Abstract
Currently, there are no studies reporting the efficacy of fisetin in premature ovarian failure (POF). In this study, using mouse and Caenorhabditis elegans models, we found that fisetin not only significantly reversed ovarian damage in POF mice, but also effectively increased C. elegans lifespan and fertility. Subsequently, we carried out 16S rRNA v3+v4 sequencing using fresh feces samples from each group of mice. Results showed that although there was no significant difference in the number of gut microbiomes between the different groups of mice, fisetin affected the diversity and distribution of gut microbiota in POF mice. Alpha and beta diversity analyses showed that in the gut of POF mice in the fisetin group, the bacterial count of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of Akkermansia was significantly decreased. Finally, flow cytometry analysis showed that the numbers of CCR9+/CXCR3+/CD4+ T lymphocytes in the peripheral blood of POF mice in the fisetin group were significantly reduced, along with the number of CD4+/interleukin (IL)-12+ cells. Therefore, our data suggested that fisetin regulates the distribution and bacterial counts of Akkermansia and uncultured_bacterium_f_Lachnospiracea in POF mice, and reduces peripheral blood CCR9+/CXCR3+/CD4+ T-lymphocyte count and IL-12 secretion to regulate the ovarian microenvironment and reduce inflammation, thus exerting therapeutic effects against POF.
Keywords: Premature ovarian failure, fisetin, gut microbiota, CCR9+/CXCR3+/CD4+ T lymphocyte, interleukin-12
Introduction
Premature ovarian failure (POF) refers to amenorrhea caused by ovarian insufficiency before the age of 40 [1-3]. POF is characterized by primary or secondary amenorrhea, accompanied by elevated blood gonadotropin, reduced oestrogen, ovarian follicle atresia, and a persistent decrease in ovarian follicle reserve function [2,4-6]. As a typical form of pathological senescence, POF severely affects pregnancy and quality of life in women [2,4-6]. However, there are no effective therapies or drugs currently available for the treatment of POF.
Fisetin (3,7,3’,4’-tetrahydroxyflavone) is a natural flavonol that is abundantly present in many fruits and vegetables and has classical anti-oxidant and anti-tumour properties [7-9]. Fisetin exhibits significant affinity with microtubule proteins and can bind to microtubules, which stabilizes its structure and function [7-9]. In our previous study, we investigated the epigenetic mechanism by which fisetin exerts an inhibitory function in renal cancer stem cells. Fisetin inhibited TET1 expression and reduced 5hmC modification in specific loci in the promoters of CCNY/CDK16 in renal cancer stem cells. This inhibition prevented gene transcription, causing cell cycle arrest and ultimately resulting in inhibition of renal cancer stem cell division [8]. However, there have been no reports so far on whether fisetin has therapeutic effects on POF in mice.
The interactions between gut microbiota and the human body have various effects, such as regulation of gut development and mucosal barrier function, regulation of nutrient intake and metabolism, promotion of immune tissue maturation, and prevention of pathogen proliferation [10,11]. Increasing numbers of studies have reported that gut microbiota (gut ecology) affects normal physiological and biochemical functions and is associated with various diseases in mammals. Guo et al. found that there were significant differences in the composition and distribution of gut microbiota between a letrozole-induced mouse model of polycystic ovary syndrome (PCOS) and healthy controls [12]. The levels of Lactobacillus, Ruminococcus, and Clostridium were lower, while the levels of Prevotella were significantly higher in the PCOS group than those in the control group [12]. Yuan et al. found significant differences between mice in the endometriosis and mock groups, where the reduction in Bacteroidetes levels was particularly significant [13]. In addition, an imbalance in gut ecology causes an abnormal increase in the blood oestrogen levels, stimulating the growth of endometriotic lesions and the pathology of cyclic bleeding [14,15]. Hence, gut microbiota appears to be close associated with the occurrence and outcome of gynaecological disorders [16].
In this study, we examined the hypothesis that fisetin regulates gut microbiota to alleviate POF in mice. Our results showed that fisetin regulated gut microbiota and decreaseed CCR9+/CXCR3+/CD4+ T-lymphocyte count and interleukin (IL-12) secretion to alleviate POF in mice.
Materials and methods
Preparation of POF mouse model
POF mouse model was established based on the method used in our previous study [1-3]. Briefly, 10-week-old female C57BL/6 mice (n=30) were purchased from Shanghai Model Organisms Center (Shanghai, China). Mice were randomly divided into three groups of ten mice each. In the fisetin intervention group, 70 mg/kg of cyclophosphamide (CTX; Sigma-Aldrich, St. Louis, USA) was injected intraperitoneally, followed by subsequent intraperitoneal injections of 20 mg/kg CTX once every two days for four continuous weeks. In addition, 100 ng/kg fisetin (Sigma-Aldrich, St. Louis, USA) was administered once every two days from the start of model construction. In the control group, 70 mg/kg of CTX (Sigma-Aldrich, St. Louis, USA) was injected intraperitoneally, followed by subsequent intraperitoneal injections of 20 mg/kg CTX once every two days for four continuous weeks. An equivalent dose of phosphate buffered saline (PBS) was administered once every two days from the start of model construction. A normal control group (WT) was also set up. This study was approved by the Ethics Committee of the Shanghai Geriatric Institute of Chinese Medicine (SHAGESYDW2016018). All animal experiments conformed to the regulations of the Ministry of Science and Technology.
Immunohistochemical staining
Immunohistochemical staining was performed according to our previously published protocol [2,3,17]. Briefly, tissue sections were blocked with blocking solution (Beyotime Biotechnology Co., Ltd., Zhejiang, China) for 30 min at 37°C, followed by incubation with primary antibody for 45 min at 37°C. After washing, slides were mounted in immunofluorescence-grade blocking solution (Sigma-Aldrich, St. Louis, USA) containing DAPI.
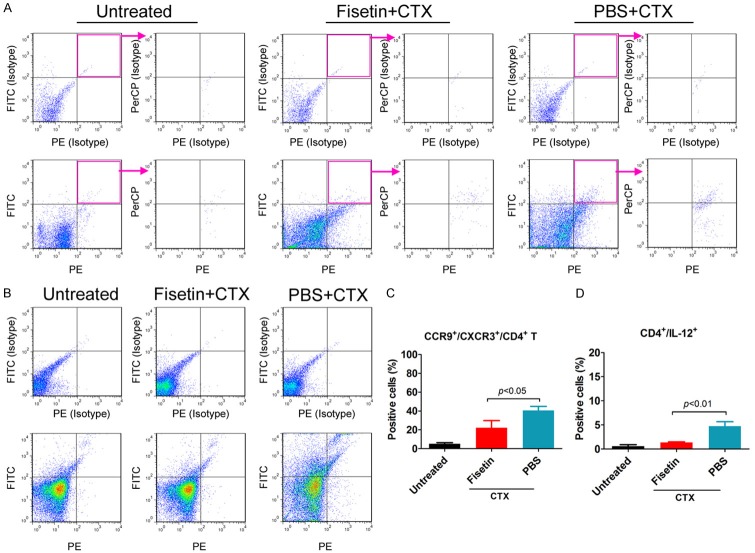
Flow cytometry (FCM) analysis
Peripheral blood mononuclear cells (PBMCs) were obtained from each group, cultured at a concentration of 1 × 106 cells/mL, and stained with primary antibodies [CD199 (CCR9) monoclonal antibody (eBioCW-1.2 (CW-1.2))-PerCP, CD183 (CXCR3) monoclonal antibody (CXCR3-173)-PE, CD4 monoclonal antibody (GK1.5)-FITC, and IL-12 p35 monoclonal antibody (27537)-PE; Invitrogen, eBioscience™, Shanghai, China] in Dulbecco’s PBS containing 10% bovine serum albumin on ice. Staining with an isotype control antibody (mouse IgG1-FITC, mouse IgG1-PE, mouse IgG1-PerCP, mouse IgG1-PE-Cyanine7 Invitrogen, eBioscience™, Shanghai, China) was performed to detect any non-specific binding. Evaluation of antibody staining by FCM was performed using FACSAria (Quanta SC, Beckman Coulter INC).
Gut microbiota analysis
Gut microbiota analysis was performed as described previously [18,19]. In brief, fresh fecal samples were collected during the final 5 days for gut microbial analysis. Bacterial genomic DNA was extracted from frozen samples stored at -80°C. The V3 and V4 regions of the 16S rRNA gene were amplified by PCR using specific bacterial primers (forward: 5’-ACTCCTACGGGAGGCAGCA-3’; reverse: 5’-GGACTACHVGGGTWTCTAAT-3’). High-throughput pyrosequencing of the PCR products was performed on an Illumina MiSeq platform at Biomarker Technologies Co. Ltd. (China). The raw paired-end reads from the original DNA fragments were merged using FLASH32 and assigned to each sample according to the unique barcodes. The UCLUST [21] in QIIME [20] (version 1.8.0) software was used to cluster sequences at 97% similarity. The tags were clustered into operational taxonomic units (OTUs). The alpha diversity index was evaluated using Mothur software (version, v.1.30). To compare the diversity index among samples, the number of sequences in each sample was standardized. Analysis treasure included OTU rank, rarefaction, and Shannon curves, and the Shannon, Chao1, Simpson, and ACE indexes were calculated. For beta diversity analysis, heatmaps of RDA-identified key OTUs, principal coordinate analysis (PcoA) [22], non-metric multi-dimensional scaling (NMDS) [23], and unweighted pair-group method with arithmetic mean (UPGMA) were performed using QIIME. Linear discriminant analysis (LDA) effect size (LEfSe) was used for the quantitative analysis of biomarkers in each group. Briefly, LEfSe analysis (at LDA threshold >4), the non-parametric factorial Kruskal-Wallis sum-rank test, and the unpaired Wilcoxon rank-sum test were performed to identify the most abundant taxa [24,25].
Construction of Caenorhabditis elegans POF model and grouping
The N2 strain of C. elegans was a gift from Shanghai Model Organisms Center (Shanghai, China). Approximately 300 C. elegans specimens in the L4 stage were placed in 12 mL of NGM culture medium containing OP50 Escherichia coli. The worms were divided equally into three groups of 10 worms/4 mL. In the fisetin intervention group, 20 μg/mL CTX and 10 ng/mL fisetin were added to the NGM culture medium. In the control group, 20 μg/mL CTX and an equivalent dose of PBS were added to the NGM culture medium. In addition, a normal control group (WT) was set up.
Hematoxylin-eosin (HE) staining
Briefly, all fresh tissues were fixed in 4% paraformaldehyde at room temperature (Sigma-Aldrich, St. Louis, USA) for 30 min [1,3]. Next, the tissues were subjected to gradient ethanol dehydration, paraffin-embedding, sectioning (thickness of 6 μm), and immersion in xylene for clearing. The tissue sections were stained with HE (Sigma-Aldrich, St. Louis, USA) before xylene (Sigma-Aldrich, St. Louis, USA) clearing and mounting with neutral resins (Sigma-Aldrich, St. Louis, USA).
C. elegans total RNA extraction and qPCR assay
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) was used to extract total RNA from C. elegans in various groups, according to the manufacturer’s instructions. After treatment with DNase I (Sigma-Aldrich, St. Louis, USA), total RNA was quantified, and reverse transcription was carried out using the ReverTra Ace-α First Strand cDNA Synthesis Kit (Toyobo. qRT-PCR was performed on the RealPlex4 Real-Time PCR Detection System (Eppendorf Co. Ltd., Hamburg, Germany). The SYBR Green Real-Time PCR Master Mix (Toyobo, Shanghai, China) was used as a fluorescent dye for nucleic acid amplification. A total of 40 amplification cycles were carried out as follows: denaturation at 95°C for 15 sec, annealing at 58°C for 30 sec, and extension at 72°C for 42 sec. The 2-ΔΔCt method was used to calculate relative gene expression, where ΔCt = Ct_genes - Ct_Act-1; ΔΔCt = ΔCt_all_groups - ΔCt_blankcontrol_group. Target gene expression levels were normalized against the 18S rRNA expression levels. The primer sequences used were as follows: Egl-17, forward: 5’-CCATTGGCAACTATTCA-3’, reverse: 5’-TTCCATCCCATTCTCC-3’; Daf-2, forward: 5’-ACCCGTGAGCAGTGTC-3’, reverse: 5’-TTGCCAACGCATTTT-3’; Act-1, forward: 5’-CTCTTGCCCCATCAACCATG-3’, reverse: 5’-CTTGCTTGGAGATCCACATC-3’.
Western blot analysis
The cells were lysed using a 2× loading lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). The proteins were separated on 12% sodium dodecyl sulfate-polyacrylamide gels and transferred onto hybrid polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After blocking with 5% (w/v) non-fat dried milk in Tris-buffered saline with Tween-20 (TBST; Beyotime Institute of Biotechnology), the membranes were washed four times (15 min each) with TBST at room temperature and incubated with primary antibodies, including rabbit anti-mouse Ki67 and AMH antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), rabbit anti-mouse p16, p-H2A.X, and GAPDH antibodies (Cell Signaling Technology, Beverly, MA, USA). After washing, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:1,000; Santa Cruz Biotechnology) for 1 h. Following washing four times (15 min each) with TBST at room temperature, the immunoreactive bands were visualized using an enhanced chemiluminescence kit (Perkin Elmer, Norwalk, CT, USA).
Evaluation of C. elegans behaviour and proliferation
Briefly, 40 C. elegans specimens at the L4 stage were collected from each group and placed in NGM culture medium containing OP50 E. coli [26-30]. The specimens were then observed under a microscope. The number of head thrashes and body bends were recorded for 1 min to calculate their frequency in C. elegans progeny. L4 stage C. elegans were randomly selected, and individual worms were added to the NGM culture medium of the experimental and control groups. The specimens were cultured at 20°C. The plates were rotated once every 12 h, and the worms were moved to NGM culture medium containing CTX until the end of the spawning period. The number of progenies in NGM culture medium from each group was calculated.
Statistical analysis
Each experiment was performed as least thrice, and GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. Data from follicular count experiment, qPCR, C. elegans study, and FCM, are expressed as mean ± standard error. Differences between groups were evaluated with Student’s t-test. A p value less than 0.05 was considered statistically significant. For gut microbiota analysis, the LDA-effect size (LEfSe) method was used for the quantitative analysis of biomarkers in each group. LEfSe analysis (at LDA threshold >4), the non-parametric factorial Kruskal-Wallis sum-rank test, and the unpaired Wilcoxon rank-sum test were performed to identify the most abundant taxa.
Results
Fisetin effectively alleviates ovarian damage in POF mice and increases the number of normal ovarian follicles
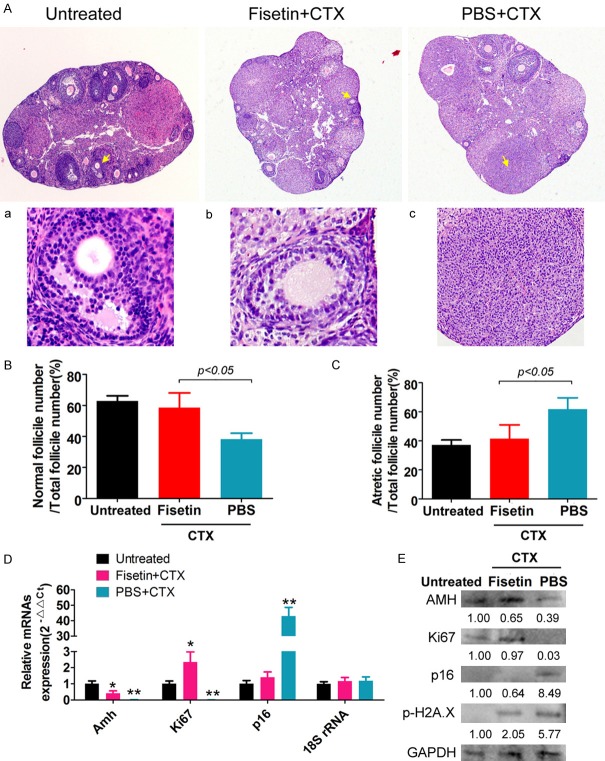
A POF model was established by intraperitoneal injection of CTX in fisetin- or PBS-treated mice. Pathological testing by HE staining showed that mature ovarian follicles disappeared, the number of atretic ovarian follicles significantly increased, and the number of quality ovarian granulosa cells significantly decreased in the ovarian tissues of POF mice in the PBS group (Figure 1A). Ovarian follicles at various stages of development were observed in the ovaries of POF mice in the fisetin group. Simultaneously, the proportion of normal ovarian follicles significantly increased while the proportion of atretic ovarian follicles significantly decreased (Figure 1B and 1C). qPCR and western blotting were used to detect the expression of proliferation- and aging-related biomarkers in each group. Results showed that the expression levels of AMH (an ovarian granulosa cell marker) and Ki67 (a cell proliferation marker) in the ovarian tissues of POF mice in the PBS group were lower significantly than those of mice in the untreated group (Figure 1D). However, the expression levels of p16 and phosphorylated H2A.X (pho-H2A.X) proteins (cell ageing markers) in the ovarian tissues of POF mice in the PBS group were significantly higher than those of mice in the untreated group (Figure 1D and 1E). However, the expression levels of p16 and pho-H2A.X in the ovaries of POF mice in the fisetin group were significantly decreased, while those of AMH and Ki67 were significantly increased (Figure 1D and 1E). These data prove that fisetin effectively alleviates ovarian damage in POF mice and increases the number of normal ovarian follicles.
Figure 1.
Fisetin effectively alleviates ovarian damage in POF mice. (A) HE staining results of the ovarian tissues in various groups of mice (magnification: 100×). (a-c) are the high magnification images (400×) of the sites indicated with yellow arrows. (B) The proportion of normal ovarian follicles in the different groups of mice. (C) The proportion of atretic ovarian follicles in the different groups of mice. Pathological testing by HE staining showed that the number of atretic ovarian follicles significantly increased, while the number of quality ovarian granulosa cells significantly decreased in the ovarian tissues of POF mice. Ovarian follicles at various stages of development appeared in the ovaries of POF mice in the fisetin group. (D) qPCR for quantitation of mRNA levels of genes related to ovarian granulosa cell proliferation and aging. **P<0.01 vs PBS+CTX group, *P<0.05 vs PBS+CTX group, as calculated by t-test. (E) Western blot analysis to detect the expression levels of proteins related to ovarian granulosa cell proliferation and aging. qPCR and western blotting results revealed that the expression levels of p16 and pho-H2A.X in the ovaries of POF mice in the fisetin group were decreased significantly, while those of AMH and Ki67 were increased significantly.
Fisetin significantly enhances C. elegans lifespan and fertility
Experimental results showed that compared with PBS, fisetin significantly increased the life span of C. elegans (Figure 2A and 2B), the frequency of head thrashes and body bends (Figure 2C, 2D, Supplementary Videos 1, 2, 3), and the number of pregnant C. elegans after CTX treatment (Figure 2E). In addition, qPCR results showed that compared with that in the PBS group, the mRNA expression of genes associated with insulin/IGF-like signaling (IIS) in C. elegans was significantly increased in the fisetin group after CTX treatment (Figure 2F). Hence, our results showed that fisetin effectively increased C. elegans lifespan and fertility via stimulating the IIS signaling pathway.
Figure 2.
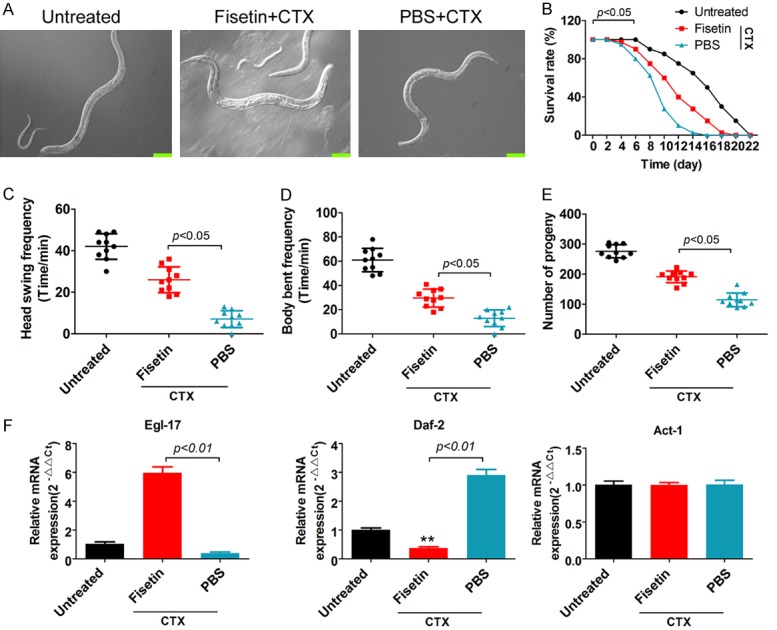
Fisetin effectively enhances C. elegans lifespan and fertility. A. Phenotypes of various groups of C. elegans. Scale bar: 100 μm. B. Life span curve: the results showed that, compared with the PBS group, the life span of C. elegans was significantly increased in the fisetin group after CTX treatment. C. Frequency of head thrashes in various groups of C. elegans: Compared with PBS, fisetin significantly increased the frequency of head thrashes after CTX treatment. D. Frequency of body bends in various groups of C. elegans: Compared with PBS, fisetin significantly increased the frequency of body bends after CTX treatment. E. Number of pregnant C. elegans in the various groups: compared with PBS, fisetin significantly increased the number of pregnant C. elegans after CTX treatment. F. qPCR for quantitation of mRNA levels of genes related to the IIS signaling pathway. **P<0.01 vs PBS+CTX group, as calculated by t-test. qPCR results showed that, compared with PBS, fisetin significantly increased the mRNA expression of genes associated with the IIS signaling pathway in C. elegans after CTX treatment.
Fisetin does not alter gut microbial counts in POF mice
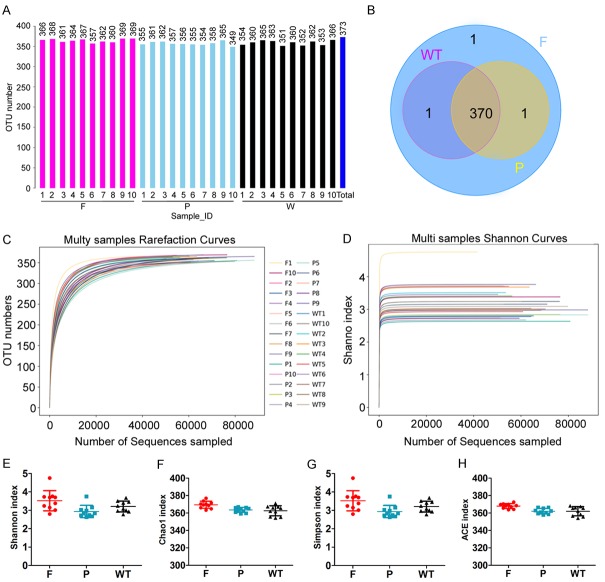
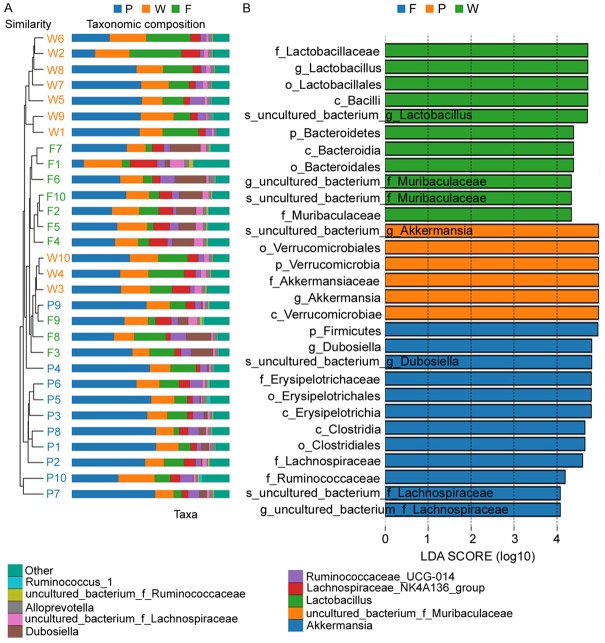
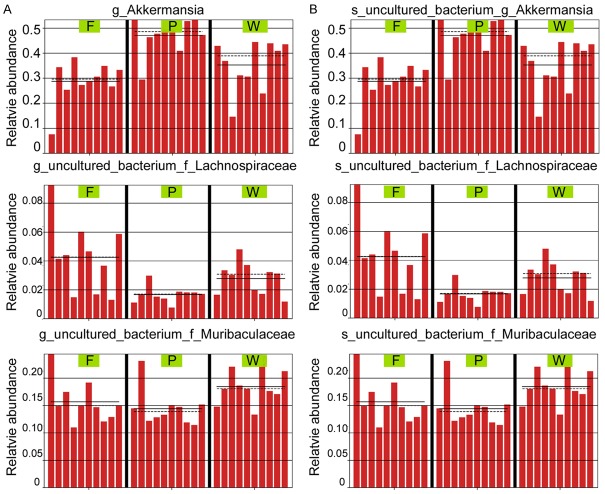
After treating the feces of mice in the normal group (W), POF mice in the fisetin group (F) and PBS group (P), bacterial 16S rRNA v3+v4 sequencing was carried out to evaluate the composition of the gut microbiota and the distribution of bacterial communities. Thirty samples were sequenced to obtain 3,255,567 reads. After the splicing of paired-end reads and filtering, a total of 2,765,085 clean tags were generated. There were at least 56,084 clean tags per sample, and 92,170 clean tags were generated on an average (Table 1). The UCLUST in QIIME (version 1.8.0) software was used to cluster tags into OTUs based on 97% sequence similarity. There were no significant differences in OTU quantity between the three groups (Figure 3A-E; Table 1). Next, we generated Venn diagrams for OTUs, OTU rank, rarefaction curves, Shannon index curves, Chao1 curves, Simpson curves, and ACE curves (Figure 3E-H; Table 2). Results showed consistent numbers of OTUs. Preliminary tests showed that fisetin intervention did not alter gut microbial counts in POF mice.
Table 1.
OUTs
| OTU_id | F | P | WT | taxonomy |
|---|---|---|---|---|
| OTU1 | 180722 | 327136 | 227626 | k__Bacteria; p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Akkermansiaceae; g__Akkermansia; s__uncultured_bacterium_g_Akkermansia |
| OTU10 | 10 | 11496 | 11 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Dubosiella; s__uncultured_bacterium_g_Dubosiella |
| OTU100 | 1095 | 1401 | 32 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU101 | 294 | 520 | 553 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU102 | 200 | 485 | 155 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus_1; s__uncultured_bacterium_g_Ruminococcus_1 |
| OTU103 | 653 | 342 | 350 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU104 | 576 | 360 | 161 | k__Bacteria; p__Fusobacteria; c__Fusobacteriia; o__Fusobacteriales; f__Fusobacteriaceae; g__Cetobacterium; s__uncultured_bacterium_g_Cetobacterium |
| OTU105 | 894 | 1198 | 1321 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU106 | 93 | 246 | 235 | k__Bacteria; p__Tenericutes; c__Mollicutes; o__Mollicutes_RF39; f__uncultured_bacterium_o_Mollicutes_RF39; g__uncultured_bacterium_o_Mollicutes_RF39; s__uncultured_bacterium_o_Mollicutes_RF39 |
| OTU107 | 362 | 185 | 302 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Tyzzerella; s__uncultured_bacterium_g_Tyzzerella |
| OTU108 | 679 | 534 | 589 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU109 | 1147 | 169 | 553 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU11 | 1798 | 4875 | 4222 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotellaceae_NK3B31_group; s__uncultured_bacterium_g_Prevotellaceae_NK3B31_group |
| OTU110 | 893 | 446 | 981 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-006; s__uncultured_bacterium_g_Lachnospiraceae_UCG-006 |
| OTU111 | 781 | 569 | 25 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU112 | 757 | 331 | 442 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU113 | 333 | 431 | 265 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_9; s__uncultured_bacterium_g_Ruminiclostridium_9 |
| OTU114 | 830 | 294 | 711 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU115 | 88 | 370 | 395 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU116 | 164 | 230 | 411 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU117 | 7 | 339 | 185 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-010; s__uncultured_bacterium_g_Ruminococcaceae_UCG-010 |
| OTU118 | 450 | 209 | 295 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU119 | 219 | 162 | 177 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU12 | 8070 | 3798 | 4800 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU120 | 431 | 163 | 399 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-001; s__uncultured_bacterium_g_Lachnospiraceae_UCG-001 |
| OTU121 | 103 | 260 | 189 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-010; s__uncultured_bacterium_g_Ruminococcaceae_UCG-010 |
| OTU122 | 158 | 257 | 237 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Family_XIII; g__[Eubacterium]_nodatum_group; s__uncultured_bacterium_g_[Eubacterium]_nodatum_group |
| OTU123 | 237 | 145 | 158 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Negativibacillus; s__uncultured_bacterium_g_Negativibacillus |
| OTU124 | 869 | 491 | 567 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU125 | 517 | 105 | 411 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU126 | 266 | 261 | 167 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU127 | 322 | 230 | 370 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Peptococcaceae; g__uncultured_bacterium_f_Peptococcaceae; s__uncultured_bacterium_f_Peptococcaceae |
| OTU128 | 342 | 113 | 480 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__[Eubacterium]_coprostanoligenes_group; s__uncultured_bacterium_g_[Eubacterium]_coprostanoligenes_group |
| OTU129 | 597 | 303 | 217 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__ASF356; s__uncultured_bacterium_g_ASF356 |
| OTU13 | 2040 | 6821 | 4164 | k__Bacteria; p__Patescibacteria; c__Saccharimonadia; o__Saccharimonadales; f__Saccharimonadaceae; g__Candidatus_Saccharimonas; s__uncultured_bacterium_g_Candidatus_Saccharimonas |
| OTU130 | 161 | 156 | 159 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Enterorhabdus; s__uncultured_bacterium_g_Enterorhabdus |
| OTU131 | 211 | 269 | 246 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Shuttleworthia; s__uncultured_bacterium_g_Shuttleworthia |
| OTU132 | 501 | 176 | 308 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU133 | 304 | 400 | 348 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU134 | 437 | 2 | 65 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-010; s__uncultured_bacterium_g_Ruminococcaceae_UCG-010 |
| OTU135 | 190 | 218 | 312 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU136 | 264 | 160 | 174 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Blautia; s__uncultured_bacterium_g_Blautia |
| OTU137 | 368 | 201 | 207 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Butyricicoccus; s__uncultured_bacterium_g_Butyricicoccus |
| OTU138 | 121 | 232 | 185 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU139 | 442 | 160 | 205 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__A2; s__uncultured_bacterium_g_A2 |
| OTU14 | 3189 | 4246 | 5318 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU140 | 462 | 197 | 565 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU141 | 456 | 304 | 291 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU142 | 288 | 157 | 153 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__UBA1819; s__uncultured_bacterium_g_UBA1819 |
| OTU143 | 25 | 283 | 77 | k__Bacteria; p__Tenericutes; c__Mollicutes; o__Mollicutes_RF39; f__uncultured_bacterium_o_Mollicutes_RF39; g__uncultured_bacterium_o_Mollicutes_RF39; s__uncultured_bacterium_o_Mollicutes_RF39 |
| OTU144 | 579 | 154 | 92 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU145 | 436 | 102 | 194 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU146 | 131 | 249 | 387 | k__Bacteria; p__Proteobacteria; c__Gammaproteobacteria; o__Betaproteobacteriales; f__Burkholderiaceae; g__Parasutterella; s__uncultured_bacterium_g_Parasutterella |
| OTU147 | 55 | 246 | 178 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU148 | 204 | 164 | 100 | k__Bacteria; p__Proteobacteria; c__Deltaproteobacteria; o__Desulfovibrionales; f__Desulfovibrionaceae; g__uncultured_bacterium_f_Desulfovibrionaceae; s__uncultured_bacterium_f_Desulfovibrionaceae |
| OTU149 | 535 | 14 | 6 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU15 | 4172 | 4640 | 7762 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__uncultured_bacterium_g_Lactobacillus |
| OTU150 | 73 | 163 | 130 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU151 | 448 | 263 | 69 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU152 | 423 | 301 | 306 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_FCS020_group; s__uncultured_bacterium_g_Lachnospiraceae_FCS020_group |
| OTU153 | 299 | 227 | 342 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU154 | 155 | 114 | 146 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU155 | 147 | 165 | 456 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU156 | 554 | 232 | 442 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU157 | 504 | 261 | 334 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU158 | 734 | 235 | 478 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU159 | 114 | 201 | 74 | k__Bacteria; p__Tenericutes; c__Mollicutes; o__Mollicutes_RF39; f__uncultured_bacterium_o_Mollicutes_RF39; g__uncultured_bacterium_o_Mollicutes_RF39; s__uncultured_bacterium_o_Mollicutes_RF39 |
| OTU16 | 7059 | 7257 | 6025 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU160 | 105 | 60 | 178 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU161 | 146 | 172 | 60 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU162 | 971 | 395 | 384 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-008; s__uncultured_bacterium_g_Lachnospiraceae_UCG-008 |
| OTU163 | 320 | 75 | 217 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU164 | 522 | 123 | 190 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU165 | 149 | 196 | 259 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Marinifilaceae; g__Butyricimonas; s__uncultured_bacterium_g_Butyricimonas |
| OTU166 | 1031 | 1603 | 505 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU167 | 218 | 50 | 167 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU168 | 221 | 22 | 279 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU169 | 83 | 68 | 251 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__uncultured_bacterium_g_Bacteroides |
| OTU17 | 2267 | 4446 | 916 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus_1; s__uncultured_bacterium_g_Ruminococcus_1 |
| OTU170 | 244 | 138 | 90 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Blautia; s__uncultured_bacterium_g_Blautia |
| OTU171 | 428 | 324 | 210 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU172 | 302 | 184 | 314 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU173 | 65 | 201 | 25 | k__Bacteria; p__Cyanobacteria; c__Melainabacteria; o__Gastranaerophilales; f__Blattella_germanica_German_cockroach; g__uncultured_bacterium_f_Blattella_germanica_German_cockroach; s__uncultured_bacterium_f_Blattella_germanica_German_cockroach |
| OTU174 | 257 | 176 | 233 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU175 | 145 | 74 | 95 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Anaerotruncus; s__uncultured_bacterium_g_Anaerotruncus |
| OTU176 | 516 | 259 | 313 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU177 | 42 | 99 | 89 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__uncultured_bacterium_f_Eggerthellaceae; s__uncultured_bacterium_f_Eggerthellaceae |
| OTU178 | 260 | 89 | 198 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus_1; s__uncultured_bacterium_g_Ruminococcus_1 |
| OTU179 | 92 | 129 | 65 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_9; s__uncultured_bacterium_g_Ruminiclostridium_9 |
| OTU18 | 4823 | 3857 | 3416 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU180 | 298 | 147 | 99 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU181 | 80 | 65 | 58 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU182 | 129 | 91 | 135 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Oscillibacter; s__uncultured_bacterium_g_Oscillibacter |
| OTU183 | 142 | 67 | 82 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU184 | 96 | 89 | 70 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Streptococcaceae; g__Streptococcus; s__uncultured_bacterium_g_Streptococcus |
| OTU185 | 64 | 99 | 81 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__uncultured_bacterium_f_Erysipelotrichaceae; s__uncultured_bacterium_f_Erysipelotrichaceae |
| OTU186 | 70 | 107 | 219 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Erysipelatoclostridium; s__uncultured_bacterium_g_Erysipelatoclostridium |
| OTU187 | 356 | 76 | 236 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU188 | 29 | 2 | 326 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU189 | 64 | 81 | 61 | k__Bacteria; p__Tenericutes; c__Mollicutes; o__Mollicutes_RF39; f__uncultured_bacterium_o_Mollicutes_RF39; g__uncultured_bacterium_o_Mollicutes_RF39; s__uncultured_bacterium_o_Mollicutes_RF39 |
| OTU19 | 1274 | 2689 | 3505 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotellaceae_UCG-001; s__uncultured_bacterium_g_Prevotellaceae_UCG-001 |
| OTU190 | 140 | 113 | 50 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__[Eubacterium]_ventriosum_group; s__uncultured_bacterium_g_[Eubacterium]_ventriosum_group |
| OTU191 | 153 | 87 | 155 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU192 | 189 | 131 | 130 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU193 | 143 | 86 | 134 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU194 | 239 | 41 | 61 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU195 | 21 | 69 | 92 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiales_vadinBB60_group; g__uncultured_bacterium_f_Clostridiales_vadinBB60_group; s__uncultured_bacterium_f_Clostridiales_vadinBB60_group |
| OTU196 | 289 | 143 | 248 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_5; s__uncultured_bacterium_g_Ruminiclostridium_5 |
| OTU197 | 170 | 36 | 55 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU198 | 182 | 104 | 233 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU199 | 84 | 72 | 101 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU2 | 45952 | 51222 | 98973 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__uncultured_bacterium_g_Lactobacillus |
| OTU20 | 699 | 6055 | 675 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Turicibacter; s__uncultured_bacterium_g_Turicibacter |
| OTU200 | 406 | 164 | 147 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_5; s__uncultured_bacterium_g_Ruminiclostridium_5 |
| OTU201 | 47 | 73 | 153 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU202 | 59 | 16 | 142 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU203 | 115 | 61 | 24 | k__Bacteria; p__Actinobacteria; c__Actinobacteria; o__Corynebacteriales; f__Nocardiaceae; g__Rhodococcus; s__bacterium_GC452011 |
| OTU204 | 455 | 254 | 304 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU205 | 252 | 14 | 15 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU206 | 101 | 44 | 35 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-005; s__uncultured_bacterium_g_Ruminococcaceae_UCG-005 |
| OTU207 | 189 | 70 | 108 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU208 | 79 | 55 | 91 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Family_XIII; g__[Eubacterium]_brachy_group; s__uncultured_bacterium_g_[Eubacterium]_brachy_group |
| OTU209 | 115 | 66 | 88 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU21 | 6617 | 9125 | 4045 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU210 | 31 | 38 | 90 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Enterorhabdus; s__uncultured_bacterium_g_Enterorhabdus |
| OTU211 | 11 | 46 | 138 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU212 | 43 | 26 | 94 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU213 | 66 | 39 | 44 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU214 | 39 | 31 | 121 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU215 | 118 | 63 | 125 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU216 | 32 | 67 | 79 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Family_XIII; g__Family_XIII_AD3011_group; s__uncultured_bacterium_g_Family_XIII_AD3011_group |
| OTU217 | 51 | 11 | 91 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU218 | 161 | 135 | 229 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU219 | 52 | 50 | 87 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Family_XIII; g__Anaerovorax; s__uncultured_bacterium_g_Anaerovorax |
| OTU22 | 10088 | 6335 | 7824 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU220 | 301 | 0 | 0 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU221 | 27 | 49 | 66 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Christensenellaceae; g__Catabacter; s__Christensenella_timonensis |
| OTU222 | 180 | 43 | 72 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU223 | 96 | 181 | 189 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Tannerellaceae; g__Parabacteroides; s__uncultured_bacterium_g_Parabacteroides |
| OTU224 | 98 | 26 | 39 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU225 | 163 | 70 | 146 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU226 | 66 | 132 | 150 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__A2; s__uncultured_bacterium_g_A2 |
| OTU228 | 82 | 60 | 32 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Enterorhabdus; s__uncultured_bacterium_g_Enterorhabdus |
| OTU229 | 28 | 66 | 49 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-005; s__uncultured_bacterium_g_Ruminococcaceae_UCG-005 |
| OTU23 | 469 | 4097 | 1749 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Faecalibaculum; s__uncultured_bacterium_g_Faecalibaculum |
| OTU231 | 1640 | 1691 | 2775 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU232 | 181 | 304 | 185 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_9; s__uncultured_bacterium_g_Ruminiclostridium_9 |
| OTU234 | 41 | 59 | 35 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Marinifilaceae; g__Butyricimonas; s__uncultured_bacterium_g_Butyricimonas |
| OTU235 | 112 | 15 | 45 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU237 | 32 | 59 | 43 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Candidatus_Soleaferrea; s__uncultured_bacterium_g_Candidatus_Soleaferrea |
| OTU239 | 231 | 54 | 88 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU24 | 5428 | 1844 | 2492 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU240 | 228 | 58 | 86 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU241 | 49 | 29 | 67 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-013; s__uncultured_bacterium_g_Ruminococcaceae_UCG-013 |
| OTU242 | 53 | 54 | 27 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU244 | 795 | 2641 | 3030 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU245 | 55 | 98 | 91 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU248 | 94 | 57 | 24 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__uncultured_bacterium_f_Erysipelotrichaceae; s__uncultured_bacterium_f_Erysipelotrichaceae |
| OTU25 | 3508 | 3405 | 3626 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU250 | 63 | 35 | 69 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU251 | 101 | 78 | 64 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU253 | 145 | 64 | 13 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU255 | 38 | 13 | 95 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU256 | 238 | 490 | 481 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU258 | 46 | 24 | 139 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU259 | 92 | 64 | 29 | k__Bacteria; p__Proteobacteria; c__Gammaproteobacteria; o__Aeromonadales; f__Aeromonadaceae; g__Aeromonas; s__uncultured_bacterium_g_Aeromonas |
| OTU26 | 5129 | 4177 | 3659 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU260 | 276 | 152 | 122 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Oscillibacter; s__uncultured_bacterium_g_Oscillibacter |
| OTU262 | 173 | 28 | 104 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU267 | 715 | 215 | 686 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU27 | 1923 | 3489 | 3732 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU270 | 296 | 50 | 10 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-001; s__uncultured_bacterium_g_Lachnospiraceae_UCG-001 |
| OTU272 | 27 | 62 | 55 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Christensenellaceae; g__Christensenellaceae_R-7_group; s__uncultured_bacterium_g_Christensenellaceae_R-7_group |
| OTU273 | 90 | 28 | 97 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU28 | 5810 | 3221 | 3670 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU284 | 76 | 60 | 33 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU288 | 45 | 65 | 36 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU29 | 1413 | 2503 | 1632 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU291 | 15 | 51 | 79 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU292 | 156 | 28 | 109 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU293 | 58 | 38 | 41 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiaceae_1; g__Candidatus_Arthromitus; s__uncultured_bacterium_g_Candidatus_Arthromitus |
| OTU294 | 77 | 68 | 61 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Marvinbryantia; s__uncultured_bacterium_g_Marvinbryantia |
| OTU297 | 107 | 25 | 259 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU3 | 11346 | 11026 | 13283 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Alloprevotella; s__uncultured_bacterium_g_Alloprevotella |
| OTU30 | 2362 | 3698 | 2913 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU304 | 140 | 44 | 111 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU31 | 2004 | 2032 | 202 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU314 | 22 | 107 | 99 | k__Bacteria; p__Tenericutes; c__Mollicutes; o__Mollicutes_RF39; f__uncultured_bacterium_o_Mollicutes_RF39; g__uncultured_bacterium_o_Mollicutes_RF39; s__uncultured_bacterium_o_Mollicutes_RF39 |
| OTU32 | 1637 | 544 | 1214 | k__Bacteria; p__Proteobacteria; c__Deltaproteobacteria; o__Desulfovibrionales; f__Desulfovibrionaceae; g__Bilophila; s__uncultured_bacterium_g_Bilophila |
| OTU323 | 1094 | 1115 | 3245 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU33 | 2570 | 3593 | 3250 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__[Eubacterium]_ruminantium_group; s__uncultured_bacterium_g_[Eubacterium]_ruminantium_group |
| OTU337 | 278 | 157 | 207 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__GCA-900066575; s__uncultured_bacterium_g_GCA-900066575 |
| OTU34 | 4113 | 1624 | 3205 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU35 | 3134 | 661 | 1991 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU357 | 480 | 492 | 270 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU36 | 1478 | 1328 | 1176 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU361 | 2649 | 1057 | 261 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU364 | 179 | 358 | 492 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU366 | 757 | 104 | 219 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU37 | 6238 | 822 | 1038 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU371 | 417 | 67 | 164 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-001; s__uncultured_bacterium_g_Lachnospiraceae_UCG-001 |
| OTU373 | 90 | 70 | 45 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Bacillales; f__Alicyclobacillaceae; g__Effusibacillus; s__Effusibacillus_pohliae |
| OTU378 | 194 | 79 | 286 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Oscillibacter; s__uncultured_bacterium_g_Oscillibacter |
| OTU38 | 974 | 934 | 893 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU39 | 1788 | 2909 | 2564 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__uncultured_bacterium_g_Bacteroides |
| OTU399 | 184 | 112 | 13 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__GCA-900066575; s__uncultured_bacterium_g_GCA-900066575 |
| OTU4 | 77346 | 1580 | 10 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Dubosiella; s__uncultured_bacterium_g_Dubosiella |
| OTU40 | 1580 | 252 | 786 | k__Bacteria; p__Deferribacteres; c__Deferribacteres; o__Deferribacterales; f__Deferribacteraceae; g__Mucispirillum; s__Mucispirillum_schaedleri_ASF457 |
| OTU400 | 423 | 164 | 363 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4B4_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4B4_group |
| OTU408 | 114 | 162 | 110 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU41 | 1384 | 2332 | 904 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiaceae_1; g__Clostridium_sensu_stricto_1; s__uncultured_bacterium_g_Clostridium_sensu_stricto_1 |
| OTU413 | 78 | 22 | 97 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU415 | 1174 | 92 | 212 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU42 | 5662 | 812 | 1352 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__[Eubacterium]_xylanophilum_group; s__uncultured_bacterium_g_[Eubacterium]_xylanophilum_group |
| OTU420 | 6 | 73 | 58 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Faecalibaculum; s__uncultured_bacterium_g_Faecalibaculum |
| OTU423 | 39 | 159 | 197 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotellaceae_NK3B31_group; s__uncultured_bacterium_g_Prevotellaceae_NK3B31_group |
| OTU426 | 54 | 50 | 98 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-006; s__uncultured_bacterium_g_Lachnospiraceae_UCG-006 |
| OTU427 | 151 | 115 | 123 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Acetatifactor; s__uncultured_bacterium_g_Acetatifactor |
| OTU428 | 115 | 69 | 68 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU43 | 1583 | 2309 | 1937 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU432 | 194 | 122 | 319 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU435 | 243 | 340 | 272 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU436 | 37 | 20 | 756 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU438 | 150 | 158 | 232 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU44 | 1671 | 1919 | 1780 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-006; s__uncultured_bacterium_g_Lachnospiraceae_UCG-006 |
| OTU449 | 356 | 261 | 601 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU45 | 1220 | 880 | 2068 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU453 | 275 | 83 | 184 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-003; s__uncultured_bacterium_g_Ruminococcaceae_UCG-003 |
| OTU46 | 873 | 1464 | 1535 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__[Eubacterium]_fissicatena_group; s__uncultured_bacterium_g_[Eubacterium]_fissicatena_group |
| OTU461 | 269 | 124 | 212 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_9; s__uncultured_bacterium_g_Ruminiclostridium_9 |
| OTU463 | 84 | 15 | 261 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU466 | 178 | 180 | 292 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU47 | 1112 | 3497 | 3989 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU470 | 232 | 60 | 298 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU478 | 385 | 158 | 280 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Intestinimonas; s__uncultured_bacterium_g_Intestinimonas |
| OTU48 | 639 | 1193 | 927 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus_1; s__uncultured_bacterium_g_Ruminococcus_1 |
| OTU485 | 155 | 9 | 57 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU486 | 948 | 861 | 350 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU487 | 218 | 211 | 332 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU489 | 220 | 209 | 358 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU49 | 1092 | 560 | 603 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU492 | 115 | 98 | 127 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU493 | 445 | 207 | 98 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnoclostridium; s__uncultured_bacterium_g_Lachnoclostridium |
| OTU495 | 252 | 64 | 178 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__[Eubacterium]_oxidoreducens_group; s__uncultured_bacterium_g_[Eubacterium]_oxidoreducens_group |
| OTU496 | 118 | 57 | 15 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnoclostridium; s__uncultured_bacterium_g_Lachnoclostridium |
| OTU5 | 10914 | 15909 | 16481 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU50 | 1484 | 1124 | 916 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU500 | 98 | 86 | 62 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU502 | 207 | 129 | 137 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU503 | 179 | 68 | 72 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU505 | 303 | 134 | 189 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Intestinimonas; s__uncultured_bacterium_g_Intestinimonas |
| OTU508 | 114 | 99 | 90 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU509 | 80 | 47 | 47 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU51 | 2189 | 1402 | 1899 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU510 | 217 | 190 | 339 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__A2; s__uncultured_bacterium_g_A2 |
| OTU52 | 597 | 1074 | 330 | k__Bacteria; p__Cyanobacteria; c__Melainabacteria; o__Gastranaerophilales; f__uncultured_bacterium_o_Gastranaerophilales; g__uncultured_bacterium_o_Gastranaerophilales; s__uncultured_bacterium_o_Gastranaerophilales |
| OTU520 | 274 | 158 | 247 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU521 | 245 | 364 | 292 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU522 | 77 | 34 | 95 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU524 | 90 | 132 | 92 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU528 | 98 | 143 | 126 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU53 | 722 | 513 | 818 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__uncultured_bacterium_g_Bacteroides |
| OTU531 | 299 | 0 | 29 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU534 | 121 | 155 | 143 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU536 | 79 | 43 | 52 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_9; s__uncultured_bacterium_g_Ruminiclostridium_9 |
| OTU54 | 2028 | 2218 | 1971 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__Muribaculum; s__Parabacteroides_sp._YL27 |
| OTU544 | 61 | 65 | 50 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU545 | 524 | 711 | 679 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU546 | 224 | 194 | 532 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU547 | 38 | 72 | 92 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Roseburia; s__uncultured_bacterium_g_Roseburia |
| OTU55 | 530 | 1109 | 1238 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Peptostreptococcaceae; g__Romboutsia; s__uncultured_bacterium_g_Romboutsia |
| OTU552 | 180 | 275 | 417 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU56 | 1780 | 3053 | 2962 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU563 | 688 | 507 | 607 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU565 | 124 | 135 | 147 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Enterorhabdus; s__uncultured_bacterium_g_Enterorhabdus |
| OTU568 | 26 | 18 | 92 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__UBA1819; s__uncultured_bacterium_g_UBA1819 |
| OTU57 | 1177 | 1160 | 1408 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU579 | 146 | 892 | 1006 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__Lactobacillus_gasseri |
| OTU58 | 984 | 509 | 674 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus_1; s__uncultured_bacterium_g_Ruminococcus_1 |
| OTU581 | 103 | 327 | 242 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU584 | 101 | 97 | 87 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU59 | 2908 | 1923 | 3017 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU593 | 61 | 109 | 31 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU596 | 392 | 52 | 64 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__[Eubacterium]_xylanophilum_group; s__uncultured_bacterium_g_[Eubacterium]_xylanophilum_group |
| OTU599 | 80 | 97 | 31 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU6 | 15510 | 17930 | 6442 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU60 | 10229 | 12249 | 10054 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU600 | 216 | 120 | 235 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU602 | 51 | 44 | 64 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU609 | 70 | 50 | 91 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU61 | 2219 | 30 | 788 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Blautia; s__uncultured_bacterium_g_Blautia |
| OTU614 | 161 | 50 | 18 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU618 | 102 | 102 | 2 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU62 | 894 | 335 | 814 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU620 | 67 | 38 | 59 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnoclostridium; s__uncultured_bacterium_g_Lachnoclostridium |
| OTU626 | 198 | 94 | 71 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU627 | 133 | 193 | 378 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU63 | 680 | 698 | 830 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU635 | 88 | 59 | 65 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_5; s__uncultured_bacterium_g_Ruminiclostridium_5 |
| OTU636 | 98 | 102 | 55 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Rikenellaceae_RC9_gut_group; s__uncultured_bacterium_g_Rikenellaceae_RC9_gut_group |
| OTU64 | 272 | 1328 | 17 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__uncultured_bacterium_g_Bacteroides |
| OTU640 | 638 | 698 | 1140 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU644 | 3945 | 113 | 0 | k__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Dubosiella; s__uncultured_bacterium_g_Dubosiella |
| OTU65 | 131 | 1061 | 353 | k__Bacteria; p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__uncultured_bacterium_g_Bifidobacterium |
| OTU652 | 259 | 323 | 590 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU658 | 243 | 289 | 708 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__uncultured_bacterium_g_Lactobacillus |
| OTU66 | 571 | 1195 | 642 | k__Bacteria; p__Cyanobacteria; c__Melainabacteria; o__Gastranaerophilales; f__Candidatus_Gastranaerophilales_bacterium_Zag_111; g__uncultured_bacterium_f_Candidatus_Gastranaerophilales_bacterium_Zag_111; s__uncultured_bacterium_f_Candidatus_Gastranaerophilales_bacterium_Zag_111 |
| OTU665 | 139 | 103 | 67 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU668 | 68 | 103 | 116 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__uncultured_bacterium_g_Bacteroides |
| OTU669 | 36 | 53 | 55 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU67 | 1431 | 1281 | 883 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU675 | 238 | 41 | 81 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU679 | 57 | 51 | 48 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU68 | 1663 | 782 | 723 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__GCA-900066575; s__uncultured_bacterium_g_GCA-900066575 |
| OTU680 | 255 | 151 | 155 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU682 | 107 | 119 | 91 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU69 | 776 | 406 | 394 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-009; s__uncultured_bacterium_g_Ruminococcaceae_UCG-009 |
| OTU691 | 42 | 27 | 70 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU693 | 180 | 236 | 146 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU694 | 74 | 24 | 74 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-006; s__uncultured_bacterium_g_Lachnospiraceae_UCG-006 |
| OTU697 | 39 | 41 | 102 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU698 | 55 | 64 | 113 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU7 | 1698 | 10259 | 8220 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__uncultured_bacterium_g_Lactobacillus |
| OTU70 | 1877 | 1138 | 1819 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU702 | 44 | 40 | 93 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU706 | 45 | 37 | 58 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU709 | 168 | 48 | 127 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU71 | 1624 | 740 | 744 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU710 | 107 | 25 | 90 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU72 | 970 | 544 | 1118 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU721 | 96 | 67 | 238 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU723 | 544 | 366 | 773 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU73 | 379 | 684 | 176 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU733 | 58 | 79 | 86 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU736 | 83 | 96 | 94 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU739 | 104 | 128 | 93 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU74 | 955 | 721 | 724 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU741 | 200 | 134 | 195 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU748 | 228 | 439 | 507 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU75 | 806 | 592 | 554 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium; s__uncultured_bacterium_g_Ruminiclostridium |
| OTU751 | 99 | 19 | 47 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-001; s__uncultured_bacterium_g_Lachnospiraceae_UCG-001 |
| OTU753 | 18 | 6 | 391 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU757 | 51 | 49 | 57 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU758 | 45 | 54 | 192 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU759 | 15 | 360 | 531 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU76 | 1076 | 1311 | 2973 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU761 | 236 | 45 | 340 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU763 | 94 | 17 | 94 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_UCG-001; s__uncultured_bacterium_g_Lachnospiraceae_UCG-001 |
| OTU77 | 110 | 819 | 509 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU78 | 436 | 377 | 1862 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU79 | 607 | 1287 | 1485 | k__Bacteria; p__Proteobacteria; c__Gammaproteobacteria; o__Betaproteobacteriales; f__Burkholderiaceae; g__Parasutterella; s__Burkholderiales_bacterium_YL45 |
| OTU8 | 9147 | 8334 | 13390 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU80 | 786 | 207 | 766 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Lachnospiraceae_NK4A136_group; s__uncultured_bacterium_g_Lachnospiraceae_NK4A136_group |
| OTU81 | 1274 | 259 | 442 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__uncultured_bacterium_f_Lachnospiraceae; s__uncultured_bacterium_f_Lachnospiraceae |
| OTU82 | 279 | 492 | 330 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminiclostridium_6; s__uncultured_bacterium_g_Ruminiclostridium_6 |
| OTU83 | 2023 | 1199 | 1182 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Intestinimonas; s__uncultured_bacterium_g_Intestinimonas |
| OTU84 | 121 | 263 | 413 | k__Bacteria; p__Proteobacteria; c__Alphaproteobacteria; o__Rhodospirillales; f__uncultured_bacterium_o_Rhodospirillales; g__uncultured_bacterium_o_Rhodospirillales; s__uncultured_bacterium_o_Rhodospirillales |
| OTU85 | 537 | 1525 | 545 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_UCG-014; s__uncultured_bacterium_g_Ruminococcaceae_UCG-014 |
| OTU86 | 772 | 548 | 236 | k__Bacteria; p__Proteobacteria; c__Alphaproteobacteria; o__Rickettsiales; f__Rickettsiaceae; g__Candidatus_Hemipteriphilus; s__secondary_symbiont_of_Sitobion_miscanthi |
| OTU87 | 852 | 682 | 1203 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU88 | 893 | 300 | 473 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Oscillibacter; s__uncultured_bacterium_g_Oscillibacter |
| OTU89 | 632 | 656 | 1021 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU9 | 3579 | 3390 | 2680 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Rikenellaceae_RC9_gut_group; s__uncultured_bacterium_g_Rikenellaceae_RC9_gut_group |
| OTU90 | 1003 | 980 | 1085 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU91 | 996 | 471 | 1283 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Marvinbryantia; s__uncultured_bacterium_g_Marvinbryantia |
| OTU92 | 59 | 576 | 115 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Atopobiaceae; g__Coriobacteriaceae_UCG-002; s__uncultured_bacterium_g_Coriobacteriaceae_UCG-002 |
| OTU93 | 247 | 196 | 642 | k__Bacteria; p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__uncultured_bacterium_g_Lactobacillus |
| OTU94 | 352 | 555 | 436 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__Alistipes; s__uncultured_bacterium_g_Alistipes |
| OTU95 | 1126 | 442 | 798 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__uncultured_bacterium_f_Ruminococcaceae; s__uncultured_bacterium_f_Ruminococcaceae |
| OTU96 | 5136 | 5810 | 15918 | k__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Muribaculaceae; g__uncultured_bacterium_f_Muribaculaceae; s__uncultured_bacterium_f_Muribaculaceae |
| OTU97 | 248 | 304 | 257 | k__Bacteria; p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Eggerthellaceae; g__Adlercreutzia; s__uncultured_bacterium_g_Adlercreutzia |
| OTU98 | 224 | 205 | 198 | k__Bacteria; p__Cyanobacteria; c__Melainabacteria; o__Gastranaerophilales; f__uncultured_bacterium_o_Gastranaerophilales; g__uncultured_bacterium_o_Gastranaerophilales; s__uncultured_bacterium_o_Gastranaerophilales |
| OTU99 | 204 | 272 | 294 | k__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcaceae_NK4A214_group; s__uncultured_bacterium_g_Ruminococcaceae_NK4A214_group |
Figure 3.
Alpha diversity analysis of the 16S rRNA v3+v4 sequencing results of the gut microbiota in the feces of mice in various groups. A. OTU statistical results of samples from various groups. B. OTU-Venn comparison results of samples from various groups. C. OTU rarefaction curve analysis results of samples from various groups. D. OTU Shannon index analysis results of samples from various groups. There were no significant differences in the number of OTUs between the three groups. E. Alpha diversity Shannon index statistics of OTUs in various groups. F. Alpha diversity Chao1 index statistics of OTUs in various groups. G. Alpha diversity Simpson index statistics of OTUs in various groups. H. Alpha diversity ACE index statistics of OTUs in various groups. The preliminary tests showed that fisetin intervention did not alter gut microbial counts in POF mice.
Table 2.
Alpha diversity
| Sample ID | OTU | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|
| F1 | 366 | 368.239 | 370 | 0.0176 | 4.7523 | 0.9998 |
| F10 | 369 | 369.668 | 369.3333 | 0.1438 | 3.3786 | 1 |
| F2 | 368 | 369.776 | 370.3333 | 0.0935 | 3.6994 | 0.9999 |
| F3 | 361 | 368.485 | 373.6667 | 0.1884 | 2.7987 | 0.9997 |
| F4 | 364 | 365.605 | 364.7895 | 0.0992 | 3.7008 | 0.9999 |
| F5 | 367 | 370.371 | 371.5833 | 0.1003 | 3.7301 | 0.9998 |
| F6 | 357 | 363.741 | 363.3333 | 0.139 | 3.1478 | 0.9996 |
| F7 | 362 | 364.106 | 366 | 0.1605 | 3.2399 | 0.9999 |
| F8 | 360 | 367.17 | 368.6364 | 0.1338 | 3.0117 | 0.9997 |
| F9 | 369 | 372.423 | 376.8571 | 0.1209 | 3.759 | 0.9998 |
| P1 | 355 | 360.549 | 363 | 0.2933 | 2.6332 | 0.9998 |
| P10 | 349 | 358.482 | 367.0714 | 0.098 | 3.758 | 0.9995 |
| P2 | 361 | 365.537 | 365.3333 | 0.2274 | 3.0253 | 0.9998 |
| P3 | 362 | 366.436 | 366.3333 | 0.2428 | 2.8388 | 0.9998 |
| P4 | 357 | 360.964 | 359.64 | 0.264 | 2.673 | 0.9998 |
| P5 | 356 | 362.862 | 365 | 0.2592 | 2.8294 | 0.9998 |
| P6 | 355 | 359.329 | 361 | 0.1832 | 3.1559 | 0.9998 |
| P7 | 354 | 357.766 | 359.2 | 0.2854 | 2.739 | 0.9997 |
| P8 | 358 | 362.32 | 362.1364 | 0.2907 | 2.7281 | 0.9998 |
| P9 | 365 | 366.481 | 366.1538 | 0.2325 | 2.9807 | 0.9999 |
| WT1 | 354 | 358.488 | 361.5 | 0.2225 | 2.7758 | 0.9998 |
| WT10 | 366 | 367.391 | 367.25 | 0.1674 | 3.2391 | 0.9999 |
| WT2 | 360 | 362.213 | 362 | 0.1198 | 3.5081 | 0.9999 |
| WT3 | 365 | 368.208 | 371.1111 | 0.1165 | 3.6781 | 0.9998 |
| WT4 | 363 | 365.394 | 367 | 0.1338 | 3.4047 | 0.9998 |
| WT5 | 351 | 355.638 | 355.3333 | 0.2165 | 2.93 | 0.9998 |
| WT6 | 360 | 366.101 | 365.913 | 0.0982 | 3.4549 | 0.9997 |
| WT7 | 352 | 354.142 | 354.3333 | 0.2106 | 3.0075 | 0.9999 |
| WT8 | 362 | 365.453 | 365.4375 | 0.2009 | 2.9697 | 0.9998 |
| WT9 | 353 | 356.592 | 355.5 | 0.2009 | 3.088 | 0.9999 |
Fisetin significantly alters the distribution and diversity of gut microbiota in POF mice
By comparing the representative sequences and microbial reference data of OTUs, every OTU can be classified into a species. Analysis at the phylum level showed that the relative abundance of Firmicutes in group F was significantly increased compared with that in group P, while the relative abundance of Verrucomicrobia was significantly decreased compared with that in group F (Figure 4A; Table 3). Analysis at the genus level showed that the relative abundance of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of Akkermansia was significantly decreased in group F compared with that in group P (Figure 4B; Table 3). Analysis at the species level showed that the relative abundance of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of uncultured_bacterium_g_Akkermans was significantly decreased in group F compared with that in group P (Figure 4C; Table 3). The Bray-Curtis algorithm analysis revealed significant differences in distribution between the microbial communities of the three groups (Figure 4D; Table 4). Hierarchical clustering analysis by UPGMA showed that the gut microbiota of mice in groups F and WT exhibited high homology and similar genetic backgrounds (Figure 5A and 5B; Table 5). In addition, integrated analysis showed that, in group P, Akkermansia was the dominant bacteria, while the levels of uncultured_bacterium_f_Lachnospiraceae were relatively low; however, opposite microbial distribution results were obtained for group F (Figure 6A; Tables 6, 7). This result was consistent with the species abundance result generated using QIIME software. LEfSe was used to identify high-dimensional biomarkers in the gut microbiota of various groups. The LDA score was set at 4.0, and species with LDA scores greater than the set value were considered to be important biomarkers (Figure 6B). As shown by cladogram and LDA analyses, the microbial count of uncultur-ed_bacterium_f_Lachnospira-ceae was significantly increased, while that of Akkermansia microorganisms was drastically reduced in the gut of group F; however, the distribution of these microbial communities was reversed in group P (Figure 7; Tables 6, 7).
Figure 4.
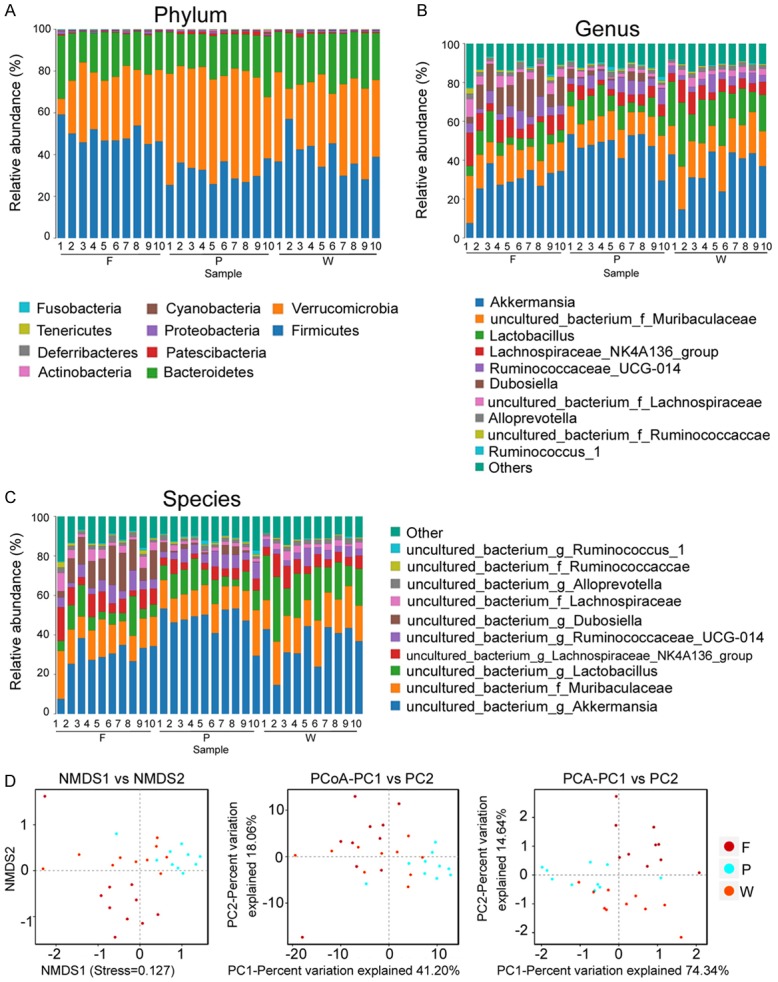
Beta diversity analysis of the 16S rRNA v3+v4 sequencing results of the gut microbiota of feces from various groups of mice. A. Community clustering analysis of OTUs in samples from various groups at the phylum level: analysis at the phylum level showed that the relative abundance of Firmicutes was significantly increased in the gut of group F compared with that in group P, while the relative abundance of Verrucomicrobia was significantly decreased compared with group F. B. Community clustering analysis of OTUs in samples from various groups at the genus level: analysis at the genus level showed that the relative abundance of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of Akkermansia was significantly decreased in group F compared with that in group P. C. Community clustering analysis of OTUs in samples from various groups at the species level: analysis at the species level showed that the relative abundance of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of uncultured_bacterium_g_Akkermans was significantly decreased in group F compared with that in group P. D. Beta diversity analysis of OTUs in samples from various groups based on unweighted UniFrac. Results revealed significant differences in community distribution between the microbial communities of the three groups.
Table 3.
Classfy
| Sample | Kindom | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|---|
| F1 | 1 | 10 | 15 | 18 | 33 | 89 | 91 |
| F2 | 1 | 10 | 15 | 19 | 34 | 91 | 93 |
| F3 | 1 | 10 | 15 | 18 | 34 | 90 | 92 |
| F4 | 1 | 10 | 15 | 19 | 35 | 92 | 94 |
| F5 | 1 | 10 | 15 | 19 | 34 | 91 | 93 |
| F6 | 1 | 10 | 15 | 18 | 34 | 89 | 91 |
| F7 | 1 | 10 | 15 | 18 | 32 | 89 | 91 |
| F8 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| F9 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| F10 | 1 | 10 | 15 | 19 | 34 | 91 | 93 |
| P1 | 1 | 10 | 15 | 19 | 34 | 90 | 92 |
| P2 | 1 | 10 | 15 | 19 | 34 | 90 | 92 |
| P3 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| P4 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| P5 | 1 | 10 | 15 | 18 | 34 | 90 | 92 |
| P6 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| P7 | 1 | 10 | 15 | 18 | 34 | 88 | 90 |
| P8 | 1 | 10 | 15 | 19 | 34 | 90 | 92 |
| P9 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| P10 | 1 | 10 | 15 | 19 | 35 | 90 | 92 |
| WT1 | 1 | 10 | 15 | 18 | 33 | 90 | 92 |
| WT2 | 1 | 10 | 15 | 18 | 33 | 89 | 91 |
| WT3 | 1 | 10 | 15 | 18 | 33 | 90 | 92 |
| WT4 | 1 | 10 | 15 | 18 | 33 | 90 | 92 |
| WT5 | 1 | 10 | 15 | 17 | 33 | 90 | 92 |
| WT6 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| WT7 | 1 | 10 | 15 | 18 | 33 | 90 | 92 |
| WT8 | 1 | 10 | 15 | 19 | 35 | 91 | 93 |
| WT9 | 1 | 10 | 15 | 18 | 34 | 90 | 92 |
| WT10 | 1 | 10 | 15 | 18 | 34 | 91 | 93 |
| Total | 1 | 10 | 15 | 19 | 35 | 92 | 94 |
Table 4.
The result of Genus ANOVA
| Genus | P (Mean) | P (Sd) | WT (Mean) | WT (Sd) | F (Mean) | F (Sd) | multiGroup (p) | multiGroup (p-corrected) | P:WT (p) | P:F (p) | WT:F (p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A2 | 0.00071 | 0.00059 | 0.00112 | 0.00037 | 0.00127 | 0.00103 | 0.21318 | 0.26866 | |||
| ASF356 | 0.00049 | 0.00034 | 0.00035 | 0.00016 | 0.00098 | 0.00078 | 0.02296 | 0.06036 | |||
| Acetatifactor | 0.00018 | 8.3E-05 | 0.00021 | 0.00013 | 0.00026 | 0.00021 | 0.45876 | 0.5147 | |||
| Adlercreutzia | 0.00044 | 0.00027 | 0.0004 | 0.00013 | 0.00041 | 0.00015 | 0.91877 | 0.91877 | |||
| Aeromonas | 9.4E-05 | 3.8E-05 | 4.7E-05 | 2.9E-05 | 0.00016 | 7.2E-05 | 0.0002 | 0.002 | ≥ 0.1 | < 0.05 | < 0.001 |
| Akkermansia | 0.47068 | 0.07268 | 0.3527 | 0.10065 | 0.28707 | 0.08508 | 0.00025 | 0.00229 | < 0.02 | < 0.001 | ≥ 0.1 |
| Alistipes | 0.00415 | 0.0024 | 0.00374 | 0.00096 | 0.00517 | 0.00149 | 0.17816 | 0.24104 | |||
| Alloprevotella | 0.0154 | 0.00641 | 0.02097 | 0.00379 | 0.01955 | 0.00723 | 0.11644 | 0.18156 | |||
| Anaerotruncus | 0.00011 | 8.6E-05 | 0.00015 | 8.1E-05 | 0.00024 | 0.00014 | 0.03213 | 0.0758 | |||
| Anaerovorax | 8.1E-05 | 0.00006 | 0.00014 | 5.9E-05 | 9.2E-05 | 9.3E-05 | 0.20167 | 0.26132 | |||
| Bacteroides | 0.00731 | 0.00344 | 0.00592 | 0.00232 | 0.00497 | 0.00174 | 0.14757 | 0.20886 | |||
| Bifidobacterium | 0.0016 | 0.00158 | 0.00056 | 0.00028 | 0.00021 | 0.00012 | 0.00662 | 0.02647 | < 0.05 | < 0.01 | ≥ 0.1 |
| Bilophila | 0.00082 | 0.00034 | 0.00197 | 0.00147 | 0.00286 | 0.00274 | 0.05624 | 0.11009 | |||
| Blautia | 0.0005 | 0.0002 | 0.00172 | 0.00139 | 0.0048 | 0.0046 | 0.00546 | 0.02284 | ≥ 0.1 | < 0.01 | < 0.05 |
| Butyricicoccus | 0.00029 | 0.00021 | 0.00034 | 0.00022 | 0.00064 | 0.00058 | 0.10623 | 0.1685 | |||
| Butyricimonas | 0.00039 | 0.0002 | 0.00045 | 0.00021 | 0.00031 | 0.00013 | 0.21607 | 0.26863 | |||
| Candidatus_Arthromitus | 5.3E-05 | 4.4E-05 | 6.5E-05 | 1.4E-05 | 9.8E-05 | 4.9E-05 | 0.04071 | 0.09362 | |||
| Candidatus_Hemipteriphilus | 0.00077 | 0.00058 | 0.00039 | 0.00033 | 0.00128 | 0.00074 | 0.00663 | 0.02539 | ≥ 0.1 | ≥ 0.1 | < 0.01 |
| Candidatus_Saccharimonas | 0.00953 | 0.00442 | 0.00663 | 0.00408 | 0.00341 | 0.00192 | 0.00342 | 0.01745 | ≥ 0.1 | < 0.01 | ≥ 0.1 |
| Candidatus_Soleaferrea | 8.5E-05 | 3.5E-05 | 6.7E-05 | 3.9E-05 | 5.9E-05 | 4.7E-05 | 0.36272 | 0.41713 | |||
| Catabacter | 6.7E-05 | 0.00006 | 0.0001 | 3.5E-05 | 4.3E-05 | 3.9E-05 | 0.02009 | 0.05961 | |||
| Cetobacterium | 0.00052 | 0.00023 | 0.00026 | 6.8E-05 | 0.00099 | 0.0003 | 0 | 7E-06 | < 0.05 | < 0.001 | < 0.001 |
| Christensenellaceae_R-7_group | 9.7E-05 | 8.2E-05 | 8.4E-05 | 5.2E-05 | 4.3E-05 | 2.5E-05 | 0.1167 | 0.17895 | |||
| Clostridium_sensu_stricto_1 | 0.00352 | 0.00223 | 0.00146 | 0.00067 | 0.00224 | 0.00074 | 0.01032 | 0.03515 | < 0.01 | ≥ 0.1 | ≥ 0.1 |
| Coriobacteriaceae_UCG-002 | 0.00084 | 0.00042 | 0.00018 | 0.00011 | 9.8E-05 | 4.4E-05 | 1E-06 | 1.2E-05 | < 0.001 | < 0.001 | ≥ 0.1 |
| Dubosiella | 0.0195 | 0.01883 | 3.4E-05 | 2.4E-05 | 0.12983 | 0.05201 | 0 | 0 | ≥ 0.1 | < 0.001 | < 0.001 |
| Effusibacillus | 0.0001 | 0.00018 | 6.5E-05 | 0.00013 | 0.00017 | 0.00035 | 0.62335 | 0.65168 | |||
| Enterorhabdus | 0.00057 | 0.00028 | 0.00068 | 0.00027 | 0.00065 | 0.0005 | 0.79595 | 0.81363 | |||
| Erysipelatoclostridium | 0.00015 | 0.0001 | 0.00033 | 0.0004 | 0.00012 | 6.8E-05 | 0.1185 | 0.17872 | |||
| Faecalibaculum | 0.00636 | 0.00523 | 0.0029 | 0.00155 | 0.00077 | 0.0003 | 0.00187 | 0.01231 | < 0.1 | < 0.01 | ≥ 0.1 |
| Family_XIII_AD3011_group | 0.00011 | 9.1E-05 | 0.00013 | 6.9E-05 | 5.6E-05 | 2.6E-05 | 0.07641 | 0.13783 | |||
| GCA-900066575 | 0.00152 | 0.00076 | 0.00156 | 0.00092 | 0.00384 | 0.00324 | 0.02141 | 0.05968 | |||
| Intestinimonas | 0.00231 | 0.0009 | 0.00269 | 0.00164 | 0.00478 | 0.00345 | 0.04559 | 0.09986 | |||
| Lachnoclostridium | 0.00045 | 0.00029 | 0.00027 | 0.0001 | 0.0011 | 0.0007 | 0.00063 | 0.00443 | ≥ 0.1 | < 0.01 | < 0.001 |
| Lachnospiraceae_FCS020_group | 0.00045 | 0.00036 | 0.00049 | 0.00013 | 0.00073 | 0.00052 | 0.19378 | 0.25468 | |||
| Lachnospiraceae_NK4A136_group | 0.04873 | 0.01641 | 0.06535 | 0.029 | 0.08475 | 0.04163 | 0.0471 | 0.10077 | |||
| Lachnospiraceae_NK4B4_group | 0.00023 | 0.00019 | 0.00062 | 0.0006 | 0.00067 | 0.00064 | 0.13038 | 0.19347 | |||
| Lachnospiraceae_UCG-001 | 0.00047 | 0.00029 | 0.00114 | 0.00068 | 0.00224 | 0.00176 | 0.00458 | 0.02219 | ≥ 0.1 | < 0.01 | < 0.1 |
| Lachnospiraceae_UCG-006 | 0.00366 | 0.00189 | 0.00467 | 0.00208 | 0.00468 | 0.00314 | 0.56423 | 0.61796 | |||
| Lachnospiraceae_UCG-008 | 0.00058 | 0.00049 | 0.00063 | 0.00045 | 0.00168 | 0.00103 | 0.0022 | 0.01263 | ≥ 0.1 | < 0.01 | < 0.01 |
| Lactobacillus | 0.09756 | 0.03877 | 0.19357 | 0.07214 | 0.08491 | 0.05631 | 0.00037 | 0.00313 | < 0.01 | ≥ 0.1 | < 0.001 |
| Marvinbryantia | 0.00076 | 0.00048 | 0.00209 | 0.00112 | 0.00193 | 0.0012 | 0.01055 | 0.03466 | < 0.02 | < 0.05 | ≥ 0.1 |
| Mucispirillum | 0.00039 | 0.00047 | 0.00125 | 0.00209 | 0.00255 | 0.00273 | 0.06836 | 0.12834 | |||
| Muribaculum | 0.00328 | 0.00134 | 0.00313 | 0.00039 | 0.00342 | 0.00073 | 0.7791 | 0.80536 | |||
| Negativibacillus | 0.00024 | 0.00024 | 0.00026 | 0.00015 | 0.00043 | 0.0003 | 0.16971 | 0.23304 | |||
| Oscillibacter | 0.00094 | 0.0003 | 0.00167 | 0.00117 | 0.00257 | 0.00171 | 0.01923 | 0.05898 | |||
| Parabacteroides | 0.00027 | 0.00013 | 0.00029 | 0.00024 | 0.00016 | 7.8E-05 | 0.1797 | 0.23959 | |||
| Parasutterella | 0.00224 | 0.00108 | 0.00297 | 0.00169 | 0.00123 | 0.00038 | 0.00995 | 0.03522 | ≥ 0.1 | ≥ 0.1 | < 0.01 |
| Prevotellaceae_NK3B31_group | 0.00743 | 0.00291 | 0.00713 | 0.0027 | 0.00305 | 0.00135 | 0.00049 | 0.00379 | ≥ 0.1 | < 0.01 | < 0.01 |
| Prevotellaceae_UCG-001 | 0.00391 | 0.0015 | 0.00558 | 0.00206 | 0.00213 | 0.00063 | 0.00011 | 0.00148 | < 0.1 | < 0.05 | < 0.001 |
| Rhodococcus | 8.4E-05 | 7.7E-05 | 3.8E-05 | 3.3E-05 | 0.00019 | 8.9E-05 | 0.00013 | 0.00153 | ≥ 0.1 | < 0.01 | < 0.001 |
| Rikenellaceae_RC9_gut_group | 0.00514 | 0.00196 | 0.00428 | 0.0018 | 0.00626 | 0.00218 | 0.10066 | 0.16247 | |||
| Romboutsia | 0.00169 | 0.00115 | 0.00202 | 0.00114 | 0.00084 | 0.00038 | 0.0305 | 0.07584 | |||
| Roseburia | 0.00395 | 0.00288 | 0.00323 | 0.00112 | 0.00732 | 0.00476 | 0.02154 | 0.05829 | |||
| Ruminiclostridium | 0.00213 | 0.0008 | 0.0048 | 0.00343 | 0.00577 | 0.00466 | 0.06001 | 0.11502 | |||
| Ruminiclostridium_5 | 0.00055 | 0.00028 | 0.00073 | 0.00033 | 0.00135 | 0.00086 | 0.00833 | 0.03066 | ≥ 0.1 | < 0.01 | < 0.05 |
| Ruminiclostridium_6 | 0.00071 | 0.0002 | 0.00055 | 0.00065 | 0.00046 | 0.00029 | 0.4272 | 0.48522 | |||
| Ruminiclostridium_9 | 0.00157 | 0.00082 | 0.00125 | 0.0006 | 0.00166 | 0.0009 | 0.46975 | 0.52068 | |||
| Ruminococcaceae_NK4A214_group | 0.0004 | 0.00021 | 0.00045 | 0.00039 | 0.00033 | 0.00016 | 0.61725 | 0.65272 | |||
| Ruminococcaceae_UCG-003 | 0.00013 | 9.8E-05 | 0.0003 | 0.00025 | 0.00048 | 0.00046 | 0.05337 | 0.10911 | |||
| Ruminococcaceae_UCG-005 | 0.00016 | 7.8E-05 | 0.00013 | 8.6E-05 | 0.00022 | 0.00011 | 0.07868 | 0.13919 | |||
| Ruminococcaceae_UCG-009 | 0.0006 | 0.0002 | 0.00062 | 0.00041 | 0.00135 | 0.00079 | 0.0046 | 0.02117 | ≥ 0.1 | < 0.01 | < 0.02 |
| Ruminococcaceae_UCG-010 | 0.00086 | 0.00068 | 0.00066 | 0.00048 | 0.00088 | 0.0004 | 0.60973 | 0.65227 | |||
| Ruminococcaceae_UCG-013 | 0.00004 | 2.5E-05 | 0.0001 | 8.6E-05 | 7.8E-05 | 4.6E-05 | 0.07933 | 0.13516 | |||
| Ruminococcaceae_UCG-014 | 0.05712 | 0.01945 | 0.04127 | 0.02049 | 0.05517 | 0.02555 | 0.2317 | 0.28422 | |||
| Ruminococcus_1 | 0.00981 | 0.00642 | 0.00443 | 0.00208 | 0.00706 | 0.0026 | 0.02674 | 0.06832 | |||
| Shuttleworthia | 0.00043 | 0.00059 | 0.00038 | 0.00026 | 0.00035 | 0.00032 | 0.91742 | 0.9275 | |||
| Streptococcus | 0.00013 | 6.7E-05 | 0.00011 | 5.1E-05 | 0.00016 | 7.3E-05 | 0.30318 | 0.35306 | |||
| Turicibacter | 0.00856 | 0.00358 | 0.00107 | 0.00058 | 0.00111 | 0.00063 | 0 | 0 | < 0.001 | < 0.001 | ≥ 0.1 |
| Tyzzerella | 0.00029 | 0.00018 | 0.00049 | 0.00027 | 0.00063 | 0.0005 | 0.09768 | 0.16047 | |||
| UBA1819 | 0.00028 | 0.00016 | 0.0004 | 0.00022 | 0.00056 | 0.00045 | 0.13449 | 0.19639 | |||
| [Eubacterium]_brachy_group | 8.3E-05 | 0.00006 | 0.00015 | 9.4E-05 | 0.00014 | 0.00012 | 0.30182 | 0.35599 | |||
| [Eubacterium]_coprostanoligenes_group | 0.00018 | 0.00016 | 0.00072 | 0.0008 | 0.0006 | 0.00039 | 0.06971 | 0.12826 | |||
| [Eubacterium]_fissicatena_group | 0.00211 | 0.0013 | 0.00235 | 0.0015 | 0.00145 | 0.00061 | 0.23596 | 0.28563 | |||
| [Eubacterium]_nodatum_group | 0.00039 | 0.0003 | 0.00038 | 0.00015 | 0.00026 | 0.0001 | 0.26487 | 0.31647 | |||
| [Eubacterium]_oxidoreducens_group | 9.8E-05 | 4.6E-05 | 0.00029 | 0.00025 | 0.00045 | 0.00038 | 0.02115 | 0.0608 | |||
| [Eubacterium]_ruminantium_group | 0.00516 | 0.00145 | 0.00506 | 0.00308 | 0.00424 | 0.00144 | 0.5765 | 0.62398 | |||
| [Eubacterium]_ventriosum_group | 0.00016 | 9.4E-05 | 7.5E-05 | 9.8E-05 | 0.00024 | 7.9E-05 | 0.00208 | 0.01276 | ≥ 0.1 | ≥ 0.1 | < 0.01 |
| [Eubacterium]_xylanophilum_group | 0.00126 | 0.00044 | 0.00229 | 0.00103 | 0.01075 | 0.00775 | 9.7E-05 | 0.00149 | ≥ 0.1 | < 0.001 | < 0.001 |
| uncultured_bacterium_f_Blattella_germanica_German_cockroach | 0.0003 | 0.00014 | 4.1E-05 | 7.1E-05 | 0.00011 | 8.7E-05 | 1.6E-05 | 0.0003 | < 0.001 | < 0.001 | ≥ 0.1 |
| uncultured_bacterium_f_Candidatus_Gastranaerophilales_bacterium_Zag_111 | 0.00187 | 0.00119 | 0.00102 | 0.00054 | 0.00095 | 0.00083 | 0.05321 | 0.11126 | |||
| uncultured_bacterium_f_Clostridiales_vadinBB60_group | 0.0001 | 0.00011 | 0.00014 | 0.00012 | 3.4E-05 | 6.1E-05 | 0.07895 | 0.13705 | |||
| uncultured_bacterium_f_Desulfovibrionaceae | 0.00024 | 0.00011 | 0.00016 | 0.00011 | 0.00036 | 0.00029 | 0.08237 | 0.13778 | |||
| uncultured_bacterium_f_Eggerthellaceae | 0.00014 | 8.1E-05 | 0.00014 | 5.6E-05 | 7.3E-05 | 4.9E-05 | 0.03097 | 0.07497 | |||
| uncultured_bacterium_f_Erysipelotrichaceae | 0.00024 | 0.00017 | 0.00017 | 5.6E-05 | 0.00027 | 0.00015 | 0.20597 | 0.26319 | |||
| uncultured_bacterium_f_Lachnospiraceae | 0.01666 | 0.00578 | 0.02775 | 0.01115 | 0.04241 | 0.02453 | 0.00467 | 0.02046 | ≥ 0.1 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_f_Muribaculaceae | 0.14418 | 0.03356 | 0.18449 | 0.03143 | 0.15664 | 0.03905 | 0.0441 | 0.09895 | |||
| uncultured_bacterium_f_Peptococcaceae | 0.00036 | 0.00018 | 0.0006 | 0.00025 | 0.00053 | 0.00037 | 0.1512 | 0.21077 | |||
| uncultured_bacterium_f_Ruminococcaceae | 0.00707 | 0.0025 | 0.00508 | 0.00227 | 0.01045 | 0.00753 | 0.05467 | 0.10934 | |||
| uncultured_bacterium_o_Gastranaerophilales | 0.00205 | 0.00135 | 0.00084 | 0.00034 | 0.00139 | 0.00053 | 0.015 | 0.0476 | < 0.02 | ≥ 0.1 | ≥ 0.1 |
| uncultured_bacterium_o_Mollicutes_RF39 | 0.00144 | 0.00082 | 0.00088 | 0.00035 | 0.00054 | 0.00019 | 0.00255 | 0.01378 | < 0.1 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_o_Rhodospirillales | 0.0004 | 0.00028 | 0.00063 | 0.00074 | 0.0002 | 0.00011 | 0.13943 | 0.20044 |
Figure 5.
Hierarchical clustering analysis of the 16S rRNA v3+v4 sequencing results of the gut microbiota of feces from various groups of mice. A. Results of UPGMA analysis showed that the gut microbiota of mice in groups F and WT exhibited high homology and similar genetic backgrounds. B. Heatmap analysis of samples from various groups based on the distance algorithm (unweighted).
Table 5.
The result of Species ANOVA
| Species | P (Mean) | P (Sd) | WT (Mean) | WT (Sd) | F (Mean) | F (Sd) | multiGroup (p) | multiGroup (p-corrected) | P::WT (p) | P::F (p) | WT::F (p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Burkholderiales_bacterium_YL45 | 0.00186 | 0.00086 | 0.00236 | 0.00137 | 0.00102 | 0.00034 | 0.01401 | 0.04542 | ≥ 0.1 | ≥ 0.1 | < 0.02 |
| Christensenella_timonensis | 6.7E-05 | 0.00006 | 0.0001 | 3.5E-05 | 4.3E-05 | 3.9E-05 | 0.02009 | 0.05901 | |||
| Effusibacillus_pohliae | 0.0001 | 0.00018 | 6.5E-05 | 0.00013 | 0.00017 | 0.00035 | 0.62335 | 0.65105 | |||
| Lactobacillus_gasseri | 0.00128 | 0.00063 | 0.00176 | 0.00226 | 0.00024 | 0.00017 | 0.05513 | 0.10797 | |||
| Mucispirillum_schaedleri_ASF457 | 0.00039 | 0.00047 | 0.00125 | 0.00209 | 0.00255 | 0.00273 | 0.06836 | 0.12599 | |||
| Parabacteroides_sp._YL27 | 0.00328 | 0.00134 | 0.00313 | 0.00039 | 0.00342 | 0.00073 | 0.7791 | 0.80479 | |||
| bacterium_GC452011 | 8.4E-05 | 7.7E-05 | 3.8E-05 | 3.3E-05 | 0.00019 | 8.9E-05 | 0.00013 | 0.00156 | ≥ 0.1 | < 0.01 | < 0.001 |
| secondary_symbiont_of_Sitobion_miscanthi | 0.00077 | 0.00058 | 0.00039 | 0.00033 | 0.00128 | 0.00074 | 0.00663 | 0.02595 | ≥ 0.1 | ≥ 0.1 | < 0.01 |
| uncultured_bacterium_f_Blattella_germanica_German_cockroach | 0.0003 | 0.00014 | 4.1E-05 | 7.1E-05 | 0.00011 | 8.7E-05 | 1.6E-05 | 0.0003 | < 0.001 | < 0.001 | ≥ 0.1 |
| uncultured_bacterium_f_Candidatus_Gastranaerophilales_bacterium_Zag_111 | 0.00187 | 0.00119 | 0.00102 | 0.00054 | 0.00095 | 0.00083 | 0.05321 | 0.11116 | |||
| uncultured_bacterium_f_Clostridiales_vadinBB60_group | 0.0001 | 0.00011 | 0.00014 | 0.00012 | 3.4E-05 | 6.1E-05 | 0.07895 | 0.13493 | |||
| uncultured_bacterium_f_Desulfovibrionaceae | 0.00024 | 0.00011 | 0.00016 | 0.00011 | 0.00036 | 0.00029 | 0.08237 | 0.13583 | |||
| uncultured_bacterium_f_Eggerthellaceae | 0.00014 | 8.1E-05 | 0.00014 | 5.6E-05 | 7.3E-05 | 4.9E-05 | 0.03097 | 0.07464 | |||
| uncultured_bacterium_f_Erysipelotrichaceae | 0.00024 | 0.00017 | 0.00017 | 5.6E-05 | 0.00027 | 0.00015 | 0.20597 | 0.26164 | |||
| uncultured_bacterium_f_Lachnospiraceae | 0.01666 | 0.00578 | 0.02775 | 0.01115 | 0.04241 | 0.02453 | 0.00467 | 0.0209 | ≥ 0.1 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_f_Muribaculaceae | 0.14418 | 0.03356 | 0.18449 | 0.03143 | 0.15664 | 0.03905 | 0.0441 | 0.0987 | |||
| uncultured_bacterium_f_Peptococcaceae | 0.00036 | 0.00018 | 0.0006 | 0.00025 | 0.00053 | 0.00037 | 0.1512 | 0.20901 | |||
| uncultured_bacterium_f_Ruminococcaceae | 0.00707 | 0.0025 | 0.00508 | 0.00227 | 0.01045 | 0.00753 | 0.05467 | 0.10934 | |||
| uncultured_bacterium_g_A2 | 0.00071 | 0.00059 | 0.00112 | 0.00037 | 0.00127 | 0.00103 | 0.21318 | 0.26718 | |||
| uncultured_bacterium_g_ASF356 | 0.00049 | 0.00034 | 0.00035 | 0.00016 | 0.00098 | 0.00078 | 0.02296 | 0.05996 | |||
| uncultured_bacterium_g_Acetatifactor | 0.00018 | 8.3E-05 | 0.00021 | 0.00013 | 0.00026 | 0.00021 | 0.45876 | 0.51337 | |||
| uncultured_bacterium_g_Adlercreutzia | 0.00044 | 0.00027 | 0.0004 | 0.00013 | 0.00041 | 0.00015 | 0.91877 | 0.91877 | |||
| uncultured_bacterium_g_Aeromonas | 9.4E-05 | 3.8E-05 | 4.7E-05 | 2.9E-05 | 0.00016 | 7.2E-05 | 0.0002 | 0.00204 | ≥ 0.1 | < 0.05 | < 0.001 |
| uncultured_bacterium_g_Akkermansia | 0.47068 | 0.07268 | 0.3527 | 0.10065 | 0.28707 | 0.08508 | 0.00025 | 0.00234 | < 0.02 | < 0.001 | ≥ 0.1 |
| uncultured_bacterium_g_Alistipes | 0.00415 | 0.0024 | 0.00374 | 0.00096 | 0.00517 | 0.00149 | 0.17816 | 0.23925 | |||
| uncultured_bacterium_g_Alloprevotella | 0.0154 | 0.00641 | 0.02097 | 0.00379 | 0.01955 | 0.00723 | 0.11644 | 0.17943 | |||
| uncultured_bacterium_g_Anaerotruncus | 0.00011 | 8.6E-05 | 0.00015 | 8.1E-05 | 0.00024 | 0.00014 | 0.03213 | 0.07551 | |||
| uncultured_bacterium_g_Anaerovorax | 8.1E-05 | 0.00006 | 0.00014 | 5.9E-05 | 9.2E-05 | 9.3E-05 | 0.20167 | 0.25968 | |||
| uncultured_bacterium_g_Bacteroides | 0.00731 | 0.00344 | 0.00592 | 0.00232 | 0.00497 | 0.00174 | 0.14757 | 0.20703 | |||
| uncultured_bacterium_g_Bifidobacterium | 0.0016 | 0.00158 | 0.00056 | 0.00028 | 0.00021 | 0.00012 | 0.00662 | 0.02705 | < 0.05 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_g_Bilophila | 0.00082 | 0.00034 | 0.00197 | 0.00147 | 0.00286 | 0.00274 | 0.05624 | 0.10789 | |||
| uncultured_bacterium_g_Blautia | 0.0005 | 0.0002 | 0.00172 | 0.00139 | 0.0048 | 0.0046 | 0.00546 | 0.02334 | ≥ 0.1 | < 0.01 | < 0.05 |
| uncultured_bacterium_g_Butyricicoccus | 0.00029 | 0.00021 | 0.00034 | 0.00022 | 0.00064 | 0.00058 | 0.10623 | 0.16643 | |||
| uncultured_bacterium_g_Butyricimonas | 0.00039 | 0.0002 | 0.00045 | 0.00021 | 0.00031 | 0.00013 | 0.21607 | 0.26725 | |||
| uncultured_bacterium_g_Candidatus_Arthromitus | 5.3E-05 | 4.4E-05 | 6.5E-05 | 1.4E-05 | 9.8E-05 | 4.9E-05 | 0.04071 | 0.09333 | |||
| uncultured_bacterium_g_Candidatus_Saccharimonas | 0.00953 | 0.00442 | 0.00663 | 0.00408 | 0.00341 | 0.00192 | 0.00342 | 0.01783 | ≥ 0.1 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_g_Candidatus_Soleaferrea | 8.5E-05 | 3.5E-05 | 6.7E-05 | 3.9E-05 | 5.9E-05 | 4.7E-05 | 0.36272 | 0.4158 | |||
| uncultured_bacterium_g_Cetobacterium | 0.00052 | 0.00023 | 0.00026 | 6.8E-05 | 0.00099 | 0.0003 | 0 | 7E-06 | < 0.05 | < 0.001 | < 0.001 |
| uncultured_bacterium_g_Christensenellaceae_R-7_group | 9.7E-05 | 8.2E-05 | 8.4E-05 | 5.2E-05 | 4.3E-05 | 2.5E-05 | 0.1167 | 0.17694 | |||
| uncultured_bacterium_g_Clostridium_sensu_stricto_1 | 0.00352 | 0.00223 | 0.00146 | 0.00067 | 0.00224 | 0.00074 | 0.01032 | 0.03591 | < 0.01 | ≥ 0.1 | ≥ 0.1 |
| uncultured_bacterium_g_Coriobacteriaceae_UCG-002 | 0.00084 | 0.00042 | 0.00018 | 0.00011 | 9.8E-05 | 4.4E-05 | 1E-06 | 1.2E-05 | < 0.001 | < 0.001 | ≥ 0.1 |
| uncultured_bacterium_g_Dubosiella | 0.0195 | 0.01883 | 3.4E-05 | 2.4E-05 | 0.12983 | 0.05201 | 0 | 0 | ≥ 0.1 | < 0.001 | < 0.001 |
| uncultured_bacterium_g_Enterorhabdus | 0.00057 | 0.00028 | 0.00068 | 0.00027 | 0.00065 | 0.0005 | 0.79595 | 0.81325 | |||
| uncultured_bacterium_g_Erysipelatoclostridium | 0.00015 | 0.0001 | 0.00033 | 0.0004 | 0.00012 | 6.8E-05 | 0.1185 | 0.17681 | |||
| uncultured_bacterium_g_Faecalibaculum | 0.00636 | 0.00523 | 0.0029 | 0.00155 | 0.00077 | 0.0003 | 0.00187 | 0.01258 | < 0.1 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_g_Family_XIII_AD3011_group | 0.00011 | 9.1E-05 | 0.00013 | 6.9E-05 | 5.6E-05 | 2.6E-05 | 0.07641 | 0.13552 | |||
| uncultured_bacterium_g_GCA-900066575 | 0.00152 | 0.00076 | 0.00156 | 0.00092 | 0.00384 | 0.00324 | 0.02141 | 0.05919 | |||
| uncultured_bacterium_g_Intestinimonas | 0.00231 | 0.0009 | 0.00269 | 0.00164 | 0.00478 | 0.00345 | 0.04559 | 0.09965 | |||
| uncultured_bacterium_g_Lachnoclostridium | 0.00045 | 0.00029 | 0.00027 | 0.0001 | 0.0011 | 0.0007 | 0.00063 | 0.00452 | ≥ 0.1 | < 0.01 | < 0.001 |
| uncultured_bacterium_g_Lachnospiraceae_FCS020_group | 0.00045 | 0.00036 | 0.00049 | 0.00013 | 0.00073 | 0.00052 | 0.19378 | 0.25299 | |||
| uncultured_bacterium_g_Lachnospiraceae_NK4A136_group | 0.04873 | 0.01641 | 0.06535 | 0.029 | 0.08475 | 0.04163 | 0.0471 | 0.10062 | |||
| uncultured_bacterium_g_Lachnospiraceae_NK4B4_group | 0.00023 | 0.00019 | 0.00062 | 0.0006 | 0.00067 | 0.00064 | 0.13038 | 0.1915 | |||
| uncultured_bacterium_g_Lachnospiraceae_UCG-001 | 0.00047 | 0.00029 | 0.00114 | 0.00068 | 0.00224 | 0.00176 | 0.00458 | 0.02267 | ≥ 0.1 | < 0.01 | < 0.1 |
| uncultured_bacterium_g_Lachnospiraceae_UCG-006 | 0.00366 | 0.00189 | 0.00467 | 0.00208 | 0.00468 | 0.00314 | 0.56423 | 0.61671 | |||
| uncultured_bacterium_g_Lachnospiraceae_UCG-008 | 0.00058 | 0.00049 | 0.00063 | 0.00045 | 0.00168 | 0.00103 | 0.0022 | 0.01291 | ≥ 0.1 | < 0.01 | < 0.01 |
| uncultured_bacterium_g_Lactobacillus | 0.09627 | 0.03827 | 0.19182 | 0.07082 | 0.08467 | 0.05625 | 0.00037 | 0.00314 | < 0.01 | ≥ 0.1 | < 0.001 |
| uncultured_bacterium_g_Marvinbryantia | 0.00076 | 0.00048 | 0.00209 | 0.00112 | 0.00193 | 0.0012 | 0.01055 | 0.03541 | < 0.02 | < 0.05 | ≥ 0.1 |
| uncultured_bacterium_g_Negativibacillus | 0.00024 | 0.00024 | 0.00026 | 0.00015 | 0.00043 | 0.0003 | 0.16971 | 0.2312 | |||
| uncultured_bacterium_g_Oscillibacter | 0.00094 | 0.0003 | 0.00167 | 0.00117 | 0.00257 | 0.00171 | 0.01923 | 0.05831 | |||
| uncultured_bacterium_g_Parabacteroides | 0.00027 | 0.00013 | 0.00029 | 0.00024 | 0.00016 | 7.8E-05 | 0.1797 | 0.23791 | |||
| uncultured_bacterium_g_Parasutterella | 0.00038 | 0.00028 | 0.00062 | 0.00037 | 0.00022 | 6.9E-05 | 0.00975 | 0.03526 | ≥ 0.1 | ≥ 0.1 | < 0.01 |
| uncultured_bacterium_g_Prevotellaceae_NK3B31_group | 0.00743 | 0.00291 | 0.00713 | 0.0027 | 0.00305 | 0.00135 | 0.00049 | 0.00387 | ≥ 0.1 | < 0.01 | < 0.01 |
| uncultured_bacterium_g_Prevotellaceae_UCG-001 | 0.00391 | 0.0015 | 0.00558 | 0.00206 | 0.00213 | 0.00063 | 0.00011 | 0.00151 | < 0.1 | < 0.05 | < 0.001 |
| uncultured_bacterium_g_Rikenellaceae_RC9_gut_group | 0.00514 | 0.00196 | 0.00428 | 0.0018 | 0.00626 | 0.00218 | 0.10066 | 0.16037 | |||
| uncultured_bacterium_g_Romboutsia | 0.00169 | 0.00115 | 0.00202 | 0.00114 | 0.00084 | 0.00038 | 0.0305 | 0.07545 | |||
| uncultured_bacterium_g_Roseburia | 0.00395 | 0.00288 | 0.00323 | 0.00112 | 0.00732 | 0.00476 | 0.02154 | 0.05786 | |||
| uncultured_bacterium_g_Ruminiclostridium | 0.00213 | 0.0008 | 0.0048 | 0.00343 | 0.00577 | 0.00466 | 0.06001 | 0.11282 | |||
| uncultured_bacterium_g_Ruminiclostridium_5 | 0.00055 | 0.00028 | 0.00073 | 0.00033 | 0.00135 | 0.00086 | 0.00833 | 0.03133 | ≥ 0.1 | < 0.01 | < 0.05 |
| uncultured_bacterium_g_Ruminiclostridium_6 | 0.00071 | 0.0002 | 0.00055 | 0.00065 | 0.00046 | 0.00029 | 0.4272 | 0.48382 | |||
| uncultured_bacterium_g_Ruminiclostridium_9 | 0.00157 | 0.00082 | 0.00125 | 0.0006 | 0.00166 | 0.0009 | 0.46975 | 0.51948 | |||
| uncultured_bacterium_g_Ruminococcaceae_NK4A214_group | 0.0004 | 0.00021 | 0.00045 | 0.00039 | 0.00033 | 0.00016 | 0.61725 | 0.65192 | |||
| uncultured_bacterium_g_Ruminococcaceae_UCG-003 | 0.00013 | 9.8E-05 | 0.0003 | 0.00025 | 0.00048 | 0.00046 | 0.05337 | 0.10906 | |||
| uncultured_bacterium_g_Ruminococcaceae_UCG-005 | 0.00016 | 7.8E-05 | 0.00013 | 8.6E-05 | 0.00022 | 0.00011 | 0.07868 | 0.13695 | |||
| uncultured_bacterium_g_Ruminococcaceae_UCG-009 | 0.0006 | 0.0002 | 0.00062 | 0.00041 | 0.00135 | 0.00079 | 0.0046 | 0.02163 | ≥ 0.1 | < 0.01 | < 0.02 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-010 | 0.00086 | 0.00068 | 0.00066 | 0.00048 | 0.00088 | 0.0004 | 0.60973 | 0.65131 | |||
| uncultured_bacterium_g_Ruminococcaceae_UCG-013 | 0.00004 | 2.5E-05 | 0.0001 | 8.6E-05 | 7.8E-05 | 4.6E-05 | 0.07933 | 0.13316 | |||
| uncultured_bacterium_g_Ruminococcaceae_UCG-014 | 0.05712 | 0.01945 | 0.04127 | 0.02049 | 0.05517 | 0.02555 | 0.2317 | 0.28286 | |||
| uncultured_bacterium_g_Ruminococcus_1 | 0.00981 | 0.00642 | 0.00443 | 0.00208 | 0.00706 | 0.0026 | 0.02674 | 0.06792 | |||
| uncultured_bacterium_g_Shuttleworthia | 0.00043 | 0.00059 | 0.00038 | 0.00026 | 0.00035 | 0.00032 | 0.91742 | 0.92729 | |||
| uncultured_bacterium_g_Streptococcus | 0.00013 | 6.7E-05 | 0.00011 | 5.1E-05 | 0.00016 | 7.3E-05 | 0.30318 | 0.35183 | |||
| uncultured_bacterium_g_Turicibacter | 0.00856 | 0.00358 | 0.00107 | 0.00058 | 0.00111 | 0.00063 | 0 | 0 | < 0.001 | < 0.001 | ≥ 0.1 |
| uncultured_bacterium_g_Tyzzerella | 0.00029 | 0.00018 | 0.00049 | 0.00027 | 0.00063 | 0.0005 | 0.09768 | 0.15831 | |||
| uncultured_bacterium_g_UBA1819 | 0.00028 | 0.00016 | 0.0004 | 0.00022 | 0.00056 | 0.00045 | 0.13449 | 0.19449 | |||
| uncultured_bacterium_g_[Eubacterium]_brachy_group | 8.3E-05 | 0.00006 | 0.00015 | 9.4E-05 | 0.00014 | 0.00012 | 0.30182 | 0.35464 | |||
| uncultured_bacterium_g_[Eubacterium]_coprostanoligenes_group | 0.00018 | 0.00016 | 0.00072 | 0.0008 | 0.0006 | 0.00039 | 0.06971 | 0.12601 | |||
| uncultured_bacterium_g_[Eubacterium]_fissicatena_group | 0.00211 | 0.0013 | 0.00235 | 0.0015 | 0.00145 | 0.00061 | 0.23596 | 0.28436 | |||
| uncultured_bacterium_g_[Eubacterium]_nodatum_group | 0.00039 | 0.0003 | 0.00038 | 0.00015 | 0.00026 | 0.0001 | 0.26487 | 0.31516 | |||
| uncultured_bacterium_g_[Eubacterium]_oxidoreducens_group | 9.8E-05 | 4.6E-05 | 0.00029 | 0.00025 | 0.00045 | 0.00038 | 0.02115 | 0.06024 | |||
| uncultured_bacterium_g_[Eubacterium]_ruminantium_group | 0.00516 | 0.00145 | 0.00506 | 0.00308 | 0.00424 | 0.00144 | 0.5765 | 0.62288 | |||
| uncultured_bacterium_g_[Eubacterium]_ventriosum_group | 0.00016 | 9.4E-05 | 7.5E-05 | 9.8E-05 | 0.00024 | 7.9E-05 | 0.00208 | 0.01304 | ≥ 0.1 | ≥ 0.1 | < 0.01 |
| uncultured_bacterium_g_[Eubacterium]_xylanophilum_group | 0.00126 | 0.00044 | 0.00229 | 0.00103 | 0.01075 | 0.00775 | 9.7E-05 | 0.00153 | ≥ 0.1 | < 0.001 | < 0.001 |
| uncultured_bacterium_o_Gastranaerophilales | 0.00205 | 0.00135 | 0.00084 | 0.00034 | 0.00139 | 0.00053 | 0.015 | 0.04701 | < 0.02 | ≥ 0.1 | ≥ 0.1 |
| uncultured_bacterium_o_Mollicutes_RF39 | 0.00144 | 0.00082 | 0.00088 | 0.00035 | 0.00054 | 0.00019 | 0.00255 | 0.01408 | < 0.1 | < 0.01 | ≥ 0.1 |
| uncultured_bacterium_o_Rhodospirillales | 0.0004 | 0.00028 | 0.00063 | 0.00074 | 0.0002 | 0.00011 | 0.13943 | 0.19859 |
Figure 6.
Significance analysis of differences in the gut microbiota of feces from various groups of mice. A. Combined UPGMA cluster tree and histogram analysis: integrated analysis showed that Akkermansia was the dominant bacteria in group P, while the levels of uncultured_bacterium_f_Lachnospiraceae were relatively low. However, opposite microbial distribution results were obtained for group F. B. LEfSe analysis of samples from various groups: the microbial count of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of Akkermansia microorganisms was drastically reduced in the gut microbiota of group F.
Table 6.
Differential genus (F vs P)
| Genus | Mean (F) | Variance (F) | Std.err (F) | Mean (P) | Variance (P) | Std.err (P) | P value | Q value |
|---|---|---|---|---|---|---|---|---|
| Blautia | 4.80E-03 | 2.12E-05 | 1.45E-03 | 4.99E-04 | 3.80E-08 | 6.17E-05 | 9.99E-04 | 7.66E-03 |
| [Eubacterium]_xylanophilum_group | 1.07E-02 | 6.01E-05 | 2.45E-03 | 1.26E-03 | 1.92E-07 | 1.39E-04 | 9.99E-04 | 7.66E-03 |
| Dubosiella | 1.30E-01 | 2.71E-03 | 1.64E-02 | 1.95E-02 | 3.55E-04 | 5.95E-03 | 9.99E-04 | 7.66E-03 |
| Mucispirillum | 2.55E-03 | 7.45E-06 | 8.63E-04 | 3.85E-04 | 2.23E-07 | 1.49E-04 | 1.90E-02 | 5.29E-02 |
| Lachnospiraceae_UCG-001 | 2.24E-03 | 3.10E-06 | 5.57E-04 | 4.66E-04 | 8.49E-08 | 9.22E-05 | 3.00E-03 | 1.72E-02 |
| [Eubacterium]_oxidoreducens_group | 4.50E-04 | 1.45E-07 | 1.20E-04 | 9.79E-05 | 2.16E-09 | 1.47E-05 | 9.99E-04 | 7.66E-03 |
| Ruminococcaceae_UCG-003 | 4.81E-04 | 2.12E-07 | 1.46E-04 | 1.29E-04 | 9.57E-09 | 3.09E-05 | 1.40E-02 | 4.44E-02 |
| Bilophila | 2.86E-03 | 7.51E-06 | 8.67E-04 | 8.17E-04 | 1.17E-07 | 1.08E-04 | 1.20E-02 | 3.94E-02 |
| [Eubacterium]_coprostanoligenes_group | 5.99E-04 | 1.54E-07 | 1.24E-04 | 1.77E-04 | 2.59E-08 | 5.09E-05 | 4.00E-03 | 1.93E-02 |
| Lachnospiraceae_NK4B4_group | 6.73E-04 | 4.14E-07 | 2.04E-04 | 2.25E-04 | 3.74E-08 | 6.11E-05 | 3.50E-02 | 7.66E-02 |
| Lachnospiraceae_UCG-008 | 1.68E-03 | 1.06E-06 | 3.26E-04 | 5.80E-04 | 2.35E-07 | 1.53E-04 | 7.99E-03 | 3.20E-02 |
| Oscillibacter | 2.57E-03 | 2.94E-06 | 5.42E-04 | 9.35E-04 | 8.82E-08 | 9.39E-05 | 4.00E-03 | 1.93E-02 |
| Ruminiclostridium | 5.77E-03 | 2.17E-05 | 1.47E-03 | 2.13E-03 | 6.46E-07 | 2.54E-04 | 9.99E-03 | 3.83E-02 |
| Marvinbryantia | 1.93E-03 | 1.43E-06 | 3.78E-04 | 7.58E-04 | 2.34E-07 | 1.53E-04 | 4.00E-03 | 1.93E-02 |
| uncultured_bacterium_f_Lachnospiraceae | 4.24E-02 | 6.02E-04 | 7.76E-03 | 1.67E-02 | 3.34E-05 | 1.83E-03 | 3.00E-03 | 1.72E-02 |
| GCA-900066575 | 3.84E-03 | 1.05E-05 | 1.02E-03 | 1.52E-03 | 5.72E-07 | 2.39E-04 | 2.30E-02 | 6.04E-02 |
| Ruminiclostridium_5 | 1.35E-03 | 7.38E-07 | 2.72E-04 | 5.51E-04 | 7.81E-08 | 8.84E-05 | 1.20E-02 | 3.94E-02 |
| Lachnoclostridium | 1.10E-03 | 4.84E-07 | 2.20E-04 | 4.51E-04 | 8.65E-08 | 9.30E-05 | 1.20E-02 | 3.94E-02 |
| Rhodococcus | 1.92E-04 | 7.90E-09 | 2.81E-05 | 8.44E-05 | 5.93E-09 | 2.43E-05 | 1.10E-02 | 3.94E-02 |
| Ruminococcaceae_UCG-009 | 1.35E-03 | 6.19E-07 | 2.49E-04 | 5.97E-04 | 4.10E-08 | 6.41E-05 | 7.99E-03 | 3.20E-02 |
| Tyzzerella | 6.33E-04 | 2.54E-07 | 1.59E-04 | 2.86E-04 | 3.38E-08 | 5.82E-05 | 2.80E-02 | 6.77E-02 |
| Anaerotruncus | 2.40E-04 | 1.95E-08 | 4.42E-05 | 1.10E-04 | 7.47E-09 | 2.73E-05 | 1.50E-02 | 4.45E-02 |
| Butyricicoccus | 6.36E-04 | 3.35E-07 | 1.83E-04 | 2.94E-04 | 4.56E-08 | 6.75E-05 | 9.79E-02 | 1.70E-01 |
| Intestinimonas | 4.78E-03 | 1.19E-05 | 1.09E-03 | 2.31E-03 | 8.04E-07 | 2.84E-04 | 2.10E-02 | 5.68E-02 |
| UBA1819 | 5.57E-04 | 1.99E-07 | 1.41E-04 | 2.77E-04 | 2.71E-08 | 5.20E-05 | 7.19E-02 | 1.37E-01 |
| ASF356 | 9.80E-04 | 6.11E-07 | 2.47E-04 | 4.88E-04 | 1.13E-07 | 1.06E-04 | 7.49E-02 | 1.37E-01 |
| Ruminococcaceae_UCG-013 | 7.84E-05 | 2.13E-09 | 1.46E-05 | 4.00E-05 | 6.16E-10 | 7.85E-06 | 2.70E-02 | 6.71E-02 |
| Cetobacterium | 9.91E-04 | 8.88E-08 | 9.42E-05 | 5.19E-04 | 5.12E-08 | 7.15E-05 | 5.00E-03 | 2.30E-02 |
| Candidatus_Arthromitus | 9.83E-05 | 2.39E-09 | 1.55E-05 | 5.30E-05 | 1.97E-09 | 1.41E-05 | 4.00E-02 | 8.36E-02 |
| Roseburia | 7.32E-03 | 2.27E-05 | 1.51E-03 | 3.95E-03 | 8.31E-06 | 9.12E-04 | 7.39E-02 | 1.37E-01 |
| A2 | 1.27E-03 | 1.07E-06 | 3.27E-04 | 7.08E-04 | 3.44E-07 | 1.85E-04 | 1.60E-01 | 2.48E-01 |
| Negativibacillus | 4.25E-04 | 9.07E-08 | 9.52E-05 | 2.37E-04 | 5.67E-08 | 7.53E-05 | 1.35E-01 | 2.14E-01 |
| Lachnospiraceae_NK4A136_group | 8.48E-02 | 1.73E-03 | 1.32E-02 | 4.87E-02 | 2.69E-04 | 5.19E-03 | 1.50E-02 | 4.45E-02 |
| Candidatus_Hemipteriphilus | 1.28E-03 | 5.40E-07 | 2.32E-04 | 7.68E-04 | 3.33E-07 | 1.83E-04 | 9.99E-02 | 1.70E-01 |
| Aeromonas | 1.55E-04 | 5.17E-09 | 2.27E-05 | 9.40E-05 | 1.44E-09 | 1.20E-05 | 1.80E-02 | 5.17E-02 |
| Lachnospiraceae_FCS020_group | 7.34E-04 | 2.69E-07 | 1.64E-04 | 4.46E-04 | 1.29E-07 | 1.13E-04 | 1.79E-01 | 2.61E-01 |
| [Eubacterium]_brachy_group | 1.35E-04 | 1.49E-08 | 3.85E-05 | 8.26E-05 | 3.57E-09 | 1.89E-05 | 2.42E-01 | 3.18E-01 |
| Effusibacillus | 1.69E-04 | 1.21E-07 | 1.10E-04 | 1.04E-04 | 3.39E-08 | 5.82E-05 | 6.79E-01 | 7.44E-01 |
| Acetatifactor | 2.60E-04 | 4.40E-08 | 6.63E-05 | 1.75E-04 | 6.88E-09 | 2.62E-05 | 2.68E-01 | 3.47E-01 |
| uncultured_bacterium_f_Ruminococcaceae | 1.05E-02 | 5.68E-05 | 2.38E-03 | 7.07E-03 | 6.25E-06 | 7.91E-04 | 2.21E-01 | 2.99E-01 |
| uncultured_bacterium_f_Peptococcaceae | 5.27E-04 | 1.35E-07 | 1.16E-04 | 3.55E-04 | 3.26E-08 | 5.71E-05 | 2.15E-01 | 2.95E-01 |
| uncultured_bacterium_f_Desulfovibrionaceae | 3.56E-04 | 8.20E-08 | 9.05E-05 | 2.42E-04 | 1.19E-08 | 3.46E-05 | 2.80E-01 | 3.53E-01 |
| [Eubacterium]_ventriosum_group | 2.36E-04 | 6.30E-09 | 2.51E-05 | 1.61E-04 | 8.92E-09 | 2.99E-05 | 5.79E-02 | 1.13E-01 |
| Ruminococcaceae_UCG-005 | 2.21E-04 | 1.13E-08 | 3.36E-05 | 1.56E-04 | 6.14E-09 | 2.48E-05 | 1.34E-01 | 2.14E-01 |
| Lachnospiraceae_UCG-006 | 4.68E-03 | 9.83E-06 | 9.91E-04 | 3.66E-03 | 3.56E-06 | 5.97E-04 | 3.86E-01 | 4.60E-01 |
| Alloprevotella | 1.96E-02 | 5.23E-05 | 2.29E-03 | 1.54E-02 | 4.11E-05 | 2.03E-03 | 1.90E-01 | 2.73E-01 |
| Alistipes | 5.17E-03 | 2.22E-06 | 4.71E-04 | 4.15E-03 | 5.74E-06 | 7.58E-04 | 2.88E-01 | 3.58E-01 |
| Rikenellaceae_RC9_gut_group | 6.26E-03 | 4.76E-06 | 6.90E-04 | 5.14E-03 | 3.82E-06 | 6.18E-04 | 2.42E-01 | 3.18E-01 |
| Streptococcus | 1.55E-04 | 5.29E-09 | 2.30E-05 | 1.30E-04 | 4.45E-09 | 2.11E-05 | 4.02E-01 | 4.68E-01 |
| Anaerovorax | 9.20E-05 | 8.72E-09 | 2.95E-05 | 8.06E-05 | 3.60E-09 | 1.90E-05 | 7.52E-01 | 7.95E-01 |
| Enterorhabdus | 6.51E-04 | 2.46E-07 | 1.57E-04 | 5.73E-04 | 7.95E-08 | 8.92E-05 | 6.50E-01 | 7.21E-01 |
| uncultured_bacterium_f_Erysipelotrichaceae | 2.71E-04 | 2.16E-08 | 4.64E-05 | 2.43E-04 | 2.94E-08 | 5.43E-05 | 7.05E-01 | 7.61E-01 |
| uncultured_bacterium_f_Muribaculaceae | 1.57E-01 | 1.53E-03 | 1.23E-02 | 1.44E-01 | 1.13E-03 | 1.06E-02 | 4.56E-01 | 5.17E-01 |
| Ruminiclostridium_9 | 1.66E-03 | 8.04E-07 | 2.84E-04 | 1.57E-03 | 6.64E-07 | 2.58E-04 | 7.80E-01 | 8.08E-01 |
| Muribaculum | 3.42E-03 | 5.29E-07 | 2.30E-04 | 3.28E-03 | 1.80E-06 | 4.24E-04 | 7.81E-01 | 8.08E-01 |
| Ruminococcaceae_UCG-010 | 8.77E-04 | 1.60E-07 | 1.27E-04 | 8.61E-04 | 4.60E-07 | 2.15E-04 | 9.57E-01 | 9.57E-01 |
| Ruminococcaceae_UCG-014 | 5.52E-02 | 6.53E-04 | 8.08E-03 | 5.71E-02 | 3.78E-04 | 6.15E-03 | 8.35E-01 | 8.44E-01 |
| Adlercreutzia | 4.12E-04 | 2.24E-08 | 4.74E-05 | 4.38E-04 | 7.06E-08 | 8.40E-05 | 8.01E-01 | 8.19E-01 |
| Lactobacillus | 8.49E-02 | 3.17E-03 | 1.78E-02 | 9.76E-02 | 1.50E-03 | 1.23E-02 | 5.63E-01 | 6.32E-01 |
| Ruminococcaceae_NK4A214_group | 3.28E-04 | 2.52E-08 | 5.02E-05 | 3.96E-04 | 4.43E-08 | 6.66E-05 | 4.32E-01 | 4.96E-01 |
| [Eubacterium]_ruminantium_group | 4.24E-03 | 2.08E-06 | 4.56E-04 | 5.16E-03 | 2.11E-06 | 4.59E-04 | 1.93E-01 | 2.73E-01 |
| Shuttleworthia | 3.50E-04 | 1.05E-07 | 1.03E-04 | 4.27E-04 | 3.52E-07 | 1.88E-04 | 7.11E-01 | 7.61E-01 |
| Butyricimonas | 3.05E-04 | 1.55E-08 | 3.94E-05 | 3.86E-04 | 3.95E-08 | 6.28E-05 | 3.16E-01 | 3.87E-01 |
| Erysipelatoclostridium | 1.15E-04 | 4.60E-09 | 2.14E-05 | 1.51E-04 | 1.03E-08 | 3.21E-05 | 3.90E-01 | 4.60E-01 |
| Ruminococcus_1 | 7.06E-03 | 6.78E-06 | 8.23E-04 | 9.81E-03 | 4.12E-05 | 2.03E-03 | 2.79E-01 | 3.53E-01 |
| Candidatus_Soleaferrea | 5.87E-05 | 2.25E-09 | 1.50E-05 | 8.46E-05 | 1.26E-09 | 1.12E-05 | 2.06E-01 | 2.87E-01 |
| [Eubacterium]_fissicatena_group | 1.44E-03 | 3.73E-07 | 1.93E-04 | 2.11E-03 | 1.69E-06 | 4.11E-04 | 1.62E-01 | 2.48E-01 |
| Bacteroides | 4.97E-03 | 3.03E-06 | 5.51E-04 | 7.31E-03 | 1.18E-05 | 1.09E-03 | 7.59E-02 | 1.37E-01 |
| uncultured_bacterium_o_Gastranaerophilales | 1.39E-03 | 2.85E-07 | 1.69E-04 | 2.05E-03 | 1.81E-06 | 4.26E-04 | 1.71E-01 | 2.53E-01 |
| [Eubacterium]_nodatum_group | 2.56E-04 | 1.08E-08 | 3.28E-05 | 3.93E-04 | 8.90E-08 | 9.43E-05 | 1.70E-01 | 2.53E-01 |
| Ruminiclostridium_6 | 4.59E-04 | 8.34E-08 | 9.13E-05 | 7.09E-04 | 4.17E-08 | 6.46E-05 | 3.40E-02 | 7.66E-02 |
| Catabacter | 4.27E-05 | 1.55E-09 | 1.24E-05 | 6.66E-05 | 3.63E-09 | 1.90E-05 | 3.20E-01 | 3.87E-01 |
| Clostridium_sensu_stricto_1 | 2.24E-03 | 5.42E-07 | 2.33E-04 | 3.51E-03 | 4.96E-06 | 7.04E-04 | 1.03E-01 | 1.72E-01 |
| Akkermansia | 2.87E-01 | 7.24E-03 | 2.69E-02 | 4.71E-01 | 5.28E-03 | 2.30E-02 | 9.99E-04 | 7.66E-03 |
| Parabacteroides | 1.59E-04 | 6.02E-09 | 2.45E-05 | 2.67E-04 | 1.72E-08 | 4.14E-05 | 3.90E-02 | 8.34E-02 |
| Parasutterella | 1.23E-03 | 1.40E-07 | 1.19E-04 | 2.24E-03 | 1.17E-06 | 3.42E-04 | 6.99E-03 | 3.06E-02 |
| Prevotellaceae_UCG-001 | 2.13E-03 | 3.98E-07 | 1.99E-04 | 3.91E-03 | 2.25E-06 | 4.74E-04 | 3.00E-03 | 1.72E-02 |
| Family_XIII_AD3011_group | 5.64E-05 | 6.94E-10 | 8.33E-06 | 1.06E-04 | 8.34E-09 | 2.89E-05 | 8.99E-02 | 1.59E-01 |
| uncultured_bacterium_f_Candidatus_Gastranaerophilales_bacterium_Zag_111 | 9.53E-04 | 6.91E-07 | 2.63E-04 | 1.87E-03 | 1.42E-06 | 3.77E-04 | 5.79E-02 | 1.13E-01 |
| uncultured_bacterium_f_Eggerthellaceae | 7.27E-05 | 2.41E-09 | 1.55E-05 | 1.43E-04 | 6.61E-09 | 2.57E-05 | 3.40E-02 | 7.66E-02 |
| uncultured_bacterium_o_Rhodospirillales | 2.01E-04 | 1.21E-08 | 3.48E-05 | 4.04E-04 | 7.89E-08 | 8.88E-05 | 4.10E-02 | 8.37E-02 |
| Romboutsia | 8.38E-04 | 1.45E-07 | 1.20E-04 | 1.69E-03 | 1.33E-06 | 3.65E-04 | 2.70E-02 | 6.71E-02 |
| Christensenellaceae_R-7_group | 4.34E-05 | 6.21E-10 | 7.88E-06 | 9.67E-05 | 6.66E-09 | 2.58E-05 | 3.50E-02 | 7.66E-02 |
| Prevotellaceae_NK3B31_group | 3.05E-03 | 1.81E-06 | 4.25E-04 | 7.43E-03 | 8.47E-06 | 9.21E-04 | 9.99E-04 | 7.66E-03 |
| uncultured_bacterium_o_Mollicutes_RF39 | 5.43E-04 | 3.60E-08 | 6.00E-05 | 1.44E-03 | 6.71E-07 | 2.59E-04 | 9.99E-04 | 7.66E-03 |
| Candidatus_Saccharimonas | 3.41E-03 | 3.67E-06 | 6.06E-04 | 9.53E-03 | 1.95E-05 | 1.40E-03 | 9.99E-04 | 7.66E-03 |
| uncultured_bacterium_f_Blattella_germanica_German_cockroach | 1.06E-04 | 7.62E-09 | 2.76E-05 | 3.03E-04 | 2.01E-08 | 4.48E-05 | 2.00E-03 | 1.41E-02 |
| uncultured_bacterium_f_Clostridiales_vadinBB60_group | 3.44E-05 | 3.73E-09 | 1.93E-05 | 1.03E-04 | 1.24E-08 | 3.51E-05 | 1.11E-01 | 1.82E-01 |
| Bifidobacterium | 2.11E-04 | 1.33E-08 | 3.65E-05 | 1.60E-03 | 2.50E-06 | 5.00E-04 | 9.99E-04 | 7.66E-03 |
| Turicibacter | 1.11E-03 | 3.92E-07 | 1.98E-04 | 8.56E-03 | 1.28E-05 | 1.13E-03 | 9.99E-04 | 7.66E-03 |
| Faecalibaculum | 7.69E-04 | 8.79E-08 | 9.37E-05 | 6.36E-03 | 2.74E-05 | 1.66E-03 | 9.99E-04 | 7.66E-03 |
| Coriobacteriaceae_UCG-002 | 9.76E-05 | 1.91E-09 | 1.38E-05 | 8.39E-04 | 1.77E-07 | 1.33E-04 | 9.99E-04 | 7.66E-03 |
Table 7.
Differential species (F vs P)
| Species | Mean (F) | Variance (F) | Std.err (F) | Mean (P) | Variance (P) | Std.err (P) | P value | Q value |
|---|---|---|---|---|---|---|---|---|
| uncultured_bacterium_g_Blautia | 4.80E-03 | 2.12E-05 | 1.45E-03 | 4.99E-04 | 3.80E-08 | 6.17E-05 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_[Eubacterium]_xylanophilum_group | 1.07E-02 | 6.01E-05 | 2.45E-03 | 1.26E-03 | 1.92E-07 | 1.39E-04 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Dubosiella | 1.30E-01 | 2.71E-03 | 1.64E-02 | 1.95E-02 | 3.55E-04 | 5.95E-03 | 2.00E-03 | 1.17E-02 |
| Mucispirillum_schaedleri_ASF457 | 2.55E-03 | 7.45E-06 | 8.63E-04 | 3.85E-04 | 2.23E-07 | 1.49E-04 | 1.10E-02 | 3.56E-02 |
| uncultured_bacterium_g_Lachnospiraceae_UCG-001 | 2.24E-03 | 3.10E-06 | 5.57E-04 | 4.66E-04 | 8.49E-08 | 9.22E-05 | 2.00E-03 | 1.17E-02 |
| uncultured_bacterium_g_[Eubacterium]_oxidoreducens_group | 4.50E-04 | 1.45E-07 | 1.20E-04 | 9.79E-05 | 2.16E-09 | 1.47E-05 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-003 | 4.81E-04 | 2.12E-07 | 1.46E-04 | 1.29E-04 | 9.57E-09 | 3.09E-05 | 1.20E-02 | 3.76E-02 |
| uncultured_bacterium_g_Bilophila | 2.86E-03 | 7.51E-06 | 8.67E-04 | 8.17E-04 | 1.17E-07 | 1.08E-04 | 8.99E-03 | 3.13E-02 |
| uncultured_bacterium_g_[Eubacterium]_coprostanoligenes_group | 5.99E-04 | 1.54E-07 | 1.24E-04 | 1.77E-04 | 2.59E-08 | 5.09E-05 | 5.00E-03 | 2.35E-02 |
| uncultured_bacterium_g_Lachnospiraceae_NK4B4_group | 6.73E-04 | 4.14E-07 | 2.04E-04 | 2.25E-04 | 3.74E-08 | 6.11E-05 | 3.60E-02 | 7.86E-02 |
| uncultured_bacterium_g_Lachnospiraceae_UCG-008 | 1.68E-03 | 1.06E-06 | 3.26E-04 | 5.80E-04 | 2.35E-07 | 1.53E-04 | 6.99E-03 | 2.74E-02 |
| uncultured_bacterium_g_Oscillibacter | 2.57E-03 | 2.94E-06 | 5.42E-04 | 9.35E-04 | 8.82E-08 | 9.39E-05 | 2.00E-03 | 1.17E-02 |
| uncultured_bacterium_g_Ruminiclostridium | 5.77E-03 | 2.17E-05 | 1.47E-03 | 2.13E-03 | 6.46E-07 | 2.54E-04 | 8.99E-03 | 3.13E-02 |
| uncultured_bacterium_g_Marvinbryantia | 1.93E-03 | 1.43E-06 | 3.78E-04 | 7.58E-04 | 2.34E-07 | 1.53E-04 | 5.99E-03 | 2.56E-02 |
| uncultured_bacterium_f_Lachnospiraceae | 4.24E-02 | 6.02E-04 | 7.76E-03 | 1.67E-02 | 3.34E-05 | 1.83E-03 | 2.00E-03 | 1.17E-02 |
| uncultured_bacterium_g_GCA-900066575 | 3.84E-03 | 1.05E-05 | 1.02E-03 | 1.52E-03 | 5.72E-07 | 2.39E-04 | 3.20E-02 | 7.33E-02 |
| uncultured_bacterium_g_Ruminiclostridium_5 | 1.35E-03 | 7.38E-07 | 2.72E-04 | 5.51E-04 | 7.81E-08 | 8.84E-05 | 1.30E-02 | 3.94E-02 |
| uncultured_bacterium_g_Lachnoclostridium | 1.10E-03 | 4.84E-07 | 2.20E-04 | 4.51E-04 | 8.65E-08 | 9.30E-05 | 9.99E-03 | 3.35E-02 |
| bacterium_GC452011 | 1.92E-04 | 7.90E-09 | 2.81E-05 | 8.44E-05 | 5.93E-09 | 2.43E-05 | 7.99E-03 | 3.00E-02 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-009 | 1.35E-03 | 6.19E-07 | 2.49E-04 | 5.97E-04 | 4.10E-08 | 6.41E-05 | 6.99E-03 | 2.74E-02 |
| uncultured_bacterium_g_Tyzzerella | 6.33E-04 | 2.54E-07 | 1.59E-04 | 2.86E-04 | 3.38E-08 | 5.82E-05 | 2.80E-02 | 6.92E-02 |
| uncultured_bacterium_g_Anaerotruncus | 2.40E-04 | 1.95E-08 | 4.42E-05 | 1.10E-04 | 7.47E-09 | 2.73E-05 | 2.10E-02 | 5.63E-02 |
| uncultured_bacterium_g_Butyricicoccus | 6.36E-04 | 3.35E-07 | 1.83E-04 | 2.94E-04 | 4.56E-08 | 6.75E-05 | 9.79E-02 | 1.64E-01 |
| uncultured_bacterium_g_Intestinimonas | 4.78E-03 | 1.19E-05 | 1.09E-03 | 2.31E-03 | 8.04E-07 | 2.84E-04 | 2.60E-02 | 6.60E-02 |
| uncultured_bacterium_g_UBA1819 | 5.57E-04 | 1.99E-07 | 1.41E-04 | 2.77E-04 | 2.71E-08 | 5.20E-05 | 6.99E-02 | 1.29E-01 |
| uncultured_bacterium_g_ASF356 | 9.80E-04 | 6.11E-07 | 2.47E-04 | 4.88E-04 | 1.13E-07 | 1.06E-04 | 8.99E-02 | 1.57E-01 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-013 | 7.84E-05 | 2.13E-09 | 1.46E-05 | 4.00E-05 | 6.16E-10 | 7.85E-06 | 3.10E-02 | 7.28E-02 |
| uncultured_bacterium_g_Cetobacterium | 9.91E-04 | 8.88E-08 | 9.42E-05 | 5.19E-04 | 5.12E-08 | 7.15E-05 | 3.00E-03 | 1.66E-02 |
| uncultured_bacterium_g_Candidatus_Arthromitus | 9.83E-05 | 2.39E-09 | 1.55E-05 | 5.30E-05 | 1.97E-09 | 1.41E-05 | 5.00E-02 | 1.04E-01 |
| uncultured_bacterium_g_Roseburia | 7.32E-03 | 2.27E-05 | 1.51E-03 | 3.95E-03 | 8.31E-06 | 9.12E-04 | 6.89E-02 | 1.29E-01 |
| uncultured_bacterium_g_A2 | 1.27E-03 | 1.07E-06 | 3.27E-04 | 7.08E-04 | 3.44E-07 | 1.85E-04 | 1.42E-01 | 2.22E-01 |
| uncultured_bacterium_g_Negativibacillus | 4.25E-04 | 9.07E-08 | 9.52E-05 | 2.37E-04 | 5.67E-08 | 7.53E-05 | 1.56E-01 | 2.40E-01 |
| uncultured_bacterium_g_Lachnospiraceae_NK4A136_group | 8.48E-02 | 1.73E-03 | 1.32E-02 | 4.87E-02 | 2.69E-04 | 5.19E-03 | 1.50E-02 | 4.27E-02 |
| secondary_symbiont_of_Sitobion_miscanthi | 1.28E-03 | 5.40E-07 | 2.32E-04 | 7.68E-04 | 3.33E-07 | 1.83E-04 | 1.10E-01 | 1.78E-01 |
| uncultured_bacterium_g_Aeromonas | 1.55E-04 | 5.17E-09 | 2.27E-05 | 9.40E-05 | 1.44E-09 | 1.20E-05 | 1.70E-02 | 4.70E-02 |
| uncultured_bacterium_g_Lachnospiraceae_FCS020_group | 7.34E-04 | 2.69E-07 | 1.64E-04 | 4.46E-04 | 1.29E-07 | 1.13E-04 | 1.60E-01 | 2.40E-01 |
| uncultured_bacterium_g_[Eubacterium]_brachy_group | 1.35E-04 | 1.49E-08 | 3.85E-05 | 8.26E-05 | 3.57E-09 | 1.89E-05 | 2.37E-01 | 3.10E-01 |
| Effusibacillus_pohliae | 1.69E-04 | 1.21E-07 | 1.10E-04 | 1.04E-04 | 3.39E-08 | 5.82E-05 | 6.71E-01 | 7.34E-01 |
| uncultured_bacterium_g_Acetatifactor | 2.60E-04 | 4.40E-08 | 6.63E-05 | 1.75E-04 | 6.88E-09 | 2.62E-05 | 2.43E-01 | 3.13E-01 |
| uncultured_bacterium_f_Ruminococcaceae | 1.05E-02 | 5.68E-05 | 2.38E-03 | 7.07E-03 | 6.25E-06 | 7.91E-04 | 2.20E-01 | 2.95E-01 |
| uncultured_bacterium_f_Peptococcaceae | 5.27E-04 | 1.35E-07 | 1.16E-04 | 3.55E-04 | 3.26E-08 | 5.71E-05 | 2.00E-01 | 2.72E-01 |
| uncultured_bacterium_f_Desulfovibrionaceae | 3.56E-04 | 8.20E-08 | 9.05E-05 | 2.42E-04 | 1.19E-08 | 3.46E-05 | 2.57E-01 | 3.22E-01 |
| uncultured_bacterium_g_[Eubacterium]_ventriosum_group | 2.36E-04 | 6.30E-09 | 2.51E-05 | 1.61E-04 | 8.92E-09 | 2.99E-05 | 6.99E-02 | 1.29E-01 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-005 | 2.21E-04 | 1.13E-08 | 3.36E-05 | 1.56E-04 | 6.14E-09 | 2.48E-05 | 1.38E-01 | 2.20E-01 |
| uncultured_bacterium_g_Lachnospiraceae_UCG-006 | 4.68E-03 | 9.83E-06 | 9.91E-04 | 3.66E-03 | 3.56E-06 | 5.97E-04 | 3.92E-01 | 4.60E-01 |
| uncultured_bacterium_g_Alloprevotella | 1.96E-02 | 5.23E-05 | 2.29E-03 | 1.54E-02 | 4.11E-05 | 2.03E-03 | 1.80E-01 | 2.52E-01 |
| uncultured_bacterium_g_Alistipes | 5.17E-03 | 2.22E-06 | 4.71E-04 | 4.15E-03 | 5.74E-06 | 7.58E-04 | 2.91E-01 | 3.55E-01 |
| uncultured_bacterium_g_Rikenellaceae_RC9_gut_group | 6.26E-03 | 4.76E-06 | 6.90E-04 | 5.14E-03 | 3.82E-06 | 6.18E-04 | 2.38E-01 | 3.10E-01 |
| uncultured_bacterium_g_Streptococcus | 1.55E-04 | 5.29E-09 | 2.30E-05 | 1.30E-04 | 4.45E-09 | 2.11E-05 | 4.27E-01 | 4.94E-01 |
| uncultured_bacterium_g_Anaerovorax | 9.20E-05 | 8.72E-09 | 2.95E-05 | 8.06E-05 | 3.60E-09 | 1.90E-05 | 7.53E-01 | 7.96E-01 |
| uncultured_bacterium_g_Enterorhabdus | 6.51E-04 | 2.46E-07 | 1.57E-04 | 5.73E-04 | 7.95E-08 | 8.92E-05 | 6.64E-01 | 7.34E-01 |
| uncultured_bacterium_f_Erysipelotrichaceae | 2.71E-04 | 2.16E-08 | 4.64E-05 | 2.43E-04 | 2.94E-08 | 5.43E-05 | 7.11E-01 | 7.69E-01 |
| uncultured_bacterium_f_Muribaculaceae | 1.57E-01 | 1.53E-03 | 1.23E-02 | 1.44E-01 | 1.13E-03 | 1.06E-02 | 4.53E-01 | 5.13E-01 |
| uncultured_bacterium_g_Ruminiclostridium_9 | 1.66E-03 | 8.04E-07 | 2.84E-04 | 1.57E-03 | 6.64E-07 | 2.58E-04 | 7.95E-01 | 8.17E-01 |
| Parabacteroides_sp._YL27 | 3.42E-03 | 5.29E-07 | 2.30E-04 | 3.28E-03 | 1.80E-06 | 4.24E-04 | 7.77E-01 | 8.12E-01 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-010 | 8.77E-04 | 1.60E-07 | 1.27E-04 | 8.61E-04 | 4.60E-07 | 2.15E-04 | 9.53E-01 | 9.53E-01 |
| uncultured_bacterium_g_Ruminococcaceae_UCG-014 | 5.52E-02 | 6.53E-04 | 8.08E-03 | 5.71E-02 | 3.78E-04 | 6.15E-03 | 8.67E-01 | 8.76E-01 |
| uncultured_bacterium_g_Adlercreutzia | 4.12E-04 | 2.24E-08 | 4.74E-05 | 4.38E-04 | 7.06E-08 | 8.40E-05 | 7.99E-01 | 8.17E-01 |
| uncultured_bacterium_g_Lactobacillus | 8.47E-02 | 3.16E-03 | 1.78E-02 | 9.63E-02 | 1.46E-03 | 1.21E-02 | 6.13E-01 | 6.86E-01 |
| uncultured_bacterium_g_Ruminococcaceae_NK4A214_group | 3.28E-04 | 2.52E-08 | 5.02E-05 | 3.96E-04 | 4.43E-08 | 6.66E-05 | 4.31E-01 | 4.94E-01 |
| uncultured_bacterium_g_[Eubacterium]_ruminantium_group | 4.24E-03 | 2.08E-06 | 4.56E-04 | 5.16E-03 | 2.11E-06 | 4.59E-04 | 1.69E-01 | 2.40E-01 |
| uncultured_bacterium_g_Shuttleworthia | 3.50E-04 | 1.05E-07 | 1.03E-04 | 4.27E-04 | 3.52E-07 | 1.88E-04 | 7.23E-01 | 7.73E-01 |
| uncultured_bacterium_g_Butyricimonas | 3.05E-04 | 1.55E-08 | 3.94E-05 | 3.86E-04 | 3.95E-08 | 6.28E-05 | 2.78E-01 | 3.43E-01 |
| uncultured_bacterium_g_Erysipelatoclostridium | 1.15E-04 | 4.60E-09 | 2.14E-05 | 1.51E-04 | 1.03E-08 | 3.21E-05 | 3.76E-01 | 4.47E-01 |
| uncultured_bacterium_g_Ruminococcus_1 | 7.06E-03 | 6.78E-06 | 8.23E-04 | 9.81E-03 | 4.12E-05 | 2.03E-03 | 2.56E-01 | 3.22E-01 |
| uncultured_bacterium_g_Candidatus_Soleaferrea | 5.87E-05 | 2.25E-09 | 1.50E-05 | 8.46E-05 | 1.26E-09 | 1.12E-05 | 1.92E-01 | 2.65E-01 |
| uncultured_bacterium_g_[Eubacterium]_fissicatena_group | 1.44E-03 | 3.73E-07 | 1.93E-04 | 2.11E-03 | 1.69E-06 | 4.11E-04 | 1.63E-01 | 2.40E-01 |
| uncultured_bacterium_g_Bacteroides | 4.97E-03 | 3.03E-06 | 5.51E-04 | 7.31E-03 | 1.18E-05 | 1.09E-03 | 8.59E-02 | 1.52E-01 |
| uncultured_bacterium_o_Gastranaerophilales | 1.39E-03 | 2.85E-07 | 1.69E-04 | 2.05E-03 | 1.81E-06 | 4.26E-04 | 1.66E-01 | 2.40E-01 |
| uncultured_bacterium_g_[Eubacterium]_nodatum_group | 2.56E-04 | 1.08E-08 | 3.28E-05 | 3.93E-04 | 8.90E-08 | 9.43E-05 | 1.68E-01 | 2.40E-01 |
| uncultured_bacterium_g_Ruminiclostridium_6 | 4.59E-04 | 8.34E-08 | 9.13E-05 | 7.09E-04 | 4.17E-08 | 6.46E-05 | 5.09E-02 | 1.04E-01 |
| Christensenella_timonensis | 4.27E-05 | 1.55E-09 | 1.24E-05 | 6.66E-05 | 3.63E-09 | 1.90E-05 | 3.33E-01 | 4.01E-01 |
| uncultured_bacterium_g_Clostridium_sensu_stricto_1 | 2.24E-03 | 5.42E-07 | 2.33E-04 | 3.51E-03 | 4.96E-06 | 7.04E-04 | 9.19E-02 | 1.57E-01 |
| uncultured_bacterium_g_Akkermansia | 2.87E-01 | 7.24E-03 | 2.69E-02 | 4.71E-01 | 5.28E-03 | 2.30E-02 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Parabacteroides | 1.59E-04 | 6.02E-09 | 2.45E-05 | 2.67E-04 | 1.72E-08 | 4.14E-05 | 5.39E-02 | 1.08E-01 |
| uncultured_bacterium_g_Parasutterella | 2.17E-04 | 4.75E-09 | 2.18E-05 | 3.82E-04 | 7.63E-08 | 8.74E-05 | 2.60E-02 | 6.60E-02 |
| Burkholderiales_bacterium_YL45 | 1.02E-03 | 1.17E-07 | 1.08E-04 | 1.86E-03 | 7.43E-07 | 2.73E-04 | 5.99E-03 | 2.56E-02 |
| uncultured_bacterium_g_Prevotellaceae_UCG-001 | 2.13E-03 | 3.98E-07 | 1.99E-04 | 3.91E-03 | 2.25E-06 | 4.74E-04 | 4.00E-03 | 1.98E-02 |
| uncultured_bacterium_g_Family_XIII_AD3011_group | 5.64E-05 | 6.94E-10 | 8.33E-06 | 1.06E-04 | 8.34E-09 | 2.89E-05 | 8.49E-02 | 1.52E-01 |
| uncultured_bacterium_f_Candidatus_Gastranaerophilales_bacterium_Zag_111 | 9.53E-04 | 6.91E-07 | 2.63E-04 | 1.87E-03 | 1.42E-06 | 3.77E-04 | 5.99E-02 | 1.17E-01 |
| uncultured_bacterium_f_Eggerthellaceae | 7.27E-05 | 2.41E-09 | 1.55E-05 | 1.43E-04 | 6.61E-09 | 2.57E-05 | 1.50E-02 | 4.27E-02 |
| uncultured_bacterium_o_Rhodospirillales | 2.01E-04 | 1.21E-08 | 3.48E-05 | 4.04E-04 | 7.89E-08 | 8.88E-05 | 3.70E-02 | 7.90E-02 |
| uncultured_bacterium_g_Romboutsia | 8.38E-04 | 1.45E-07 | 1.20E-04 | 1.69E-03 | 1.33E-06 | 3.65E-04 | 2.90E-02 | 6.98E-02 |
| uncultured_bacterium_g_Christensenellaceae_R-7_group | 4.34E-05 | 6.21E-10 | 7.88E-06 | 9.67E-05 | 6.66E-09 | 2.58E-05 | 3.30E-02 | 7.38E-02 |
| uncultured_bacterium_g_Prevotellaceae_NK3B31_group | 3.05E-03 | 1.81E-06 | 4.25E-04 | 7.43E-03 | 8.47E-06 | 9.21E-04 | 2.00E-03 | 1.17E-02 |
| uncultured_bacterium_o_Mollicutes_RF39 | 5.43E-04 | 3.60E-08 | 6.00E-05 | 1.44E-03 | 6.71E-07 | 2.59E-04 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Candidatus_Saccharimonas | 3.41E-03 | 3.67E-06 | 6.06E-04 | 9.53E-03 | 1.95E-05 | 1.40E-03 | 2.00E-03 | 1.17E-02 |
| uncultured_bacterium_f_Blattella_germanica_German_cockroach | 1.06E-04 | 7.62E-09 | 2.76E-05 | 3.03E-04 | 2.01E-08 | 4.48E-05 | 4.00E-03 | 1.98E-02 |
| uncultured_bacterium_f_Clostridiales_vadinBB60_group | 3.44E-05 | 3.73E-09 | 1.93E-05 | 1.03E-04 | 1.24E-08 | 3.51E-05 | 1.07E-01 | 1.76E-01 |
| Lactobacillus_gasseri | 2.44E-04 | 2.99E-08 | 5.46E-05 | 1.28E-03 | 3.96E-07 | 1.99E-04 | 2.00E-03 | 1.17E-02 |
| uncultured_bacterium_g_Bifidobacterium | 2.11E-04 | 1.33E-08 | 3.65E-05 | 1.60E-03 | 2.50E-06 | 5.00E-04 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Turicibacter | 1.11E-03 | 3.92E-07 | 1.98E-04 | 8.56E-03 | 1.28E-05 | 1.13E-03 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Faecalibaculum | 7.69E-04 | 8.79E-08 | 9.37E-05 | 6.36E-03 | 2.74E-05 | 1.66E-03 | 9.99E-04 | 1.04E-02 |
| uncultured_bacterium_g_Coriobacteriaceae_UCG-002 | 9.76E-05 | 1.91E-09 | 1.38E-05 | 8.39E-04 | 1.77E-07 | 1.33E-04 | 9.99E-04 | 1.04E-02 |
Figure 7.
Significance analysis of differential gut microbial communities in the feces from various groups of mice. As shown by cladogram and LDA analyses, the microbial count of uncultured_bacterium_f_Lachnospiraceae was significantly increased, while that of Akkermansia microorganisms was drastically reduced in the gut microbiota of mice in Group F.
Fisetin alters the expression of 16S functional genes and metabolic signaling pathways in the gut microorganisms in POF mice
Differential analysis of KEGG metabolic pathways was used to analyze the differences and changes in the functional genes of microbial communities in the metabolic pathways between different groups, thereby evaluating metabolic changes in different gut samples in response to environmental changes. Results showed that, compared with that in group P, the gut microbiota in group F showed increased carbohydrate and nucleotide metabolism, but decreased lipid and amino acid metabolism, thereby promoting membrane transport activation (environmental information processing), cell motility (cellular processes), and translation (genetic information processing) (Figure 8A; Table 8).
Figure 8.
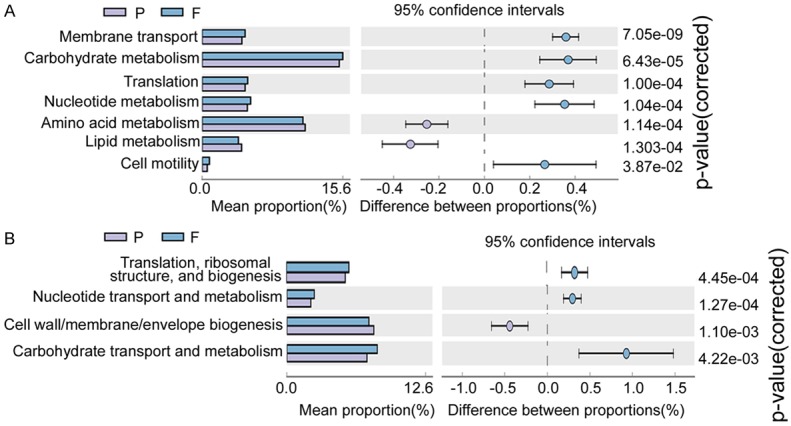
16S functional gene prediction analysis of gut microbiota in various groups of mice. A. KEGG functional prediction analysis showed that, compared with that in Group P, the gut microbiota in Group F could increase both carbohydrate and nucleotide metabolism, while decreasing both lipid and amino acid metabolism. B. COG functional prediction analysis showed that compared with that in Group P, the abundance of proteins associated with transcription, translation, ribosomal structure and biogenesis, carbohydrate transport and metabolism, and nucleotide transport and metabolism was significantly increased in Group F.
Table 8.
KEGG (P vs F)
| Class 1 | Class 2 | P: mean rel.freq.(%) | P: std.dev.(%) | F: mean rel.freq.(%) | F: std.dev.(%) | p-values | p-values (corrected) | Difference between means | 95.0% lower CI | 95.0% upper CI |
|---|---|---|---|---|---|---|---|---|---|---|
| Metabolism | Carbohydrate metabolism | 15.2158 | 0.098465 | 15.5841 | 0.1462 | 1.20E-05 | 6.43E-05 | -0.368309 | -0.493 | -0.243608866 |
| Metabolism | Lipid metabolism | 4.37768 | 0.092 | 4.05142 | 0.146 | 4.24E-05 | 0.00013 | 0.326259 | 0.20377 | 0.448746455 |
| Metabolism | Metabolism of cofactors and vitamins | 6.40804 | 0.025488 | 6.24375 | 0.04993 | 6.28E-07 | 6.75E-06 | 0.164293 | 0.12404 | 0.204544501 |
| Metabolism | Energy metabolism | 7.0241 | 0.041369 | 6.89513 | 0.08907 | 0.001766 | 0.003302 | 0.128965 | 0.05808 | 0.199851964 |
| Metabolism | Nucleotide metabolism | 5.03781 | 0.109697 | 5.39101 | 0.14845 | 2.65E-05 | 0.000104 | -0.3532 | -0.4833 | -0.223129628 |
| Metabolism | Biosynthesis of other secondary metabolites | 1.40308 | 0.008529 | 1.38291 | 0.01267 | 0.001143 | 0.002457 | 0.020177 | 0.00937 | 0.030980301 |
| Metabolism | Amino acid metabolism | 11.4485 | 0.069283 | 11.1952 | 0.11124 | 3.43E-05 | 0.000114 | 0.253385 | 0.16031 | 0.346457558 |
| Metabolism | Metabolism of terpenoids and polyketides | 2.09511 | 0.029918 | 2.02984 | 0.0807 | 0.043072 | 0.063866 | 0.065272 | 0.00241 | 0.128130574 |
| Metabolism | Xenobiotics biodegradation and metabolism | 2.36127 | 0.067271 | 2.17755 | 0.09547 | 0.000225 | 0.00057 | 0.183717 | 0.10126 | 0.266173915 |
| Metabolism | Metabolism of other amino acids | 2.05794 | 0.01307 | 2.06663 | 0.01359 | 0.183913 | 0.22595 | -0.008688 | -0.0219 | 0.004520898 |
| Metabolism | Glycan biosynthesis and metabolism | 3.10912 | 0.040491 | 2.91094 | 0.06041 | 4.72E-07 | 6.76E-06 | 0.198184 | 0.14672 | 0.249643975 |
| Genetic Information Processing | Translation | 4.76588 | 0.096159 | 5.05097 | 0.11761 | 2.80E-05 | 0.0001 | -0.285099 | -0.3918 | -0.17841167 |
| Metabolism | Global and overview maps | 13.5316 | 0.056769 | 13.5306 | 0.151 | 0.984286 | 1.085239 | 0.001082 | -0.1166 | 0.118814514 |
| Human Diseases | Drug resistance | 0.93973 | 0.015243 | 0.88032 | 0.04027 | 0.001492 | 0.003056 | 0.059412 | 0.028 | 0.090828372 |
| Environmental Information Processing | Membrane transport | 4.44104 | 0.051621 | 4.79906 | 0.06255 | 1.64E-10 | 7.05E-09 | -0.358021 | -0.415 | -0.301081011 |
| Environmental Information Processing | Signal transduction | 3.44014 | 0.094492 | 3.2234 | 0.10309 | 0.000203 | 0.000546 | 0.216738 | 0.11875 | 0.31472534 |
| Cellular Processes | Cell motility | 0.5768 | 0.098952 | 0.84214 | 0.29143 | 0.025219 | 0.038729 | -0.265346 | -0.491 | -0.039666622 |
| Genetic Information Processing | Folding, sorting and degradation | 2.28493 | 0.022624 | 2.21118 | 0.05424 | 0.002683 | 0.004438 | 0.073744 | 0.03108 | 0.116409292 |
| Genetic Information Processing | Transcription | 0.22556 | 0.00285 | 0.23609 | 0.00715 | 0.001518 | 0.002967 | -0.010534 | -0.0161 | -0.004929328 |
| Genetic Information Processing | Replication and repair | 4.20832 | 0.071461 | 4.44943 | 0.09408 | 1.19E-05 | 7.31E-05 | -0.241111 | -0.3243 | -0.157943165 |
| Organismal Systems | Endocrine system | 0.50868 | 0.005102 | 0.49429 | 0.00524 | 1.39E-05 | 6.65E-05 | 0.014387 | 0.00926 | 0.019511202 |
| Environmental Information Processing | Signaling molecules and interaction | 0 | 0 | 0 | 0 | 1 | 1.075 | 0 | 0 | 0 |
| Cellular Processes | Cell growth and death | 0.71639 | 0.007562 | 0.72397 | 0.01043 | 0.095905 | 0.128873 | -0.007584 | -0.0167 | 0.001498704 |
| Cellular Processes | Transport and catabolism | 0.64595 | 0.010187 | 0.5868 | 0.01182 | 1.53E-09 | 3.29E-08 | 0.059149 | 0.0482 | 0.070095597 |
| Organismal Systems | Circulatory system | 0.00053 | 0.000248 | 0.00075 | 0.00035 | 0.129507 | 0.168751 | -0.000227 | -0.0005 | 7.38E-05 |
| Organismal Systems | Development | 0 | 0 | 0 | 0 | 1 | 1.04878 | 0 | 0 | 0 |
| Cellular Processes | Cellular community | 0 | 0 | 0 | 0 | 1 | 1.02381 | 0 | 0 | 0 |
| Organismal Systems | Immune system | 0.11342 | 0.002376 | 0.12145 | 0.00272 | 3.25E-06 | 2.33E-05 | -0.008032 | -0.0106 | -0.005497639 |
| Organismal Systems | Environmental adaptation | 0.16438 | 0.002798 | 0.17464 | 0.00853 | 0.005696 | 0.009071 | -0.010257 | -0.0168 | -0.003666729 |
| Organismal Systems | Nervous system | 0.38693 | 0.007892 | 0.37309 | 0.01956 | 0.072761 | 0.100927 | 0.013842 | -0.0015 | 0.02917844 |
| Organismal Systems | Sensory system | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Human Diseases | Endocrine and metabolic diseases | 0.12607 | 0.004953 | 0.13412 | 0.00453 | 0.00208 | 0.003578 | -0.008049 | -0.0128 | -0.003345654 |
| Organismal Systems | Excretory system | 0.11442 | 0.008281 | 0.08404 | 0.01532 | 0.000132 | 0.000377 | 0.030383 | 0.01792 | 0.042849674 |
| Organismal Systems | Digestive system | 0.01355 | 0.002889 | 0.01664 | 0.0038 | 0.069293 | 0.099319 | -0.003087 | -0.0064 | 0.000272377 |
| Human Diseases | Neurodegenerative diseases | 0.20071 | 0.002501 | 0.18878 | 0.00417 | 2.58E-06 | 2.22E-05 | 0.011936 | 0.00848 | 0.015393269 |
| Human Diseases | Substance dependence | 0.10587 | 0.010866 | 0.07204 | 0.019 | 0.000363 | 0.00082 | 0.033834 | 0.01822 | 0.049448215 |
| Human Diseases | Infectious diseases: Bacterial | 0.74233 | 0.002251 | 0.74307 | 0.00606 | 0.737421 | 0.834451 | -0.000741 | -0.0055 | 0.003982918 |
| Human Diseases | Infectious diseases: Parasitic | 0.10874 | 0.010226 | 0.07877 | 0.01618 | 0.000277 | 0.000661 | 0.029962 | 0.01638 | 0.04354289 |
| Human Diseases | Infectious diseases: Viral | 0.00122 | 0.000353 | 0.00103 | 0.00036 | 0.250759 | 0.299517 | 0.000198 | -0.0002 | 0.000549562 |
| Human Diseases | Cancers: Overview | 0.94709 | 0.012065 | 0.8997 | 0.03345 | 0.001984 | 0.003554 | 0.047393 | 0.02139 | 0.073396642 |
| Human Diseases | Cancers: Specific types | 0.0935 | 0.003377 | 0.09216 | 0.00775 | 0.641648 | 0.745699 | 0.001345 | -0.0048 | 0.007470511 |
| Human Diseases | Immune diseases | 0.05753 | 0.001452 | 0.06283 | 0.00216 | 1.59E-05 | 6.82E-05 | -0.005301 | -0.0071 | -0.003461283 |
| Human Diseases | Cardiovascular diseases | 0.00017 | 8.21E-05 | 0.00025 | 0.00012 | 0.136062 | 0.172079 | -7.41E-05 | -0.0002 | 2.59E-05 |
Through clusters of orthologous groups of proteins (COG) analysis, the distribution and abundance of homologous protein families in microorganisms were analyzed. Results showed that, compared with that in group P, the abundance of proteins associated with transcription, translation, ribosomal structure and biogenesis, and carbohydrate and nucleotide transport and metabolism was significantly increased in group F. Meanwhile, the abundance of proteins associated with cell wall/membrane/envelope biogenesis in cellular processes and signaling was significantly decreased (Figure 8B; Table 9). Thus, our results suggested that fisetin significantly affects the metabolic pathways of the gut microbiota in POF mice.
Table 9.
COG (P vs F)
| Class 1 | Class 2 | P: mean rel.freq.(%) | P: std.dev.(%) | F: mean rel.freq.(%) | F: std.dev.(%) | p-values | p-values (corrected) | Difference between means | 95.0% lower CI | 95.0% upper CI |
|---|---|---|---|---|---|---|---|---|---|---|
| INFORMATION STORAGE AND PROCESSING | RNA processing and modification | 0.04424 | 0.00378 | 0.0325 | 0.00631 | 0.00025419 | 0.000489 | 0.01174 | 0.0065 | 0.016973591 |
| INFORMATION STORAGE AND PROCESSING | Chromatin structure and dynamics | 0.04419 | 0.00408 | 0.03105 | 0.0071 | 0.00025742 | 0.00046 | 0.01313 | 0.00729 | 0.018970949 |
| METABOLISM | Energy production and conversion | 4.57045 | 0.08251 | 4.64817 | 0.11357 | 0.11569553 | 0.131472 | -0.0777 | -0.1767 | 0.02126776 |
| CELLULAR PROCESSES AND SIGNALING | Cell cycle control, cell division, chromosome partitioning | 0.96205 | 0.03834 | 1.09534 | 0.05938 | 4.13E-05 | 0.000172 | -0.1333 | -0.1834 | -0.083179818 |
| METABOLISM | Amino acid transport and metabolism | 7.15645 | 0.03858 | 7.28732 | 0.04532 | 3.87E-06 | 4.83E-05 | -0.1309 | -0.1726 | -0.089104438 |
| METABOLISM | Nucleotide transport and metabolism | 2.19879 | 0.08924 | 2.49334 | 0.11736 | 1.53E-05 | 0.000127 | -0.2946 | -0.3983 | -0.190771936 |
| METABOLISM | Carbohydrate transport and metabolism | 7.28615 | 0.28753 | 8.21234 | 0.70532 | 0.00337815 | 0.004223 | -0.9262 | -1.4798 | -0.372551875 |
| METABOLISM | Coenzyme transport and metabolism | 3.84124 | 0.0295 | 3.74354 | 0.0239 | 5.37E-07 | 1.34E-05 | 0.09769 | 0.07102 | 0.124367872 |
| METABOLISM | Lipid transport and metabolism | 2.81747 | 0.04787 | 2.58496 | 0.10658 | 5.52E-05 | 0.000197 | 0.23252 | 0.14803 | 0.317007595 |
| INFORMATION STORAGE AND PROCESSING | Translation, ribosomal structure and biogenesis | 5.39996 | 0.14566 | 5.72952 | 0.16261 | 0.00026715 | 0.000445 | -0.3296 | -0.4826 | -0.176553788 |
| INFORMATION STORAGE AND PROCESSING | Transcription | 7.58899 | 0.12905 | 8.07005 | 0.35014 | 0.00245464 | 0.003409 | -0.4811 | -0.7537 | -0.208450554 |
| INFORMATION STORAGE AND PROCESSING | Replication, recombination and repair | 6.10104 | 0.06951 | 6.32243 | 0.1059 | 8.84E-05 | 0.000246 | -0.2214 | -0.3111 | -0.131655 |
| CELLULAR PROCESSES AND SIGNALING | Cell wall/membrane/envelope biogenesis | 7.8961 | 0.11425 | 7.45362 | 0.27131 | 0.00070138 | 0.001096 | 0.44248 | 0.22886 | 0.656102178 |
| CELLULAR PROCESSES AND SIGNALING | Cell motility | 1.57647 | 0.05978 | 1.42295 | 0.0702 | 0.00010096 | 0.000229 | 0.15352 | 0.08883 | 0.218210194 |
| CELLULAR PROCESSES AND SIGNALING | Posttranslational modification, protein turnover, chaperones | 3.4028 | 0.04029 | 3.23856 | 0.10484 | 0.00095795 | 0.001409 | 0.16425 | 0.08237 | 0.246128913 |
| METABOLISM | Inorganic ion transport and metabolism | 5.25735 | 0.08135 | 4.88141 | 0.15717 | 2.05E-05 | 0.000128 | 0.37595 | 0.24898 | 0.502910122 |
| METABOLISM | Secondary metabolites biosynthesis, transport and catabolism | 1.47389 | 0.05604 | 1.28654 | 0.09765 | 0.00018346 | 0.000382 | 0.18735 | 0.10704 | 0.267664261 |
| POORLY CHARACTERIZED | General function prediction only | 12.5919 | 0.14023 | 12.321 | 0.28875 | 0.02503819 | 0.029807 | 0.27085 | 0.03973 | 0.501972433 |
| POORLY CHARACTERIZED | Function unknown | 8.85463 | 0.24438 | 8.23404 | 0.452 | 0.00281298 | 0.003701 | 0.62059 | 0.25285 | 0.988323449 |
| CELLULAR PROCESSES AND SIGNALING | Signal transduction mechanisms | 5.65728 | 0.08178 | 5.71257 | 0.21303 | 0.48169017 | 0.523576 | -0.0553 | -0.2217 | 0.111075864 |
| CELLULAR PROCESSES AND SIGNALING | Intracellular trafficking, secretion, and vesicular transport | 2.86423 | 0.10006 | 2.4761 | 0.18932 | 9.53E-05 | 0.000238 | 0.38813 | 0.23469 | 0.541571855 |
| CELLULAR PROCESSES AND SIGNALING | Defense mechanisms | 2.39035 | 0.07462 | 2.70361 | 0.14814 | 7.10E-05 | 0.000222 | -0.3133 | -0.4324 | -0.194074878 |
| CELLULAR PROCESSES AND SIGNALING | Extracellular structures | 0 | 0 | 0 | 0 | 1 | 1.041667 | 0 | 0 | 0 |
| CELLULAR PROCESSES AND SIGNALING | Nuclear structure | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| CELLULAR PROCESSES AND SIGNALING | Cytoskeleton | 0.02398 | 0.0013 | 0.019 | 0.0021 | 2.25E-05 | 0.000112 | 0.00498 | 0.00322 | 0.006738383 |
Fisetin significantly decreases CCR9+/CXCR3+/CD4+ T-lymphocyte counts and IL-12 levels in POF mice
Immunofluorescence staining and FCM analysis showed that the peripheral blood CCR9+/CXCR3+/CD4+ T-lymphocyte counts (Figure 9A and 9C) as well as the IL-12+/CD4+ T-lymphocyte counts were significantly lower in the fisetin group compared to those in the PBS group (Figure 9B and 9D). Hence, our results suggested that fisetin significantly decreased CCR9+/CXCR3+/CD4+ T-lymphocyte count and IL-12 secretion in POF mice.
Figure 9.
Analysis of peripheral blood CCR9+/CXCR3+/CD4+ T-lymphocyte counts and IL-12 levels in various groups of mice. A and C. FCM analysis showed that the peripheral blood CCR9+/CXCR3+/CD4+ T-lymphocyte count of mice in the fisetin group was significantly lower than that of mice in the control group; B and D. FCM analysis showed that the peripheral blood CD4+/IL-12+ cell count of mice in the fisetin group was significantly lower than that of mice in the control group.
Discussion
Currently, an increasing number of studies have found that POF is a multi-factorial disease [1-4,6,31]. Several factors, such as genetic factors, infection and inflammation, mental health condition, and hormone levels contribute towards the development of POF [1-4,6]. Ovum maturation is an extremely complex process and requires the participation of hormones, growth factors, plasmin, and other substances [1-4,6,31]. The ovaries, which provide a natural microenvironment for ovum maturation, consist of granulosa, stromal, ovarian epithelial, and vascular endothelial cells [1-4,6,31]. The ovaries require large amounts of nutrients to function properly [1-4,6,31]. In the presence of endocrine and metabolic abnormalities in the body, the ovarian microenvironment is affected [1-4,6,31]. Therefore, maintaining an overall healthy endocrine and metabolic state is extremely important for the maintenance of a healthy ovarian microenvironment. Some studies have reported that fisetin, as a flavonol present in fruits and vegetables, possesses anti-oxidant, anti-inflammatory, and anti-aging effects in addition to its anti-cancer properties [7-9]. However, the exact mechanisms by which it exerts its effects are currently unknown. In this study, we investigated whether orally adminstered fisetin undergoes digestion and absorption in the gastrointestinal tract, and whether the gut microbiota carries out metabolic-biochemical reactions using fisetin as a raw material. However, whether fisetin affects the composition and metabolic pathways of the gut microbiota remains unknown.
In this study, we aimed to investigate: 1) whether fisetin exhibits therapeutic efficacy against POF, 2) whether fisetin exerts its effect in POF mice via regulating the gut microbiota, and 3) the mechanism by which fisetin regulates the gut microbiota to alleviate POF. To achieve this, we first established POF mouse and C. elegans models by injection of CTX, followed by fisetin treatment. Our previous studies have confirmed that POF is typically characterized by pathological aging of the ovarian tissue [1,17,32]. C. elegans is a suitable animal model to study aging [32]. Therefore, in this study, we still selected C. elegans to evaluate the anti-aging function of fisetin. Results of pathological and molecular analyses suggested that fisetin could reverse ovarian damage and atresia in the ovarian follicles of mice and prevent premature senescence and reduction of ovarian function in C. elegans. Subsequently, we carried out 16S rRNA v3+v4 sequencing of fresh feces samples of mice in various groups to assess the composition of gut microbiota and differences in the distribution of specific flora. The results showed that fisetin treatment did not alter gut microbial counts in POF mice, but affected the composition and distribution of gut microbiota. Alpha and beta diversity analyses showed that the bacterial count of uncultured_bacterium_f_Lachnospiraceae was significantly increased in the gut microbiota of POF mice in the fisetin group, but that of Akkermansia was significantly reduced. Routy and Zitvogel previously reported that the efficacy of PD-1 treatment was higher by 25-40% in some cancer patients containing Akkermansia in their gut compared to those lacking these gut microorganisms [33,34]. Their results suggested that Akkermansia microorganisms may help mediate the immunoregulatory function of PD-1. Further, an in-depth study showed that when experimental mice were orally administered Akkermansia microorganisms, the recruitment of CCR9+/CXCR3+/CD4+ T lymphocytes increased, which suppressed IL-12-dependent PD-1 blockade, thereby increasing the cytotoxic effects of PD-1 antagonists [33,34]. Hence, it suggested that Akkermansia microorganisms could effectively increase the activity of immune cells. In POF, certain immune cells, such as CD8+ T cells and M1 macrophages are in an overactivated state. In our study, we found that when fisetin alleviates POF symptoms in POF mice, it also simultaneously decreases the number of Akkermansia microorganisms in the gut of mice. These results indicate that fisetin may decrease Akkermansia counts to reduce the counts and activity of CCR9+/CXCR3+/CD4+ T lymphocytes in POF mice. Subsequently, the counts of CCR9+/CXCR3+/CD4+ T lymphocytes and CD4+/IL-12+ cells in the PBMCs of various groups of mice were measured. Interestingly, CCR9+/CXCR3+/CD4+ T-lymphocyte and CD4+/IL-12+ counts were significantly decreased in POF mice in the fisetin group (Figure 10), which are consistent with the findings of Routy and Zitvogel [33,34].
Figure 10.
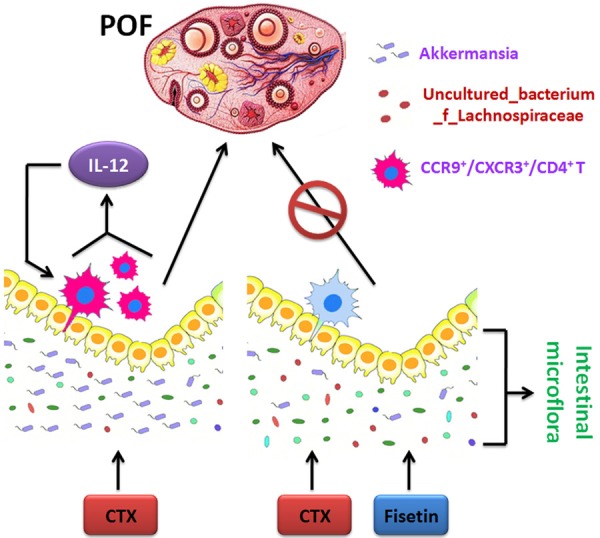
Fisetin alleviates POF in mice by regulating gut microbiota via decreasing CCR9+/CXCR3+/CD4+ T-lymphocyte count and IL-12 secretion.
In summary, in this study, we showed that fisetin alleviates POF through novel pharmacological mechanisms, including regulating the distribution and counts of Akkermansia and uncultured_bacterium_f_Lachnospiracea in POF mice to reduce peripheral blood CCR9+/CXCR3+/CD4+ T-lymphocyte counts and IL-12 secretion. This in turn results in the regulation of ovarian microenvironment and reduction of inflammation, which have therapeutic effects on POF.
Acknowledgements
This work was supported by grant from the National Natural Science Foundation of China (No. 81973899). And, grant from Shanghai Natural Science Foundation (No. 16ZR1434000). And, grant from the projects sponsored by the development fund for Shanghai talents (2017054). And, grant from the projects sponsored by the fund for Xinglin talents of Shanghai University of TCM (201707081). And, grant from the graduate student innovation training project of Shanghai University of Traditional Chinese Medicine (Y201860).
Disclosure of conflict of interest
None.
Abbreviations
- POF
Premature ovarian failure
- Fisetin
3,7,3’,4’-tetrahydroxyflavone
- FCM
Flow cytometry
- PBMCs
Peripheral blood mononuclear cells
- DPBS
Dulbecco’s phosphate-buffered saline
- LEfSe
LDA-effect size
- PCOA
Principal coordinates analysis
- PCA
Principal component analysis
- NMDS
Non-metric multi-dimensional scaling
- UPGMA
Unweighted pair-group method with arithmetic mean
- LDA
Line discriminant analysis
- COG
Clusters of orthologous groups of proteins
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
References
- 1.Xiong Y, Liu T, Wang S, Chi H, Chen C, Zheng J. Cyclophosphamide promotes the proliferation inhibition of mouse ovarian granulosa cells and premature ovarian failure by activating the lncRNA-Meg3-p53-p66Shc pathway. Gene. 2017;596:1–8. doi: 10.1016/j.gene.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Liu T, Wang S, Li Q, Huang Y, Chen C, Zheng J. Telocytes as potential targets in a cyclophosphamide-induced animal model of premature ovarian failure. Mol Med Rep. 2016;14:2415–2422. doi: 10.3892/mmr.2016.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu T, Huang Y, Zhang J, Qin W, Chi H, Chen J, Yu Z, Chen C. Transplantation of human menstrual blood stem cells to treat premature ovarian failure in mouse model. Stem Cells Dev. 2014;23:1548–1557. doi: 10.1089/scd.2013.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, Huang Y, Guo L, Cheng W, Zou G. CD44+/CD105+ human amniotic fluid mesenchymal stem cells survive and proliferate in the ovary long-term in a mouse model of chemotherapy-induced premature ovarian failure. Int J Med Sci. 2012;9:592–602. doi: 10.7150/ijms.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T, Qin W, Huang Y, Zhao Y, Wang J. Induction of estrogen-sensitive epithelial cells derived from human-induced pluripotent stem cells to repair ovarian function in a chemotherapy-induced mouse model of premature ovarian failure. DNA Cell Biol. 2013;32:685–698. doi: 10.1089/dna.2013.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu T, Li Q, Wang S, Chen C, Zheng J. Transplantation of ovarian granulosalike cells derived from human induced pluripotent stem cells for the treatment of murine premature ovarian failure. Mol Med Rep. 2016;13:5053–5058. doi: 10.3892/mmr.2016.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhtar E, Adhami VM, Sechi M, Mukhtar H. Dietary flavonoid fisetin binds to beta-tubulin and disrupts microtubule dynamics in prostate cancer cells. Cancer Lett. 2015;367:173–183. doi: 10.1016/j.canlet.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Si Y, Liu J, Shen H, Zhang C, Wu Y, Huang Y, Gong Z, Xue J, Liu T. Fisetin decreases TET1 activity and CCNY/CDK16 promoter 5hmC levels to inhibit the proliferation and invasion of renal cancer stem cell. J Cell Mol Med. 2019;23:1095–1105. doi: 10.1111/jcmm.14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SC, Kim YH, Son SW, Moon EY, Pyo S, Um SH. Fisetin induces Sirt1 expression while inhibiting early adipogenesis in 3T3-L1 cells. Biochem Biophys Res Commun. 2015;467:638–644. doi: 10.1016/j.bbrc.2015.10.094. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, Xia H, Liu Z, Cui B, Liang P, Xi L, Jin J, Ying X, Wang X, Zhao X, Li W, Jia H, Lan Z, Li F, Wang R, Sun Y, Yang M, Shen Y, Jie Z, Li J, Chen X, Zhong H, Xie H, Zhang Y, Gu W, Deng X, Shen B, Xu X, Yang H, Xu G, Bi Y, Lai S, Wang J, Qi L, Madsen L, Wang J, Ning G, Kristiansen K, Wang W. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 11.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y, Qi Y, Yang X, Zhao L, Wen S, Liu Y, Tang L. Association between polycystic ovary syndrome and gut microbiota. PLoS One. 2016;11:e0153196. doi: 10.1371/journal.pone.0153196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan M, Li D, Zhang Z, Sun H, An M, Wang G. Endometriosis induces gut microbiota alterations in mice. Hum Reprod. 2018;33:607–616. doi: 10.1093/humrep/dex372. [DOI] [PubMed] [Google Scholar]
- 14.Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, Gajer P, Ravel J, Goedert JJ. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker JM, Al-Nakkash L, Herbst-Kralovetz MM. Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas. 2017;103:45–53. doi: 10.1016/j.maturitas.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 16.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, Backhed F, Isolauri E, Salminen S, Ley RE. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ai A, Xiong Y, Wu B, Lin J, Huang Y, Cao Y, Liu T. Induction of miR-15a expression by tripterygium glycosides caused premature ovarian failure by suppressing the Hippo-YAP/TAZ signaling effector Lats1. Gene. 2018;678:155–163. doi: 10.1016/j.gene.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Xu P, Wang J, Hong F, Wang S, Jin X, Xue T, Jia L, Zhai Y. Melatonin prevents obesity through modulation of gut microbiota in mice. J Pineal Res. 2017;62 doi: 10.1111/jpi.12399. [DOI] [PubMed] [Google Scholar]
- 19.Ji ZH, Ren WZ, Gao W, Hao Y, Gao W, Chen J, Quan FS, Hu JP, Yuan B. Analyzing the innate immunity of NIH hairless mice and the impact of gut microbial polymorphisms on Listeria monocytogenes infection. Oncotarget. 2017;8:106222–106232. doi: 10.18632/oncotarget.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 22.Sakaki T, Takeshima T, Tominaga M, Hashimoto H, Kawaguchi S. Recurrence of ICA-PCoA aneurysms after neck clipping. J Neurosurg. 1994;80:58–63. doi: 10.3171/jns.1994.80.1.0058. [DOI] [PubMed] [Google Scholar]
- 23.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR, Hashsham SA, Tiedje JM, Stanton TB. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A. 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polanska UM, Edwards E, Fernig DG, Kinnunen TK. The cooperation of FGF receptor and Klotho is involved in excretory canal development and regulation of metabolic homeostasis in Caenorhabditis elegans. J Biol Chem. 2011;286:5657–5666. doi: 10.1074/jbc.M110.173039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Kwon G, Lim YH. Elucidating the mechanism of weissella-dependent lifespan extension in caenorhabditis elegans. Sci Rep. 2015;5:17128. doi: 10.1038/srep17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu C, Hwang W, Jeong DE, Ryu Y, Ha CM, Lee SV, Liu L, He ZM. Genetic inhibition of an ATP synthase subunit extends lifespan in C. elegans. Sci Rep. 2018;8:14836. doi: 10.1038/s41598-018-32025-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo Y, Kingsley S, Walker G, Mondoux MA, Tissenbaum HA. Metabolic shift from glycogen to trehalose promotes lifespan and healthspan in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2018;115:E2791–E2800. doi: 10.1073/pnas.1714178115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaletsky R, Murphy CT. The role of insulin/IGF-like signaling in C. elegans longevity and aging. Dis Model Mech. 2010;3:415–419. doi: 10.1242/dmm.001040. [DOI] [PubMed] [Google Scholar]
- 31.Lee DH, Pei CZ, Song JY, Lee KJ, Yun BS, Kwack KB, Lee EI, Baek KH. Identification of serum biomarkers for premature ovarian failure. Biochim Biophys Acta Proteins Proteom. 2019;1867:219–226. doi: 10.1016/j.bbapap.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Liu T, Liu Y, Huang Y, Chen J, Yu Z, Chen C, Lai L. miR-15b induces premature ovarian failure in mice via inhibition of alpha-Klotho expression in ovarian granulosa cells. Free Radic Biol Med. 2019;141:383–392. doi: 10.1016/j.freeradbiomed.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Routy B, Gopalakrishnan V, Daillere R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15:382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 34.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.