Abstract
Class 1 integrons (c1-integrons) are associated with multidrug resistance in diarrheagenic Escherichia coli (DEC). However, little is known about gene cassettes located within these c1-integrons, particularly truncated c1-integrons, in DEC strains. Therefore, the aims of the present study were to reveal the relationship between antimicrobial resistance and the presence of truncated c1-integrons in DEC isolates derived from human stool samples in Japan. A total of 162 human stool-derived DEC isolates from Japan were examined by antimicrobial susceptibility testing, PCR-based gene detection, and next-generation sequencing analyses. Results showed that 44.4% (12/27) of c1-integrons identified in the DEC isolates harbored only intI1 (an element of c1-integrons) and were truncated by IS26, Tn3, or IS1-group insertion sequences. No difference in the frequency of antimicrobial resistance was recorded between intact and truncated c1-integron-positive DEC isolates. Isolates containing intact/truncated c1-integrons, particularly enteroaggregative E. coli isolates, were resistant to a greater number of antimicrobials than isolates without c1-integrons. aadA and dfrA were the most prevalent antimicrobial resistance genes in the intact/truncated c1-integrons examined in this study. Therefore, gene cassettes located within these intact/truncated c1-integrons may only play a limited role in conferring antimicrobial resistance among DEC. However, DEC harboring truncated c1-integrons may be resistant to a greater number of antimicrobials than c1-integron-negative DEC, similar to strains harboring intact c1-integrons.
1. Introduction
Gene cassettes located within class 1 integrons (c1-integrons) may play an important role in diarrheagenic Escherichia coli (DEC) strains. DEC are generally classified into five categories (enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enteroaggregative E. coli (EAEC), and enteroinvasive E. coli) on the basis of their virulence traits [1]. Among the categories, EPEC and EAEC are known for their high prevalence in both community and/or clinical settings [2, 3]. EAEC strains display higher rates of resistance to several antibiotics when compared with that of other DEC pathotypes [4, 5]. c1-integrons are a major source of antibiotic resistance genes and contain three main elements: an integrase gene (intI), a primary recombination site (attI), and a strong promoter. c1-integrons capture gene cassettes conferring resistance to antibiotics via IntI-catalyzed recombination between the attI recombination site and a 59-bp element called attC present on the gene cassettes (Figure 1(a)) [6]. c1-integron-harboring bacterial strains generally show higher rates of antibiotic resistance than those without c1-integrons [7, 8]. Moreover, the presence of c1-integrons contributes to multidrug resistance (MDR), defined as resistance to three or more classes of antimicrobials, in Enterobacteriaceae [9]. While little is known about gene cassettes located within intact c1-integrons in DEC strains [6], even less is known about genes found in truncated c1-integrons. Therefore, further research is needed to evaluate gene cassettes in truncated c1-integrons.
Figure 1.
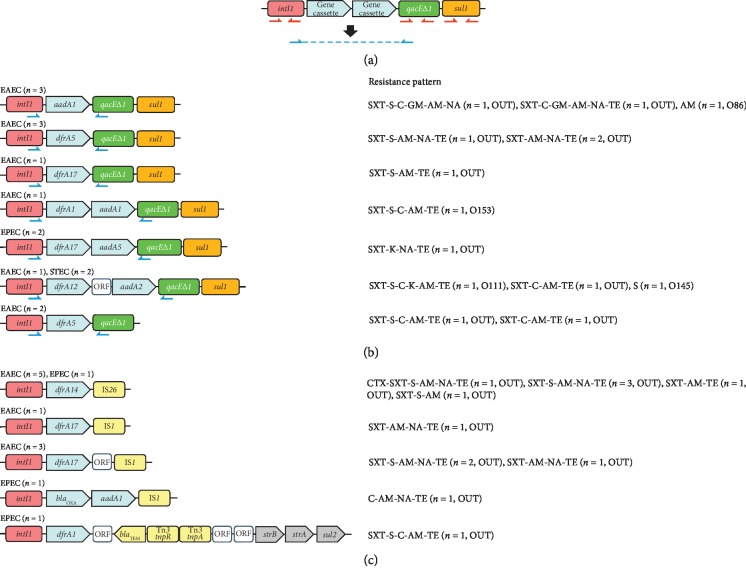
(a) General structure of class 1 integrons (c1-integrons). The red arrows show the positions of primers used for detection of intI1, qacEΔ1, and sul1. The blue arrows show the positions of primers used for sequencing. P indicates the promoter. (b) Intact c1-integron cassette arrays that were confirmed by sequencing of PCR products, along with the corresponding resistance patterns. (c) Truncated c1-integron cassette arrays that were confirmed by next-generation sequencing analysis, along with the corresponding resistance patterns. EAEC: enteroaggregative Escherichia coli; EPEC: enteropathogenic E. coli; STEC: Shiga toxin-producing E. coli; CTX: cefotaxime; SXT: sulfamethoxazole-trimethoprim; (S) streptomycin; (C) chloramphenicol; (K) kanamycin; AM: ampicillin; NA: nalidixic acid; TE: tetracycline; aadA1: aminoglycoside resistance gene; dfrA: dihydrofolate reductase gene (trimethoprim resistance); OUT: O-serogroup-untypeable. All insertion sequences designated “IS1” belong to the IS1 family.
There is also a lack of information about the role of truncated c1-integrons in the antimicrobial resistance of DEC. Previous work has shown that truncated c1-integrons are involved in the dissemination of antimicrobial resistance genes such as blaSHV-12 and blaVIM-7 in bacteria other than DEC, including Enterobacter cloacae [10] and Pseudomonas aeruginosa [11], respectively. Thus, truncated c1-integron cassettes should be evaluated in DEC. However, it is difficult to investigate gene arrays in truncated c1-integron cassettes because repeat sequences, insertion sequences (IS), and transposons can result in truncation of the genes, inhibiting amplification reactions [12]. As such, the aims of the present study were to reveal the relationship between antimicrobial resistance and the presence of truncated c1-integrons in DEC isolates derived from human stool samples in Japan using both conventional sequencing and next-generation sequencing (NGS) analyses.
2. Materials and Methods
2.1. Bacterial Strains
A total of 162 DEC isolates, consisting of 40 EAEC, 37 EPEC, 83 STEC, and two ETEC, were examined. All DEC isolates, except for 51 of the STEC isolates, were obtained from stool samples collected from asymptomatic carriers and patients with gastrointestinal symptoms at the Kawasaki City Institute for Public Health, Japan, from 2012 to 2014. The remaining 51 STEC isolates were collected from outpatients at several hospitals in Kawasaki between April 2012 and December 2014 (Table 1). The 40 EAEC isolates were also examined in our previous study of antimicrobial resistance patterns [13]. The ETEC and EPEC isolates were identified by PCR-based assays using primers targeting eae [14] and elt and est (primers ELT−1/−2, ESH−1/−2, and ESP−1/−2; Takara Biomedicals, Kusatsu, Japan). The 83 STEC isolates were identified using stx-targeting PCR primers EVC−1/−2 (Takara Biomedicals) or using a Loopamp Verotoxin Typing Kit (Eiken Chemical Co., Tokyo, Japan). For all of the DEC isolates, O-serotyping was conducted using a slide agglutination method with 43 commercially available O-antisera (Denka Seiken Co., Tokyo, Japan).
Table 1.
Diarrheagenic Escherichia coli strains used in this study (n = 162).
| Pathogenic categories | No. of strains | Origin | O-serogroup | Isolation year |
|---|---|---|---|---|
| EAEC∗ | 40 | Symptomatic patient (n = 17) | 86 (n = 1), 111 (n = 1), 125 (n = 1), 126 (n = 1), 127 (n = 2), 153 (n = 1), OUT (n = 10) | 2012–2014 |
| Asymptomatic carrier (n = 23) | 44 (n = 1), 55 (n = 1), 86 (n = 1), 126 (n = 2), OUT (n = 18) | 2012–2014 | ||
| EPEC | 37 | Symptomatic patient (n = 12) | 55 (n = 1), 114 (n = 1), 164 (n = 1), OUT (n = 9) | 2012–2014 |
| Asymptomatic carrier (n = 25) | 15 (n = 1), 63 (n = 1), 124 (n = 2), 125 (n = 1), 145 (n = 1), 167 (n = 1), OUT (n = 18) | 2013-2014 | ||
| STEC | 83 | Symptomatic patient (n = 68) | 26 (n = 3), 103 (n = 3), 111 (n = 4), 145 (n = 2), 157 (n = 54), 165 (n = 1), 186 (n = 1) | 2012–2014 |
| Asymptomatic carrier (n = 15) | 26 (n = 3), 157 (n = 12) | 2012–2014 | ||
| ETEC | 2 | Asymptomatic carrier (n = 2) | 148 (n = 1), 169 (n = 1) | 2013-2014 |
∗EAEC: enteroaggregative E. coli; EPEC: enteropathogenic E. coli; STEC: Shiga toxin-producing E. coli; ETEC: enterotoxigenic E. coli; OUT: O-serogroup untypeable.
2.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility profiles were determined using the disc diffusion method with BD Sensi-Discs (Becton Dickinson, Tokyo, Japan) according to the guidelines outlined in Clinical and Laboratory Standards Institute documents M02-A13 and M100-S28 [15, 16]. The following 14 antibiotic discs were used: cefotaxime (30 μg), norfloxacin (10 μg), sulfamethoxazole-trimethoprim (23.75 μg, 1.25 μg), streptomycin (10 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), kanamycin (30 μg), gentamicin (10 μg), ampicillin (10 μg), fosfomycin (50 μg), nalidixic acid (30 μg), tetracycline (30 μg), imipenem (10 μg), and meropenem (10 μg).
2.3. Detection of c1-Integrons
DNA template was extracted from each isolate using a QIAamp DNA Stool Mini Kit (Qiagen GmbH, Hilden, Germany). In general, the 5ʹ-conserved segment (5ʹCS) of c1-integrons contains intI1, while the 3ʹ-conserved segment (3ʹ-CS) contains both qacEΔ1 and sul1 (Figure 1(a)). In this study, the presence of a c1-integron was confirmed by three independent amplifications of intI1, qacEΔ1, and sulI (Figure 1(a)) via PCR-based assays [17].
2.4. Amplification and Sequencing of Gene Cassette Regions
The isolates from which genes in the 5ʹ-CS and 3ʹ-CS regions could be amplified were classed as containing intact c1-integrons. These isolates were then subjected to PCR using the 5ʹCS/3ʹ-CS primers, followed by Sanger sequencing of the resulting amplicons to determine the sequence of the region between intI1 and qacEΔ1 in intact c1-integrons (Figure 1(a)) [17]. Acquired resistance genes within each c1-integron were analyzed using the ResFinder platform (http://genomicepidemiology.org/), while similarity searches were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) [18, 19]. The primers used for PCR analyses are described in Table 2.
Table 2.
Primers used in this study.
| Target gene | Primer direction | Nucleotide sequence (5′–3′) | Amplicon size (bp) | Reference number |
|---|---|---|---|---|
| intI1 | F | CAGTGGACATAAGCCTGTTC | 160 | 15 |
| R | CCCGAGGCATAGACTGTA | |||
| sul1 | F | CGGCGTGGGCTACCTGAACG | 433 | 15 |
| R | GCCGATCGCGTGAAGTTCCG | |||
| qacEΔ1 | F | ATCGCAATAGTTGGCGAAGT | 250 | 15 |
| R | GAAGCTTTTGCCCATGAAGC | |||
| Class 1 gene cassette | F | GGCATCCAAGCAGCAAGC | Variable | 15 |
| R | AAGCAGACTTGACCTGAT | |||
| aggR | F | GTATACACAAAAGAAGGAAGC | 254 | 10 |
| R | ACAGAATCGTCAGCATCAGC | |||
| eae | F | GCTTAGTGCTGGTTTAGGAT | 591 | 10 |
| R | CTCTGCAGATTAACCTCTGC |
2.5. Next-Generation Sequencing
Integrons from which only intI1 could be amplified (i.e., missing qacEΔ1 and sulI) were classed as truncated integrons. Isolates harboring truncated c1-integrons were subjected to next-generation sequencing analysis. DNA extraction from the strains was carried out as described previously [20]. A short insert size (approximately 0.5 kb) paired-end library was constructed using a Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA), followed by whole-genome sequencing using the Illumina NextSeq 500 platform with a 300-cycle NextSeq 500 Reagent Kit v2 (2 × 150 mer). The extracted contigs were validated by comparison against the whole-genome sequence database GenEpid-J [21] and by using the ResFinder and VirulenceFinder tools available from the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/).
2.6. Detection of Extended-Spectrum β-Lactamase (ESBL) Genes
Isolates that showed resistance to cefotaxime during antimicrobial susceptibility testing were further examined for the presence of ESBL genes (blaCTX-M, blaTEM, and blaSHV) by PCR, as described previously [22, 23]. Primers used for sequencing of blaTEM were designed in this study: TEMseq-F, 5ʹ-GGTGCGGTATTATCCCGTGT-3ʹ; TEMseq-R, 5ʹ-TTGTTGCCGGGAAGCTAGAG-3ʹ. The resulting PCR amplicons were sequenced and the nucleotide and deduced amino acid sequences were compared with entries in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/), as well as with those described on the β-lactamase classification website (http://www.lahey.org/Studies/, accessed February 2017), to determine the β-lactamase gene subtype.
2.7. Statistical Analyses
Statistical analyses were performed using Fisher's exact test. A p-value of <0.05 was considered statistically significant.
2.8. Ethical Approval
This study was performed in accordance with the guidelines of the Ethics Regulations Related to Medical Research Involving Human Subjects at the Kawasaki City Institute for Public Health under permit number 28-2.
3. Results
3.1. Susceptibility to Antimicrobial Agents
Of the 162 DEC isolates, 64 were resistant to at least one of the antibiotics tested, with 32 isolates showing MDR (Table 3). As shown in Table 4, the highest prevalence of antimicrobial resistance was associated with ampicillin (50 isolates, 30.9%), followed by tetracycline (39 isolates, 24.1%) and sulfamethoxazole-trimethoprim (28 isolates, 17.3%). Furthermore, EAEC isolates showed resistance to a greater number of antimicrobial agents than EPEC or STEC isolates. MDR phenotypes were more frequently associated with DEC (p < 0.001), EAEC (p < 0.0011), and EPEC isolates (p=0.002) harboring intact/truncated c1-integrons than with c1-integron-negative isolates (Table 4).
Table 3.
Number of antibiotics to which the strains with/without integrons showed resistance.
| Presence of integrons | Intact/truncated integron and pathotype | No. of strains | Number (%) of antibiotics to which each strain showed resistance† | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | One | Two | Three | Four | Five | Six | |||
| Strains with integrons | Intact integron | ||||||||
| EAEC∗ | 11 | 0 | 1 | 0 | 0 | 5 | 3 | 2 | |
| EPEC∗ | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | |
| STEC∗ | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| ETEC∗ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 15 | 1 (7%) | 2 (13%) | 0 | 0 | 6 (40%) | 3 (20%) | 3 (20%) | |
| Truncated integron | |||||||||
| EAEC | 9 | 0 | 0 | 0 | 1 | 2 | 5 | 1 | |
| EPEC | 3 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | |
| STEC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| ETEC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 12 | 0 | 0 | 0 | 2 (16.7) | 3 (25.0) | 6 (50.0) | 1 (8.3) | |
| Strains without integrons | EAEC | 20 | 6 | 6 | 5 | 1 | 1 | 1 | 0 |
| EPEC | 32 | 25 | 2 | 2 | 1 | 1 | 0 | 1 | |
| STEC | 81 | 66 | 6 | 7 | 1 | 1 | 0 | 0 | |
| ETEC | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| Subtotal | 135 | 97 (71.6) | 15 (11.1) | 14 (10.4) | 3 (2.2) | 3 (2.2) | 2 (1.5) | 1 (0.7) | |
|
| |||||||||
| Total | 162 | 98 (60.5) | 17 (10.5) | 14 (8.6) | 5 (3.1) | 12 (7.4) | 11 (6.8) | 5 (3.1) | |
†A total of 14 antimicrobials were tested (ampicillin, sulfamethoxazole-trimethoprim, tetracycline, nalidixic acid, streptomycin, chloramphenicol, gentamicin, cefotaxime, norfloxacin, kanamycin, ciprofloxacin, fosfomycin, imipenem, and meropenem).
Table 4.
Number (%) of antibiotic-resistant diarrheagenic Escherichia coli isolates in Japan, with/without class 1 integrons.
| Antibiotics | EAEC∗ | EPEC∗ | STEC∗ | ETEC∗ | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Integron + (n = 20) | Integron − (n = 20) | Total (n = 40) | Integron + (n = 5) | Integron − (n = 32) | Total (n = 37) | Integron + (n = 2) | Integron − (n = 81) | Total (n = 83) | Integron + (n = 0) | Integron − (n = 2) | Integron + (n = 27) | Integron − (n = 135) | |
| Ampicillin | 20 (100)‡ | 13 (65.0) | 33 (82.5) | 3 (60.0)‡ | 4 (12.5) | 7 (18.9) | 1 (50.0) | 8 (9.9) | 9 (10.8) | 0 | 1 (50.0) | 24 (88.9)‡ | 26 (19.3) |
| Sulfamethoxazole-trimethoprim | 18 (90.0)‡ | 3 (15.0) | 21 (52.5) | 3 (60.0)‡ | 1 (3.1) | 4 (10.8) | 1 (50.0)‡ | 1 (1.2) | 2 (2.4) | 0 | 1 (50) | 22 (81.5)‡ | 6 (4.4) |
| Tetracycline | 18 (90.0)‡ | 3 (15.0) | 21 (52.5) | 3 (60.0)‡ | 4 (12.5) | 7 (18.9) | 1 (50.0) | 9 (11.1) | 10 (12.0) | 0 | 1 (50.0) | 22 (81.5)‡ | 17 (12.6) |
| Nalidixic acid | 13 (65.0)‡ | 3 (15.0) | 16 (40.0) | 2 (40.0) | 3 (9.4) | 5 (13.5) | 0 | 0 | 0 | 0 | 2 (100) | 15 (55.6)‡ | 8 (5.9) |
| Streptomycin | 11 (55.0)‡ | 3 (15.0) | 14 (35.5) | 2 (40.0) | 2 (6.3) | 4 (10.8) | 2 (100)‡ | 6 (7.4) | 8 (9.6) | 0 | 1 (50.0) | 15 (55.6)‡ | 12 (8.9) |
| Chloramphenicol | 6 (30.0) | 1 (5.0) | 7 (17.5) | 2 (40.0) | 2 (6.3) | 4 (10.8) | 1 (50.0)‡ | 1 (1.2) | 2 (2.4) | 0 | 0 | 9 (33.3)‡ | 4 (3.0) |
| Gentamicin | 2 (9.52) | 0 | 2 (5.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (7.4)‡ | 0 |
| Cefotaxime | 1 (10.0) | 2 (10.0) | 3 (7.5) | 0 | 2 (6.3) | 2 (5.4) | 0 | 0 | 0 | 0 | 0 | 1 (3.7) | 4 (3.0) |
| Norfloxacin | 1 (5.0) | 0 | 1 (2.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (3.7) | 0 |
| Kanamycin | 0 | 0 | 0 | 1 (20.0) | 1 (3.1) | 2 (5.4) | 1 (50.0) | 2 (2.5) | 3 (3.6) | 0 | 0 | 2 (7.4) | 3 (2.2) |
| Ciprofloxacin, fosfomycin, imipenem, or meropenem | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Phenotype of multidrug resistance† | 19 (95.0)‡ | 2 (10.0) | 21 (52.5) | 4 (80.0)‡ | 3 (9.4) | 7 (18.9) | 1 (50.0) | 2 (2.5) | 3 (3.6) | 0 | 1 (50.0) | 24 (88.9)‡ | 8 (5.9) |
| β-lactamase genes detected | bla CTX-M-14 (n = 1), blaTEM-1 (n = 1) | bla CTX-M-14 (n = 1), blaCTX-M-15 (n = 1) | bla CTX-M-14 (n = 2), blaCTX-M-15 (n = 1), blaTEM-1 (n = 1) | bla OXA-1 (n = 1) | bla CTX-M-14 (n = 1), blaTEM-1 (n = 1) | bla CTX-M-14 (n = 1), blaTEM-1 (n = 1) blaOXA-1 (n = 1) | bla CTX-M-14 (n = 1), blaTEM-1(n = 1) blaOXA-1(n = 1) | bla CTX-M-14 (n = 2), blaCTX-M-15 (n = 1), blaTEM-1 (n = 1) | |||||
†Defined as resistance to three or more classes of antimicrobials. ‡Denotes significantly higher rate of resistance to antibiotics for class 1 integron-positive isolates compared with class 1 integron-negative isolates (p > 0.05).
3.2. Frequency of c1-Integrons
EAEC isolates were more likely to harbor intact/truncated c1-integrons (50.0%) compared with EPEC (13.5%), STEC (2.4%), and ETEC (0%) isolates (Tables 3 and 4). Within DEC pathotypes EAEC, EPEC, and STEC isolates containing c1-integrons had significantly higher rates of resistance to seven specific drugs compared with c1-integron-negative isolates (Tables 3 and 4).
3.3. Identification of Gene Cassette Arrays within c1-Integrons
NGS analysis revealed that the 3ʹ-CS regions of the 12 isolates harboring truncated c1-integrons were truncated by the insertion of IS26 (n = 6), insertion sequences belonging to the IS1 group (n = 5), or by transposon Tn3 (n = 1) (Figures 1(b) and 1(c)). No differences in the frequency of antibiotic resistance or in the number of antibiotics to which isolates showed resistance were observed between isolates harboring intact and truncated c1-integrons (Table 3).
Overall, 7.5% (3/40) of EAEC isolates and 2.7% (1/37) of EPEC isolates contained ESBL genes, none of which were identified in the STEC or ETEC isolates. Importantly, none of the ESBL genes detected in the current study (three blaCTX-M-14 and one blaCTX-M-15; Table 4) were located within the c1-integron cassettes.
3.4. Nucleotide Sequences
All sequence data for c1-integrons amplified using primers 5ʹCS/3ʹ-CS have been deposited in the GenBank database under accession numbers LC380541–LC380554 and LC383355. Raw sequence reads have been deposited in the DNA Data Bank of Japan Sequence Read Archive under Biosample IDs SAMD00117734–SAMD00117745 (Run IDs DRR129827–DRR129838) (Supplementary ).
4. Discussion
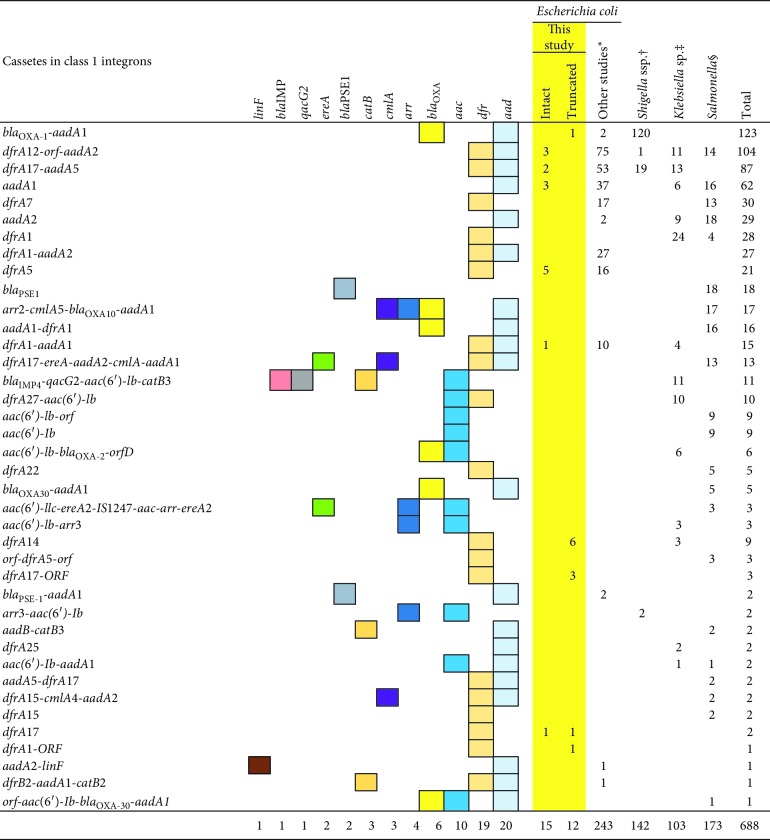
Based on the results of this study, DEC harboring truncated c1-integrons may be resistant to a greater number of antimicrobials than c1-integron-negative DEC. Structures of c1-integrons from strains in this study were compared with those from other Enterobacteriaceae (Table 5) [24–32]. Dominant resistant genes aadA (conferring resistance to aminoglycoside) and/or dfrA (conferring resistance to trimethoprim) within intact/truncated c1-integrons from strains examined in this study are also the major genes in other c1-integrons of other Enterobacteriaceae strains (Table 5). Only “dfrA17,” “dfrA17-ORF,” or “dfrA1-ORF” were unique gene cassettes from strains in the present study. In contrast, seven of the ten sequence patterns of cassette-borne antimicrobial and related genes in intact/truncated c1-integrons from strains studied herein have also been identified in other strains from other countries (Table 5), suggesting a worldwide circulation of the c1-integrons among Enterobacteriaceae. The cassette-borne genes identified in the present study suggest that gene cassettes within intact/truncated c1-integrons play a limited role in determining the antimicrobial resistance of Enterobacteriaceae. This indicates that the majority of antimicrobial resistance genes, except for aadA and dfrA, in DEC isolates are not cassette-borne and are located outside intact/truncated c1-integrons. Intact/truncated c1-integrons are generally associated with mobile genetic elements like transposons [10, 33, 34], which are major reservoirs of antimicrobial resistance genes. Subsequently, like strains with intact c1-integrons, DEC strains containing truncated c1-integrons might be resistant to a greater number of antimicrobials than strains without c1-integrons, as observed in the present study.
Table 5.
Comparison between this study and other studies of gene cassettes within class 1 integron.
|
∗Kang et al. (Korea) and Heir et al. (Norway). †Yang et al. (China). ‡Chang et al. (Taiwan), Li et al. (China) and Chowdhury et al. (Argentina, Chile, Uruguay, and Australia). §Peirano et al. (Brazil), Zhang et al. (China), and Krauland et al. (United States, Canada, Argentina, Australia, Belgium, South Africa, Spain, Italy, Denmark, and Taiwan).
The high rates of resistance genes in EAEC isolates may be attributed to the presence of intact/truncated c1-integrons and may be promoted in animal production environments. The results of the present study align with those of previous reports showing that EAEC strains display higher rates of resistance to several antibiotics compared with other DEC pathotypes [4, 5]. In addition, significantly higher resistance rates were observed among c1-integron-positive EAEC compared with the other three DEC pathotypes (Tables 3 and 4). Moreover, our results showed that the antimicrobial resistance patterns of intact/truncated c1-integron-positive EAEC isolates were similar to those of Japanese E. coli isolates originating from livestock, particularly broiler chickens, although previous studies have reported that EAEC isolates from humans are characteristically divergent from those from animals [35–37]. Therefore, the high prevalence of intact/truncated c1-integrons incorporating resistance genes among EAEC in the current study suggests that these isolates may be derived from meat or meat products.
5. Conclusion
Regardless of whether the integrons were intact or truncated, c1-integron-positive DEC isolates examined in the current study were more frequently resistant to antibiotics than integron-negative isolates even through intact/truncated c1-integrons may only play a limited role in conferring antimicrobial resistance among DEC isolates. Thus, truncated c1-integrons may also be involved in the acquisition of antimicrobial resistance genes by DEC, particularly EAEC. Continuous surveillance is therefore required to better monitor cassette-borne resistance genes in DEC in clinical and related fields.
Acknowledgments
This study was supported in part by a Grant-in-Aid from the Japan Society for the Promotion of Sciences, KAKENHI (http://www.jsps.go.jp/english/index.html) (grant no. 19K10590), and grants from the Japan Agency for Medical Research and Development, AMED (https://www.amed.go.jp/en/index.html) (grant nos. JP19fk0108103 and JP19fk0108033). The authors are grateful to Ms. Anzawa, Mr. Sasaki, Ms. Kojima, and Ms. Homma of the Kawasaki City Institute for Public Health and Ms. Doi and Ms. Yamada of the National Institute of Infectious Diseases, for their assistance. The authors thank Tamsin Sheen, PhD, from Edanz Group, (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Contributor Information
Akiko Kubomura, Email: kubomurak@gmail.com.
Koichi Murakami, Email: kmuraka@nih.go.jp.
Data Availability
The authors declare that raw data generated in this project are available from the corresponding authors upon reasonable request.
Disclosure
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
Supplementary Materials
Supplementary Table 1: strains tested and accession numbers of DNA sequences deposited in the DDBJ/GenBank/EMBL database.
References
- 1.Nataro J. P., Kaper J. B. Diarrheagenic Escherichia coli. Clinical Microbiology Reviews. 1998;11(1):142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochoa T. J., Contreras C. A. Enteropathogenic E. coli (EPEC) infection in children. Current Opinion in Infectious Diseases. 2011;24(5):478–483. doi: 10.1097/qco.0b013e32834a8b8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnenberg M. S. Enterobacteriaceae. In: Mandell G., Douglas Jr. R., Bennett J. E., editors. Principles and Practice of Infectious Diseases. Philadelphia, PA, USA: Elsevier; 2015. pp. 2503–2517. [Google Scholar]
- 4.Ochoa T. J., Molina M., Lanata C. F., et al. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. The American Journal of Tropical Medicine and Hygiene. 2009;81(2):296–301. doi: 10.4269/ajtmh.2009.81.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Chen X., Zheng S., et al. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China. Clinical Microbiology and Infection. 2014;20(1):52–58. doi: 10.1111/1469-0691.12188. [DOI] [PubMed] [Google Scholar]
- 6.Domingues S., da Silva G. J., Nielsen K. M. Integrons. Mobile Genetic Elements. 2012;2(5):211–223. doi: 10.4161/mge.22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L.-T., Wan L.-H., Song X.-H., Xiong Y., Jin S.-J., Zhou L.-M. Relevance of class 1 integrons and extended-spectrum β-lactamases in drug-resistant Escherichia coli. Molecular Medicine Reports. 2013;8(4):1251–1255. doi: 10.3892/mmr.2013.1626. [DOI] [PubMed] [Google Scholar]
- 8.Phongpaichit S., Wuttananupan K., Samasanti W. Class 1 integrons and multidrug resistance among Escherichia coli isolates from human stools. The Southeast Asian Journal of Tropical Medicine and Public Health. 2008;39(2):279–287. [PubMed] [Google Scholar]
- 9.Leverstein-van Hall M. A., Blok H. E. M., Donders A. R. T., Paauw A., Fluit A. C., Verhoef J. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. The Journal of Infectious Diseases. 2003;187(2):251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 10.Chen C. M., Yu W. L., Huang M., et al. Characterization of IS26-composite transposons and multidrug resistance in conjugative plasmids from Enterobacter cloacae. Microbiology and Immunology. 2015;59(9):516–525. doi: 10.1111/1348-0421.12289. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Toleman M. A., Bennett P. M., Jones R. N., Walsh T. R. Complete sequence of p07-406, a 24, 179-base-pair plasmid harboring the blaVIM-7 metallo- -lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrobial Agents and Chemotherapy. 2008;52(9):3099–3105. doi: 10.1128/aac.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunde M. Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. Journal of Antimicrobial Chemotherapy. 2005;56(6):1019–1024. doi: 10.1093/jac/dki377. [DOI] [PubMed] [Google Scholar]
- 13.Kubomura A., Misaki T., Homma S., Matsuo C., Okabe N. Phenotypic and molecular characterization of enteroaggregative Escherichia coli isolated in Kawasaki, Japan. Japanese Journal of Infectious Diseases. 2017;70(5):507–512. doi: 10.7883/yoken.jjid.2016.387. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K., Seto K., Yatsuyanagi J., et al. Presence of the genes regarding adherence factors of Escherichia coil isolates and a consideration of the procedure for detection of diarrheagenic strain. Journal of the Japanese Association for Infectious Diseases. 2002;76(11):911–920. doi: 10.11150/kansenshogakuzasshi1970.76.911. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standard Institute. M02-A13: Performance Standards for Antimicrobial Disk Susceptibility Tests. 13th. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 16.Clinical and Laboratory Standard Institute. M100-S. 28. Performance Standards for Antimicrobial Susceptibility Testing. 28th. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 17.Karczmarczyk M., Abbott Y., Walsh C., Leonard N., Fanning S. Characterization of multidrug-resistant Escherichia coli isolates from animals presenting at a university veterinary hospital. Applied and Environmental Microbiology. 2011;77(20):7104–7112. doi: 10.1128/aem.00599-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zankari E., Hasman H., Cosentino S., et al. Identification of acquired antimicrobial resistance genes. Journal of Antimicrobial Chemotherapy. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Akiba M., Sekizuka T., Yamashita A., et al. Distribution and relationships of antimicrobial resistance determinants among extended-spectrum-cephalosporin-resistant or carbapenem-resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrobial Agents and Chemotherapy. 2016;60(5):2972–2980. doi: 10.1128/aac.01950-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki S., Ohnishi M., Kawanishi M., Akiba M., Kuroda M. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. The Lancet Infectious Diseases. 2016;16(3):284–285. doi: 10.1016/s1473-3099(16)00008-6. [DOI] [PubMed] [Google Scholar]
- 22.Shibata N., Kurokawa H., Doi Y., et al. PCR classification of CTX-M-type-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrobial Agents and Chemotherapy. 2006;50(2):791–795. doi: 10.1128/aac.50.2.791-795.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi T., Kurokawa H., Shibata N., Shibayama K., Arakawa Y. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiology Letters. 2000;184(1):53–56. doi: 10.1016/s0378-1097(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 24.Kang H. Y., Jeong Y. S., Oh J. Y., et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. Journal of Antimicrobial Chemotherapy. 2005;55(5):639–644. doi: 10.1093/jac/dki076. [DOI] [PubMed] [Google Scholar]
- 25.Yang H., Pan Y., Hu L., et al. Antimicrobial resistance patterns and characterization of integrons in clinical isolates of Shigella from China. Canadian Journal of Microbiology. 2014;60(4):237–242. doi: 10.1139/cjm-2013-0893. [DOI] [PubMed] [Google Scholar]
- 26.Chang C. Y., Fang Y. T., Tsai S. M., Chang L. L., Yu W. L. Characterization of class 1 integrons and gene cassettes in clinical isolates of Klebsiella pneumoniae from Taiwan. Diagnostic Microbiology and Infectious Disease. 2009;65(2):214–216. doi: 10.1016/j.diagmicrobio.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Li B., Hu Y., Wang Q., et al. Structural diversity of class 1 integrons and their associated gene cassettes in Klebsiella pneumoniae isolates from a hospital in China. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0075805.e75805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy Chowdhury P., Ingold A., Vanegas N., et al. Dissemination of multiple drug resistance genes by class 1 integrons in Klebsiella pneumoniae isolates from four countries: a comparative study. Antimicrobial Agents and Chemotherapy. 2011;55(7):3140–3149. doi: 10.1128/aac.01529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peirano G., Agersø Y., Aarestrup F. M., dos Reis E. M., dos Prazeres Rodrigues D. Occurrence of integrons and antimicrobial resistance genes among Salmonella enterica from Brazil. Journal of Antimicrobial Chemotherapy. 2006;58(2):305–309. doi: 10.1093/jac/dkl248. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Shi L., Li L., et al. Identification and characterization of class 1 integron resistance gene cassettes among salmonella strains isolated from healthy humans in China. Microbiology and Immunology. 2004;48(9):639–645. doi: 10.1111/j.1348-0421.2004.tb03473.x. [DOI] [PubMed] [Google Scholar]
- 31.Krauland M. G., Marsh J. W., Paterson D. L., Harrison L. H. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerging Infectious Diseases. 2009;15(3):388–396. doi: 10.3201/eid1503.081131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heir E., Lindstedt B. A., Leegaard T. M., Gjernes E., Kapperud G. Prevalence and characterization of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class 1 integron-located lincosamide resistance gene. Annals of Clinical Microbiology and Antimicrobials. 2004;3(12) doi: 10.1186/1476-0711-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naas T., Mikami Y., Imai T., Poirel L., Nordmann P. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. Journal of Bacteriology. 2001;183(1):235–249. doi: 10.1128/jb.183.1.235-249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partridge S. R., Tsafnat G., Coiera E., Iredell J. R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiology Reviews. 2009;33(4):757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang R., Gu D. X., Huang Y. L., Chan E. W., Chen G. X., Chen S. Comparative genetic characterization of enteroaggregative Escherichia coli strains recovered from clinical and non-clinical settings. Scientific Reports. 2016;6:p. 24321. doi: 10.1038/srep24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uber A. P., Trabulsi L. R., Irino K., et al. Enteroaggregative Escherichia coli from humans and animals differ in major phenotypical traits and virulence genes. FEMS Microbiology Letters. 2006;256(2):251–257. doi: 10.1111/j.1574-6968.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 37.National Veterinary Assay Laboratory Ministry of Agriculture Forestry Fisheries. A report on the Japanese veterinary antimicrobial resistance monitoring system: 2012 to 2013. 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: strains tested and accession numbers of DNA sequences deposited in the DDBJ/GenBank/EMBL database.
Data Availability Statement
The authors declare that raw data generated in this project are available from the corresponding authors upon reasonable request.