical and pathological characteristics of the patients are shown (Table 1). The TNM staging was determined by the criteria established by the International Union Against Cancer (UICC) in 2009.
Table 1.
Clinicopathological characteristics of patients with non-small cell lung cancer
| Characteristics | No. of patients |
|---|---|
| Age (year) | |
| ≥ 60 | 34 |
| < 60 | 27 |
| Gender | |
| Male | 45 |
| Female | 16 |
| Subtype | |
| AD | 25 |
| SC | 28 |
| Lymphatic metastasis | |
| N0 | 17 |
| N1 | 13 |
| N2 | 22 |
| N3 | 1 |
| Distal metastasis | |
| M0 | 52 |
| M1 | 1 |
| Invasion | |
| Tis & T1-T3 | 46 |
| T4 | 7 |
| TNM stage | |
| 0 & I & II | 24 |
| III & IV | 29 |
Abbreviations: AD, adenocarcinoma; SCC, squamous cell carcinoma.
Each patient was informed and was fully aware of this research. Consent was received by written agreement. The study was conducted according to the rules of the Medical Ethics Commission of Soochow University. All methods were carried out in accordance with relevant guidelines and regulations.
Cell culture
The human lung cell lines A549, BEAS-2B, H1299, and H460 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). A549 cells were cultured in DMEM, and BEAS-2B, H1299, and H460 were cultured in RPMI 1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2. The cells were passaged at ~70% confluence.
Total RNA extraction and reverse transcription
Total RNA was extracted from tissues and cell samples with the Total RNA Isolation Kit according to manufacturer’s instructions (Ambion, Carlsbad, CA, USA). The purity and concentration were measured with a spectrophotometer 2000. Purified RNA was stored at -80°C for use in experiments.
Complementary DNA was synthesized using random primers and the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA).
Quantitative reverse transcription polymerase chain reaction
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using SYBR-Green Premix Ex Taq (Takara Bio, Nojihigashi, Kusatsu, Japan) and monitored by Roche Light Cycler480 (Roche, USA) according to the manufacturer’s instructions. The PCR reaction protocol was 95°C 3 min, followed by 40 cycles of 95°C 30 s and 60°C 50 s. The divergent primer sequences of hsa_circ_0037515, hsa_circ_0037516, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, used as internal standard for normalization), were synthesized by Generay Biotech Ltd. (Shanghai, China). All experiments were conducted in triplicate. The primers for qRT-PCR are shown in Table 2. The data were analyzed using the comparative cycle threshold (ΔCt) method. All results are expressed as the mean ± SD.
Table 2.
qRT-PCR primer sequences
| Primer name | Primer F (5’-3’) | Primer R (5’-3’) |
|---|---|---|
| ABCA3 | GACCACGACTTTCAAAATGC | CGCAGAGTGTTCTCCACGCA |
| hsa_circ_0037515 | AGCACCTTCTTCAGCAAAGGA | ACACAGAAGAGCAGGGTCATG |
| hsa_circ_0037516 | TGTCGTGCAGGAGAAGGAAAG | TGATGGTCACCGTCAGTCTCT |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC |
Abbreviations: ABCA3, ATP binding cassette subfamily A member 3; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Sequencing of qRT-PCR products
qRT-PCR products of hsa_circ_0037515 and hsa_circ_0037516 in NSCLC tumor and adjacent lung tissues were cloned using T vector (Transgene, China). Sanger sequencing was performed by Saiyin (Shanghai, China).
Polymerase chain reaction (PCR)
cDNA and genomic DNA of lung tissues from two randomly selected patients were used in PCR. The reactions for PCR were performed using 2× Es Taq MasterMix (Dye) (Cwbiotech, China), according to the manufacturer’s instructions. The divergent primers were as described above (Table 2), and convergent primers are shown in Table 3.
Table 3.
PCR convergent primer sequences
| Primer name | Convergent primer F (5’-3’) | Convergent primer R (5’-3’) |
|---|---|---|
| GAPDH | ACCACAGTCCATGCCATCAC | CCACCTGGTGCTCAGTGTAG |
| hsa_circ_0037515 | ATGATGGGGCTCAGCAGC | GACACAGAAGAGCAGGGTCAT |
| hsa_circ_0037516 | GACGGTGACCATCAAGAGGTT | CTTCTCCTGCACGACAGCAC |
Abbreviations: PCR, polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Statistical analysis
GraphPad Prism 5.0 (GraphPad Software, LaJolla, CA, USA) software was used to generate graphs of the results. The hsa_circ_0037515 and hsa_circ_0037516 expression differences between NSCLC and non-tumorous tissues were evaluated by the paired sample t test. Correlation analysis of the two circRNAs was performed by Statistical Product and Service Solutions (SPSS). Differences between cancer subtypes were analyzed with the independent t-test. R 3.2.1 was used to construct ROC curves. A value of P < 0.05 was considered statistically significant.
Results
Amplification of hsa_circ_0037515 and hsa_circ_0037516 in NSCLC tissues
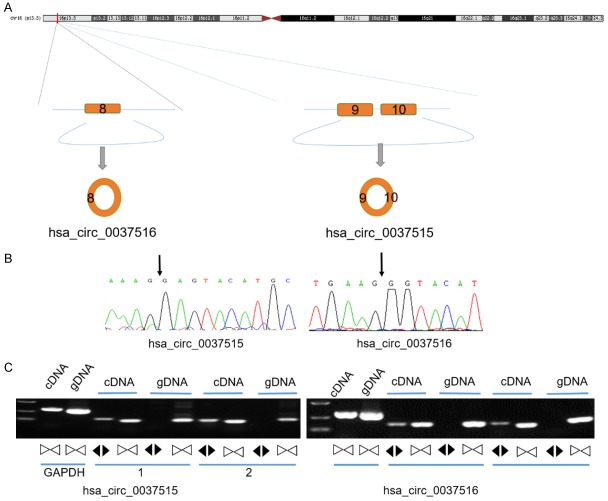
Using CircBase database, we evaluated whether the ABCA3 gene can be spliced to form hsa_circ_0037515 and hsa_circ_0037516. Hsa_circ_0037515 is predicted to be produced at an ABCA3 gene locus containing exon 9 and 10, while Hsa_circ_0037516 is predicted to be produced at a locus containing exon 8 (Figure 1A). To verify that these circRNAs are expressed in NSCLC tissues, first we used melting curve analysis to demonstrate that amplified products were single peaks, which indicates that there were no primer dimers or non-specific amplification (data not shown). Second, the qRT-PCR products of these two circRNAs in NSCLC tissues were sequenced, and the results confirmed the backsplice junctions, which were completely consistent with the predictions from CircBase database (Figure 1B). In order to exclude genomic contamination, we designed convergent primers for linear RNA. The results showed that the circRNAs were amplified from cDNA, but not from genomic DNA of NSCLC tissues using divergent primers (Figure 1C). Therefore, these results confirm the structure and extragenomic composition of these circRNAs in NSCLC tissues.
Figure 1.
Identification of hsa_circ_0037515 and hsa_circ_0037516 in samples of NSCLC tissues. A. The genomic location of hsa_circ_0037515 and hsa_circ_0037516. These circRNAs are predicted to be derived from exons 8-10 of the ABCA3 gene. B. Sanger sequencing of the qPCR products of hsa_circ_0037515 and hsa_circ_0037516. Results from the splice junctions region are shown. C. RT-PCR products with divergent primers in the gDNA of NSCLC tissues from two randomly selected patients (coded as 1 and 2 in the figure). The black and white arrowheads represent the direction of the primers. White arrowheads represent convergent primer, and black arrowheads represent divergent primer that designed for circRNA. Abbreviations: ABCA3, ATP binding cassette subfamily A member 3; NSCLC, non-small cell lung cancer.
Assessment of hsa_circ_0037515 and hsa_circ_0037516 expression levels in lung cancer cell lines and tissues
To evaluate the potential significance of hsa_circ_0037515 and hsa_circ_0037516 in NSCLC, we measured their expression levels in lung cancer cell lines and tissues by RT-qPCR. As shown in Figure 2A, the expression levels of both circRNAs were significantly lower in A549, H1299, and H460 lung cancer cell lines than in BEAS-2B normal lung cells. Consistently, the expression levels were lower in NSCLC tissues compared to adjacent non-tumorous tissues (P < 0.001; Figure 2B). Moreover, reduced expression levels of the ABCA3 gene were observed in lung cancer cell lines and tissues compared to normal lung cells and tissues (P < 0.001; Figure 2C). Therefore, the levels of hsa_circ_0037515, hsa_circ_0037516, and ABCA3 tend to be reduced in NSCLC.
Figure 2.
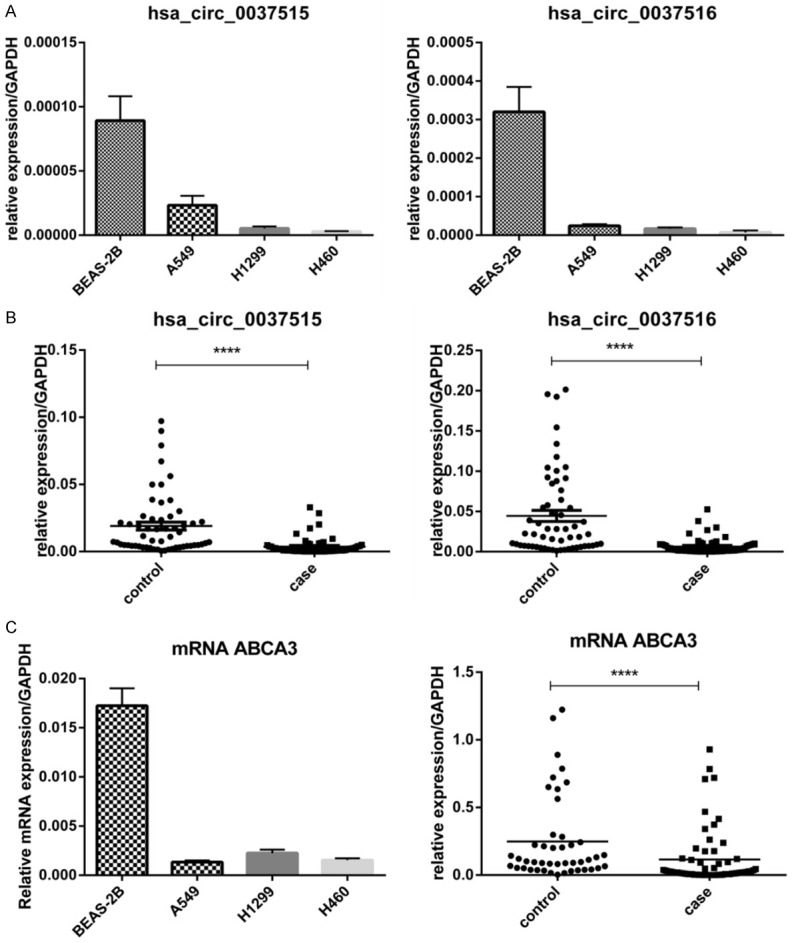
The expression levels of candidate circRNAs. Results were obtained by performing RT-qPCR in triplicate. RNA expression levels were normalized to expression of the reference gene GAPDH. A. The expression of circRNAs in A549, H1299, and H460 lung cancer cells were compared to the expression in BEAS-2B normal lung cells. Results represent the mean ± SD. B. The average expression of circRNAs in 61 pairs of NSCLC tissues (case) and adjacent lung tissues (control). C. The expression of ABCA3 mRNA in lung cancer cells (left panel) and tissues (right panel). ****P < 0.001. Abbreviations: ABCA3, ATP binding cassette subfamily A member 3; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NSCLC, non-small cell lung cancer.
Levels of hsa_circ_0037515 and hsa_circ_0037516 are correlated but cannot be used to distinguish NSCLC subtypes
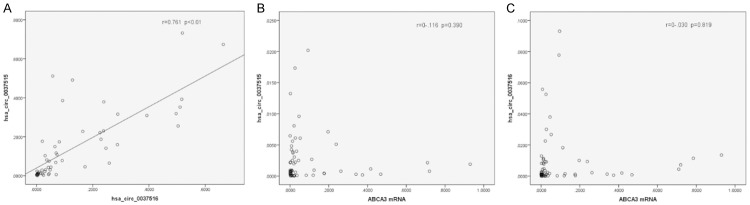
Next, we examined our data for potential correlations between the expression of the two circRNAs. The results showed that hsa_circ_0037515 and hsa_circ_0037516 expression is consistent, with a Pearson correlation coefficient of -0.745 (P < 0.01) (Figure 3A). However, there were no significant differences in expression levels among different cancer subtypes (Table 4). The expression of neither of these circRNAs correlated with the expression of ABCA3 (Figure 3B, 3C). Therefore, these two circRNAs may be useful indicators of NSCLC but cannot distinguish between cancer subtypes.
Figure 3.
Correlation between hsa_circ_0037515, hsa_circ_0037516, and ABCA3 expression levels in NSCLC tissues. Expression level differences between NSCLC and adjacent tissues were plotted for the 61 paired samples. A. hsa_circ_0037515 vs. hsa_circ_0037516 was plotted. The best fit line is shown. B. hsa_circ_0037515 vs. ABCA3 was plotted. C. hsa_circ_0037516 vs. ABCA3 was plotted. Abbreviations: ABCA3, ATP binding cassette subfamily A member 3; NSCLC, non-small cell lung cancer.
Table 4.
The relative expression of hsa_circ_0037515 and hsa_circ_0037516 in SSC and AD
| Subtype | 2-ΔΔCt ± SD | P value | |
|---|---|---|---|
| hsa_circ_0037515 | AD | 0.25 ± 0.24 | 0.77 |
| SSC | 0.22 ± 0.50 | ||
| hsa_circ_0037516 | AD | 0.48 ± 0.97 | 0.10 |
| SSC | 0.16 ± 0.24 |
Abbreviations: AD, adenocarcinoma; SCC, squamous cell carcinoma.
Potential diagnostic values of hsa_circ_0037515 and hsa_circ_0037516 in NSCLC cancer
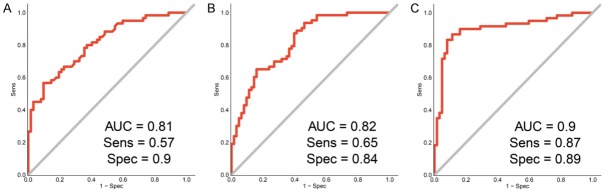
Given the differential expression of ABCA3, hsa_circ_0037515, and hsa_circ_0037516 between NSCLC and non-cancer tissues (Figure 2), we evaluated their potential diagnostic values by using the ROC analysis as a comprehensive index to reflect the sensitivity and specificity of continuous variables. For the ABCA3 mRNA, the sensitivity was 0.57, the specificity was 0.95, and the area under the ROC curve (AUC) was 0.79 (Figure S1), which indicates moderate diagnostic potential. However, the AUC of hsa_circ_0037515 was 0.81, with a sensitivity of 0.57 and a specificity of 0.9 (Figure 4A), and the AUC of hsa_circ_0037516 was 0.82, with a sensitivity of 0.65 and a specificity of 0.84 (Figure 4B). To improve their diagnostic potential, we evaluated the predictive value of combining the two circRNAs. The results show that the AUC was 0.9, with a sensitivity of 0.87 and a specificity of 0.89 (Figure 4C). These results suggest that each of the circRNAs can be used to predict NSCLC on their own, but that the predictive value is best for the two markers in combination.
Figure 4.
ROC curve analysis of the expression of hsa_circ_0037515 and hsa_circ_0037516 in NSCLC versus control tissues. A. ROC curve of hsa_circ_0037515. B. ROC curve of hsa_circ_0037516. C. ROC curve of hsa_circ_0037515 and hsa_circ_0037516 combined. Abbreviations: ROC, receiver operating characteristic; NSCLC, non-small cell lung cancer; AUC, area under the ROC curve.
Discussion
CircRNAs are distinctive single-stranded closed circular RNA molecules that are found in a variety of organisms. In 1979, circRNAs were isolated from several eukaryotes [19]. At first, they were thought to be byproducts of splicing with no functional consequence. However, it has become increasingly clear that circRNAs can modulate the expression of sets of genes involved in important biological processes. The primary mechanism of circRNAs is thought to involve competitive binding of miRNAs [20]. With the development of RNA sequencing technology and bioinformatics, an increasing number of circRNAs has been discovered [21], and the circRNA database is expanding rapidly [22]. This database contains numerous circRNA genomic and mature RNA sequences of different species, as well as circRNAs relating to various diseases [23]. Currently, more than 20,000 different circRNAs have been discovered in eukaryotes [24]. Furthermore, emerging research demonstrates that circRNAs can play important roles in cancer development. For example, Zhong et al. found that the circRNA MYLK is a competing endogenous RNA that promotes bladder cancer progression by regulating the VEGFA/VEGFR2 signaling pathway [25]. Additionally, several circRNAs have been identified as potential biomarkers in NSCLC [6,13].
In this study, we assessed the potential roles of two circRNAs residing in the ABCA3 locus (hsa_circ_0037515 and hsa_circ_0037516) in 61 paired NSCLC samples. We chose to evaluate circRNAs within this gene because of its role in pulmonary surfactant homeostasis and lung disease [14,15]. Our results demonstrate that both circRNAs are expressed on cancerous and non-cancerous lung tissues, but that they are downregulated in NSCLC, which indicates that their expression level may negatively correlate with lung cancer development. The expression of ABCA3 was also decreased in NSCLC, which might account in part for the decrease in these circRNAs; however, the decreases were larger for the circRNAs, raising the possibility that their expression may be dependent on additional factors.
To evaluate the potential of ABCA3 mRNA, hsa_circ_0037515, and hsa_circ_0037516 as biomarkers, we analyzed their diagnostic effectiveness. Each had moderate predictive value (AUC 0.79-0.84); however, the value was improved for the two circRNAs in combination (AUC 0.9). These results indicate that hsa_circ_0037515 and hsa_circ_0037516 have the potential to serve as novel diagnostic markers for NSCLC with high accuracy, specificity, and sensitivity. Interestingly, high ABCA3 expression levels have been shown to correlate with worse disease-free and overall survival in tumors from 89 patients with NSCLC [18]. We did not have sufficient information to assess the prognostic potential of hsa_circ_0037515 and hsa_circ_0037516 in our study; however, future studies to determine their prognostic potential would be valuable.
In addition to their diagnostic potential, these circRNAs are likely to add mechanistic insight into the regulatory mechanisms of NSCLC and other cancers. Currently, the detailed molecular mechanisms of these two circRNAs are unknown, but given their genomic location and similarity in expression pattern, they could potentially influence the function of ABCA3. Future investigations to reveal functional roles for these RNAs would be valuable. Furthermore, verification of our findings with a larger and more diverse set of tissue samples will be an important future endeavor.
Acknowledgements
The study was supported by research grants from the National Natural Science Foundation of China (81872417, 81572923 and 81871866), the Natural Science Foundation of Shanxi Province (2019SF-033), Project of Tangdu Hospital, the Fourth Military Medical University (2018 Key Talents), the Priority Academic Program Development of Jiangsu Higher Education Institutions of China (PAPD), and the Suzhou City Science and Technology Program (grant number SYS201419).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, Chirieac LR, D’Amico TA, Demmy TL, Ganti AKP, Govindan R, Grannis FW, Jahan T, Jahanzeb M, Johnson DH, Kessinger A, Komaki R, Kong FM, Kris MG, Krug LM, Le QT, Lennes IT, Martins R, O’Malley J, Osarogiagbon RU, Otterson GA, Patel JD, Pisters KM, Reckamp K, Riely GJ, Rohren E, Simon GR, Swanson SJ, Wood DE, Yang SC. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Mao F, Shen TY, Luo QQ, Ding ZP, Qian LQ, Huang J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu SB, Yang XS, Li XL, Wang JL, Gao Y, Shang RZ, Sun W, Dou KF, Li HM. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Li C, Tan CL, Liu XB. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 6.Cui JG, Li W, Liu GH, Chen XJ, Gao XL, Lu HL, Lin D. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47:810–821. doi: 10.1080/21691401.2019.1575847. [DOI] [PubMed] [Google Scholar]
- 7.Shen L, Hu YY, Lou JW, Yin S, Wang WL, Wang YY, Xia Y, Wu W. CircRNA-0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting miR-107. Mol Med Rep. 2019;19:3923–3932. doi: 10.3892/mmr.2019.10011. [DOI] [PubMed] [Google Scholar]
- 8.Su CY, Han Y, Zhang HT, Li Y, Yi L, Wang XJ, Zhou SJ, Yu DP, Song XY, Xiao N, Cao XQ, Liu ZD. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhong ZY, Lv MX, Chen JX. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SJ, Li TW, Zhao QF, Xiao BX, Guo JM. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 12.Wang CD, Tan SY, Liu WR, Lei Q, Qiao WL, Wu YP, Liu XQ, Cheng W, Wei YQ, Peng Y, Li WM. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18:134. doi: 10.1186/s12943-019-1061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Fraipont F, Gazzeri S, Cho WC, Eymin B. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet. 2019;10:390. doi: 10.3389/fgene.2019.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beers MF, Mulugeta S. The biology of the ABCA3 lipid transporter in lung health and disease. Cell Tissue Res. 2017;367:481–493. doi: 10.1007/s00441-016-2554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kröner C, Wittmann T, Reu S, Teusch V, Klemme M, Rauch D, Hengst M, Kappler M, Cobanoglu N, Sismanlar T. Lung disease caused by ABCA3 mutations. Thorax. 2017;72:213–220. doi: 10.1136/thoraxjnl-2016-208649. [DOI] [PubMed] [Google Scholar]
- 16.Schimanski S, Wild PJ, Treeck O, Horn F, Sigruener A, Rudolph C, Blaszyk H, Klinkhammer-Schalke M, Ortmann O, Hartmann A, Schmitz G. Expression of the lipid transporters ABCA3 and ABCA1 is diminished in human breast cancer tissue. Horm Metab Res. 2010;42:102–109. doi: 10.1055/s-0029-1241859. [DOI] [PubMed] [Google Scholar]
- 17.Aberuyi N, Rahgozar S, Khosravi Dehaghi Z, Moafi A, Masotti A, Paolini A. The translational expression of ABCA2 and ABCA3 is a strong prognostic biomarker for multidrug resistance in pediatric acute lymphoblastic leukemia. Onco Targets Ther. 2017;10:3373–3380. doi: 10.2147/OTT.S140488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overbeck TR, Arnemann J, Waldmann-Beushausen R, Trümper L, Schöndube FA, Reuter-Jessen K, Danner BC. ABCA3 phenotype in non-small cell lung cancer indicates poor outcome. Oncology. 2017;93:270–278. doi: 10.1159/000477619. [DOI] [PubMed] [Google Scholar]
- 19.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 20.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am J Cancer Res. 2018;8:1142–1156. [PMC free article] [PubMed] [Google Scholar]
- 24.Geng YT, Jiang JT, Wu CP. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong ZY, Huang MG, Lv MX, He YF, Duan CZ, Zhang LY, Chen JX. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.