Abstract
Pilocytic astrocytoma is a low-grade glial neoplasm of the central nervous system (CNS) that tends to occur in the pediatric population and less commonly presents in adults. Hereditary pilocytic astrocytoma is often associated with germline genetic alterations in the tumor suppressor NF1, the gene responsible for the syndrome neurofibromatosis type 1. Sporadic pilocytic astrocytoma frequently harbors somatic alterations in BRAF, with rare pilocytic astrocytomas containing alterations in FGFR1 and NTRK2. NTRK2 encodes for the protein tropomyosin receptor kinase B (TrkB), which is a neurotrophin receptor with high affinity for Brain-Derived Neurotrophic Factor (BDNF), and plays a role in several physiological functions of neurons, including cell survival and differentiation. In this report, we describe a novel PML-NTRK2 gene fusion occurring in an adult sporadic pilocytic astrocytoma and review the biology and implications of specific NTRK2 mutations occurring in CNS neoplasms.
1. Introduction
Pilocytic astrocytoma, World Health Organization (WHO) grade I, is the most common low-grade glial neoplasm found in the pediatric population and is infrequent in adults. Pilocytic astrocytoma can occur in the setting of hereditary disorders, such as neurofibromatosis type 1 (NF1), or sporadically in the absence of any tumor predisposition syndrome. The most common (observed in 15-20% overall) intracranial neoplasm in pediatric NF1 patients is the optic pathway glioma, which usually is classified as pilocytic astrocytoma and typically presents in children younger than 7 years of age [1–3]. NF1 results from germline mutations in the NF1 tumor suppressor gene, and pilocytic astrocytoma associated with NF1 additionally contain biallelic inactivation of NF1 and loss of expression of the NF1 protein product (neurofibromin) [4]. Sporadically occurring pilocytic astrocytoma frequently contains somatic alterations in the BRAF gene, which encodes for a serine/threonine kinase also involved in the RAS/MAPK/ERK signaling pathway [5]. Unlike the loss-of-function mutations found in NF1, BRAF alterations predominantly occur as an oncogenic gene fusion product, KIAA1549-BRAF [5]. Outside of the frequent alterations occurring in NF1 and BRAF genes, very rare genetic alterations in FGFR1 and NTRK2 have been reported [6]. To our knowledge, NTRK2 (the gene that encodes for the neurotrophin receptor TrkB) alterations have been described in only eight cases of low-grade circumscribed gliomas, with half (n = 4) being associated with pilocytic astrocytomas, which are resultant from gene fusions with partners including QKI, NACC2, and KANK1 (Table 1) [6, 7]. Additionally, a single case of a low-grade diffuse glioma has been reported with an NTRK2 gene fusion (Table 1) [8]. NTRK2 fusion partners in nonpilocytic astrocytoma low-grade gliomas include QKI, SLMAP, TLE4, NAV1, and BCR [8–11]. Here, we describe a patient where a novel PML-NTRK2 gene fusion was identified in an adult sporadic pilocytic astrocytoma. The PML gene itself is a transcription factor that is associated with promyelocytic leukemia, and such alterations have not been reported in pilocytic astrocytoma. In addition to expanding the landscape of NTRK2 mutations occurring in the setting of pilocytic astrocytoma, we review the biological and therapeutic implications of altered TrkB signaling in low-grade glial neoplasms.
Table 1.
Summary of reported NTRK2 gene fusion alterations in low-grade neuroepithelial tumors. PA = pilocytic astrocytoma; GG = ganglioglioma; DNT = dysembryoplastic neuroepithelial tumor; LGG-NOS = low-grade glioma not otherwise specified.
| 5′ fusion gene | 3′ fusion gene | Number of cases | Histology | Reference |
|---|---|---|---|---|
| QKI | NTRK2 | 2 | PA | Jones DT, 2013 |
| NACC2 | NTRK2 | 1 | PA | Jones DT, 2013 |
| KANK1 | NTRK2 | 1 | PA | Lopez GY, 2019 |
| PML | NTRK2 | 1 | PA | Current patient |
| SLMAP | NTRK2 | 1 | GG | Qaddoumi I, 2016 |
| TLE4 | NTRK2 | 1 | GG | Prabhkaran N, 2018 |
| NAV1 | NTRK2 | 1 | DNT | Qaddoumi I, 2016 |
| QKI | NTRK2 | 1 | LGG-NOS | Solomon JP, 2019 |
| BCR | NTRK2 | 1 | Diffuse LGG-NOS | Jones KA, 2019 |
2. Case Presentation
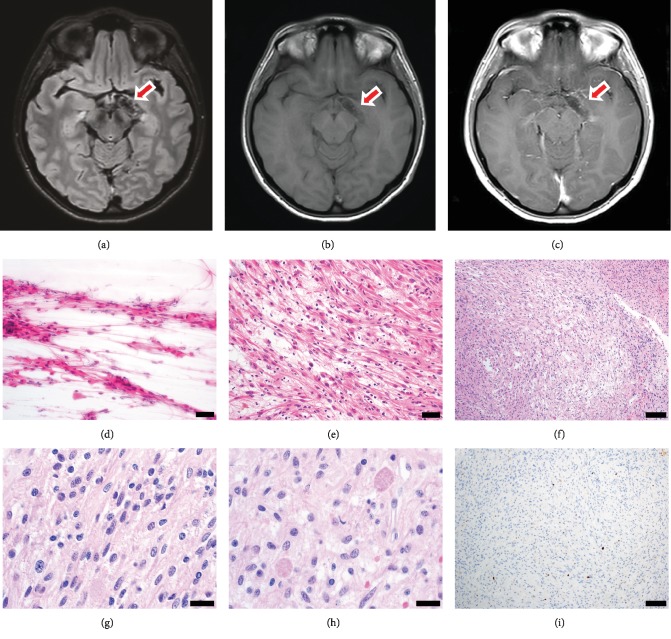
The patient is a 33-year-old woman with past medical history of hypothyroidism and gastroesophageal reflux disease, who has had chronic headaches refractory to medical management. A maxillofacial computed tomography (CT) evaluation for possible sinusitis noted an abnormal curvilinear “calcification” involving the left suprasellar cistern and mesial temporal region. Follow-up brain magnetic resonance imaging (MRI) demonstrated an abnormal T1-weighted hypointense, T2-weighted/FLAIR hyperintense, nonenhancing lesion in the area of the left suprasellar cistern and mesial temporal lobe, with mass effect over the optic tract (Figures 1(a)–1(c)). The radiographic impression included a wide differential including vascular malformation or neoplasm. She subsequently underwent left frontotemporal craniotomy and orbital osteotomy for excision of the lesion.
Figure 1.
Pilocytic astrocytoma radiographic and histopathologic features. Brain magnetic resonance imaging demonstrates T2/FLAIR curvilinear hyperintensity (a) and T1 hypointensity (b) without contrast enhancement (c). Hematoxylin and eosin-stained intraoperative cytological smear preparation (d) and frozen section (e) demonstrating an astrocytoma with thin elongated piloid (“hair-like”) cellular processes (scale bar = 50 microns). (f) Permanent sections showed a piloid astrocytoma with compact areas and adjacent slightly looser areas (scale bar = 100 microns). (g) The astrocytic nuclei have mild pleomorphism and are elongated, slightly irregular, and hyperchromatic (scale bar = 20 microns). (h) Abundant eosinophilic granular bodies are found throughout the astrocytoma (scale bar = 20 microns). (i) The Ki-67 proliferative index is low (<1%) overall (scale bar = 100 microns).
Hematoxylin and eosin- (H&E-) stained slides of intraoperative cytologic smear preparation and frozen section tissue demonstrated a glial neoplasm with piloid features (Figures 1(d) and 1(e)). Permanent H&E-stained sections showed multiple fragments of a solid-growing, biphasic, glial neoplasm characterized by compact piloid areas comprised of bipolar cells with bland, ovoid to elongate nuclei adjacent to more loosely arranged areas comprised of cells with angular and hyperchromatic nuclei in a fibrillary background (Figures 1(f)–1(g)). Eosinophilic granular bodies were frequently noted throughout the neoplasm, with occasional scattered Rosenthal fibers (Figure 1(h)). There were occasional hyalinized vessels as well as vessels cuffed by small lymphocytes. There was no necrosis. Abundant fresh hemorrhage and clusters of hemosiderin-laden macrophages were present. Mitotic activity was not identified, and the Ki-67 proliferative index was less than 1% overall (Figure 1(i)). Neurofilament highlighted a predominantly solid growth pattern (not shown). The overall histopathological features are diagnostic for pilocytic astrocytoma, WHO grade I. Ancillary fluorescence in situ hybridization (FISH) was negative for BRAF duplication or rearrangement.
The clinically validated UW-OncoPlex [12] next-generation sequencing (NGS) assay was used to examine 262 cancer-associated genes in the neoplastic tissue. Average target coverage for the tested sample was 577-fold, with no single-nucleotide variants (SNVs), insertion-deletion (indel), or structural mutations identified in other pilocytic astrocytoma-related genes including BRAF, NF1, and FGFR1. Also of note, there were no alterations detected for IDH1, IDH2, TP53, or ATRX. A novel translocation between PML and NTRK2 is identified by multiple bioinformatics pipeline programs CREST [13] and BreakDancer [14], with approximate genomic breakpoints of HG19 chr9:g.87467299 with chr15:g.7431663 and chr15:g.74316451 with chr9:g.87467483. BLAT (BLAST-like alignment tool) analysis of the consensus sequence mapped uniquely to PML and NTRK2 was employed, and split-read sequences were readily identified in the sequencing BAM file using the Integrative Genomics Viewer [15, 16] (IGV, Broad Institute, Cambridge, MA, USA) (Figure 2). The consensus-read data indicates that this rearrangement occurs within NTRK2 intron 14, which is upstream of the kinase domain, and PML intron 3. While at the DNA level, the functional consequences of this rearrangement are not known, the newly juxtaposed exons are predicted to be in-frame for both the NTRK2-PML and PML-NTRK2 rearrangements, if the splicing within the fusion gene products are not disrupted, suggesting that the genomic rearrangement may be a balanced translocation. Other glial neoplasms that have been identified to harbor NTRK1, NTRK2, or NTRK3 fusions have been described with similarly structured rearrangements [17]. The clinically validated FusionPlex (ArcherDx, Inc., Boulder, CO, USA) NGS analysis using a custom 114-gene solid tumor panel with RNA extracted from the tumor detected a fusion between genes PML (5′ partner) and NTRK2 (3′ partner). Two isoforms of fusion transcripts in-frame were detected, and both had exon 3 of PML (NM_002675.3) at the 5′ end with one isoform having exon 16 of NTRK2 (NM_006180.3) at the 3′ end and the other isoform having exon 15 of NTRK2 (NM_006180.3) at the 3′ end. For confirmation of the novel fusions discovered by NGS analysis, RT-PCR analysis was also performed for both 5′PML-NTRK2 3′ fusion products and potential reciprocal 5′NTRK2-PML 3′ fusion products. By RT-PCR, the two in-frame fusion isoforms of 5′PML-NTRK2 3′ were detected, while there were no detectable reciprocal 5′NTRK2-PML 3′ fusion products (Figure 2(c)). So while DNA analysis indicates that the PML and NTRK2 rearrangement may result in balanced fusion products, at the RNA level, only 5′NTRK2-PML 3′ fusion products were detectable.
Figure 2.
Pilocytic astrocytoma PML-NTRK2 gene fusion. (a) Illustrations of the breakpoints in NTRK2 on chromosome 9 and PML on chromosome 15, and associated transcripts, as determined by DNA next-generation sequencing (NGS). (b) Illustration of in-frame PML-NTRK2 gene fusion product confirmed with FusionPlex RNA NGS. (c) RT-PCR analysis validating PML-NTRK2 in-frame fusions F1 and F2. F1 = fusion transcript 1 with exon 3 of PML (5′ end partner) and exon 16 of NTRK2 (3′ end partner); F2 = fusion transcript 2 with exon 3 of PML (5′ end partner) and exon 15 of NTRK2 (3′ end partner). There are no detectable reciprocal 5′-NTRK2-PML-3′ fusion products.
The patient has had 26 months of clinical follow-up and is currently without symptoms or radiographic evidence of disease.
3. Discussion
Pilocytic astrocytomas are typically associated with alterations in either NF1 or BRAF, and less commonly in FGFR1. Rare somatic NTRK2 gene fusions have been previously described in pediatric low-grade neuroepithelial tumors, including pilocytic astrocytoma and ganglioglioma (summarized in Table 1), but the incidence in adults has not been described. In the current case, there was a lack of genetic alterations in NF1 and BRAF, but rather this pilocytic astrocytoma harbored a novel NTRK2 fusion with interesting implications for biology and therapeutics. TRK inhibitors have been proposed as therapeutic targets in neoplasms with activating NTRK fusions [18, 19] with promising results in early-stage clinical evaluations [20, 21]. To our knowledge, this is the first report of a NTRK2 gene fusion product in a pilocytic astrocytoma that has PML-NTRK2.
Located on chromosome 9 and spanning more than 350,000 base pairs (bp), NTRK2 encodes for numerous transcripts with precise developmental and tissue-specific precision [22–24]. The initial 5′ exons exhibit complex splicing patterns with varied transcriptional start sites while also forming an internal ribosomal entry site (IRES) to the translational start site while subsequent exons encode for the extracellular domain containing several IG-like domains and a neurotrophin binding site. A single exon encodes for the transmembrane domain, which is shared between multiple transcripts, followed by several exons encoding intracellular domains with the most 3′ exons encoding for the kinase domain. To date, known NTRK2 fusions observed in low-grade neuroepithelial tumors (summarized in Table 1), as well as diffuse intrinsic pontine glioma and other higher-grade gliomas [17], have resulted in fusions containing exons encoding for the TrkB kinase domain, postulating constitutive kinase activation and subsequent downstream signaling as the oncogenic driver. Additional gliomas with unspecified histological classification or grading have been reported as well [25].
Similar to previously published TRK fusions, the PML-NTRK2 fusion described here results in an NTRK2 fusion harboring the TrkB kinase domain, which is of interest biologically for downstream signaling (Figure 3). Additionally, this fusion contains the first three exons of PML encoding the TRIpartite motif responsible for binding DNA. PML fusions have been well characterized in other cancer types, particularly the PML-RAR alpha oncofusion in acute promyelocytic leukemia, and are typically associated with loss-of-function mutations in the PML protein, leading to subsequent loss of tumor suppressor roles.
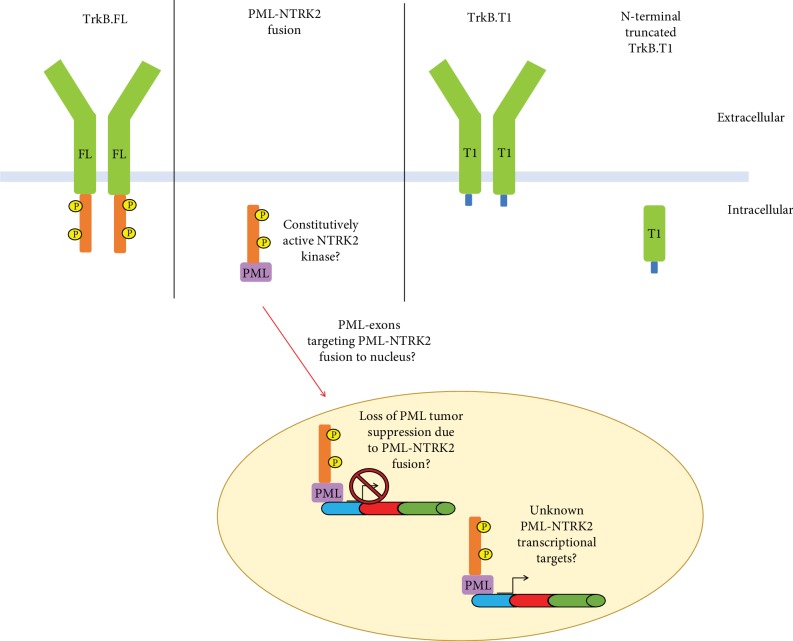
Figure 3.
Predicted TrkB receptor outcomes encoded by NTRK2 fusions in pediatric low-grade glioma. FL = full-length variant; KD = kinase domain; T1 = known alternative splice variant; P=phosphorylation site.
Given NTRK2's uniquely complex splice patterns, it is also of interest to note that the fusions occur downstream of exons that may still allow for multiple kinase-deficient NTRK2 transcripts. In addition to the full-length receptor tyrosine kinase, TrkB, several alternatively spliced NTRK2variants have been discovered, including TrkB.T1, TrkB-Shc, and N-terminal-truncated-TrkB.T1 (Figure 3) [22]. Once thought to be a dominant-negative/decoy receptor due to its lack of a kinase domain, one truncated splice variant, TrkB.T1, shares the same extracellular and transmembrane domains and first 12 intracellular amino acids as other variants [26]. Lacking a kinase domain, TrkB.T1 has a specific C-terminal sequence of 11 amino acids that is 100% conserved across species from rodents to humans [22, 24, 26, 27], and structure-function analysis confirms that this sequence is required for TrkB.T1 activity [28], supporting an evolutionary role beyond a dominant negative. This alternatively spliced receptor variant has been previously shown to alter Ca2+ signaling in glial cells in response to BDNF signaling [29] and mediate signal transduction in vitro [30]. Additionally, TrkB.T1 has been shown to regulate neuronal [31] and astrocytic morphology via rho GTPases [32] and outgrowth of filopodia in neuroblastoma cells [33] and contribute to glioma cell proliferation in vitro [32, 34]. The NTRK2 fusion point observed here (chr9:g.87467299) is within an intron just downstream of the TrkB.T1 alternate exon (which terminates at chr9:87430621) which contains multiple poly-A sites, regulatory elements, and a stop codon, highlighting the potential that while TrkB kinase activation is not possible, an intact form of truncated TrkB.T1 (NM_001007097), among other NTRK2 transcripts, may also be functionally expressed in conjunction with this fusion.
Basic scientific and clinical investigation surrounding Trks' precise role in cancer has long been hindered due to nonspecific of pan-Trk antibodies and kinase inhibitors alike. Current clinical trials are also plagued by nonspecificity when it comes to exploring Trks. For example, the small-molecule LOXO-01 has been designed to block the ATP binding site of Trks (including TrkA, TrkB, and TrkC) and has shown promising outcomes in clinical trials for other cancer types, but as this drug lacks specificity, its mechanism of action remains unclear [19]. The combined use of pan-Trk detection reagents for research and pan-Trk inhibitors in clinical trials may offer marginal insight into their biology. Further investigation is needed in order to hone in on precise signaling relevant to tumor biology in an effort to avoid off-target effects from multi-Trk inhibition, especially those involving the pediatric nervous system, as neurotoxic damage could have lifelong consequences. Due to the nonspecificity of the current pan-Trk kinase inhibitors, additional research confirming the kinase activity of the observed NTRK1, NTRK2, or NTRK3 fusions is warranted. Future exploration into the role of alternative splicing of Trk receptors, and NTRK2 specifically, will be paramount to understanding tumor biology and subsequently designing appropriate therapeutics and diagnostic approaches.
Acknowledgments
Pattwell S.S. was partially supported by the National Cancer Institute of the NIH under award number 5R21CA223531, as well as the Jacobs Foundation Research Fellowship.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Lewis R. A., Gerson L. P., Axelson K. A., Riccardi V. M., Whitford R. P. von Recklinghausen neurofibromatosis. II. Incidence of optic gliomata. Ophthalmology. 1984;91(8):929–935. doi: 10.1016/s0161-6420(84)34217-8. [DOI] [PubMed] [Google Scholar]
- 2.Listernick R., Ferner R. E., Liu G. T., Gutmann D. H. Optic pathway gliomas in neurofibromatosis-1: controversies and recommendations. Annals of Neurology. 2007;61(3):189–198. doi: 10.1002/ana.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez F. J., Lim K. S., Bowers D., Eberhart C. G. Pathological and molecular advances in pediatric low-grade astrocytoma. Annual Review of Pathology: Mechanisms of Disease. 2013;8:361–379. doi: 10.1146/annurev-pathol-020712-164009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutmann D. H., McLellan M. D., Hussain I., et al. Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Research. 2013;23(3):431–439. doi: 10.1101/gr.142604.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins V. P., Jones D. T., Giannini C. Pilocytic astrocytoma: pathology, molecular mechanisms and markers. Acta Neuropathologica. 2015;129(6):775–788. doi: 10.1007/s00401-015-1410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones D. T. W., Hutter B., Jäger N., et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nature Genetics. 2013;45(8):927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López G. Y., Perry A., Harding B., Li M., Santi M. CDKN2A/B loss is associated with anaplastic transformation in a case of NTRK2 fusion-positive pilocytic astrocytoma. Neuropathology and Applied Neurobiology. 2019;45(2):174–178. doi: 10.1111/nan.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones K. A., Bossler A. D., Bellizzi A. M., Snow A. N. BCR-NTRK2 fusion in a low-grade glioma with distinctive morphology and unexpected aggressive behavior. Molecular Case Studies. 2019;5(2, article a003855) doi: 10.1101/mcs.a003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prabhakaran N., Guzman M. A., Navalkele P., Chow-Maneval E., Batanian J. R. NovelTLE4-NTRK2fusion in a ganglioglioma identified by array-CGH and confirmed by NGS: potential for a gene targeted therapy. Neuropathology. 2018;38(4):380–386. doi: 10.1111/neup.12458. [DOI] [PubMed] [Google Scholar]
- 10.Qaddoumi I., Orisme W., Wen J., et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathologica. 2016;131(6):833–845. doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon J. P., Linkov I., Rosado A., et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Modern Pathology. 2020;33(1):38–46. doi: 10.1038/s41379-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritchard C. C., Salipante S. J., Koehler K., et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. The Journal of Molecular Diagnostics. 2013;16(1):56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Mullighan C. G., Easton J., et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nature Methods. 2011;8(8):652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K., Wallis J. W., McLellan M. D., et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J. T., Thorvaldsdóttir H., Winckler W., et al. Integrative genomics viewer. Nature Biotechnology. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson J. T., Thorvaldsdóttir H., Wenger A. M., Zehir A., Mesirov J. P. Variant review with the integrative genomics viewer. Cancer Research. 2017;77(21):e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G., Diaz A. K., Paugh B. S., et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nature Genetics. 2014;46(5):444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amatu A., Sartore-Bianchi A., Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1(2, article e000023) doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doebele R. C., Davis L. E., Vaishnavi A., et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discovery. 2015;5(10):1049–1057. doi: 10.1158/2159-8290.CD-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagasubramanian R., Wei J., Gordon P., Rastatter J. C., Cox M. C., Pappo A. Infantile fibrosarcoma with NTRK3-ETV6 fusion successfully treated with the tropomyosin-related kinase inhibitor LOXO-101. Pediatric Blood & Cancer. 2016;63(8):1468–1470. doi: 10.1002/pbc.26026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drilon A., Nagasubramanian R., Blake J. F., et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion–positive solid tumors. Cancer Discovery. 2017;7(9):963–972. doi: 10.1158/2159-8290.CD-17-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luberg K., Wong J., Weickert C. S., Timmusk T. Human TrkB gene: novel alternative transcripts, protein isoforms and expression pattern in the prefrontal cerebral cortex during postnatal development. Journal of Neurochemistry. 2010;113(4):952–964. doi: 10.1111/j.1471-4159.2010.06662.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawara A., Liu X. G., Ikegaki N., et al. Cloning and chromosomal localization of the human TRK-B tyrosine kinase receptor gene (NTRK2) Genomics. 1995;25(2):538–546. doi: 10.1016/0888-7543(95)80055-q. [DOI] [PubMed] [Google Scholar]
- 24.Stoilov P., Castren E., Stamm S. Analysis of the human TrkB gene genomic organization reveals novel TrkB isoforms, unusual gene length, and splicing mechanism. Biochemical and Biophysical Research Communications. 2002;290(3):1054–1065. doi: 10.1006/bbrc.2001.6301. [DOI] [PubMed] [Google Scholar]
- 25.Gatalica Z., Xiu J., Swensen J., Vranic S. Molecular characterization of cancers with NTRK gene fusions. Modern Pathology. 2019;32(1):147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 26.Fenner B. M. Truncated TrkB: beyond a dominant negative receptor. Cytokine & Growth Factor Reviews. 2012;23(1-2):15–24. doi: 10.1016/j.cytogfr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Haapasalo A., Sipola I., Larsson K., et al. Regulation of TRKB surface expression by brain-derived neurotrophic factor and truncated TRKB isoforms. Journal of Biological Chemistry. 2002;277(45):43160–43167. doi: 10.1074/jbc.M205202200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao L., Sheng A. L., Huang S. H., et al. Mechanism underlying activity-dependent insertion of TrkB into the neuronal surface. Journal of Cell Science. 2009;122(17):3123–3136. doi: 10.1242/jcs.047712. [DOI] [PubMed] [Google Scholar]
- 29.Rose C. R., Blum R., Pichler B., Lepier A., Kafitz K. W., Konnerth A. Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature. 2003;426(6962):74–78. doi: 10.1038/nature01983. [DOI] [PubMed] [Google Scholar]
- 30.Baxter G. T., Radeke M. J., Kuo R. C., et al. Signal transduction mediated by the truncated trkB receptor isoforms, trkB.T1 and trkB.T2. The Journal of Neuroscience. 1997;17(8):2683–2690. doi: 10.1523/JNEUROSCI.17-08-02683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartmann M., Brigadski T., Erdmann K. S., et al. Truncated TrkB receptor-induced outgrowth of dendritic filopodia involves the p75 neurotrophin receptor. Journal of Cell Science. 2004;117(24):5803–5814. doi: 10.1242/jcs.01511. [DOI] [PubMed] [Google Scholar]
- 32.Ohira K., Kumanogoh H., Sahara Y., et al. A truncated tropo-myosine-related kinase B receptor, T1, regulates glial cell morphology via rho GDP dissociation inhibitor 1. The Journal of Neuroscience. 2005;25(6):1343–1353. doi: 10.1523/JNEUROSCI.4436-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haapasalo A., Saarelainen T., Moshnyakov M., et al. Expression of the naturally occurring truncated trkB neurotrophin receptor induces outgrowth of filopodia and processes in neuroblastoma cells. Oncogene. 1999;18(6):1285–1296. doi: 10.1038/sj.onc.1202401. [DOI] [PubMed] [Google Scholar]
- 34.Ohira K., Homma K. J., Hirai H., Nakamura S., Hayashi M. TrkB-T1 regulates the RhoA signaling and actin cytoskeleton in glioma cells. Biochemical and Biophysical Research Communications. 2006;342(3):867–874. doi: 10.1016/j.bbrc.2006.02.033. [DOI] [PubMed] [Google Scholar]