Summary
Robust production of terminally differentiated cells from self-renewing resident stem cells is essential to maintain proper tissue architecture and physiological functions, especially in high-turnover tissues. However, the transcriptional networks that precisely regulate cell transition and differentiation are poorly understood in most tissues. Here, we identified Sox100B, a Drosophila Sox E family transcription factor, as a critical regulator of adult intestinal stem cell differentiation. Sox100B is expressed in stem and progenitor cells and required for differentiation of enteroblast progenitors into absorptive enterocytes. Mechanistically, Sox100B regulates the expression of another critical stem cell differentiation factor, Sox21a. Supporting a direct control of Sox21a by Sox100B, we identified a Sox21a intronic enhancer that is active in all intestinal progenitors and directly regulated by Sox100B. Taken together, our results demonstrate that the activity and regulation of two Sox transcription factors are essential to coordinate stem cell differentiation and proliferation and maintain intestinal tissue homeostasis.
Keywords: Sox100B, Sox21a, Drosophila, intestinal stem cells, differentiation
Graphical Abstract

Highlights
-
•
Sox100B is expressed in progenitor cells in the adult intestine
-
•
Sox100B is required for stem cell differentiation
-
•
Sox100B is required for Sox21a expression
-
•
Sox100B directly controls the activity of a Sox21a intronic enhancer
In this article, Biteau and colleagues show that the transcription factor Sox100B is specifically expressed in stem cells and progenitors in the adult Drosophila intestinal epithelium. They demonstrate that it is essential for cell differentiation in the absorptive lineage and directly regulates the expression of the differentiation factor Sox21a.
Introduction
The proper maintenance of tissue homeostasis is essential for their normal architecture and physiological functions, especially in high-turnover tissues, such as intestinal epithelium. In most tissues, this is achieved by their resident stem cells, which are capable of self-renewing and differentiating into a variety of cell types within tissues. To answer the fundamental question of how tissue homeostasis is properly maintained, it is critical to identify the genetic networks that control stem cell proliferation and differentiation. Although proliferation has been extensively studied over the decades, mechanisms by which progressive and robust differentiation is achieved in vivo remain less understood in many lineages.
The adult Drosophila intestinal epithelium provides a genetically tractable experimental system to examine molecular mechanisms regulating stem cell activities (Biteau et al., 2011, Miguel-Aliaga et al., 2018). The adult midgut epithelium is actively maintained by multipotent intestinal stem cells (ISCs), which self-renew to maintain a stable stem cell population and give rise to post-mitotic progenitors committed to one of two distinct cell lineages: diploid secretary enteroendocrine cells (EEs) and polyploid absorptive enterocytes (ECs) (Micchelli and Perrimon, 2006, Ohlstein and Spradling, 2006). In the EC lineage, ISCs turn on the Notch signaling in the daughter cells termed enteroblasts (EBs) that are committed to differentiation into the absorptive fate. EBs then go through several rounds of endo-replication and finally differentiate into Pdm1-positive ECs (Ohlstein and Spradling, 2007). To maintain the secretory lineage, ISCs give rise to Prospero-positive pre-EE daughter cells (Biteau and Jasper, 2014, Zeng and Hou, 2015). A number of signaling pathways and transcription factors have been implicated in regulating ISC differentiation, including Delta/Notch (Bardin et al., 2010, Kapuria et al., 2012, Ohlstein and Spradling, 2007), JAK/STAT92E (Beebe et al., 2010, Jiang et al., 2009), escargot (Korzelius et al., 2014, Loza-Coll et al., 2014), Sox21a (Chen et al., 2016, Zhai et al., 2015, Zhai et al., 2017), GATAe (Okumura et al., 2016), and Pdm1 (Korzelius et al., 2014). However, our understanding of the transcriptional network involved in the control of EB differentiation remains incomplete.
Sox (Sry-related HMG Box) family transcription factors are important regulators of cell fate specification and cell differentiation during development and in multiple adult stem cell populations (Kamachi and Kondoh, 2013, Lefebvre et al., 2007, Sarkar and Hochedlinger, 2013, She and Yang, 2015). Sox21a, a Drosophila Sox B gene, is specifically expressed in ISCs and EBs and plays important roles in regulating ISC proliferation and EB differentiation into EC, both at homeostasis and under stress conditions (Chen et al., 2016, Meng and Biteau, 2015, Zhai et al., 2015, Zhai et al., 2017). However, how ISC- and EB-specific Sox21a expression pattern is established remains unknown. Here, we investigated the expression and function of another Sox family transcription factor, the Sox E factor Sox100B, and found that it is required for ISC differentiation into the EC lineage. We show that Sox100B is required for both Sox21a protein expression and the activity of a transcriptional enhancer located in the first intron of the Sox21a gene. Our identification of Sox100B binding sites in this intronic enhancer strongly supports the notion that Sox21a is a direct Sox100B target gene. Our results identify an essential player in the transcriptional network that regulates the complex process of stem cell differentiation in the adult Drosophila intestine.
Results
Sox100B Is Expressed in ISCs and EBs in the Adult Intestine
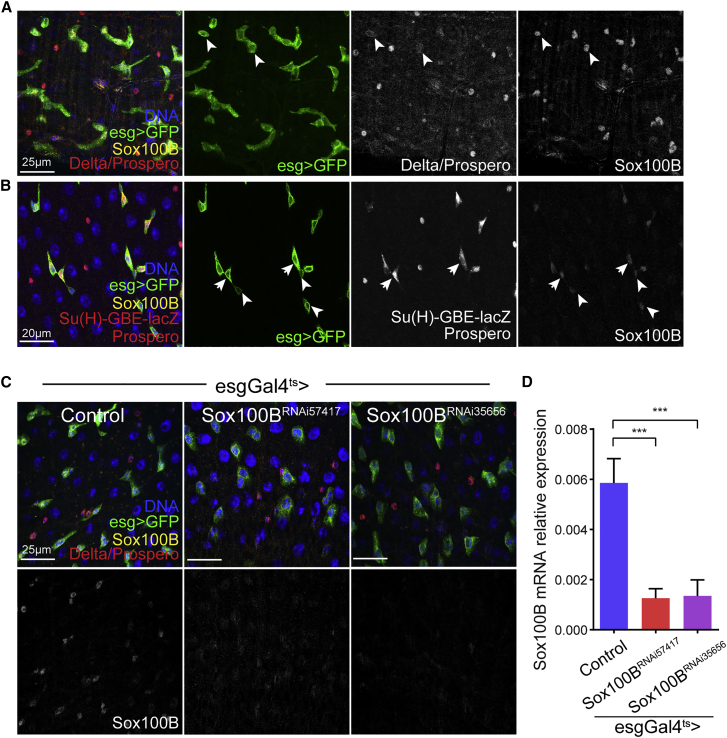
We previously found that the Sox family transcription factor Sox21a is specifically expressed in the progenitor cells, ISCs and EBs, in the adult intestine (Meng and Biteau, 2015). To investigate whether other Sox family transcription factors are involved in regulating the adult ISC lineage, we first asked whether other Sox family transcription factors are expressed in the fly intestinal epithelium. Sox100B, which encodes the sole Sox E group transcription factor in Drosophila, shows high mRNA levels in both larval midgut and adult midgut among different tissues (http://flyatlas.org, Figure S1A). In the adult intestine, ISCs and EBs are labeled by the expression of the transcription factor escargot (esg), while terminally differentiated ECs and EEs are identified by the expression of transcription factors Pdm1 and Prospero, respectively (Micchelli and Perrimon, 2006, Ohlstein and Spradling, 2006). Using a polyclonal antibody directed against full-length Sox100B protein (Loh and Russell, 2000), we found that Sox100B protein is expressed in small diploid cells in the adult intestine, overlapping with esgGal4-driven GFP expression, strongly suggesting that Sox100B is expressed in ISCs and EBs (54 Sox100B-positive cells among 155 esg-positive cells counted across 5 guts) (Figures 1A and 1B). We confirmed the identity of these Sox100B-positive cells using specific markers: ISCs express the Notch ligand Delta (Figure 1A, indicated by arrowheads) and EBs show high activity for the Notch reporter GBE-Su(H)-LacZ (Figure 1B, indicated by arrows) (Ohlstein and Spradling, 2007). We detected Sox100B protein expression in both ISCs and EBs, but not in fully differentiated ECs (no polyploid nuclei stain strongly positive for Sox100B across all our samples) and rarely in pre-EEs/EEs (15/47 Prospero-positive cells show low levels of Sox100B staining across 5 guts) (Figures 1A and 1B). Consistent with the mostly ISC/EB-specific expression pattern for Sox100B, both Sox100B protein and mRNA expression levels were strongly reduced when two independent RNAi constructs directed against Sox100B (Sox100BTRiP lines, Bloomington 57417 and 35656) were expressed specifically in the ISCs/EBs using the esgGal4ts driver (Figures 1C and 1D). Thus, our data demonstrate that Sox100B is expressed in ISCs and EBs in the adult midgut, confirming a recent study that reported expression of a Sox100B-GFP genomic BAC construct in intestinal progenitors (Doupe et al., 2018).
Figure 1.
Sox100B Is Expressed in ISCs and EBs in the Adult Intestine
(A and B) Representative confocal images of Sox100B expression in the adult posterior midgut showing that Sox100B is expressed in both Delta-positive ISCs––indicated by arrowheads in (A)––and EBs which have high Notch reporter activity––indicated by arrows in (B). Sox100B is not expressed in mature ECs and only marginally detected in a subset of EEs. ISC, intestinal stem cell; EB, enteroblast; EE, enteroendocrine; EC, enterocyte.
(C) Sox100B antibody staining is strongly reduced when two independent Sox100BRNAi (Bloomington TRiP stocks 57417 and 35656) were specifically expressed in the ISCs and EBs using the esgGal4ts driver for 6 days.
(D) qRT-PCR data demonstrates that Sox100B mRNA expression is strongly reduced in Sox100BRNAi -expressing ISCs and EBs after 7days knocking down. Sox100B mRNA expression is normalized to actin5c expression.
In (A–C) UAS-GFP expression driven by the esgGal4 labels both ISCs and EBs (green), DNA is stained by Hoechst (blue), Delta/Prospero/β-gal antibody staining (red) labels ISCs, mature EEs, and GBE-lacZ-positive EBs, respectively (Delta, membrane staining; Prospero, nuclear staining; GBE-lacZ, cytoplasmic staining). Scale bars, 25 μm (A and C) and 20 μm (B). In (D), values are presented as averages ± SEM of four independent biological replicates per condition; p values are calculated using unpaired two-tailed Student's t test; ∗∗∗p < 0.001.
Sox100B Is Required for Terminal Differentiation but Dispensable for ISC Proliferation
To investigate the function of Sox100B in ISCs and EBs, we generated Sox100B deletion alleles using the CRISPR/Cas9 method (Gratz et al., 2014). We designed two gRNAs targeting the third exon of Sox100B and isolated two independent deletion alleles, Sox100Bd1 and Sox100Bd2 (Figure S1B). Both alleles were confirmed by sequencing the Sox100B locus, which revealed that 62% and 65% of the endogenous Sox100B coding sequence were deleted in the Sox100Bd1 and Sox100Bd2 alleles, respectively. Both Sox100Bd1/d1 and Sox100Bd2/d2 homozygous mutants die as pharate adults, consistent with the adult-lethal phenotype for a previously reported Sox100B null allele which was generated by an imprecise excision using a P element insertion in the neighboring gene discs overgrown (dco) (Nanda et al., 2009). We found that both alleles are lethal in trans with the deficiency line Df(3R) tll-e that covers the entire Sox100B locus (Figure S1C) strongly suggesting that the lethality of Sox100Bd1/d1 and Sox100Bd2/d2 homozygous mutants is not due to off-target mutations induced by the gRNAs but instead due to the disruption of endogenous Sox100B protein and function. To validate the lack of Sox100B protein expression in these mutants, we stained larval testes where pigment cells express high levels of Sox100B (Nanda et al., 2009) and found no Sox100B protein expression in Sox100Bd1/d1 homozygous mutants (Figure S1D, indicated by arrows). Altogether, these data establish that Sox100Bd1 and Sox100Bd2 are null or at least strong loss-of-function alleles.
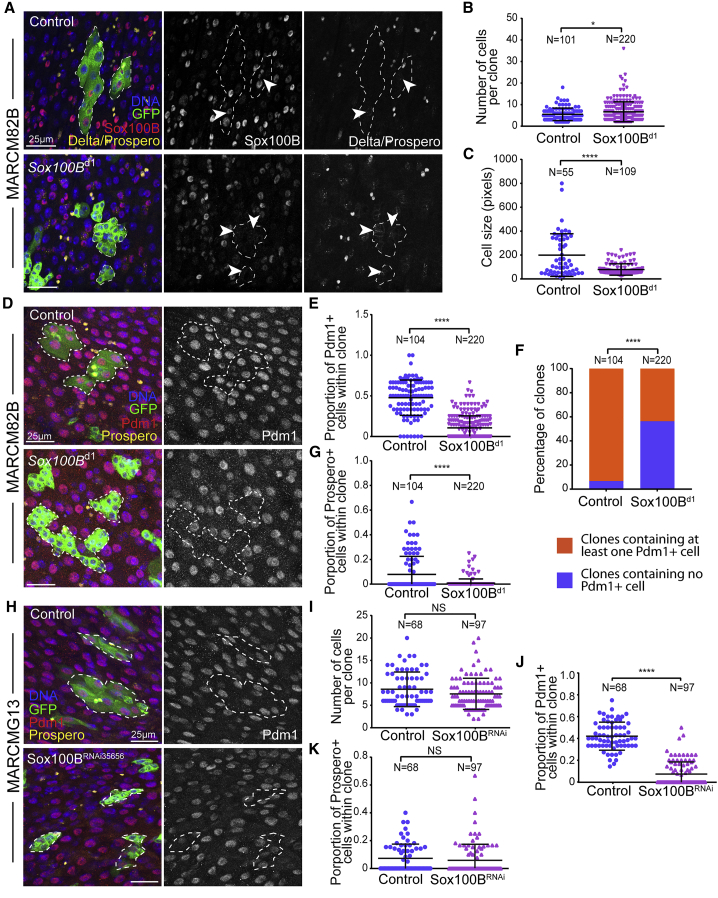
To characterize the role of Sox100B in the adult intestine, we first generated Sox100Bd1 homozygous mutant clones using the Mosaic Analysis with a Repressible Cell Marker (MARCM)method (Lee and Luo, 1999). This method allows to permanently label individual ISCs and their progeny with GFP expression and quantify cell number and cell identity within clones, to monitor ISC proliferation and differentiation. In wild-type control clones, Sox100B protein was detected only in small diploid cells, but not in polyploid ECs and Prospero-positive EEs (Figure 2A), consistent with our previously described ISC/EB-specific Sox100B expression pattern (Figures 1A and 1B). No Sox100B protein was detected in Sox100Bd1 homozygous mutant clones (Figure 2A), further validating that Sox100Bd1 allele disrupts endogenous Sox100B protein expression.
Figure 2.
Sox100B Is Required for Proper ISC Differentiation
(A and B) Representative confocal images of 7-day-old MARCM clones in the adult posterior midgut showing that Sox100Bd1 homozygous mutant ISCs maintain Delta expression––indicated by arrowheads in (A)––and the ability to form multicellular clones. Note that absence of Sox100B antibody staining in the Sox100Bd1 homozygous mutant clones further validates the mutant allele. Number of cells per clone is plotted in (B) showing that Sox100B is not essential for ISC proliferation. In (A) Delta and Prospero staining are separated by cellular localization (Delta, membrane staining; Prospero, nuclear staining).
(C) Cell size distribution in Sox100Bd1 homozygous mutant clones is affected compared with control clones. Cell shape within clones is outlined by membrane Arm staining and sizes of individual cell represented by pixels in the outlined area is plotted.
(D–G) Clonal analysis of Sox100Bd1 mutant 7-day-old MARCM clones using differentiation markers shows that proper ISC differentiation to ECs and EEs is impaired. Representative confocal images are shown in (D). Numbers of Pdm1-positive ECs and Prospero-positive EEs are counted in individual clones and the ratios to total cell number in each clone are plotted in (E) and (G), respectively. Distributions of total clones categorized by whether containing Pdm1-positive ECs in control and Sox100Bd1 mutant clones is shown in (F).
(H–K) Sox100BRNAi-expressing 5-day-old MARCM clones shows normal clone size (I) but impaired EC differentiation (J), confirming that Sox100B is not required for ISC proliferation but is required for EC differentiation. EE differentiation appears to be not affected in Sox100BRNAi-expressing MARCM clones (K).
In (A), (D), and (H), GFP expression labels MARCM clones. Scale bars, 25 μm. In (B), (C), (E), (G), and (I)–(K), values are presented as averages ± SEM; all p values are calculated using unpaired two-tailed Student's t test; ∗p < 0.05, ∗∗∗∗p < 0.0001. In (F), the p value is calculated using the chi-squared test (p < 10−16).
We then asked whether Sox100B is required for ISC maintenance or proliferation. We observed that strong Delta-expressing ISCs were present in Sox100Bd1 mutant clones 9 days after clone induction (Figure 2A, indicated by arrowhead), showing that Sox100B is not required for ISC maintenance. Importantly, Sox100Bd1 homozygous mutant clones contain multiple cells and grow over time to a slightly larger clone size compared with controls (control: 5.5 ± 0.3, N = 104; Sox100Bd1/d1: 6.7 ± 0.3, N = 200, Figures 2A and 2B), indicating that Sox100B is not required for ISC proliferation. We did, however, notice that most Sox100Bd1 clones do not contain large cells with large nuclei and strong DNA staining (Figure 2A), a typical feature of the fully differentiated large polyploid ECs. We thus quantified the size of cells within clones, using Armadillo staining to outline cell membranes (Figure S2A) and found that cells in Sox100Bd1 clones show a significantly reduced size distribution compared with control clones (Figure 2C), suggesting that proper EC differentiation may be disrupted in Sox100Bd1 mutant clones. Fully differentiated ECs and EEs are identified by their expression of transcription factors Pdm1 and Prospero, respectively. As expected, nearly 50% of cells in control clones show strong Pdm1 expression, while most cells in Sox100Bd1 clones are negative for Pdm1 staining (Figures 2D and 2E). Moreover, almost 60% of Sox100Bd1 clones do not contain any Pdm1-positive ECs (only 8% in control clones) (Figure 2F), demonstrating that Sox100B is required for proper EC differentiation. We also quantified the proportion of Prospero-positive EEs in these clones and found that the number of EEs within Sox100Bd1 mutant clones is slightly reduced compared with controls (Figure 2G). To support the defects observed in Sox100Bd1 mutant lineages, we generated MARCM clones expressing the Sox100BRNAi35656 construct, which strongly suppresses Sox100B expression (Figures 1C, 1D, and 2H). We found that knocking down Sox100B does not affect clone size distribution (Figure 2I) but causes a strong reduction in the number of Pdm1-positive ECs (Figures 2H and 2J), confirming that Sox100B is required for EC differentiation but does not significantly affect ISC proliferation. Of note, we observed no significant difference in Prospero-positive EEs in these conditions (Figure 2K).
Because of the observed differentiation defect, we asked whether EBs are properly specified and whether the activity of the Notch signaling pathway, a known driver of EB differentiation, is affected in EBs by the loss or reduction of Sox100B. However, we found no change in expression of the Notch reporter GBE-Su(H)-LacZ reporter in the intestine when Sox100BRNAi35656 is expressed in ISCs and EBs, using the esgGal4ts driver (Figure S2E). We also show that comparable numbers of GBE-Su(H)-LacZ-positive EBs can be detected in wild-type or Sox100B null MARCM clones (Figure S2F). These data support the notion that Sox100B is required for EB differentiation, after their commitment to the EC lineage and independently of the Notch signaling pathway.
We next investigated further the effect of Sox100B on ISC proliferation. To this end, we complemented our clonal analysis by knocking down Sox100B specifically in ISCs using the esgGal4ts; GBEGal80 (ISC-Gal4ts) driver. Confirming our clone size analysis, we found that ISC proliferation was not inhibited when Sox100B was knocked down, but rather we observed overall increased ISC proliferation possibly due to feedback regulation to ISCs when proper differentiation is affected (Figure S2B). Similar results were observed when knocking down Sox100B in ISCs and EBs using the esgGal4ts driver (Figure S2C). In addition, we found that Sox100B was also not required for ISC proliferation when flies were challenged with the chemical stressor DSS (dextran sulfate sodium), a treatment that strongly induces ISC proliferation (Figure S2D).
Finally, we recently found that overexpression of the transcription factor zfh2 specifically in EBs is sufficient to drive their activation and growth (Rojas Villa et al., 2019). Further confirming that Sox100B is essential for EB early differentiation, knocking down Sox100B blocks zfh2-mediated EB growth (Figure S3). In addition, we found that long-term knockdown of Sox100B in EBs does not cause uncontrolled ISC proliferation (Figure S2G), as opposed to Sox21a knockdown, which causes accumulation of activated and tumorigenic EBs (Chen et al., 2016, Zhai et al., 2015).
Overall, our data establish that Sox100B is not required for ISC maintenance or proliferation but is essential for proper early cell differentiation in the EC lineage.
Sox100B Is Required for Sox21a Expression in Both ISCs and EBs
Several signaling pathways and transcription factors involved in the EC differentiation process have been identified, including Delta/Notch (Bardin et al., 2010, Kapuria et al., 2012, Ohlstein and Spradling, 2007), JAK/STAT92E (Beebe et al., 2010, Jiang et al., 2009), Escargot (Korzelius et al., 2014, Loza-Coll et al., 2014), Sox21a (Chen et al., 2016, Zhai et al., 2015, Zhai et al., 2017), GATAe (Okumura et al., 2016), and Pdm1 (Korzelius et al., 2014, Tang et al., 2018). The functional requirement of Sox21a in EC differentiation and the ISC/EB-specific Sox21a expression pattern prompted us to test whether Sox21a expression is affected in the Sox100Bd1 mutant clones. Interestingly, we found that cells in the Sox100Bd1 mutant clones show reduced Sox21a protein expression compared with neighboring Sox21a-positive non-clonal cells (Figures 3A and 3B). Consistent with this observation, we performed a blind analysis based on Sox21a antibody staining and found that Sox21a expression was strongly reduced in ISCs/EBs when Sox100B was knocked down using the esgGal4ts driver (Figures 3C and 3D). Importantly, Sox100Bd1 mutant cells and Sox100BRNAi-expressing cells showed strong GFP expression (Figures 3A and 3C), which argues against the possibility that the reduced Sox21a expression we observed is due to a global reduction of gene expression. To examine the requirement of Sox100B for Sox21a protein expression in a cell-type-specific manner, we quantified the Sox21a staining intensity specifically in Sox100BRNAi-expressing ISCs and in Sox100BRNAi-expressing EBs and found reduced Sox21a levels in both ISCs and EBs compared with control cells (Figures 3E and 3F). In addition, we found that Sox21a mRNA expression in the intestine is strongly reduced when Sox100BRNAi is expressed in all ISCs/EBs (Figure 3G). Noteworthy, escargot and Delta mRNA expression levels were induced under the same condition, confirming that the reduction of Sox21a mRNA expression is not due to premature loss of progenitor cells in the epithelium; rather, suggesting that Sox100BRNAi expression causes a moderate increase in the number of progenitor cells, ISCs and EBs, with low Sox21a levels (Figure 3G).
Figure 3.
Sox100B Is Required for Sox21a Expression in ISCs and EBs
(A and B) Representative confocal images of 9-day-old MARCM clones showing that Sox21a expression is reduced in Sox100Bd1 homozygous mutant clones. Expression level of Sox21a is quantified based on Sox21a antibody staining and relative Sox21a intensity is calculated by comparing Sox21a intensity within Sox100Bd1 MARCM clones to that of the nearest Sox21a-positive cell outside clones, and quantification is shown in (B). In (A), Delta and Prospero staining are separated by cellular localization (Delta, membrane staining; Prospero, nuclear staining).
(C and D) Representative images of Sox21a antibody staining in wild-type- and Sox100BRNAi- expressing posterior midguts, illustrating the reduced Sox21a expression after 7day Sox100B knockdown. Category scoring of endogenous Sox21a expression is performed and quantification is shown in (D).
(E and F) Sox21a intensity quantification in individual cells specifically in ISCs (E) and EBs (F) confirms that Sox100B is required for Sox21a expression in both ISCs and EBs.
(G) qRT-PCR data demonstrates that Sox21a mRNA expression is strongly reduced in the intestine, while escargot and Delta mRNA expression are slightly induced after ISC/EB-specific Sox100B knockdown for 7 days. All mRNA expression is normalized to actin5c expression. Four independent biological replicates were analyzed per condition.
In (D), p values are calculated using Fisher's exact test (p < 10−5 versus control); the number of guts scored for each condition is indicated above the bar. In (B) and (E)–(G), values are presented as averages ± SEM; p values are calculated using unpaired two-tailed Student's t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We have previously shown that Sox21a expression is induced in ISCs and EBs in response to tissue damage induced by feeding flies with chemical stressors DSS or reactive oxygen species-generating compound Paraquat (Meng and Biteau, 2015). We therefore asked whether Sox100B is required for stress-induced Sox21a expression. Indeed, we found that the induction of Sox21a by Paraquat treatment was strongly impaired when Sox100B was knocked down in ISCs and EBs (Figure S4A). Furthermore, activation of stress-sensing signaling pathways, such as Jun-N-terminal kinase (JNK) and epidermal growth factor receptor (EGFR)/Ras is sufficient to induce Sox21a expression in the absence of environmental stressors (Meng and Biteau, 2015, Zhai et al., 2017). Similarly, we found that JNKK/Hep- and RasV12-induced Sox21a expression was strongly impaired when Sox100BRNAi was also expressed in esg-positive cells (Figure S4B).
Altogether, our data demonstrate that Sox100B is required for optimal Sox21a expression in both ISCs and EBs under homeostatic conditions or in response to stress or tissue injury. To further confirm that Sox100B is upstream of Sox21a, we asked whether, conversely, manipulating Sox21a may affect Sox100B. We found that Sox100B expression is not reduced, but rather slightly induced, in the intestine of Sox21a homozygous null mutant animals (Figure S4C). This supports the notion that Sox100B is upstream of Sox21a in the EB differentiation process.
Sox100B Is Necessary and Sufficient for the Activity of the Sox21a First-Intron Enhancer
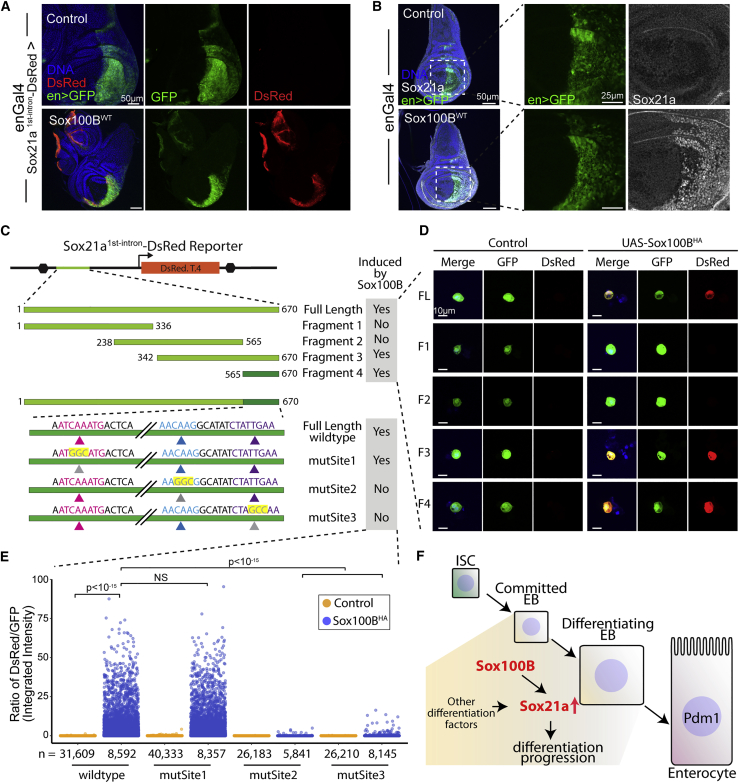
Our results support the notion that Sox21a is transcriptional target gene of Sox100B. Therefore, we examined the Sox21a genomic locus to identify potential transcriptional enhancers that could be directly regulated by Sox100B. Although Sox21a mRNA and protein have been shown to be specifically expressed in intestinal progenitors (Chen et al., 2016, Meng and Biteau, 2015), the molecular mechanism underlying this ISC/EB-specific expression pattern remains unclear. By screening potential enhancers and generating in vivo reporters (Figure 4A), we identified that combining the first intron of Sox21a with a minimal promoter is sufficient to drive the expression of a dsRed reporter gene in an ISC/EB-specific expression pattern in the adult intestine. In the intestine of animals carrying this reporter, dsRed expression overlaps exactly with esg-driven GFP expression (153 Sox100B-positive cells among 153 esg-positive cells, including 65/65 Delta-positive ISCs, counted across 4 guts) (Figures 4B and 4C). Of note, dsRed expression is undetectable in polyploid ECs or Prospero-positive EEs (Figure 4D).
Figure 4.
Characterization of a Sox100B Activity Reporter
(A) Schematic diagram showing the generation of Sox21a1st intron dsRed reporter line. The entire first intron of Sox21a (shown in green) was cloned into dsRed reporter construct to drive dsRed expression in vivo.
(B and C) Representative confocal images of posterior midgut showing that Sox21a1st intron dsRed expression overlaps with esgGal4-driven GFP expression. Zoom in images of the outlined region is shown in (C).
(D) Representative confocal images of posterior midguts illustrating that Sox21a1st intron dsRed reporter expression is almost abolished in 5 day Sox100BRNAi-expressing ISC/EBs. Delta and Prospero staining are separated by cellular localization (Delta, membrane staining; Prospero, nuclear staining).
(E) Quantification of a blind scoring experiment based on dsRed intensity. The number of guts scored for each condition is indicated above the bar, and p values are calculated using Fisher's exact test (p < 10−5 versus control). ∗∗∗∗p < 0.0001.
(F) Representative confocal images of L3 stage larval testes showing that Sox21a1st intron dsRed expression completely overlaps with Sox100B protein expression and is Sox100B dependent in the testis pigment cells. The expression of Sox100B and Sox21a1st intron dsRed are both absent in Sox100Bd1 homozygous null mutant testes.
Interestingly, we found that the Sox21a first-intron dsRed reporter is expressed in other tissues where Sox100B has been reported to be expressed (Loh and Russell, 2000, Nanda et al., 2009). For example, we observed strong dsRed expression in larval testis pigment cells (Figure 4F) and larval hindgut boundary cells (Figure S5C), suggesting that the Sox21a first intron could serve as a reporter of Sox100B activity. To test this idea, we first examined whether Sox21a first-intron dsRed reporter expression is dependent on Sox100B. We found that dsRed reporter expression is completely absent in the intestinal epithelium when Sox100B was knocked down in ISCs and EBs using the esgGal4ts driver (Figures 4D and 4E). Similarly, the reporter activity is lost in larval intestinal progenitors and larval testes in Sox100Bd1/d1 homozygous mutants (Figures 4F and S5A).
In addition to Sox100B, we and others have previously shown that the transcription factors Fos and Stat92E are involved in regulating Sox21a expression (Meng and Biteau, 2015, Zhai et al., 2015). We found that Sox21a first-intron dsRed reporter expression is not affected when Fos or Stat92E was knocked down (Figure S5D). In addition, we did not find any evidence that Sox21a itself is required, as reporter expression was not affected when Sox21a was knocked down (Figure S5D). Thus, our data strongly support the idea that this Sox21a first-intron dsRed reporter is specifically under the control of Sox100B.
Next, we asked whether Sox100B is sufficient to induce ectopic Sox21a first-intron dsRed reporter expression. To this end, we expressed Sox100B using the engrailed-Gal4 driver and a UAS-Sox100B construct that allows strong overexpression (Figure S5B). The Sox21a first-intron dsRed reporter is normally not detected in larval epidermis, hindgut ECs, or wing imaginal discs (Figures 5A and S5C). However, we found strong dsRed expression in the engrailed-positive domain in all these tissues when Sox100B is ectopically expressed (Figures 5A and S5C), demonstrating that Sox100B is sufficient to induce reporter activity. Remarkably, ectopic Sox21a protein expression was also detected in the Sox100B-expressing posterior domain of the larval wing imaginal disc (Figure 5B), strongly supporting the hypothesis that Sox21a is a direct transcriptional target gene of Sox100B.
Figure 5.
Sox100B Directly Regulates the Activity of the Sox21a Intronic Enhancer
(A) Representative images showing that 24 h Sox100B overexpression is sufficient to induce ectopic Sox21a1st intron dsRed in L3 stage larval wing imaginal disc.
(B) Representative confocal images showing that 24 h Sox100B overexpression is sufficient to induce ectopic Sox21a protein expression in larval wing imaginal disc.
(C–E) (C) Schematic diagram representing the experimental design to identify putative Sox100B binding sites in the first intron of Sox21a using Drosophila S2 cells. A series of reporter constructs using different fragments of the full-length first intron (fragments 1–4) identified a 106 bp region that is necessary and sufficient to mediate Sox100B-induced dsRed reporter expression. Then site-directed mutagenesis of three candidate sites (sites 1–3) in the mapped region were constructed and tested, revealing that both site 2 and site 3 are necessary for reporter induction. Representative images of S2 carrying the different fragments are shown in (D). Quantification of the dsRed signal, normalized to GFP expression, is shown in (E) for the full-length fragments carrying individual mutations of the predicted Sox binding sites. In (E), n represents the number of transfected cells (GFP-positive cells) analyzed; p values were calculated using unpaired two-tailed Student's t test.
(F) A proposed scheme depicting the role of Sox100B-Sox21a transcriptional cascade in the process of EB to EC differentiation. Sox100B is required for proper EB differentiation progression by regulating Sox21a and potentially other differentiation factors.
To identify potential Sox100B binding sites within the Sox21a first intron, we conducted a series of experiments in Drosophila S2 cells where the Sox21a first-intron dsRed reporter is not expressed but can be induced by co-transfection of an HA-tagged Sox100B construct (Figures 5C–5E). We generated multiple reporter constructs using different fragments of the first intron and examined individually their inducibility by Sox100B. This assay identified a 106 bp region in the Sox21a first intron that is necessary and sufficient to mediate Sox100B-overexpression-induced reporter expression in S2 cells (Figures 5C and 5D). Based on the consensus binding motif for Sox proteins 5ʹ-(A/T) (A/T)CAA(A/T)G-3ʹ (Harvey et al., 1994), we identified three putative Sox100B binding sites within this fragment. Thus, we performed site-directed mutagenesis of these sites individually (CAA mutated to GGC) and found that both the second and third binding sites are required for optimal reporter induction (Figures 5C and 5E). Interestingly, these two binding sites are physically spaced close to each other in a manner that is highly similar to binding sites observed in mammalian Sox9 target genes (Kamachi and Kondoh, 2013). Altogether, our in vivo and in vitro data strongly support our conclusion that Sox21a is a Sox100B target gene, at least partially regulated by direct binding of Sox100B to the 3′ region of its first intron.
Discussion
In this work, we show that the Sox E family transcription factor Sox100B is expressed in ISCs and EBs in the adult Drosophila intestine, consistent with a recent report using a GFP-tagged genomic BAC construct for Sox100B (Doupe et al., 2018). Recently, another Sox family transcription factor Sox21a has already been shown to be expressed specifically in ISCs and EBs (Chen et al., 2016, Meng and Biteau, 2015, Zhai et al., 2015). These two Sox transcription factors do not simply share an overlapping expression pattern, as demonstrated by our work, but Sox100B is required for proper Sox21a expression in both cell types. Our data altogether led us to propose a transcriptional cascade in which Sox21a is a Sox100B target gene. The overlapping expression pattern is not surprising, given the fact that Sox factors are commonly co-expressed in several tissues. For example, Drosophila B Group Sox factors SoxNeuro and Dichaete are co-expressed in part of the neuroectoderm during early development of the CNS (Overton et al., 2002). However, we present evidence showing a direct transcriptional regulation between different Sox factors in the somatic stem cell lineage in Drosophila.
Previous studies by us and others have shown that transcription factors, such as Stat92E and AP-1 factor Fos are involved in regulating Sox21a expression at basal and stress conditions (Chen et al., 2016, Meng and Biteau, 2015, Zhai et al., 2015). At basal condition, Stat92E has been implicated in regulating Sox21a expression, and the second intron of Sox21a alone is sufficient to drive gene expression in ISCs and EBs (Zhai et al., 2015); however, whether this intronic enhancer is directly regulated by Stat92E has not been addressed. Here, we identified that the first intron of Sox21a is also sufficient to direct endogenous Sox21a expression pattern. Interestingly, this intronic enhancer is not dependent on Stat92E, suggesting that parallel signal inputs from both Sox21a introns act together to robustly control the ISC/EB-specific Sox21a expression pattern. In support of this model, we found that the first intronic enhancer is specifically responsive to Sox100B. This model is consistent with our observation that Sox21a expression is strongly reduced but not absent in Sox100B mutant clones, suggesting that parallel inputs mediated by the second intron via other factors, such as Stat92E contribute to Sox21a expression even in the absence of Sox100B. This model also partially accounts for the notion that depleting Sox100B does not inhibit ISC proliferation while depleting Sox21a strongly inhibits ISC proliferation (Meng and Biteau, 2015), and it is likely that residual Sox21a expression in Sox100B loss-of-function conditions allows ISC proliferation. In response to stress, Sox21a expression is strongly induced which involves multiple stress-sensing signaling pathways and factors, such as JNK, EGFR, AP-1 factor Fos, and Stat92E (Meng and Biteau, 2015, Zhai et al., 2015, Zhai et al., 2017). We showed here that Sox100B is required for stress-, JNK-, and RasV12-induced Sox21a expression but that Sox100B overexpression in ISCs and EBs is not sufficient to induce Sox21a expression. Our data suggest a model where Sox100B provides cell-type specificity and allows Sox21a expression in intestinal progenitors, ISCs and EBs, while other pathways control Sox21a induction during the differentiation process or in response to stresses (Figure 5F). This raises an interesting question of whether Sox100B directly interacts with stress-sensing and differentiation pathways to ensure that Sox21a expression is stress inducible and gradually increases during differentiation. As an example of such a mechanism, mammalian Sox9 has been shown to physically interact with AP-1 factors to co-activate target gene expression in developing chondrocytes (He et al., 2016). We anticipate that the identification of Sox100B-interacting cofactors will provide a better view regarding the mechanism(s) by which Sox100B cooperates with stress-sensing signaling and other differentiation pathways to precisely adapt stem cell activities to tissue demands.
The last decade has witnessed significant advances in understanding the mechanisms by which ISC activities are regulated in response to infection, tissue injury, and during aging, with a strong focus on ISC proliferation (Guo et al., 2016, Jiang et al., 2016, Li and Jasper, 2016, Liu and Jin, 2017). In contrast, a well-defined progressive differentiation process is still inadequately understood. Sox100B has been recently implicated in regulating acute gut regeneration in response to pathogenic bacteria Pseudomonas entomophila (Lan et al., 2018); however, the exact role of Sox100B in this process has not been characterized. Here we found that Sox100B is functionally required for robust stem cell differentiation, as a strong reduction in the EC lineage and a reduction to a lesser extent in the EE lineage were observed in the Sox100B mutant clones. We further showed that the expression of a critical differentiation regulator Sox21a is reduced in Sox100B mutants, establishing a transcriptional cascade during the ISC differentiation process. However, in preliminary experiments, we found that Sox21a overexpression alone is not sufficient to rescue the differentiation defects of the Sox100B mutants (data not shown), suggesting that other Sox100B targets, in addition to Sox21a, are required for proper differentiation. Further identification of Sox100B transcriptional target genes is needed to fully decipher the role of Sox100B during the differentiation process.
Interestingly, Sox9, the mammalian counterpart of Drosophila Sox100B, is expressed in ISCs and differentiated Paneth cells at the bottom of the crypts (Blache et al., 2004, Formeister et al., 2009). In Sox9 knockout intestine, Paneth cells are found missing and crypt hyperplasia is widely observed (Bastide et al., 2007, Mori-Akiyama et al., 2007). We also observed a mild but significant increase of ISC proliferation in Sox100B loss-of-function conditions, which could be due to disruption of normal differentiation process, since it has been well documented that blocking differentiation process by genetically manipulating Notch and Sox21a causes strong pro-mitotic feedback regulation on ISC proliferation (Chen et al., 2016, Patel et al., 2015, Zhai et al., 2015). In addition to Sox9, several other Sox family factors are expressed in the mammalian intestine, and their expression pattern and function remain elusive (Blache et al., 2004). The transcriptional regulatory pattern between different Sox factors in the intestine could be conserved from flies to mammals, and such possibility needs to be further examined.
Experimental Procedures
Drosophila Stocks and Culture
The following strains were obtained from the Bloomington Drosophila Stock Center: w1118, FRT82B, FRTG13, enGal4,UAS-EGFP (Bl25752), Df(3R) tll-e/TM6B (Bl5415), UAS-RasV12, UAS-Sox100BRNAi(TRiP) (Bl57417 and Bl35656), UAS-Sox21aRNAi(TriP) (Bl31902 and Bl53991), UAS-Stat92ERNAi(TRiP) (Bl35600 and Bl31318), UAS-zfh2 (Bl56545), and Sox21aJC2 null mutant (Bl68154). UAS-FosRNAi (VDRC10813) were obtained from the Vienna Drosophila RNAi Center. The line esgGal4NP5130 was provided by S. Hayashi, esgGal4ts,Su(H)GBEGal80 was provided by H. Jasper, DeltaGal4 and Su(H)GBEGal4 by S. Hou, Su(H)GBELacZ by S. Brand, and UAS-Hep by M. Mlodzik.
All flies were raised on standard yeast and molasses-based food, at 25°C and 65% humidity, on a 12 h light/dark cycle, unless otherwise indicated.
Conditional Expression of UAS-Linked Transgenes
The esgGal4, DeltaGal4, and Su(H)GBEGal4 drivers were combined with a ubiquitously expressed temperature-sensitive Gal80 inhibitor (tubGal80ts). Crosses and flies were kept at 18°C (permissive temperature), 3- to 5-day-old females were then shifted to 29°C for the indicated periods of time to allow expression of the transgenes before analysis or additional treatment. For these experiments, control animals are progeny of crosses with w1118 flies.
Generation of Sox100B Deletion Mutant and Overexpression Construct
To generate Sox100B deletion allele, the following two gRNAs are designed using the CRISPR Optimal Target Finder with maximum stringency (http://tools.flycrispr.molbio.wisc.edu/targetFinder/) (Gratz et al., 2014). gRNA1 forward: CTTCGGTGCGGGGTCACCCAGTGT, gRNA1 reverse: AAACACACTGGGTGACCCCGCACC; gRNA2 forward: CTTCGATGACGTTATCCGCCACTA, gRNA2 reverse: AAACTAGTGGCGGATAACGTCATC. A mix of two gRNAs was injected into the following fly line: y sc v; attP40{nos-Cas9}/Cyo in the lab to generate deletion mutant alleles. Two independent mutant alleles were confirmed by sequencing the Sox100B locus and were named Sox100Bd1 and Sox100Bd2 in this study (see Figure S1B).
The UAS-Sox100BWT and UAS-Sox100BHA plasmids were constructed by amplifying the entire coding sequence of Sox100B using the following primers forward: 5′-AAAGCGGCCGCATGAGTGACAGTTCCAGCTC-3′ and Reverse (WT): 5′-AAATCTAGATTAGGGATTGACATAAGTGT-3′ or Reverse (HA) 5′-AAATCTAGAGGGATTGACATAAGTGTAAG-3′ and cloning these fragments in the pUAST or pUAST-HA vectors respectively, using NotI and XbaI sites. Vectors were verified by sequencing, and transgenic animals carrying the UAS-Sox100B construct were generated by Genetic Services using a standard P element transgenesis protocol. An insertion on the second chromosome was used for all the presented experiments.
Generation of Sox21a First-Intron dsRed In Vivo Reporter Line
The following primers were used to amplify the whole first intron of Sox21a: forward: 5′-AAAGCATGCAGACAATTAATACAGGTAAGCT-3′; reverse: 5′-AAACTCGAGCTCTGAAATGCCAACGGAAATG-3′, which were then cloned into pB-ARE-DsRedT.4 plasmid to replace the 4XARE sequence (Chatterjee and Bohmann, 2012). Transgenic flies were generated using the following fly line (y1w67c23;;P{CaryP} attP2) by BestGene.
Mosaic Analysis with a Repressible Cell Marker Clones
Positively marked clones were generated by somatic recombination using the following MARCM stocks: MARCM82B (hsFlp, UAS-GFP; tubGal4; FRT82B; tubGal80) and MARCMG13 (hsFlp, UAS-CD8GFP; FRTG13; tubGal80; tubGal4). Five- to 7-day-old mated female flies were heat shocked for 45–60 min at 37°C to induce somatic recombination. Flies were transferred to 25°C and clones were observed 7–9 days after induction. Only isolated ISC clones in the posterior midgut were included in our analysis. The number of clones analyzed, from at least four guts, is indicated in the figures. The different genetic conditions were compared using unpaired two-tailed Student's t test or Pearson's chi-squared test, as mentioned in the figure legend.
Analysis of Gene Expression in the Gut
Total RNA from eight dissected guts from young mated females was extracted using RNeasy Mini Kit (QIAGEN), according to manufacturer’s instructions. cDNA was synthesized using an oligo-dT primer. Real-time PCR was performed on a Bio-Rad iQ5 detection system using the following primers: Sox100B forward 5′-TCACCACTGCGGTTATGAAG-3′; Sox100B reverse 5′-GGGCTTCTTATCACTGTCCTTTA-3′; Sox21a forward 5′-GCCGAGTGGAAATTACTCACCGAA-3′, Sox21a reverse 5′-TGCGACGTGGTCGATACTTGTAGT-3′; Delta forward 5′-AGGCTTGTACTGCAACCAGGATCT-3′, Delta reverse 5′-TGAGCACTTTCTCCTCGCACATCT-3′; escargot forward 5′-GCCGCAGGATTTGTGCGTAAAGAA-3′, escargot reverse 5′-ATGACCCTGCTGATTGATGGTCCT-3′; actin5c forward 5′-CTCGCCACTTGCGTTTACAGT-3′, actin5c reverse 5′-TCCATATCGTCCCAGTTGGTC-3′. Relative expression was calculated using the ΔΔCt method and normalized to actin5c levels. All qPCR experiments were performed using four independent biological replicates. To determine significant differences in gene expression, p values were calculated using unpaired two-tailed Student's t test.
Phenotype Analysis
To score the intensity of Sox21a immunostaining in the midgut, individual intestines were mounted, randomized, and attributed blindly to one of the following categories––weak expression, normal expression, and high expression––based on Sox21a immunostaining. For scoring the intensity of Sox21a first-intron dsRed reporter, individual intestines were mounted, randomized, and attributed blindly to one of the following categories––weak/no expression and normal expression––based on dsRed intensity. Results of these scoring experiments were analyzed using RStudio and p values were calculated using Fischer's exact test. The number of intestines scored for each category is indicated in the figure.
S2 Cell Reporter Assay
To test Sox21a dsRed reporter inducibility by Sox100B, Drosophila S2 cells were transiently transfected with Sox21a dsRed reporter constructs, pActin-Gal4, pUAS-GFP, and either a pUAST for control condition or a pUAST-Sox100BHA for experimental condition using the Effectene Transfection Reagent (QIAGEN, cat. no. 301425) following the manufacturer's suggested protocol. GFP-positive cells were selected, GFP and dsRed expression were then examined and measured either by confocal microscopy or by a Celigo Cytometer. Reporter expression in individual cells was calculated as the ratio of dsRed integrated intensity to GFP integrated intensity and plotted using RStudio. For our cytometer-based quantification of reporter activity, significant differences were determined using an unpaired two-tailed Student's t test. Mutagenesis of individual putative Sox binding sites (CAA mutated to GGC) were designed and performed using a Q5 site-directed mutagenesis kit (NEB E0554S).
Lethality Assay
To examine the adult-lethal phenotype of Sox100B mutant alleles, Sox100Bd1 and Sox100Bd2 heterozygous mutant flies over TM6 were crossed to a deficiency line Df(3R) tll-e/TM6 in which the whole Sox100B coding sequence was deleted. One- to 2-day-old adult progenies from the above-mentioned crosses were collected, sorted based on their sex and genotype, and counted.
DSS and Paraquat Treatments
For all stress experiments, young mated females were cultured on standard food. Flies were starved for 6 h in empty vials and re-fed with a 5% sucrose (AMRESCO) solution with or without 5 mM Paraquat (Sigma-Aldrich) or 4% DSS (Sigma-Aldrich, 9–20 kDa). Flies were then dissected at the indicated time points for immunocytochemistry.
Immunocytochemistry and Microscopy
Intact fly intestines were dissected and fixed as described previously (Meng and Biteau, 2015). The Sox100B antibody was kindly provided by Steve Russell (1:1,000 dilution), and Pdm1 antibody was kindly provided by Xiaohang Yang and Cai Yu (1:1,000 dilution). Sox21a antibody was generated in the lab (1:5,000 dilution). The anti-Delta (C594.9B; 1:100 dilution), anti-Armadillo (N2 7A1; 1:100 dilution), anti-Prospero (MR1A; 1:250 dilution), anti-β-galactosidase (40-1a; 1:100 dilution) were obtained from the Developmental Studies Hybridoma Bank and the Anti-phospho-Histone H3 (06-570; 1:2,000 dilution) from Millipore. Fluorescent secondary antibodies were obtained from Jackson Immunoresearch. Hoechst 33258 was used to stain DNA.
Confocal images were collected using a Leica SP5 confocal system and processed using the Leica software, Fiji, and Adobe Photoshop CC.
For all quantifications of cell numbers, cell proportion or signal intensity, the data are represented as average ± SEM and p values are calculated using an unpaired two-tailed Student's t test unless stated otherwise.
Author Contributions
The project was designed by F.W.M and supervised by B.B. Experiments were performed and analyzed by F.W.M and S.R.V. F.W.M and B.B wrote the manuscript.
Acknowledgments
We thank Taylor McKenty for her technical assistance with the generation of Sox100Bd1 and Sox100Bd2 mutant alleles, Nicole Gorski and Sreejith Perinthottathil for their help with S2 cell reporter assay. This work was funded in part by a New Scholar in Aging Award from the Ellison Medical Foundation to B.B. (AG-NS-0990-13), the National Institute of General Medical Sciences (5R01GM108712 to B.B.) and the Goodman Dissertation Fellowship to F.M. (University of Rochester).
Published: February 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.01.003.
Supplemental Information
References
- Bardin A.J., Perdigoto C.N., Southall T.D., Brand A.H., Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide P., Darido C., Pannequin J., Kist R., Robine S., Marty-Double C., Bibeau F., Scherer G., Joubert D., Hollande F. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K., Lee W.C., Micchelli C.A. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C.E., Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Jasper H. Slit/robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 2014;7:1867–1875. doi: 10.1016/j.celrep.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J.N., Clevers H., Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N., Bohmann D. A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One. 2012;7:e34063. doi: 10.1371/journal.pone.0034063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu N., Huang H., Cai T., Xi R. A feedback amplification loop between stem cells and their progeny promotes tissue regeneration and tumorigenesis. Elife. 2016;5:e14330. doi: 10.7554/eLife.14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe D.P., Marshall O.J., Dayton H., Brand A.H., Perrimon N. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Proc. Natl. Acad. Sci. U S A. 2018;115:12218–12223. doi: 10.1073/pnas.1719169115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formeister E.J., Sionas A.L., Lorance D.K., Barkley C.L., Lee G.H., Magness S.T. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S.J., Ukken F.P., Rubinstein C.D., Thiede G., Donohue L.K., Cummings A.M., O'Connor-Giles K.M. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Lucchetta E., Rafel N., Ohlstein B. Maintenance of the adult Drosophila intestine: all roads lead to homeostasis. Curr. Opin. Genet. Dev. 2016;40:81–86. doi: 10.1016/j.gde.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey V.R., Lovell-Badge R., Goodfellow P.N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Ohba S., Hojo H., McMahon A.P. AP-1 family members act with Sox9 to promote chondrocyte hypertrophy. Development. 2016;143:3012–3023. doi: 10.1242/dev.134502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Tian A., Jiang J. Intestinal stem cell response to injury lessons from Drosophila. Cell Mol. Life Sci. 2016;73:3337–3349. doi: 10.1007/s00018-016-2235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- Kapuria S., Karpac J., Biteau B., Hwangbo D., Jasper H. Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLoS Genet. 2012;8:e1003045. doi: 10.1371/journal.pgen.1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzelius J., Naumann S.K., Loza-Coll M.A., Chan J.S., Dutta D., Oberheim J., Glasser C., Southall T.D., Brand A.H., Jones D.L. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014;33:2682–2967. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q., Cao M., Kollipara R.K., Rosa J.B., Kittler R., Jiang H. FoxA transcription factor Fork head maintains the intestinal stem/progenitor cell identities in Drosophila. Dev. Biol. 2018;433:324–343. doi: 10.1016/j.ydbio.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible neurotechnique cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Dumitriu B., Penzo-Mendez A., Han Y., Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Jasper H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis. Models. Mech. 2016;9:487–499. doi: 10.1242/dmm.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Jin L.H. Tissue-resident stem cell activity: a view from the adult Drosophila gastrointestinal tract. Cell Commun. Signal. 2017;15:33. doi: 10.1186/s12964-017-0184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh S.H.Y., Russell S. A Drosophila group E Sox gene is dynamically expressed in the embryonic alimentary canal. Mech. Dev. 2000;93:185–188. doi: 10.1016/s0925-4773(00)00258-6. [DOI] [PubMed] [Google Scholar]
- Loza-Coll M.A., Southall T.D., Sandall S.L., Brand A.H., Jones D.L. Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor Escargot. EMBO J. 2014;33:2983–2996. doi: 10.15252/embj.201489050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F.W., Biteau B. A sox transcription factor is a critical regulator of adult stem cell proliferation in the Drosophila intestine. Cell Rep. 2015;13:906–914. doi: 10.1016/j.celrep.2015.09.061. [DOI] [PubMed] [Google Scholar]
- Micchelli C.A., Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Jasper H., Lemaitre B. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics. 2018;210:357–396. doi: 10.1534/genetics.118.300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y., van den Born M., van Es J.H., Hamilton S.R., Adams H.P., Zhang J., Clevers H., de Crombrugghe B. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Nanda S., DeFalco T.J., Loh S.H., Phochanukul N., Camara N., Van Doren M., Russell S. Sox100B, a Drosophila group E Sox-domain gene, is required for somatic testis differentiation. Sex Dev. 2009;3:26–37. doi: 10.1159/000200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Okumura T., Takeda K., Kuchiki M., Akaishi M., Taniguchi K., Adachi-Yamada T. GATAe regulates intestinal stem cell maintenance and differentiation in Drosophila adult midgut. Dev. Biol. 2016;410:24–35. doi: 10.1016/j.ydbio.2015.12.017. [DOI] [PubMed] [Google Scholar]
- Overton P.M., Meadows L.A., Urban J., Russell S. Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during CNS development in Drosophila. Dev. Biol. 2002;129:4219–4228. doi: 10.1242/dev.129.18.4219. [DOI] [PubMed] [Google Scholar]
- Patel P.H., Dutta D., Edgar B.A. Niche appropriation by Drosophila intestinal stem cell tumours. Nat. Cell Biol. 2015;17:1182–1192. doi: 10.1038/ncb3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Villa S.E., Meng F.W., Biteau B. zfh2 controls progenitor cell activation and differentiation in the adult Drosophila intestinal absorptive lineage. PLoS Genet. 2019;15:e1008553. doi: 10.1371/journal.pgen.1008553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Z.Y., Yang W.X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015;94:547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Tang X., Zhao Y., Buchon N., Engstrom Y. The POU/Oct Transcription factor nubbin controls the balance of intestinal stem cell maintenance and differentiation by isoform-specific regulation. Stem Cell Reports. 2018;10:1565–1578. doi: 10.1016/j.stemcr.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Hou S.X. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142:644–653. doi: 10.1242/dev.113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Boquete J.P., Lemaitre B. A genetic framework controlling the differentiation of intestinal stem cells during regeneration in Drosophila. PLoS Genet. 2017;13:e1006854. doi: 10.1371/journal.pgen.1006854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Kondo S., Ha N., Boquete J.P., Brunner M., Ueda R., Lemaitre B. Accumulation of differentiating intestinal stem cell progenies drives tumorigenesis. Nat. Commun. 2015;6:10219. doi: 10.1038/ncomms10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.