Abstract
Background: Triple-negative breast cancer (TNBC) is an aggressive breast cancer subtype. G protein coupled receptor (GPER), the key player in the intercellular signaling communication, has been verified to participate in tumorigenesis. The present study aims to explore the effects of GPER on cell proliferation, invasion and EMT through CD151/miR-199a-3p bio-axis in TNBC cells. Methods: Total proteins were isolated from TNBC cell lines and GPER expression was determined using western blot assay. CCK-8 assay was used to detect cell viability after being treated with GPER activation. Western blotting and immunofluorescence were applied to measure the level of proteins associated with cell proliferation, angiogenesis and EMT, as well as the Hippo signal pathway. The level of miR-199a-3p and transfection efficiency were evaluated by reverse transcriptase quantitative PCR (RT-qPCR) after being transfected with miR-199a-3p mimics. Cell migration and invasion of TNBC cells were assessed by wound healing and transwell assays. Moreover, luciferase reporter assay was conducted to verify the relationship between CD151 and miR-199a-3p. Results: GPER activation treatment suppressed MDA-MB-231 cell viability, proliferation, migration, invasion, angiogenesis and EMT process. The expression of E-cadherin was increased, but N-cadherin, Vimentin, VEGFA, AngII and CD151 were decreased after GPER activation treatment. Conversely, inhibition of GPER indeed up-regulated CD151 expression. In addition, overexpression of miR-199a-3p supressed cell proliferation, migration, invasion and angiogenesis, as well as EMT process and the Hippo signal pathway. Conclusion: Collectively, the activation of GPER inhibits cells proliferation, invasion and EMT of triple-negative breast cancer via CD151/miR-199a-3p bio-axis. This study provides a novel intervention target for the treatment of breast cancer cells and a fresh idea for the clinical therapy of breast cancer.
Keywords: Triple-negative breast cancer, G protein coupled receptor, miRNA-199a-3p, CD151, hippo pathway
Introduction
Breast cancer is an extraordinarily diverse disease, including manifestations, morphology, molecular structure, and response to treatment. According to different criteria, breast cancer can be divided into different subtypes, among which triple negative breast cancer (TNBC) is a special clinical pathological subtype. TNBC refers to breast cancer with negative expressions of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2), expressing cytokeratin 5/6 (CK5/6) and/or either epidermal growth factor receptor (EGFR) [1]. As an aggressive breast cancer subtype, TNBC is genetically heterogeneous, which challenges the identification of clinically effective molecular makers and treatments.
Estrogens regulates breast cancer progression mainly by binding to and activating the estrogen receptor (ER) α and ERβ to regulate the expression of genes related to cell proliferation, migration and viability [2]. The G protein estrogen receptor (GPR30/GPER) can also mediate the action of estrogens in both normal and malignant cell contexts [3]. Further, ligand-activated GPER induces a network of signal transduction pathways including epidermal growth factor receptor (EGFR), intracellular cyclic AMP, calcium mobilization, MAPK and PI3K [4]. It has been reported that GPER was detected in tissues of 132 patients with TNBC and its expression level was negatively correlated with high-level tumors, showing that the lower the expression level of GPER was, the worse the prognosis became [5,6]. These all suggest that GPER is a positive prognostic factor in TNBC.
Epithelial-mesenchymal transition (EMT) process mainly includes the decrease of intercellular connection and adhesion, the enhancement of cell vitality and the changes of various related molecules, and it can be determined by the loss of E-cadherin along with the up-regulation of N-cadherin, Fibronectin and Vimentin [7]. Previous studies have also reported that the activation of GPER inhibits EMT, migration and angiogenesis of TNBC cells via NF-κB signaling pathway. More importantly, the role of GPER has been highlighted in nervous, cardiovascular and immune systems as well as the inflammatory processes [8,9]. For instance, GPER was shown to be involved in thymic atrophy and thymocyte apoptosis induced by estrogens and GPER agonist G-1 in a knockout mice in vivo [10]. Interestingly, GPER expression has been associated with poor clinical-pathological features in breast, endometrial and ovarian cancer patients.
MicroRNAs (miRNAs), about 18~22 nucleotides, are small non-coding RNA molecules [11]. They regulate the expression of targeted genes by directly binding the 3’-untranslated regions (3’-UTR) of corresponding messenger RNAs (mRNAs) [12]. miRNAs participate in the pathogenesis of various biological behaviors, such as suppressing or promoting tumors. As a tumor suppressive factor, miRNA-199a-3p (miR-199a-3p) is down-regulated in multiple cancer tissues and cells, including hepatocellular carcinoma [13], osteosarcoma [14] and papillary thyroid carcinoma [15]. Highly expressed in hair follicles and in some tumor cells, miR-199a-3p participated in tumor progression. However, it is significantly under expressed in hepatocellular carcinoma and bladder cancer and regulates cell proliferation and migration. In addition, miR-199a-3p promotes cell proliferation and survival of endothelial cells as well as breast cancer cells [16]. CD151, also known as GP-27, MER-2, PETA-3, SFA-1 or Tspan-24, can be expressed in many cell types and considered to comprise molecular facilitators [17]. The mRNA and protein levels of CD151 are highly expressed in breast cancer, colon cancer and hepatocellular carcinoma [18]. Moreover, studies have shown that the expression change of CD151 is markedly correlated with the growth process, invasion and migration of cancers [19]. Other studies have reported that CD151 is highly expressed in ER positive and TNBC cells and can promote the proliferation, invasion and migration of breast cancer cells through targeted binding with miR-124 [20]. Therefore, this study aims to explore whether the activation of GPER in TNBC cells can suppress the process of TNBC cells by inhibiting the expression of CD151 binding to miR-199a-3p.
It still remains unclear that whether the activation of GPER inhibits cells proliferation, invasion and EMT of triple-negative breast cancer via CD151/miR-199a-3p bio-axis, thus, more researches are needed. The regulatory role of GPER in the expression of miR-199a-3p/CD151 are also investigated to reveal the possible internal molecular mechanisms and signaling pathways. This finding will provide new theoretical basis for in-depth exploration of the breast cancer treatment.
Materiel and methods
Cell culture and treatment
Three TNBC cell lines (HCC1806, HCC1937, MDA-MB-231) and normal breast epithelial cell lines (HMEC-184) were cultured in RPMI 1640 media (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin solution (Gibco). Cultures were maintained in a humidified incubator with 5% CO2 at 37°C.
17β-Estradiol (E2) was purchased from Sigma-Aldrich, and solubilized in ethanol. G-1(1-[4-(-6-bromobenzol [1,3] diodo-5-yl)-3a,4,5,9b tetrahidro3H5cyclopenta[c]quinolin-8yl]-ethanone) was obtained from Tocris Bioscience (Bristol, UK), which was solubilized in ethanol. G-1 and E2 inducers have been reported to belong to the GPER agonists for up-regulating GPER expression. Cultured in regular growth medium, MDA-MB-231 cells were switched to medium without serum and phenol red for 24 h, and then treated with E2 (10 nM) for 6 h and 8 h or with G-1 (1 μM) for 24 h and 48 h. Experiments Grouping, Control, G-1 (24 h), G-1 (48 h), E2 (6 h) and E2 (8 h) groups.
Cell transfection
Cell transfection was performed to up-regulate the expression of miR-199a-5p in MDA-MB-231 cells. miR-199a-5p mimics and its negative control (NC) were both designed and synthesized by GenePharma Corporation (Shanghai, China). The plasmids along with miR-199a-5p mimics or scramble were transfected into MDA-MB-231 cells with Lipofectamine 2000 reagents (Invitrogen, USA) according to the manufacturer’s instructions. Transfection efficiency was confirmed with qRT-PCR.
Cell viability assay
Cell viability after corresponding treatments was assessed using cell counting kit-8 (CCK-8) assay (Beyotime Biotechnology, Shanghai, China). Briefly, after with or without treatment, MDA-MB-231 cells were seeded into 96-well plate (Thermo Fisher Scientific, Inc.) with cell concentration of 1 × 104 cells/well. Then, 10 μl CCK-8 solution was added into each well and the cells were maintained in humidified incubator for 1 h at 37°C. Then, the absorbance at 490 nm was measured using a microplate reader (Bio-Tek Instruments, USA). Cell viability (%) was calculated using the average absorbance of different treatment groups/average absorbance of control groups × 100%.
Wound healing assay and transwell assay
Cells were put into a 6-well plate. After cells received different treatments, a 200-μl sterile pipette tip was used to create scratches. Subsequently, cells were washed twice with PBS and placed in DMEM without FBS. Photographs were captured at 0 h and after 24 h using a microscope (Carl Zeiss), and the data was analysed with Image pro-plus software.
Transwell membranes was coated with Matrigel (BD Biosciences, San Jose, CA). 1 × 104 cells in 150 μl serum-free medium were added to the upper chamber, while the medium in the lower chamber was kept with 10% FBS. After 24 h, the cells in the top well was removed. The bottom cells were fixed with 95% ethanol, stained with 0.1% crystal violet. Additionally, photographs were taken in three independent fields for each well.
Immunofluorescence assay
MDA-MB-231 cells were seeded at 10% confluence onto small glass coverslips placed in 24-well plates. After different treatments were performed, the coverslips were removed, washed with phosphate-buffered saline (PBS) three times, and fixed with 4% paraformaldehyde in PBS for 20 minutes. Pushing through the cytomembrane (0.1% Triton, 0.1% sodium citrate for 10 minutes) and blocking in 5% goat serum for 1 h were followed by incubation with VEGFA and Ang II primary antibodies at 4°C overnight. After washing with PBS, cells were incubated with a 1:500 dilution of a fluorescent tag (Alexa Fluor 488; Thermo Fisher Scientific, Inc.) and conjugated with secondary antibodies in dark for 30 minutes. Next, cells were treated with DAPI (1:10,000, Invitrogen) for 5 minutes, and then covered with an antifade mounting medium and placed onto microscope slides. Finally, representative images were captured by Olympus FV1000 Digital laser scanning microscopy.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted with TRIZOL reagent (Invitrogen) in accordance with the manufacturer’s instructions. qRT-PCR was conducted to measure the expression level of miR-199a-3p in MDA-MB-231 cells after relevant treatment or transfection. The cDNA was synthesized using the PrimeScript RT reagent Kit (TaKaRa). Furthermore, quantitative PCR was performed using SYBR Premix Ex Taq (TaKaRa) to detect the expression of miR-199a-3p and U6. The expression of U6 was used as internal control. All experiments were performed in triplicate. The data was analysed using 2-ΔΔCt method.
Western blotting
Western blotting was employed to assess the protein expression of genes involved in cell proliferation and angiogenesis, EMT process and hippo pathways in MDA-MB-231 cells after relevant transfection or treatment. Briefly, total proteins were isolated using RIPA lysis buffer from MDA-MB-231 cells and then quantified using the BCA protein assay kit (Beyotime Biotechnology, Shanghai, China). Then, proteins were electrophoresed in polyacrylamide gels and transferred onto PVDF membranes (Millipore, USA), which were incubated with relevant antibodies. All primary antibodies were purchased from Abcam Biotechnology, Inc., and the information of catalog was as follows: Anti-GPER antibody (ab39742), Anti-VEGFA antibody (ab46154), Anti-p21 antibody (ab109199), Anti-Ang II antibody (ab236317), Anti-CDK2 antibody (ab235941), Anti-CD151 (ab33315), Anti-Cyclin E1 antibody (ab3927), Anti-E-cadherin antibody (ab15148), Anti-N-cadherin antibody (ab18203), Anti-LATS1 antibody (ab70561), Anti-YAP1 antibody (ab56701) and Anti-Vimentin antibody (ab8978). After primary antibodies incubation overnight at 4°C, membranes were washed with Tris-Buffered Saline Tween 20 (TBST) and then incubated with Goat anti-Rabbit (or anti-Mouse) IgG H&L (HRP) at room temperature for 1 h. After washing with TBST again, the signals were visualized with an enhanced chemiluminescence (ECL) reagents (GE Healthcare). All data were analyzed and quantified using the Image-J software. The relative amount of proteins was normalized to GAPDH.
Luciferase reporter assay
Luciferase activity was measured using the Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer’s instructions. A fragment of the CD151-3’ untranslated region (UTR) containing the miR-199-3p predicted seed region (wild-type; WT) was amplified from the cDNA of cells and inserted into pGL-3 plasmids (Promega Corporation). These constructs were transfected into MDA-MB-231 cells, using Lipofectamine 2000 reagent (Invitrogen) with or without miR-199-3p, conforming to the manufacturer’s protocol. For normalization, they were co-transfected with Renilla luciferase plasmid (Genechem, Shanghai, China). The luciferase activity was defined from the ratio of Firefly Luciferase activity to Renilla.
Statistical analysis
All experiments were repeated at least three times. All data were presented as mean ± standard deviation (SD) and statistical analyses were performed using Graphpad Prism 6.0 statistical software (Graphpad, San Diego, CA, USA). Differences between two groups were analyzed using student’s t-test and among three or more groups were analyzed by a One-way analysis of variance (ANOVA). P<0.05 was considered statistically significant.
Results
Expression of GPER and the effects of its activation on cell viability in TNBC cells
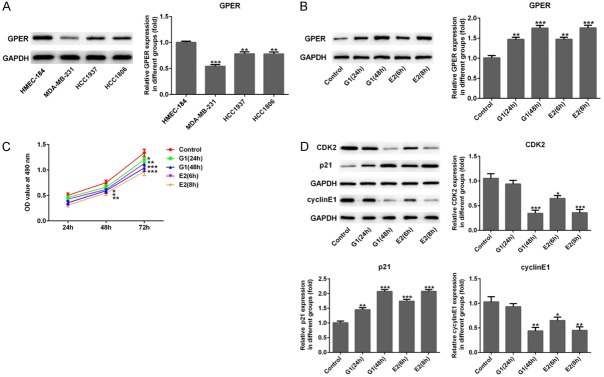
Firstly, we assessed the GPER expression in three TNBC cell lines (HCC1806, HCC1937 and MDA-MB-231) and the normal breast epithelial cell line (HMEC-184) using western blotting. Figure 1A displayed that GPER was decreased significantly in three TNBC cells when compared with HMEC-184 cells, in particular, the downward trend was most significant in MDA-MB-231 cells (P<0.001). Therefore, MDA-MB-231 cells were chosen for subsequent experiments. After being treated with E2 (10 nM) for 6 h and 8 h or with G-1 (1 μM) for 24 h and 48 h, GPER expression was elevated sharply in MDA-MB-231 cells in a time-dependent manner (Figure 1B, P<0.01 or P<0.001). In addition, Figure 1C presented that E2 or G-1 treatment obviously reduced MDA-MB-231 cell viability with dependence of time. Western blotting showed that the protein expression of CDK2 and Cyclin E1 was notably decreased and p21 was significantly increased after E2 or G-1 treatment in time dependence (Figure 1D, P<0.05, P<0.01 or P<0.001). Considering the effects of G-1 or E2 on GPER expression and cell vitality, G-1 (24 h) and E2 (8 h) were chosen for further experiments.
Figure 1.
Expression of GPER and the effects of its activation on cell viability of MDA-MB-231 cells. A. Western blotting indicated that the relative GPER expression in three TNBC cell lines (HCC1806, HCC1937 and MDA-MB-231) was downregulated significantly when compared with the normal breast epithelial cell line (HMEC-184). **P<0.01 and ***P<0.001 versus HMEC-184. B. The GPER expression in MDA-MB-231 cells was remarkably elevated using western blot after G-1 (1 μM for 24 h and 48 h) and E2 (for 6 h and 8 h) treatment. C. After G-1 (1 μM for 24 h and 48 h) or E2 (10 nM for 6 h and 8 h) treatment, the viability of MDA-MB-231 cells was decreased using cell counting kit-8 (CCK-8) assay. D. After G-1 (1 μM for 24 h and 48 h) or E2 (10 nM for 6 h and 8 h) treatment, western blotting showed that the protein expression of CDK2 and Cyclin E1 were downregulated but p21 was increased significantly. *P<0.05, **P<0.01 and ***P<0.001 versus control.
GPER suppressed cell migration, invasion, EMT and angiogenesis of MDA-MB-231 cells
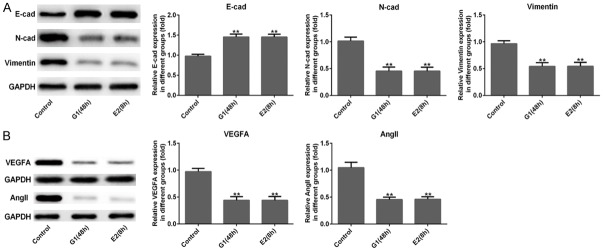
The effects of GPER on cell migration and invasion of MDA-MB-231 cells were measured using wound healing and transwell assays. Figure 2A showed that the relative migration of MDA-MB-231 cells remarkably decreased both in G-1 (48 h) and E2 (8 h) treatment groups compared to control group (P<0.001). Similarly, the relative invasive cell rate sharply decreased in G-1 (48 h) and E2 (8 h) groups (Figure 2B, P<0.001). The results of western blotting revealed that the protein levels of N-cad and Vimentin related to EMT process was significantly reduced and E-cad was obviously increased in MDA-MB-231 cells after G-1 (48 h) and E2 (8 h) treatments (Figure 3A, P<0.01). Additionally, the result of Figure 3B showed that G-1 or E2 (GPER agonists) prominently reduced the protein expression of VEGFA and Ang II related to angiogenesis in MDA-MB-231 cells (P<0.001). Moreover, the result of immunofluorescence staining also illustrated that the expression of VEGFA and Ang II (stained green) was noticeably decreased after G-1 (48 h) and E2 (8 h) treatments in MDA-MB-231 cells when compared with control group (Figure 4A and 4B). These above findings indicated that GPER (treated with GPER agonists, G-1 or E2) inhibited migration, invasion, EMT and angiogenesis of MDA-MB-231 cells.
Figure 2.
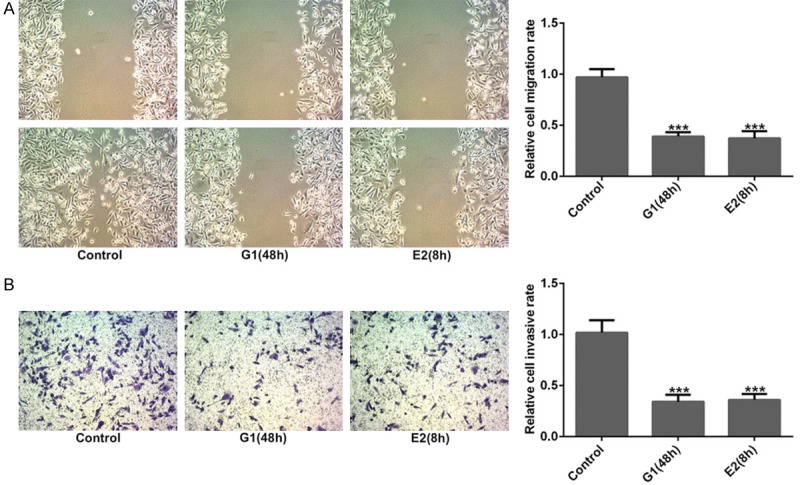
Activation of GPER suppressed cell migration and invasion. After G-1 (1 μM for 48 h) or E2 (10 nM for 8 h) treatment, the relative migration (A) and invasion (B) of MDA-MB-231 cells were reduced sharply when compared with the control group from the results of wound healing and transwell assays. ***P<0.001 versus control.
Figure 3.
Activation of GPER inhibited cell epithelial-mesenchymal transition (EMT) and angiogenesis. (A) Western blotting showed that the protein expression levels of E-cad was increased significantly with a striking reduce in the expression of N-cad and Vimentin, (B) as well as the VEGFA and Ang II expressions were decreased notably in MDA-MB-231 cells after G-1 (1 μM for 48 h) or E2 (10 nM for 8 h) treatment when compared with the control group. **P<0.01 versus control.
Figure 4.
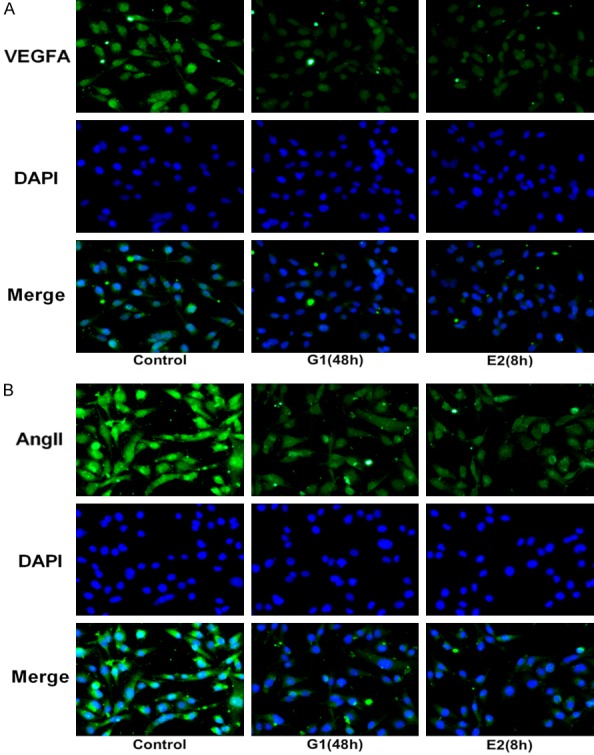
Activation of GPER suppressed the protein expression related to angiogenesis. Representative images of immunofluorescence staining indicated that the expressions of VEGFA (A) and Ang II (B) in MDA-MB-231 cells were downregulated after G-1 (1 μM for 48 h) or E2 (10 nM for 8 h) treatment when compared with the control group. Magnification, × 200.
GPER regulated the levels of CD151 and miR-199a-3p expression in MDA-MB-231 cells
Firstly, Figure 5A displayed that CD151 expression was obviously reduced in MDA-MB-231 cells after G-1 (48 h) and E2 (8 h) treatments (P<0.01). To investigate the effects of GPER on CD151 and miR-199a-3p expression, G15 (100 nm, GPER inhibitor) was used to dispose MDA-MB-231 cells for 6 h after G-1 (48 h) and E2 (8 h) treatments. Subsequently, the result of western blotting showed that G15 remarkably up-regulated the CD151 expression in G-1 (48 h) and E2 (8 h) groups (Figure 5B, P<0.05), and the result of qRT-PCR showed that G-1 (48 h) or E2 (8 h) treatments distinctly increased the expression of miR-199a-3p which was significantly reduced in both G-1+G15 and E2+G15 groups when compared with control group in MDA-MB-231 cells (Figure 5C, P<0.05, P<0.01 or P<0.001). Meanwhile, the results of the online bioinformatics tool TargetScan suggested that CD151 was predicted to be a possible target gene of miR-199a-3p. A luciferase reporter assay was conducted to verify this prediction. As shown in Figure 5D, the 3’-UTR of the gene CD151 was showed to contain the binding sequences for miR-199a-3p, suggesting that CD151 may be a downstream target gene of miR-199a-3p. Luciferase activity was significantly reduced in the CD151 3’UTR WT group transfected with miR-199a-3p (P<0.01), whereas there was no variation in the mutant-type CD151 3’UTR. These results implied that GPER could regulate CD151 and miR-199a-3p expressions in MDA-MB-231 cells and there was a targeting relationship between CD151 and miR-199a-3p.
Figure 5.
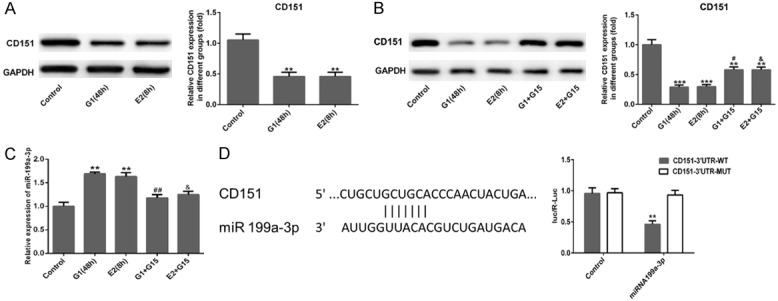
Activation of GPER suppressed CD151 expression and elevated miR-199a-3p. (A) The relative expression of CD151 in MDA-MB-231 cells was downregulated significantly after G-1 (1 μM for 48 h) or E2 (10 nM for 8 h) treatment, and (B) their expression were elevated after G15 (100 nM for 6 h) treatment following G-1 (1 μM for 48 h) or E2 (10 nM for 8 h) treatment using western blot assay. (C) qRT-PCR was used to measure the expression level of miR-199a-3p in MDA-MB-231 cells in different groups. (D) The TargetScan database revealed the putative binding site of CD151 in the 3’-UTR of miR-199a-3p. The relative luciferase activity was reduced when cells were co-transfected with miR-199a-3p and CD151-3’-UTR (WT). ***P<0.001 vs. 3’-UTR (MUT). **P<0.01 and ***P<0.001 versus control; #P<0.05 and ##P<0.01 versus G1 (48 h); &P<0.05 versus E2 (8 h). 3’-UTR, 3’-untranslated region; WT, wild type; MUT, mutant.
miR-199a-3p suppressed cell proliferation, migration, invasion, EMT and angiogenesis of MDA-MB-231 cells
To confirm the effects of miR-199a-3p on cell proliferation, migration, invasion, EMT and angiogenesis, miR-199a-3p mimic was transfected into MDA-MB-231 cells. Figure 6A showed that miR-199a-3p mimic transfection distinctly increased the expression level of miR-199a-3p in MDA-MB-231 cells (P<0.001). The results of Figure 6B presented that compared to control and mimic NC groups, cell proliferation was obviously increased in miR-199a-3p mimic group (Figure 6B, P<0.001), indicating that miR-199a-3p might play anti-cancer role in TNBC cells. In response, Figure 6C and 6D presented that the relative migration and invasion of MDA-MB-231 cells were both significantly reduced in miR-199a-3p mimic group (P<0.001), compared to control and mimic NC groups. The result of immunofluorescence showed that miR-199a-3p mimic notably attenuated VEGFA and Ang II expression (stained green) in MDA-MB-231 cells (Figure 7A and 7B). Similarly, the result of western blotting showed that miR-199a-3p mimic remarkably reduced N-cad and Vimentin expression levels and up-regulated E-cad expression in MDA-MB-231 cells (Figure 8A, P<0.01). These results further suggested that miR-199a-3p also participated in cell proliferation, migration, invasion, EMT and angiogenesis of MDA-MB-231 cells.
Figure 6.
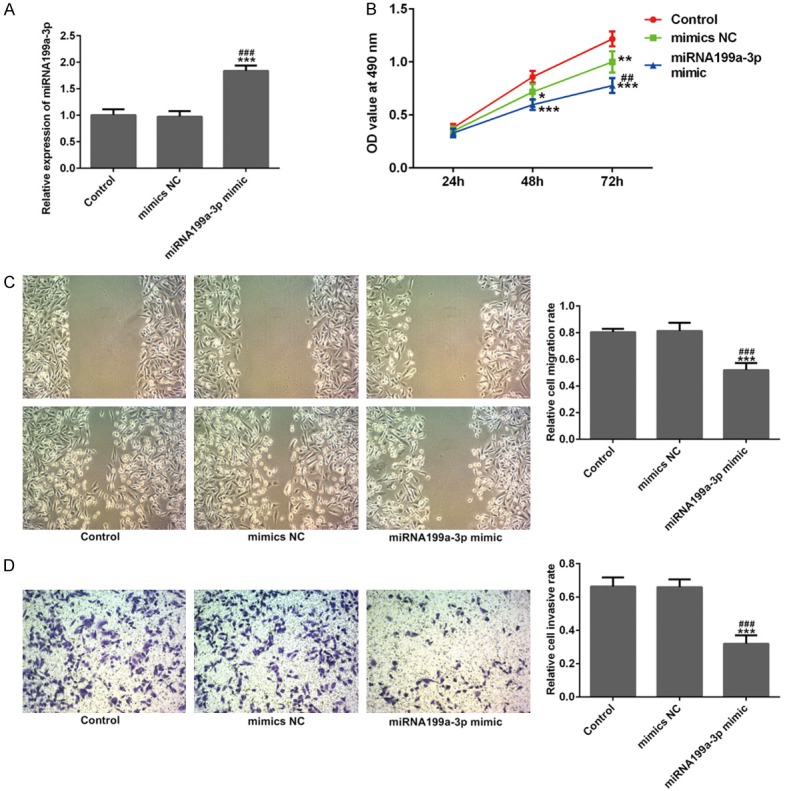
miR-199a-3p suppressed cell viability, migration and invasion. A. After miR-199a-3p mimic transfection, the relative expression level of miR-199a-3p was overexpressed in MDA-MB-231 cells using quantitative RT-PCR. B-D. After miR-199a-3p mimic transfection, relative cell viability, migration and invasion of MDA-MB-231 cells were inhibited significantly from the results of CCK-8, wound healing and Transwell assays. ***P<0.001 versus control; ###P<0.001 versus mimic NC.
Figure 7.
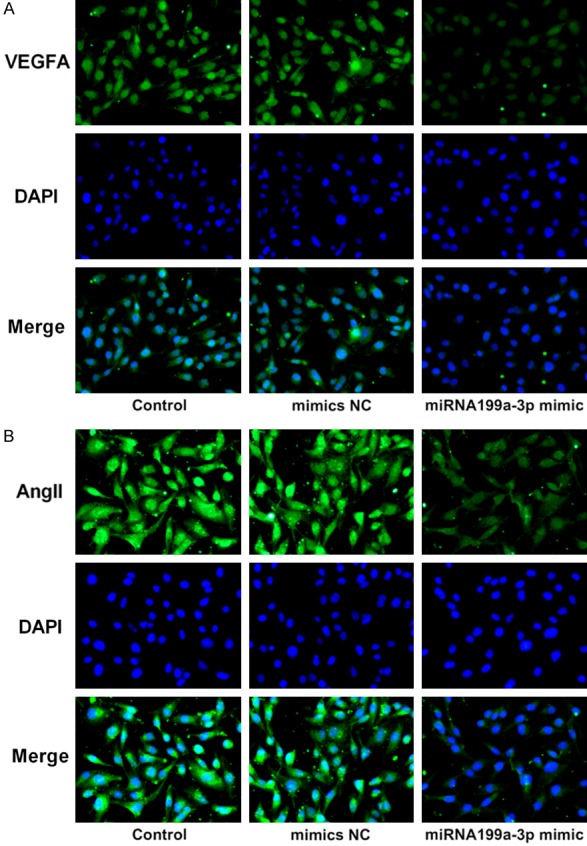
miR-199a-3p s suppressed the protein expression related to angiogenesis. Representative images of immunofluorescence staining indicated the expression of VEGFA (A) and Ang II (B) were downregulated in MDA-MB-231 cells after miR-199a-3p mimic transfection. Magnification, × 200.
Figure 8.
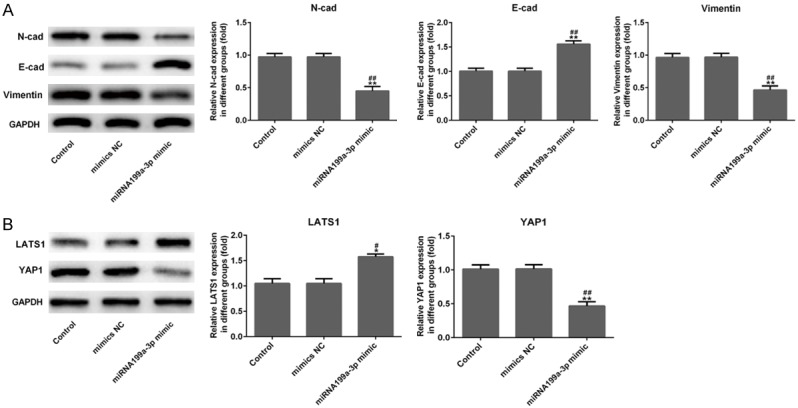
miR-199a-3p inhibited cell epithelial-mesenchymal transition (EMT) and regulated Hippo signaling pathway. A. Results of western blotting showed that the protein expression levels of N-cad and Vimentin were decreased distinctly in MDA-MB-231 cells were evaluated using after miR-199a-3p mimic transfection with a marked increase in E-cad expression. B. After miR-199a-3p mimic transfection in MDA-MB-231 cells, overexpression of miR-199a-3p increased the protein expression of LATS1 and suppressed the YAP1 expression, which are the key factors in the Hippo pathway. *P<0.05 and **P<0.01 versus control; #P<0.05 and ##P<0.01 versus mimic NC.
miR-199a-3p inactivated Hippo signaling pathway in MDA-MB-231 cells by upregulating LATS1 and inhibiting YAP1
Finally, we evaluated the effect of miR-199a-3p mimic on Hippo signaling pathway using western blotting. Figure 8B showed that miR-199a-3p mimic remarkably up-regulated the expression of LATS1 (P<0.05), while miR-199a-3p mimic distinctly down-regulated the expression of YAP1 in MDA-MB-231 cells (P<0.01). These findings suggested that miR-199a-3p might suppress Hippo signaling pathway in TNBC cells by up-regulating LATS1 and inhibiting YAP1.
Discussion
In this research, we mainly revealed the inhibiting effects of activation of GPER and overexpression of miR-199a-3p on TNBC cell proliferation, migration, invasion and angiogenesis, as well as EMT process. GPER was under-expressed in TNBC cells, especially in MDA-MB-231 cells. The expression level of and CD151 was decreased and miR-199a-3p was increased in MDA-MB-231 cells after GPER overexpression with G-1 or E2 treatment. Further, overexpression of miR-199a-3p, the same as activation of GPER, significantly suppressed cell proliferation, migration, invasion, angiogenesis and EMT processes in MDA-MB-231 cells through inhibiting Hippo signal pathway. Meanwhile, there is a targeting correlation between miR-199a-3p and CD151 regulating the progression of TNBC cells.
Breast cancer is one of the most commonly diagnosed cancers in women, but there is no targeted therapies for TNBC at present; Chemotherapy regimens, along with their adverse effects, remain the mainstay of treatment in most TNBC patients. Thus, finding a targeted biological agent for TNBC is supposed to make a paradigm shift in the treatment of these patients [21]. GPER is expressed extensively in TNBC clinical specimens and positively associated with high recurrence of TNBCs [22]. In addition, GPER mediates a specific gene signature related to cell growth, migration and angiogenesis in estrogen-sensitive tumors [23-27]. In consistent with previous studies, we found that GPER expression was lowly prevalent in TNBC cell lines, and E2 or G-1 treatment significantly up-regulated GPER expression and inhibited cell proliferation, migration and invasion of MDA-MB-231 cells (Figures 1 and 2).
In solid tumors, vascular endothelial growth factor (VEGF)-A and its receptor are involved in carcinogenesis, invasion and distant metastasis as well as tumor angiogenesis. Contrary to the cell-autonomous effects of most oncogenes, high expression of VEGFA suggests that tumors may also select for genetic alterations that mediate tumor-stromal interactions, and Ang II promotes inflammation by inducing the production of adhesion molecules, inflammatory cytokines and reactive oxygen species [28]. Research has shown that tumor progression is closely related to the expression of genes involved in endothelial function and angiogenesis, including the VEGF receptors and Ang II [29]. Similarly, we observed that activation of GPER inhibited the protein expression of VEGFA and Ang II in MDA-MB-231 cells (Figure 3B). This finding was further confirmed by data in the Figure 4.
Recent advances in the study of microRNAs indicate their important roles in regulating cellular activities, such as proliferation, morphogenesis, apoptosis, and differentiation, by regulating the expression of various target genes [30]. Previous findings suggested that miR-199a-3p targeting of caveolin-2 might have a crucial role in breast cancer tumor progression, making it a potential candidate for intervention in cancer [16]. CD151 was highly expressed in breast cancer as a potential tumor biomarker and CD151 overexpression was independently associated with poor OS in invasive breast cancer [18,31]. In our study, GPER agonist remarkably inhibited CD151 expression and up-regulated miR-199a-3p level, which was reversed by the GPER inhibitor (G15) (Figure 5). This indicated that CD151 and miR-199a-3p might be regulated by GPER, which consistent with that CD151 overexpression was found to be significantly associated with absence of estrogen receptor in previous research [31]. Moreover, TargetScan analysis and Luciferase reporter assay suggested that CD151 was a possible target gene of miR-199a-3p. Subsequently, we found that miR-199a-3p mimic obviously reduced cell proliferation, migration, invasion and angiogenesis, as well as EMT process in MDA-MB-231 cells, which were similar to those of activating GPER (Figures 6 and 7).
Over the past decade, the Hippo pathway has been shown to play a critical role in controlling organ size via regulating cell proliferation and apoptosis [32]. As a major downstream effector of Hippo pathway, it is not surprising that Yes-associated protein (YAP) functions as an oncogene, and since its expression is strikingly increased in perivascular cells, which are believed to own the properties of cancer stem cells [33]. The core kinase cascade of the Hippo pathway consists of upstream Ste20-like protein kinases (STKs) (commonly known as MST1/2) and the large tumor suppressor kinases 1/2 (LATS1/2) [34]. LATS1/2 kinase can directly phosphorylate and inactivate YAP and the Hippo signaling cascade is regulated by numerous upstream signals which include G-protein coupled receptors (GPCRs) [35], Wnt signaling [36], as well as microRNAs [37]. In this research, we found that overexpression of miR-199a-3p distinctly inhibited the Hippo pathway via up-regulating LATS1 and down-regulating YAP1 in MDA-MB-231 cells (Figure 8).
Conclusion
The present study revealed that the exact anti-tumor effects of GPER and miR-199a-3p overexpression on TNBC cells. Briefly, activation of GPER suppressed TNBC MDA-MB-231 cell proliferation, migration, invasion and angiogenesis, as well as EMT process, by regulating miR-199a-3p/CD151 axis, inactivating Hippo signaling pathway. This finding may provide new theoretical basis for deeply exploring the targeted therapies for Triple-Negative Breast Cancer.
Acknowledgements
We thank Director Mei-long Hu in the Chemoradiotherapy Department of the First Hospital Affiliated to Wenzhou Medical College, and Director Hui Dai in the Hangzhou Tumor Hospital, for their inspiration in article, suggestions and supports of experimental work. This work is supported by the Jiangsu Provincial Medical Youth Talent, the Project of Invigorating Health Care through Science, Technology and Education (QNRC2016661); Project of Jiangsu provincial Six Talent Peaks in China (WSN-057) and Jiangsu Provincial Talent on Maternal and Child Health (FRC201759).
Disclosure of conflict of interest
None.
References
- 1.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 2.Jia M, Dahlman-Wright K, Gustafsson JA. Estrogen receptor alpha and beta in health and disease. Best Pract Res Clin Endocrinol Metab. 2015;29:557–568. doi: 10.1016/j.beem.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 4.Maggiolini M, Picard D. The unfolding stories of GPR30, a new membrane-bound estrogen receptor. J Endocrinol. 2010;204:105–114. doi: 10.1677/JOE-09-0242. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Wei W, Jiang GM, Liu H, Wei WD, Yang X, Wu YM, Liu H, Wong CK, Du J, Wang HS. Activation of GPER suppresses epithelial mesenchymal transition of triple negative breast cancer cells via NF-kappaB signals. Mol Oncol. 2016;10:775–788. doi: 10.1016/j.molonc.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang S, Chen Z, Jiang G, Zhou Y, Liu Q, Su Q, Wei W, Du J, Wang H. Activation of GPER suppresses migration and angiogenesis of triple negative breast cancer via inhibition of NF-kappaB/IL-6 signals. Cancer Lett. 2017;386:12–23. doi: 10.1016/j.canlet.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 8.De Marco P, Bartella V, Vivacqua A, Lappano R, Santolla MF, Morcavallo A, Pezzi V, Belfiore A, Maggiolini M. Insulin-like growth factor-I regulates GPER expression and function in cancer cells. Oncogene. 2013;32:678–688. doi: 10.1038/onc.2012.97. [DOI] [PubMed] [Google Scholar]
- 9.Albanito L, Sisci D, Aquila S, Brunelli E, Vivacqua A, Madeo A, Lappano R, Pandey DP, Picard D, Mauro L, Ando S, Maggiolini M. Epidermal growth factor induces G protein-coupled receptor 30 expression in estrogen receptor-negative breast cancer cells. Endocrinology. 2008;149:3799–3808. doi: 10.1210/en.2008-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Zhang A, Li Y, Zhang K, Han L, Du W, Yan W, Li R, Wang Y, Wang K, Pu P, Jiang T, Jiang C, Kang C. MiR-24 regulates the proliferation and invasion of glioma by ST7L via beta-catenin/Tcf-4 signaling. Cancer Lett. 2013;329:174–180. doi: 10.1016/j.canlet.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Zhang W, Yan W, Han L, Zhang K, Shi Z, Zhang J, Wang Y, Li Y, Yu S, Pu P, Jiang C, Jiang T, Kang C. The putative tumor suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in glioma. Carcinogenesis. 2012;33:2276–2282. doi: 10.1093/carcin/bgs261. [DOI] [PubMed] [Google Scholar]
- 13.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, Tian W, Zhang Q, Wang C, Zhang Q, Zhuang SM, Zheng L, Liang A, Tao W, Cao X. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Duan Z, Choy E, Harmon D, Liu X, Susa M, Mankin H, Hornicek F. MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol Cancer Ther. 2011;10:1337–1345. doi: 10.1158/1535-7163.MCT-11-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minna E, Romeo P, De Cecco L, Dugo M, Cassinelli G, Pilotti S, Degl’Innocenti D, Lanzi C, Casalini P, Pierotti MA, Greco A, Borrello MG. miR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget. 2014;5:2513–2528. doi: 10.18632/oncotarget.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–2836. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Wang F, Li Q, Zhang H, Cui Y, Ma C, Zhu J, Gu X, Sun Z. CD151 knockdown inhibits osteosarcoma metastasis through the GSK-3beta/beta-catenin/MMP9 pathway. Oncol Rep. 2016;35:1764–1770. doi: 10.3892/or.2015.4517. [DOI] [PubMed] [Google Scholar]
- 18.Yang XH, Richardson AL, Torres-Arzayus MI, Zhou P, Sharma C, Kazarov AR, Andzelm MM, Strominger JL, Brown M, Hemler ME. CD151 accelerates breast cancer by regulating alpha 6 integrin function, signaling, and molecular organization. Cancer Res. 2008;68:3204–3213. doi: 10.1158/0008-5472.CAN-07-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novitskaya V, Romanska H, Dawoud M, Jones JL, Berditchevski F. Tetraspanin CD151 regulates growth of mammary epithelial cells in three-dimensional extracellular matrix: implication for mammary ductal carcinoma in situ. Cancer Res. 2010;70:4698–4708. doi: 10.1158/0008-5472.CAN-09-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han ZB, Yang Z, Chi Y, Zhang L, Wang Y, Ji Y, Wang J, Zhao H, Han ZC. MicroRNA-124 suppresses breast cancer cell growth and motility by targeting CD151. Cell Physiol Biochem. 2013;31:823–832. doi: 10.1159/000350100. [DOI] [PubMed] [Google Scholar]
- 21.Nakhjavani M, Palethorpe HM, Tomita Y, Smith E, Price TJ, Yool AJ, Pei JV, Townsend AR, Hardingham JE. Stereoselective anti-cancer activities of ginsenoside Rg3 on triple negative breast cancer cell models. Pharmaceuticals (Basel) 2019;12 doi: 10.3390/ph12030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiman J, Peralta EA, Louis S, Kamel O. Biology of the estrogen receptor, GPR30, in triple negative breast cancer. Am J Surg. 2013;206:698–703. doi: 10.1016/j.amjsurg.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Pandey DP, Lappano R, Albanito L, Madeo A, Maggiolini M, Picard D. Estrogenic GPR30 signalling induces proliferation and migration of breast cancer cells through CTGF. EMBO J. 2009;28:523–532. doi: 10.1038/emboj.2008.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filardo EJ, Graeber CT, Quinn JA, Resnick MB, Giri D, DeLellis RA, Steinhoff MM, Sabo E. Distribution of GPR30, a seven membrane-spanning estrogen receptor, in primary breast cancer and its association with clinicopathologic determinants of tumor progression. Clin Cancer Res. 2006;12:6359–6366. doi: 10.1158/1078-0432.CCR-06-0860. [DOI] [PubMed] [Google Scholar]
- 25.Smith HO, Leslie KK, Singh M, Qualls CR, Revankar CM, Joste NE, Prossnitz ER. GPR30: a novel indicator of poor survival for endometrial carcinoma. Am J Obstet Gynecol. 2007;196:386, e381–389. doi: 10.1016/j.ajog.2007.01.004. discussion 386 e389-311. [DOI] [PubMed] [Google Scholar]
- 26.Smith HO, Arias-Pulido H, Kuo DY, Howard T, Qualls CR, Lee SJ, Verschraegen CF, Hathaway HJ, Joste NE, Prossnitz ER. GPR30 predicts poor survival for ovarian cancer. Gynecol Oncol. 2009;114:465–471. doi: 10.1016/j.ygyno.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivacqua A, Romeo E, De Marco P, De Francesco EM, Abonante S, Maggiolini M. GPER mediates the Egr-1 expression induced by 17beta-estradiol and 4-hydroxitamoxifen in breast and endometrial cancer cells. Breast Cancer Res Treat. 2012;133:1025–1035. doi: 10.1007/s10549-011-1901-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhan Y, Brown C, Maynard E, Anshelevich A, Ni W, Ho IC, Oettgen P. Ets-1 is a critical regulator of Ang II-mediated vascular inflammation and remodeling. J Clin Invest. 2005;115:2508–2516. doi: 10.1172/JCI24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 30.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon MJ, Park S, Choi JY, Oh E, Kim YJ, Park YH, Cho EY, Kwon MJ, Nam SJ, Im YH, Shin YK, Choi YL. Clinical significance of CD151 overexpression in subtypes of invasive breast cancer. Br J Cancer. 2012;106:923–930. doi: 10.1038/bjc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zygulska AL, Krzemieniecki K, Pierzchalski P. Hippo pathway-brief overview of its relevance in cancer. J Physiol Pharmacol. 2017;68:311–335. [PubMed] [Google Scholar]
- 33.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, Flores F, Yu FX, Halder G, Guan KL. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL, Attisano L. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Mitamura T, Watari H, Wang L, Kanno H, Kitagawa M, Hassan MK, Kimura T, Tanino M, Nishihara H, Tanaka S, Sakuragi N. microRNA 31 functions as an endometrial cancer oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer. 2014;13:97. doi: 10.1186/1476-4598-13-97. [DOI] [PMC free article] [PubMed] [Google Scholar]