Abstract
Introduction: Burkitt’s lymphoma (BL) is a rare and highly aggressive B cell non-Hodgkin lymphoma. High toxicity of chemotherapy for BL treatment causes morbidity and mortality. Many miRNAs have been used as biomarkers for early detection or therapy targets for tumors. However, the roles of miR-21 and miR-155 in Burkitt’s lymphoma remain unclear. Methods: We collected 15 blood samples from patients with Burkitt’s lymphoma and evaluated the expression of miR-21 and miR-155. Then, we knocked down miR-21 and miR-155 expression in Burkitt’s lymphoma cell lines and assessed cell proliferation, cell cycle, and apoptosis. Furthermore, we detected the activation of PI3K/AKT pathway by qPCR and western blot. Finally, we predicted the target genes of miR-21 and miR-155 by publicly available databases. Results: The expression of miR-21 and miR-155 in blood samples from patients with Burkitt’s lymphoma were significantly upregulated. Knockdown of miR-21 and miR-155 significantly suppressed cell proliferation, and resulted in S phase arrest and cell apoptosis. The knockdown of miR-21 and miR-155 inhibited the activation of the PI3K/AKT pathway. We found that the target genes of miR-21 and miR-155 were C1RL and TCAP. Conclusion: miR-21 and miR-155 promote the progression of Burkitt’s lymphoma through PI3K/AKT signaling by targeting C1RL and TCAP. Our findings will provide a novel biomarker and therapeutic strategies for Burkitt’s lymphoma.
Keywords: miR-21, miR-155, Burkitt’s lymphoma, PI3K/AKT, C1RL, TCAP
Introduction
Burkitt’s lymphoma (BL) is a rare and highly aggressive B cell non-Hodgkin lymphoma (NHL) originating from germinal center B cells [1]. In malaria-endemic areas, BL is the most frequent childhood cancer and the fastest growing human tumor [2]. Currently, the most common therapeutic strategy for BL is chemotherapy. However, the high toxicity of chemotherapy causes morbidity and mortality [3]. Therefore, it is urgent to find novel potential approaches for treatment of BL.
MicroRNAs are 21-23 nucleotide long, non-coding RNAs that regulate gene expression posttranscriptionally by degradation of its mRNA and suppression of expression of its target genes [4,5]. They are involved in various physiologic and pathologic processes, such as cell differentiation, proliferation, cell cycle, apoptosis, inflammation, and metabolism [6-8]. Dysregulation of miRNA expression result in many kinds of cancer [9,10]. Thus, many miRNAs have been used as biomarkers for early detection or therapy targets for tumors [11].
MicroRNA-21 (miR-21) has been demonstrated to regulate cardiac hypertrophy, cardiac fibrosis, and cardiac muscle contractility [12,13]. It has also been implicated in cell proliferation, division, and apoptosis. For example, miR-21 was overexpressed in gastric cancer, glioma, cervical cancer, and non-small cell lung cancer and can enhance cell proliferation, invasion and migration [14-16]. Inhibition of miR-21 resulted in arrest in the G1 phase and increased apoptosis rate in esophageal cancer [17]. MiR-155 is primarily upregulated in activated B cells and T cells and in the inflammation of monocytes and macrophages [18-20]. It regulates the development and function of immune cells [21,22]. Its dysregulation is also related to cancers [18]. MiR-155 is overexpressed in colorectal cancer and can promote cell proliferation and invasion [23,24]. Expression of miR-155 is elevated in hepatocellular carcinoma, and miR-155 can promote cell cycle arrest, cell proliferation and inhibit apoptosis [25]. miR-155 was also reported to suppress epithelial mesenchymal transition, cell proliferation, invasion and migration in human Caski cervical cancer cells [26]. In gastric cancer, decreasing the expression of miR-155-5p is associated with advanced tumor grade and metastasis [27].
In hematopoietic malignancy, the first microRNAs identified were miR15 and miR16-1, which were associated with the pathogenesis of B cell chronic lymphocytic leukemia [28]. Many other miRNAs were also reported in the pathogenesis of the most frequent forms of lymphoma, such as miR15, miR17HG, miR-21, miR-155, miR34A, and miR125B (28, 29) [29]. MiR-21 and miR-155 expression were significantly higher in NK-cell lymphoma [30]. Serum miR-21 and miR-155 were significantly elevated in patients with B-lymphoma and associated with advanced disease stage [31,32]. MiR-155 expression was significantly higher in chronic lymphocytic leukemia, acute myeloid leukemia, and Waldenström’s macroglobulinemia [33]. However, the roles of miR-21 and miR-155 in Burkitt’s lymphoma remain unclear.
The present study investigated the expression of miR-21 and miR-155 in Burkitt’s lymphoma tissues and cell lines. Furthermore, the roles and mechanisms in cell proliferation, cell cycle, and apoptosis after knockdown of miR-21 and miR-155 were examined. Finally, their target genes were predicted and evaluated. We found that miR-21 and miR-155 promote the progression of Burkitt’s lymphoma through PI3K/AKT signaling by targeting C1RL and TCAP. Thus, our findings will provide novel therapeutic strategies for Burkitt’s lymphoma.
Materials and methods
Cell culture
Daudi, Raji and U-937 cell lines were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 mg/mL streptomycin in a 5.0% CO2 incubator at 37°C.
siRNA transfection
Raji cells were seeded into 96-well plates at a density of 5×104/mL cells, and then transfected with miR-21 inhibitor, miR-155 inhibitor or negative control through Lipofectamine 2,000 according to the manufacturer’s instruction. After 48 hours, cells were used for further experiments.
Patient samples
Peripheral blood mononuclear cells (PBMCs) were isolated from lymphoma patients or healthy donors by density centrifugation, respectively. The expression of miR-21 and miR-155 were analyzed by qPCR. Ethical protocols for using lymphoma patient samples were approved by the committee of Qingdao Hiser Hospital Affiliated to Qingdao University.
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted from PBMCs and cell lines by using a RNA extraction kit, and then reverse-transcribed into cDNA by RevertAid First Strand cDNA Synthesis Kit according to the manufacturer’s instruction (Thermo, Shanghai, China). qPCR was used to access the mRNA level of corresponding genes. The expression of actin was used as a control. The 2-ΔΔCt method was used to calculate the relative mRNA levels of the indicated genes. All primers were as follows: MiR-21 RT: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA-3’; MiR-21 forward: 5’-CCAtagcttatcagactga-3’; MiR-21 reverse: 5’-GCAGGGTCCGAGGTATTC-3’; MiR-155- RT: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCCC-3’; MiR-155 forward: 5’-ATTaaTgcTaaTcgTgaTag-3’; miR-155 reverse: 5’-GCAGGGTCCGAGGTATTC-3’; PTEN forward: 5’-ACCCACCACAGCTAGAACTT-3’; PTEN reverse: 5’-GGGAATAGTTACTCCCTTTTTGTC-3’; PI3K forward: 5’-GCAGTTTTGGAAGCAGTCACA-3’; PI3K reverse: 5’-ATTCAGTTCAATTGCAGAAGGAG-3’; AKT forward: 5’-TGCATCCTGGTCCTGTCTTCC-3’; AKT reverse: 5’-CCTAAGCCCCTGGTGACAGAT-3’; SHIP-1 forward: 5’-GTCCTGGTTTCTCTCCAAGGG-3’; SHIP-1 reverse: 5’-ACGATGCGGATGTTCCAGAG-3’; mTOR forward: 5’-CTTAGAGGACAGCGGGGAAG-3’; mTOR reverse: 5’-TGGTTTCCTCATTCCGGCTC-3’; actin forward: 5’-CTCGCTTCGGCAGCACA-3’; actin reverse: 5’-AACGCTTCACGAATTTGCGT-3’.
Cell proliferation assay
The MTT assay and CCK-8 assay were performed to examine the effect of miR-21 and miR-155 on the cell proliferation of Raji cells. Raji cells at a density of 5×104/mL cells were seeded into 96-well plates, and then transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control for 48 hours. At various time points (0 h, 24 h, 48 h, 72 h, 96 h), 10 μL MTT solution or 10 μL CCK-8 solution was added into Raji cells for another 4 hours. Then, the absorbance in each well was measured at 490 nm using a microplate reader.
BrdU incorporation
Raji cells were transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control for 24 hours. BrdU was added into the culture medium in the dark for another 24 hours. Then, cells were incubated with anti-BrdU and the secondary antibody for 1 hour, respectively. Flow cytometry was accessed to detect the cell growth of Raji cells.
Cell cycle analysis
Flow cytometry was performed to explore the effect of miR-21 and miR-155 on the cell cycle of Raji cells. After transfection with miR-21 inhibitor, miR-155 inhibitor or negative control for 48 hours, Raji cells were collected, washed and then fixed with 70% ethanol overnight at 4°C. Raji cells were washed again and incubated with 2 μL RNAse for 30 min at room temperature. Then, 5 μL PI Staining Solution was added for another 30 min in the dark at room temperature. Flow cytometry was performed to analyze the cell cycle change and data were analyzed using the Flowjo software.
Cell apoptosis assay
Cell apoptosis assay was performed by Annexin V-FITC/PI apoptosis detection kit according to the manufacturer’s instruction. After transfected with miR-21 inhibitor, miR-155 inhibitor or negative control for 48 hours, cells were collected and incubated with 5 μL Annexin V-FITC and 10 μL PI Staining Solution in the dark for 15 min at room temperature. Then, flow cytometry was performed to analyze the cell apoptosis rate and data were analyzed using the Flowjo software.
Western blot
RIPA lysis buffer (Beyotime, Shanghai, China) was used to lyse Raji cells transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control. Then, protein extracts were obtained and protein concentration was measured by using a bicinchonininc acid (BCA, Beyotime, Shanghai, China) assay. SDS-PAGE was used to separate the protein extracts, then the protein extracts were transferred to a polyvinylidene fluoride (PVDF) membrane. The PVDF membranes were blocked with 5.0% non-fat milk (BD, San Jose, CA, USA) for 1 hour at room temperature and incubated with antibodies against PTEN, PI3K, AKT, mTOR, SHIP-1, C1RL, TCAP and Actin at 4°C overnight. Then, the membranes were incubated with secondary antibodies for 1 hour at room temperature. Actin was used as a loading control. The blots were visualized using enhanced chemiluminescence (Millipore, Massachusetts, USA).
Statistical analysis
All statistical analyses were performed with SPSS 18.0 (SPSS, Chicago, USA). Data were represented as mean ± standard deviation based on at least three repeats. Group difference was assessed using Student’s t-test. Association was identified by Pearson correlation analysis. P<0.05 was considered significant.
Results
MiR-21 and miR-155 were upregulated in Burkitt’s lymphoma blood samples and cell lines
To evaluate the potential roles of miR-21 and miR-155 in Burkitt’s lymphoma, we first quantified the expression of miR-21 and miR-155 in 15 blood samples of patients with Burkitt’s lymphoma at the mRNA level. Comparing with healthy volunteers, miR-21, and miR-155 were overexpressed in each blood samples, up to 6 fold and 11 fold respectively (P<0.05, Figure 1A).
Figure 1.
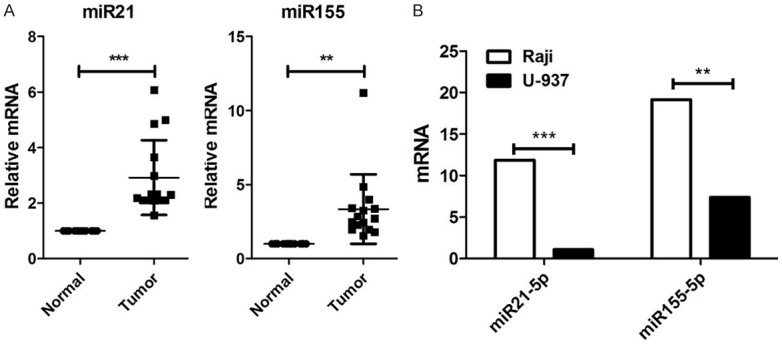
MiR-21 and miR-155 were upregulated in Burkitt’s lymphoma blood samples and cell lines. A. Relative expression levels of miR-21 and miR-155 in blood samples of lymphoma patients (n=15) and healthy donors (n=15) was assessed by qPCR. Data are presented as mean ± SD of fold change. B. Relative expression levels of miR-21, miR-155 in Raji and U-937 lymphoma cell lines. Data are representative of three independent experiments and are shown as the mean ± SD. (**P<0.01, ***P<0.001, Student’s t-test).
Furthermore, we detected the expression of miR-21 and miR-155 in two lymphoma cell lines. The results showed that miR-21-5p and miR-155-5p were highly expressed in Burkitt’s lymphoma cell Raji, in comparison to human histiocytic lymphoma cells U937 (P<0.05, Figure 1B). Taken together, our results demonstrated that miR-21 and miR-155 are upregulated in Burkitt’s lymphoma blood samples and cell lines.
Knockdown of miR-21 and miR-155 suppress cell proliferation
To investigate the biologic functions of miR-21 and miR-155 in the development of Burkitt’s lymphoma, we knocked down miR-21 and miR-155 in Raji cells. The results showed that expression of miR-21 and miR-155 were significantly inhibited (P<0.05, Figure 2A). Then, cell proliferation was assessed by MTT and BrdU incorporation assay. BrdU positive cells were dramatically reduced after transfection with miR-21 and miR-155 inhibitor (P<0.05, Figure 2B). Similarly, after transfection with miR-21 and miR-155 inhibitor for 48 h, 72 h, and 96 h, cell proliferation was significantly suppressed (P<0.05, Figure 2C). Overall, these data indicate that knockdown of miR-21 and miR-155 suppress cell proliferation.
Figure 2.
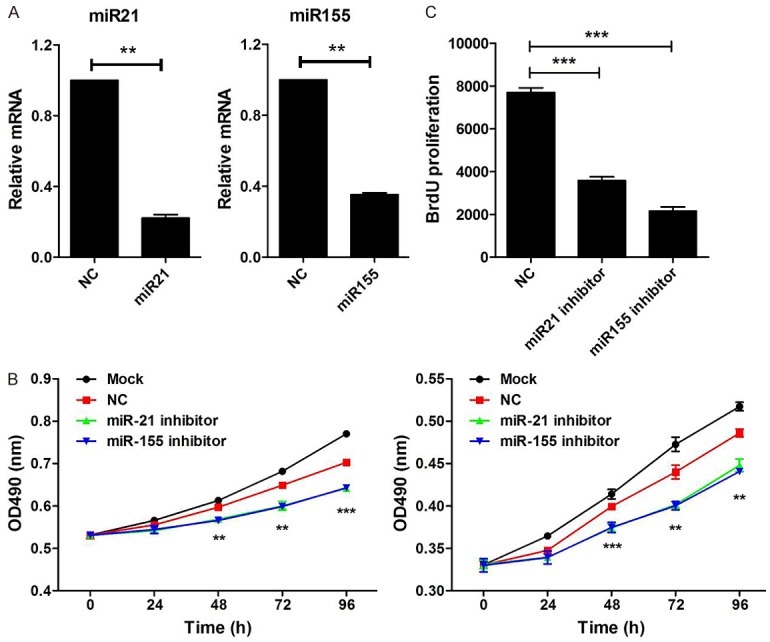
Knockdown of miR-21 and miR-155 suppress cell proliferation. A. Raji cells were transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control (NC) for 48 hours. The expression of miR-21 and miR-155 were measured by qPCR, wh normalized to human actin (n=3). B. Raji cells were transfected with miR-21 inhibitor, miR-155 inhibitor or negative control (NC) for 24 hours. Then, BrdU was added and incubated for another 24 hours. Cell proliferation was analyzed by flow cytometry. C. Raji cells were transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control (NC) for 48 hours. Then, the CCK-8 assay (right) was performed to examine cell proliferation at various time points (0 h, 24 h, 48 h, 72 h, 96 h). Results are shown as absorbance at 490 nm. Data are representative of three independent experiments and shown as the mean ± SD. (**P<0.01, ***P<0.001, Student’s t-test).
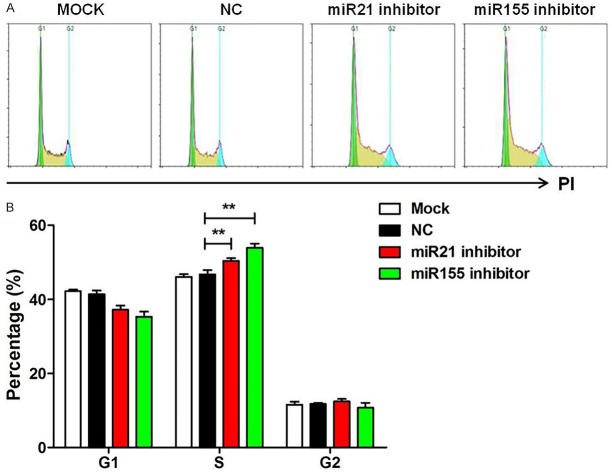
Knockdown of miR-21 and miR-155 induced S phase cell cycle arrest
We further explored the contribution of miR-21 and miR-155 to the cell cycle. MiR-21 and miR-155 were inhibited in Raji cells and cell cycle assay was performed. Compared with control group, Raji cells were significantly arrested at S phase after treatment with miR-21 and miR-155 inhibitor (P<0.05, Figure 3A and 3B).
Figure 3.
Knockdown of miR-21 and miR-155 induced S phase cell cycle arrest. (A) Flow cytometry was performed to detect changesin cell cycle in Raji cells transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control (NC) for 48 hours. (B) The percentage of cells in G1, S and G2 phase was obtained as at (A). Data are representative of three independent experiments and shown as the mean ± SD. (**P<0.01, Student’s t-test).
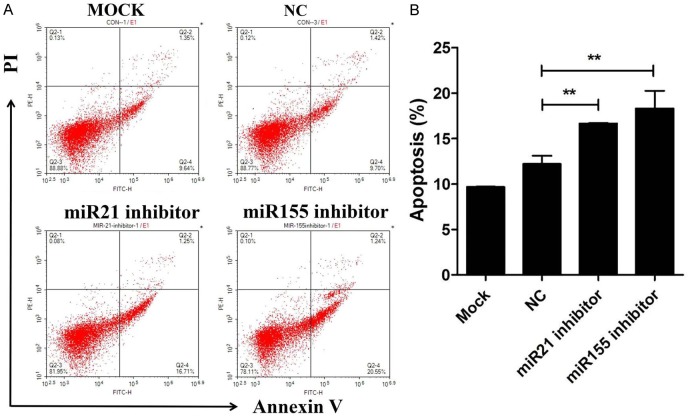
Knockdown of miR-21 and miR-155 promoted cell apoptosis
To further exploit the role of miR-21 and miR-155 in cell apoptosis, Annexin V/PI staining was performed. The results showed that knockdown of miR-21 and miR-155 promoted cell apoptosis (Figure 4A and 4B). Taken together, these results indicate that miR-21 and miR-155 promote progression of Burkitt’s lymphoma.
Figure 4.
Knockdown of miR-21 and miR-155 promoted cell apoptosis. (A) The cell apoptosis was assayed by flow cytometry in Raji cells transfected with miR-21 inhibitor, miR-155 inhibitor, or negative control (NC) for 48 hours. The percentage of apoptotic cells is shown in (B). Data are representative of three independent experiments and shown as the mean ± SD. (**P<0.01, Student’s t-test).
MiR-21 and miR-155 promoted Burkitt’s lymphoma via PI3K/AKT signaling pathway
It was reported that miR-21 accelerates hepatocyte proliferation through the PI3K/AKT/mTOR pathway by targeting PTEN [34,35]. Thus, we detected the expression of PTEN, PI3K and AKT in Raji cells after treatment with miR-21 inhibitor. The expression of PTEN, PI3K and AKT were significantly decreased after the knockdown of miR-21 at the mRNA level and protein level (Figure 5A left and Figure 5B left).
Figure 5.
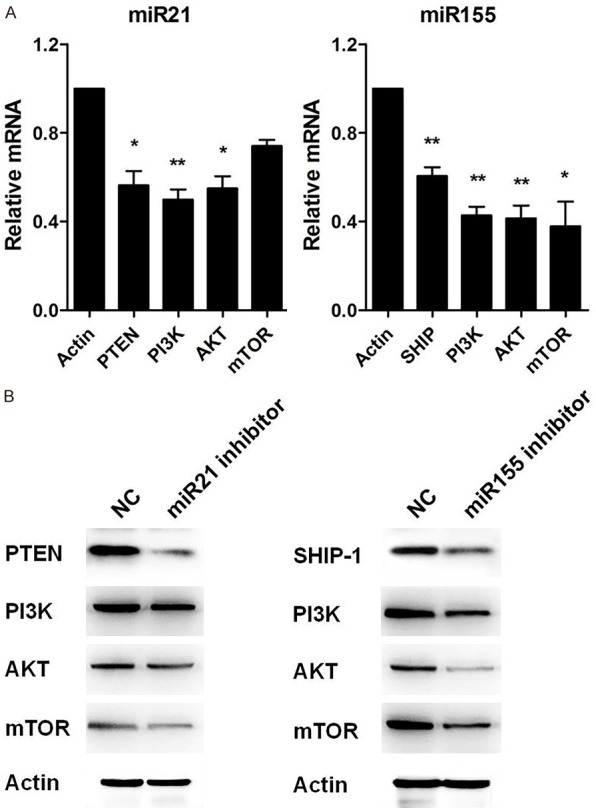
miR-21 and miR-155 promoted Burkitt’s lymphoma via PI3K/AKT signaling pathway. A. qPCR was performed to measure the mRNA levels of PTEN, PI3K, AKT, and mTOR in Raji cells transfected with miR-21 inhibitor or miR-155 inhibitor for 48 hours. B. Western blot was used to detect the protein levels of PTEN, PI3K, AKT, and mTOR in Raji cells transfected with miR-21 inhibitor or miR-155 inhibitor for 48 hours. Actin was used as a control. Data are representative of three independent experiments and shown as the mean ± SD. (*P<0.05, **P<0.01, Student’s t-test).
MiR-155 involved in oxLDLinduced autophagy by PI3K/AKT/mTOR pathway [36]. Triptolide inhibited Fibril-induced microglial activation by the miR-155-5p/SHIP1 Pathway pathway [37]. We next detected the expression of SHIP-1, PI3K, AKT and mTOR after treatment with miR-155 inhibitor. The expression of SHIP-1, PI3K, AKT and mTOR were significantly decreased after knockdown of miR-155 at mRNA level and protein level (Figure 5A right and Figure 5B right). These data illustrate that miR-21 and miR-155 acted through the PI3K/AKT signaling pathway in Burkitt’s lymphoma.
C1RL and TCAP are the targets of miR-21 and miR-155
In order to clarify the mechanisms of miR-21 and miR-155 in the progression of Burkitt’s lymphoma, we used publicly available databases TargetScan 6.2 and miRanda to search the target genes of miR-21 and miR-155 in Raji cells. Among them, the 3’-UTR of C1RL and TCAP were conserved for miR-21 and miR-155 binding. To verify the role of miR-21 and miR-155 in C1RL and TCAP, we detected the expression of C1RL and TCAP after treatment with miR-21 and miR-155 inhibitor. We found that expression of C1RL and TCAP proteins were significantly decreased after the knockdown of miR-21 and miR-155 (Figure 6).
Figure 6.
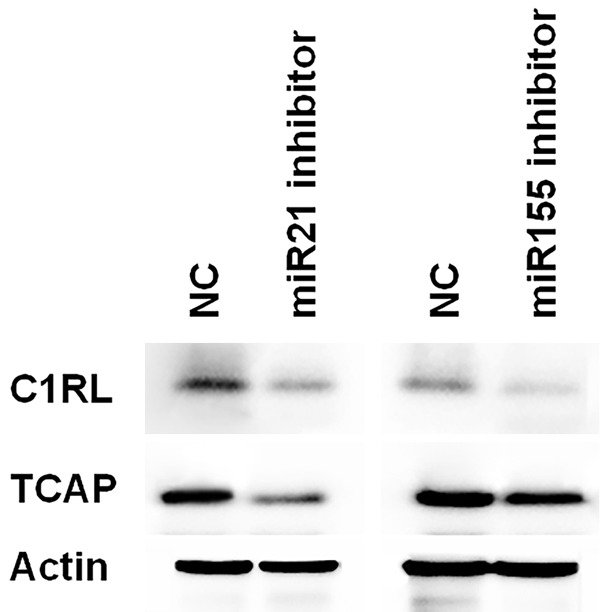
C1RL and TCAP are targets of miR-21 and miR-155. The protein levels of C1RL and TCAP was further detected by western blot in Raji cells transfected with miR-21 inhibitor or miR-155 inhibitor for 48 hours. Actin was used as a control. Data are representative of three independent experiments.
Discussion
In this study, we found that miR-21 and miR-155 were overexpressed in blood samples of Burkitt’s lymphoma. Knockdown of miR-21 and miR-155 significantly inhibited the cell growth, resulted in S phase arrest and cell apoptosis in Burkitt’s lymphoma cells. Furthermore, knockdown of miR-21 and miR-155 significantly suppressed the expression of PI3K and AKT at mRNA and protein levels. In addition, C1RL and TCAP are the target genes of miR-21 and miR-155 according to publicly available databases and knockdown experiments.
Uncontrolled proliferation of cancer cells is a hallmarker of cancer [38]. Previous studies reported that miR-21 and miR-155 were overexpressed and enhanced cell growth in various cancers, such as gastric cancer, glioma, cervical cancer, and non-small cell lung cancer [14-16,23,24]. In this study, we found that miR-21 and miR-155 were overexpressed in the blood samples of Burkitt’s lymphoma, and related to proliferation of Burkitt’s lymphoma cell Raji.
The PI3K/AKT pathway is known for promoting cell growth and inhibiting apoptosis in most hematologic cancers including diffuse large B cell lymphoma, mantle cell lymphoma and T-acute lymphoblastic leukemia [39-41]. MiR-21 accelerates hepatocyte proliferation via PI3K/AKT/mTOR pathway by targeting PTEN [34,35], and miR-155 involved in oxLDLinduced autophagy by PI3K/AKT/mTOR pathway [36]. Our results showed that miR-21 and miR-155 promote the progression of Burkitt’s lymphoma by the PTEN/PI3K/AKT and SHIP-1/PI3K/AKT/mTOR signaling pathways, respectively.
Tumor progression is a complex process including cell growth, migration, invasion, metastasis, colony formation, and adhesion [42,43]. Although we here report that miR-21 and miR-155 promote the progress of Burkitt’s lymphoma, their relationship to survival of patients with this disease, their roles in cell migration, invasion, metastasis, colony formation, and adhesion, and the mechanisms of action in the progress of Burkitt’s lymphoma still need to be further investigated. In addition, we only predicted the target genes of miR-21 and miR-155, but the roles of C1RL and TCAP in the progression of Burkitt’s lymphoma and their interaction are still unknown.
Taken together, miR-21 and miR-155 promote the progression of Burkitt’s lymphoma by PI3K/AKT signaling by targeting C1RL and TCAP, and they could be novel candidate genes to diagnose patients with Burkitt’s lymphoma.
Disclosure of conflict of interest
None.
References
- 1.Burkitt D. A sarcoma involving the jaws in African children. Br J Surg. 1958;46:218–223. doi: 10.1002/bjs.18004619704. [DOI] [PubMed] [Google Scholar]
- 2.Molyneux EM, Rochford R, Griffin B, Newton R, Jackson G, Menon G, Harrison CJ, Israels T, Bailey S. Burkitt’s lymphoma. Lancet. 2012;379:1234–1244. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- 3.Roland S, Young RM, Michele C, Sameer J, Wenming X, Meili Z, George W, Shaffer AL, Hodson DJ, Eric B. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong Q, Wang W, Li P. Regulator role of HPV E7 protein on miR-21 expression in cervical carcinoma cells and its functional implication. Int J Clin Exp Pathol. 2014;8:15808–15813. [PMC free article] [PubMed] [Google Scholar]
- 5.Han Y, Xu GX, Lu H, Yu DH, Ren Y, Wang L, Huang XH, Hou WJ, Wei ZH, Chen YP, Cao YG, Zhang R. Dysregulation of miRNA-21 and their potential as biomarkers for the diagnosis of cervical cancer. Int J Clin Exp Pathol. 2015;8:7131–7139. [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 8.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 9.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 10.Meng F, Francis H, Alpini G. Non-coding RNAs in human liver malignancies: critical regulators for cancer stemness? Translational Gastrointestinal Cancer. 2012;1:1–4. [Google Scholar]
- 11.Yan LX, Liu YH, Xiang JW, Wu QN, Xu LB, Luo XL, Zhu XL, Liu C, Xu FP, Luo DL, Mei P, Xu J, Zhang KP, Chen J. PIK3R1 targeting by miR-21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int J Oncol. 2016;48:471–484. doi: 10.3892/ijo.2015.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong S, Ma W, Hao B, Hu F, Yan L, Yan X, Wang Y, Chen Z, Wang Z. MicroRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int J Clin Exp Pathol. 2014;7:565–574. [PMC free article] [PubMed] [Google Scholar]
- 13.Da Costa Martins PA, De Windt LJ. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res. 2012;93:563–572. doi: 10.1093/cvr/cvs013. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clinica Chimica Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 16.Luo G, Luo W, Sun X, Lin J, Wang M, Zhang Y, Luo W, Zhang Y. MicroRNA21 promotes migration and invasion of glioma cells via activation of Sox2 and βcatenin signaling. Mol Med Rep. 2016;15:187. doi: 10.3892/mmr.2016.5971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Wu YR, Qi HJ, Deng DF, Luo YY, Yang SL. MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal cancer. Tumour Biol. 2016;37:12061–12070. doi: 10.1007/s13277-016-5074-2. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229:545–550. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 19.Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, Ortiz M, Nacken W, Sorg C, Vogl T, Roth J, Gabrilovich DI. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang S, Wang RH, Akagi K, Kim KA, Martin BK, Cavallone L Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer. Haines DC, Basik M, Mai P, Poggi E, Isaacs C, Looi LM, Mun KS, Greene MH, Byers SW, Teo SH, Deng CX, Sharan SK. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat Med. 2011;17:1275–1282. doi: 10.1038/nm.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collazo MM, Paraiso KH, Park MY, Hazen AL, Kerr WG. Lineage extrinsic and intrinsic control of immunoregulatory cell numbers by SHIP. Eur J Immunol. 2012;42:1785–1795. doi: 10.1002/eji.201142092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas C, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan YC, Li T, Han YD, Zhang HY, Lin H, Zhang B. MicroRNA-155 enhances the activation of Wnt/β-catenin signaling in colorectal carcinoma by suppressing HMG-box transcription factor 1. Mol Med Rep. 2016;13:2221–2228. doi: 10.3892/mmr.2016.4788. [DOI] [PubMed] [Google Scholar]
- 24.He B, Gao SQ, Huang LD, Huang YH, Zhang QY, Zhou MT, Shi HQ, Song QT, Shan YF. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep. 2015;11:2355–2359. doi: 10.3892/mmr.2014.2994. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Wang W, Li X, He S, Yao J, Wang X, Zhang D, Sun X. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int J Oncol. 2016;48:2425–2434. doi: 10.3892/ijo.2016.3465. [DOI] [PubMed] [Google Scholar]
- 26.Lei C, Wang Y, Huang Y, Yu H, Huang Y, Wu L, Huang L. Up-regulated miR-155 reverses the epithelial-mesenchymal transition induced by EGF and increases chemo-sensitivity to cisplatin in human Caski cervical cancer cells. PLoS One. 2012;7:e52310. doi: 10.1371/journal.pone.0052310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zare A, Alipoor B, Omrani MD, Zali MR, Malekpour Alamdari N, Ghaedi H. Decreased miR-155-5p, miR-15a, and miR-186 expression in gastric cancer is associated with advanced tumor grade and metastasis. Iran Biomed J. 2019;23:338–343. doi: 10.29252/.23.5.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrie CH. MicroRNAs and lymphomagenesis: a functional review. Br J Haematol. 2013;160:571–581. doi: 10.1111/bjh.12157. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang ZG, Zhang JA, Luo HL, Liu GB, Lu YB, Ge NH, Zheng BY, Li RX, Chen C, Wang X, Liu YQ, Liu FH, Zhou Y, Cai XZ, Chen ZW, Xu JF. The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol. 2017;90:264–272. doi: 10.1016/j.molimm.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Z, Sun R, Zhao HJ, Fu D, Zhong HJ, Weng XQ, Qu B, Zhao Y, Wang L, Zhao WL. MiR-155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol Cancer. 2019;18:54. doi: 10.1186/s12943-019-0977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caivano A, Rocca FL, Simeon V, Girasole M, Dinarelli S, Laurenzana I, Stradis AD, Luca LD, Trino S, Traficante A. MicroRNA-155 in serum-derived extracellular vesicles as a potential biomarker for hematologic malignancies - a short report. Cell Oncol. 2017;40:1–7. doi: 10.1007/s13402-016-0300-x. [DOI] [PubMed] [Google Scholar]
- 34.Bai YN, Yu ZY, Luo LX, Jiang Y, Xia QJ, Yong Z. MicroRNA-21 accelerates hepatocyte proliferation in vitro via PI3K/Akt signaling by targeting PTEN. Biochem Biophys Res Commun. 2014;443:802–807. doi: 10.1016/j.bbrc.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 35.Hong L, Han Y, Zhang Y, Zhang H, Zhao Q, Wu K, Fan D. MicroRNA-21: a therapeutic target for reversing drug resistance in cancer. Expert Opin Ther Targets. 2013;17:1073–1080. doi: 10.1517/14728222.2013.819853. [DOI] [PubMed] [Google Scholar]
- 36.Yin S, Yang S, Pan X, Ma A, Ma J, Pei H, Dong Y, Li S, Li W, Bi X. MicroRNA155 promotes oxLDLinduced autophagy in human umbilical vein endothelial cells by targeting the PI3K/Akt/mTOR pathway. Mol Med Rep. 2018;18:2798–2806. doi: 10.3892/mmr.2018.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Zheng C, Zhang Y, Xing C, Cai W, Li R, Chen J, Duan Y. Triptolide inhibits preformed fibril-induced microglial activation by targeting the microRNA155-5p/SHIP1 pathway. Oxid Med Cell Longev. 2019;2019:6527638. doi: 10.1155/2019/6527638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhu F, Guo H, Bates PD, Zhang S, Zhang H, Nomie KJ, Li Y, Lu L, Seibold KR, Wang F, Rumball I, Cameron H, Hoang NM, Yang DT, Xu W, Zhang L, Wang M, Capitini CM, Rui L. PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia. 2019;33:2898–2911. doi: 10.1038/s41375-019-0489-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao Y, Xu L, Wang J, Zhang L, Hou N, Xu J, Wang L, Yang S, Chen Y, Xiong L, Zhu J, Fan W, Xu J. ROR1 associates unfavorable prognosis and promotes lymphoma growth in DLBCL by affecting PI3K/Akt/mTOR signaling pathway. Biofactors. 2019;45:416–426. doi: 10.1002/biof.1498. [DOI] [PubMed] [Google Scholar]
- 41.Padgaonkar A, Rechkoblit O, Vasquez-Del Carpio R, Pallela V, Venkata Subbaiah D, Cosenza SC, Baker SJ, Ramana Reddy MV, Aggarwal A, Reddy EP. Targeting protein kinase CK2 and CDK4/6 pathways with a multi-kinase inhibitor ON108110 suppresses pro-survival signaling and growth in mantle cell lymphoma and T-acute lymphoblastic leukemia. Oncotarget. 2018;9:37753–37765. doi: 10.18632/oncotarget.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 43.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]