Abstract
Urban alteration of neutral and adaptive evolutionary processes is still underexplored. Using a genome-wide SNP dataset, we investigated (i) urban-induced modifications of population demography, genetic diversity and population structure and (ii) signature of divergent selection between urban and forest populations in the ant species, Temnothorax nylanderi. Our results did not reveal an impact of urbanization on neutral processes since we observed: (i) analogous genetic diversity among paired urban/forest sites and two control populations; (ii) weak population genetic structure explained neither by habitat (urban versus forest) nor by geography; (iii) a remarkably similar demographic history across populations with an ancestral growth followed by a recent decline, regardless of their current habitat or geographical location. The micro-geographical home range of ants may explain their resilience to urbanization. Finally, we detected 19 candidate loci discriminating urban/forest populations and associated with core cellular components, molecular function or biological process. Two of these loci were associated with a gene ontology term that was previously found to belong to a module of co-expressed genes related to caste phenotype. These results call for transcriptomics analyses to identify genes associated with ant social traits and to infer their potential role in urban adaptation.
Keywords: RADseq, urban adaptation, population genomics, social insects, random forest, triploid
1. Background
Urbanization alters natural habitats [1–3], leading to the extirpation of native species and the establishment of non-native species, and promoting biotic homogenization [4–6]. Nevertheless, some species are able to persist in urban landscapes providing the unique opportunity to assess the consequences of urbanization on both neutral and adaptive evolutionary processes [7]. Habitat modification resulting from urbanization involves habitat loss and fragmentation. These changes may induce a rapid decline of genetic diversity and an increase of population differentiation in species with small effective population size whose dispersal is hampered by the new habitat configuration [8–10]. Environmental changes resulting from urbanization may also impose new selective pressures, ultimately leading to genotypic and phenotypic differences between urban and non-urban populations [11–13]. Several studies reported urban-induced shifts in phenotypic traits, including life history, morphology, behaviour, physiology and reproductive traits. Although mechanisms underlying such phenotypic changes are unknown for most organisms, some common garden experiments disentangled phenotypic plasticity from genetic adaptation to urban-induced environmental changes [14,15]. Because such an approach allows study of only a restricted number of traits that are relatively straightforward to measure, difficult-to-measure but potentially informative traits may be overlooked [16]. Genomic data may help to overcome this limitation and to identify genomic signatures of local adaptation. Only a small number of studies have profited from genome-wide data to search for the footprints of divergent selection between urban and non-urban populations (but see [17–19]). In addition, they focused on a single urban area although replication over multiple urban areas is required to understanding urban adaptation [7,20].
Ants are frequently used as bio-indicators in ecological studies and monitoring programmes: they respond rapidly to environmental changes and have extensively adapted to a wide range of environments [21,22]. Studies on the consequences of urbanization on the genetic variability of ant species are rare (but see [23,24]), generally investigating the community level and reporting contrasted effects on species assemblages [25]. Here, using a replicated design over three cities, we explored (i) urban-induced modifications of genetic diversity, population structure and demography; (ii) signature of divergent selection between urban and forest populations in Temnothorax nylanderi, a forest leaf litter-dwelling, cavity nesting, small-sized predatory ant found in small colonies of 50–200 workers. An absence of genetic structure in Western Europe was previously suggested for this species based on one allozyme and two mitochondrial genes [26]. However, single-locus (such as mtDNA) inference of spatial genetic differentiation is not reliable [27] and further instigation based on genome-wide loci is warranted. To this end, we generated a genome-wide SNP dataset for 96 colonies belonging to eight localities (12 colonies per locality). Colonies were sampled in wooded parts of historical urban parks (e.g. the Jardin des Plantes of Paris acquired its present form in the seventeenth century, around 400 generations before present considering a generation time of 1 year) of three highly populated French cities: Bordeaux (BorC), Lyon (LyoC) and Paris (ParC). Forest localities (BorF, LyoF and ParF) were chosen to achieve a paired study design including three replicates, each consisting of one urban and one forest locality in close geographical proximity (30–50 km) but functionally disconnected. Two additional forest localities (Bretagne, Bre and Villefranche, Vil) were included to provide a better coverage of the T. nylanderi geographical distribution (figure 1). Because of the tiny size of T. nylanderi (ca 3 mm), we randomly pooled 50 workers per colony to obtain a suitable amount of DNA. We followed a single-digestion RADseq protocol [28], individually barcoding each colony. Temnothorax nylanderi colonies are predominantly monogynous and monoandrous [29]: pooling several workers from a colony results in sequencing three alleles per locus, since workers carry the allele from the male and one of the two alleles from the queen. Barcoding a colony is therefore analogous to barcoding a triploid genotype. Popular pipelines performing de novo assembly and SNP calling for RADseq data cannot handle triploid individuals. To circumvent this problem, we split the bioinformatics analysis of raw fastq reads into two steps: (i) we first built loci using the denovo_map pipeline implemented in STACKS [30]; (ii) we artificially built a reference sequence using the assembled loci and mapped reads back using BWA v0.7.15 [31]. Triploid genotypes were called after local realignment using GATK v3.8-1-0 [32].
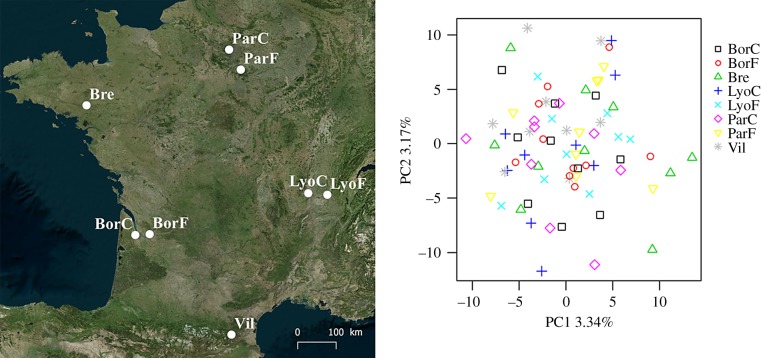
Figure 1.
Sampled localities and individual genotypes plotted in the space of the first two axes of a principal coordinate axis (PCA). Colonies were sampled in wooded parts of urban parks (BorC, LyoC and ParC) and in forest localities (BorF, LyoF, ParF, Bre and Vil). (Online version in colour.)
To test the hypothesis of urban-induced reductions of population size, genetic diversity and gene flow, we estimated within-population diversity, in all sampled localities, by calculating the observed heterozygosity (HO) using the gametic homozygosity concept of Moody et al. [33], the expected heterozygosity (HE) and multilocus FIS values corrected for the ploidy level following Hardy [34]. We also investigated the genetic structure by: (i) a principal component analysis (PCA) on triploid genotype frequencies; (ii) a non-metric multidimensional scaling (NMDS) on the matrix of FST pairwise distances. Finally, for each locality, we investigated the variation of the scaled mutation rate (Θ, the product of the effective population size Ne and the mutation rate per generation µ) using the stairwayplot software [35] and the abc-skyline method of [36]. To investigate the potential genomic signature of local adaptation to urban landscape in T. nylanderi, we applied two complementary random forest (RF) variable selection procedures, implemented in the packages VSURF [37] and Boruta [38], by feeding genotypes as discriminating variables for each pair of urban/forest localities.
Full details on the methodology for this study are in electronic supplementary material, S1.
2. Results and discussion
(a). Locus assembly and data filtering
Following [39], the optimal set of STACKS's parameters resulted in m3, M3 and n3 (electronic supplementary material, S2). After implementing a newly developed pipeline for building triploid genotypes in non-model organisms, we obtained the final filtered dataset composed of 5728 polymorphic loci and 10 723 SNP for 75 colonies, with an average coverage of 47X and 4.9% of missing data.
(b). Genetic diversity and population structure
Expected (HE) and observed heterozygosity (HO) averaged across loci ranged, respectively, from 0.216 to 0.235 and from 0.217 to 0.243 (table 1). We observed similar values between each pair of forest (F) and urban (C) sites. Multilocus FIS corrected for the ploidy level were slightly positive within each sampled site and ranged from 0 (BorC) to 0.046 (LyoF), suggesting that populations within each site are at Hardy–Weinberg equilibrium.
Table 1.
Estimates of genetic diversity. For each site, geographical coordinates, the number of sampled colonies (N), expected (HE) and observed (HO) heterozygosity and FIS values are provided.
| locality | site | latitude | longitude | N | HE | HO | FIS |
|---|---|---|---|---|---|---|---|
| Bor | BorC | 44°51'16.60″ N | 0°35'17.21″ O | 10 | 0.224 | 0.231 | 0 |
| BorF | 44°52'23.61″ N | 0° 8'15.25″ O | 11 | 0.216 | 0.217 | 0.028 | |
| Lyo | LyoC | 45°46'43.87″ N | 4°51'20.35″ E | 9 | 0.229 | 0.230 | 0.034 |
| LyoF | 45°44'40.28″ N | 5°27'23.36″ E | 9 | 0.230 | 0.228 | 0.046 | |
| Par | ParC | 48°50'39.39″ N | 2°21'43.45″ E | 9 | 0.235 | 0.243 | 0.003 |
| ParF | 48°25'42.14″ N | 2°43'50.81″ E | 9 | 0.235 | 0.237 | 0.029 | |
| Bre | Bre | 47°40'37.84″ N | 2° 7'30.27″ O | 9 | 0.230 | 0.233 | 0.027 |
| Vil | Vil | 42°35'9.77″ N | 2°26'13.07″ E | 9 | 0.233 | 0.234 | 0.032 |
Principal component analyses revealed a lack of structure (figure 1), as colonies were not separated on the basis of their origin for any of the first 20 axes (representing 48% of the total variance). Consistently, both the global FST (FST = 0.028) and the pairwise FST values suggest that most of the observed genetic variance is partitioned within sites. Indeed, none of the pairwise FST values exceeded 0.039 (LyoF versus Bre), despite many comparisons being significant at the 0.05 level (electronic supplementary material, S3). The lack of genetic structure driven by habitat type was confirmed in two out of three forest versus urban comparisons by the NDMS (electronic supplementary material, S4), the only exception being Lyon (in agreement with the pairwise FST, electronic supplementary material, S3). An absence of genetic structure in Western Europe was previously suggested for this species based on one allozyme and two mitochondrial genes [26]. Here we confirmed this finding, strengthened not only by genome-wide data, but also by the finer spatial sampling. The dispersal flight of sexuals over large distances and/or the passive transport of established colonies in acorns and hazelnuts by vertebrates including humans [40] could allow high level of gene flow and may explain this absence of contemporary genetic structure.
(c). Demographic history and adaptation
We inferred the demographic history of the eight populations using stairwayplot (figure 2) and the abc-skyline. After removing missing data, an average of 2800 polymorphic loci were available for each population, the majority of which have one SNP only (approx. 90% of the loci). The inferred variation of Θ through time was substantially unchanged whether the singletons were included or not in the case of the stairwayplot (figure 2). The results reveal an initial growth followed by a recent decline for all populations analysed, regardless of their current habitat. This suggests that the observed demographic history is the result of events independent from the contemporary urbanization process. The lack of a robust estimation of the genome-wide molecular rate hampers a flawless dating of the observed demography. We therefore preferred to report estimates of Θ and scaled time. However, we note that using the evolutionary rate proposed for the whole ant group by Romiguier et al. [41], the expansion and the following decline (figure 2) would have occurred around approximately 50 000 and approximately 20 000 years B.P. respectively, further strengthening the idea that the historical demography of T. nylanderi was not affected by urbanization. We further check the robustness of the stairwayplot, the performance of which has been recently questioned for datasets of less than approximately 10 000 SNPs [42], by running the abc-skyline (electronic supplementary material, S4). Results were in agreement, suggesting that the recent decline is not an artefact of the inferential methods but the result of real biological processes shared among all sampled sites regardless of the habitat type.
Figure 2.
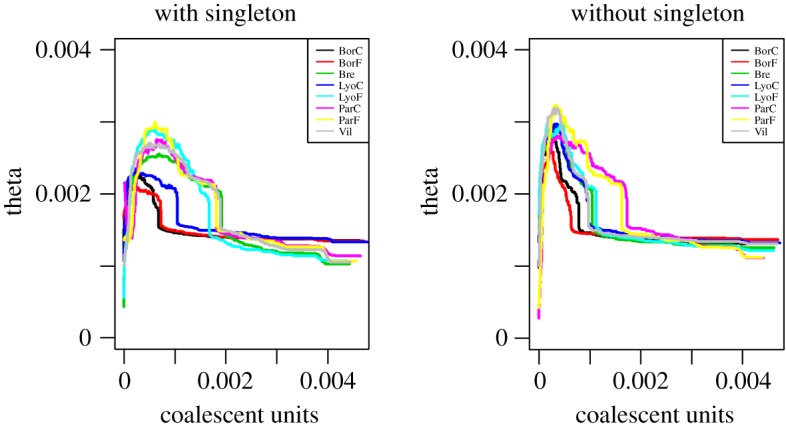
Maximized composite likelihood of Θ through time obtained through the stairwayplot method on the folded SFS. The inferred effective size Θ is plotted, from left to right, from present time (0) to the past (in scaled units of time). (Online version in colour.)
The demographic signature and the absence of contemporary genetic structure are compatible with the scenario of rapid post-glacial recolonization from southern refuges hosting genetically homogeneous populations previously invoked in this species [26]. However, the causes of the decline observed in all populations remain to be further explored. A recent increase in connectivity may result in an apparent decrease of Θ in populations analysed with unstructured models such as the stairwayplot [43]. This explanation would be consistent with the low population differentiation found, but the causes of such a recent increase in connectivity remain to be elucidated.
The two selection procedures identified 19 common SNPs that discriminate urban and forest paired sites (three for Par and eight for both Bor and Lyo, electronic supplementary material, S5). To validate RF results, following [44], we further computed FST at the selected SNPs between urban and forest paired sites and found an approximately 10 times increase compared to values based on the whole SNP dataset, with all three comparisons becoming significant. These SNPs are therefore efficient in discriminating forest versus urban landscape and could be considered candidates for being under divergent selection. We recorded five loci with significant homologies after BLASTn searches (table 2). The Blat2GO annotation tool identified a protein kinase C, involved in catalysis of a molecular reaction, from two different candidate loci found in Bor and Lyo, strongly suggesting the role of this protein family in urban adaptation. An uncharacterized protein recognized as an integral component of membrane and a serine/arginine repetitive matrix protein involved in biological processes related to mRNA processing were also identified (table 2). The mRNA processing GO term associated with the locus carrying the SNP 827337 (table 2) was previously found to belong to a module of co-expressed genes correlated with caste phenotype [45]. The genetic toolkit hypothesis posits that conserved sets of genes and gene pathways involved in core physiological processes have been repeatedly used in the evolution of complex social behaviour [46]. Recent studies identified modules of co-expressed genes whose up- and downregulation is associated with phenotypic traits related to social behaviour (e.g. caste, worker sterility) and species ecology (e.g. invasiveness) [45,47]. Further transcriptomics analyses will help to identify genes related to social traits associated with adaptation to urban habitat.
Table 2.
Candidate loci discriminating urban and forest sites and corresponding functional annotation (Gene Ontology GO).
| paired urban–forest sites | SNP IDs | BLASTn E-values | description of significant hits | GO IDs | GO names |
|---|---|---|---|---|---|
| BorC–BorF | 775198 | no significant similarity found | — | — | — |
| BorC–BorF | 782672 | 1.00 × 10−7 | protein kinase C | GO:0004697 | protein kinase C activity |
| BorC–BorF | 855369 | no significant similarity found | — | — | — |
| BorC–BorF | 1416915 | no significant similarity found | — | — | — |
| BorC–BorF | 1471596 | no significant similarity found | — | — | — |
| BorC–BorF | 1640863 | no significant similarity found | — | — | — |
| BorC–BorF | 2343564 | 6.00 × 10−20 | ultra-conserved locus | no GO found | no GO found |
| BorC–BorF | 2627927 | no significant similarity found | — | — | — |
| LyoC–LyoF | 280520 | no significant similarity found | — | — | — |
| LyoC–LyoF | 395140 | no significant similarity found | — | — | — |
| LyoC–LyoF | 813323 | 4.00 × 10−22 | protein kinase C | GO:0004697 | protein kinase C activity |
| LyoC–LyoF | 827337 | 4.00 × 10−56 | serine/arginine repetitive matrix protein | GO:0006397 | mRNA processing |
| LyoC–LyoF | 1567225 | no significant similarity found | — | — | — |
| LyoC–LyoF | 2136348 | 3.00 × 10−58 | uncharacterized protein | GO:0016021 | integral component of membrane |
| LyoC–LyoF | 2602998 | no significant similarity found | — | — | — |
| LyoC–LyoF | 3297929 | no significant similarity found | — | — | — |
| ParC–ParF | 808830 | no significant similarity found | — | — | — |
| ParC–ParF | 865740 | no significant similarity found | — | — | — |
| ParC–ParF | 2841848 | no significant similarity found | — | — | — |
3. Conclusion
Recent empirical research on urban dweller species has shown that urbanization may increase genetic differentiation among populations through a reduction of functional connectivity and an increase of genetic drift [8,9,18,48,49]. Our results, substantially strengthened by the congruence among the replicated landscapes, support a different scenario and call for a species-specific null model in urbanization studies. Despite living in fragmented patches of habitat, colonies of T. nylanderi are apparently genetically not isolated. The demographic reconstruction suggests that this is the consequence of past demographic events rather than urbanization. Moreover, a recent increase in connectivity is possibly ongoing, contributing to further hinder the effects of urbanization on genetic patterns. Indeed, with the intensification of transport networks, human-mediated dispersal contributes, in some species like ants, to gene flow by transgressing dispersal barriers [50,51].
Discrepancies between observed and expected consequences of urbanization on neutral evolutionary processes were previously described in studies on ant communities structure, suggesting that biotic homogenization has not taken place in this group at the European scale [52].
Models of natural habitat islands isolated within an urban matrix that reduces functional connectivity are often used to describe patterns of vertebrate diversity. Our results provide evidence, at another organizational level of biodiversity (within-species genetic diversity), that these models may not be relevant for small species exploiting micro-habitats [22,25]. Such ecological and/or life-history requirements drive species-specific patterns of landscape functional connectivity. Species-centred approaches offer considerable promise to predict species sensitivity to human-induced landscape alteration [53,54].
Despite the predominant influence of the demographic history, we found evidence of divergent selection at 19 loci; among them, four are known to be involved in core cellular components, molecular functions or biological processes. Although recent transcriptomics studies in ants (e.g. [45,47]) identified conserved functional genomic units involved in social behaviour traits, their potential role in adaptation to urban habitats was not investigated. Group living, by allowing workers to manipulate the environment of the developing larvae, could buffer environmental variations between urban and forest environments [55,56]. Nevertheless, we found signatures of divergent selection between urban and forest sites, potentially linked to sociality (table 2). This is in agreement with recent phenotypic studies that suggest rapid phenotypic differentiation in urban populations in response to heavy metal pollution in T. nylanderi [57], rapid temperature increases in T. curvispinosus [14,58] and human food inputs [59], indicating that further transcriptomic studies under controlled conditions are warranted.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to the Genotoul platform (http://bioinfo.genotoul.fr/) for computing resources. We thank Nicolas Navarro.
Ethics
All applicable national and/or institutional guidelines for the collection of animals were followed.
Data accessibility
Home-made code to reproduce the analysis of population structure (PCA, Fst and bootstrap confidence interval, NMDS) and the filtered vcf with one SNP per locus are deposited in Dryad and they are accessible with this temporary review link: https://datadryad.org/stash/share/9y3WVtfMO1YPoszoIvaoHlOGFiaaW7U6EPqJIUvq3Zs.
Authors' contributions
C.D. and M.M. designed the study; A.K., R.P., C.D., M.M., B.K. and P.A.E. performed fieldwork; A.K., S.M. and C.D. analysed the data; A.K., S.M. and C.D. drafted the manuscript and all authors contributed to the final version. All authors agree to be held accountable for the content therein and approve the final version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by ANR grant no. ANTEVO ANR-12-JSV7-0003-01 and by the ARP EVOL from the Ecole Pratique des Hautes Etudes.
References
- 1.McKinney ML. 2002. Urbanization, biodiversity, and conservation: the impacts of urbanization on native species are poorly studied, but educating a highly urbanized human population about these impacts can greatly improve species conservation in all ecosystems. BioScience 52, 883–890. ( 10.1641/0006-3568(2002)052[0883:UBAC]2.0.CO;2) [DOI] [Google Scholar]
- 2.Pickett STA, Cadenasso ML, Grove JM, Nilon CH, Pouyat RV, Zipperer WC, Costanza R. 2001. Urban ecological systems: linking terrestrial ecological, physical, and socioeconomic components of metropolitan areas. Annu. Rev. Ecol. Syst. 32, 127–157. ( 10.1146/annurev.ecolsys.32.081501.114012) [DOI] [Google Scholar]
- 3.Shochat E, Warren PS, Faeth SH, McIntyre NE, Hope D. 2006. From patterns to emerging processes in mechanistic urban ecology. Trends Ecol. Evol. 21, 186–191. ( 10.1016/j.tree.2005.11.019) [DOI] [PubMed] [Google Scholar]
- 4.McKinney ML. 2006. Urbanization as a major cause of biotic homogenization. Biol. Conserv. 127, 247–260. ( 10.1016/j.biocon.2005.09.005) [DOI] [Google Scholar]
- 5.Devictor V, Julliard R, Couvet D, Lee A, Jiguet F. 2007. Functional homogenization effect of urbanization on bird communities. Conserv. Biol. 21, 741–751. ( 10.1111/j.1523-1739.2007.00671.x) [DOI] [PubMed] [Google Scholar]
- 6.Holway DA, Suarez AV. 2006. Homogenization of ant communities in mediterranean California: the effects of urbanization and invasion. Biol. Conserv. 127, 319–326. ( 10.1016/j.biocon.2005.05.016) [DOI] [Google Scholar]
- 7.Johnson MTJ, Munshi-South J. 2017. Evolution of life in urban environments. Science 358, eaam8327 ( 10.1126/science.aam8327) [DOI] [PubMed] [Google Scholar]
- 8.Björklund M, Ruiz I, Senar JC. 2010. Genetic differentiation in the urban habitat: the great tits (Parus major) of the parks of Barcelona city. Biol. J. Linn. Soc. 99, 9–19. ( 10.1111/j.1095-8312.2009.01335.x) [DOI] [Google Scholar]
- 9.Noël S, Ouellet M, Galois P, Lapointe F-J. 2007. Impact of urban fragmentation on the genetic structure of the eastern red-backed salamander. Conserv. Genet. 8, 599–606. ( 10.1007/s10592-006-9202-1) [DOI] [Google Scholar]
- 10.Wandeler P, Funk SM, Largiadèr CR, Gloor S, Breitenmoser U. 2003. The city-fox phenomenon: genetic consequences of a recent colonization of urban habitat. Mol. Ecol. 12, 647–656. ( 10.1046/j.1365-294X.2003.01768.x) [DOI] [PubMed] [Google Scholar]
- 11.Alberti M. 2015. Eco-evolutionary dynamics in an urbanizing planet. Trends Ecol. Evol. 30, 114–126. ( 10.1016/j.tree.2014.11.007) [DOI] [PubMed] [Google Scholar]
- 12.Alberti M, Marzluff J, Hunt VM. 2017. Urban driven phenotypic changes: empirical observations and theoretical implications for eco-evolutionary feedback. Phil. Trans. R. Soc. B 372, 20160029 ( 10.1098/rstb.2016.0029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelletier F, Coltman DW. 2018. Will human influences on evolutionary dynamics in the wild pervade the Anthropocene? BMC Biol. 16, 7 ( 10.1186/s12915-017-0476-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond SE, Chick LD, Perez A, Strickler SA, Zhao C. 2018. Evolution of plasticity in the city: urban acorn ants can better tolerate more rapid increases in environmental temperature. Conserv. Physiol. 6, 30 ( 10.1093/conphys/coy030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RA, Chick LD, Yilmaz AR, Diamond SE. 2019. Evolution, not transgenerational plasticity, explains the adaptive divergence of acorn ant thermal tolerance across an urban–rural temperature cline. Evol. Appl. 12, 1678–1687. ( 10.1111/eva.12826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudman SM, et al. 2018. What genomic data can reveal about eco-evolutionary dynamics. Nat. Ecol. Evol. 2, 9–15. ( 10.1038/s41559-017-0385-2) [DOI] [PubMed] [Google Scholar]
- 17.Perrier C, Lozano del Campo A, Szulkin M, Demeyrier V, Gregoire A, Charmantier A. 2018. Great tits and the city: distribution of genomic diversity and gene–environment associations along an urbanization gradient. Evol. Appl. 11, 593–613. ( 10.1111/eva.12580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munshi-South J. 2012. Urban landscape genetics: canopy cover predicts gene flow between white-footed mouse (Peromyscus leucopus) populations in New York City. Mol. Ecol. 21, 1360–1378. ( 10.1111/j.1365-294X.2012.05476.x) [DOI] [PubMed] [Google Scholar]
- 19.Harris SE, Munshi-South J. 2017. Signatures of positive selection and local adaptation to urbanization in white-footed mice (Peromyscus leucopus). Mol. Ecol. 26, 6336–6350. ( 10.1111/mec.14369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson JL, Brady SP, Wang IJ, Spear SF. 2016. Navigating the pitfalls and promise of landscape genetics. Mol. Ecol. 25, 849–863. ( 10.1111/mec.13527) [DOI] [PubMed] [Google Scholar]
- 21.Andersen AN, Fisher A, Hoffmann BD, Read JL, Richards R. 2004. Use of terrestrial invertebrates for biodiversity monitoring in Australian rangelands, with particular reference to ants. Austral Ecol. 29, 87–92. ( 10.1111/j.1442-9993.2004.01362.x) [DOI] [Google Scholar]
- 22.Pacheco R, Vasconcelos HL. 2007. Invertebrate conservation in urban areas: ants in the Brazilian Cerrado. Landsc. Urban Plan. 81, 193–199. ( 10.1016/j.landurbplan.2006.11.004) [DOI] [Google Scholar]
- 23.Menke SB, Booth W, Dunn RR, Schal C, Vargo EL, Silverman J. 2010. Is it easy to be urban? Convergent success in urban habitats among lineages of a widespread native ant. PLoS ONE 5, e9194 ( 10.1371/journal.pone.0009194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto J, Uchida K, Takami Y. 2013. Colonization and persistence of urban ant populations as revealed by joint estimation of kinship and population genetic parameters. J. Hered. 104, 639–648. ( 10.1093/jhered/est041) [DOI] [PubMed] [Google Scholar]
- 25.Menke SB, Guénard B, Sexton JO, Weiser MD, Dunn RR, Silverman J. 2011. Urban areas may serve as habitat and corridors for dry-adapted, heat tolerant species; an example from ants. Urban Ecosyst. 14, 135–163. ( 10.1007/s11252-010-0150-7) [DOI] [Google Scholar]
- 26.Pusch K, Seifert B, Foitzik S, Heinze J. 2006. Distribution and genetic divergence of two parapatric sibling ant species in Central Europe. Biol. J. Linn. Soc. 88, 223–234. ( 10.1111/j.1095-8312.2006.00618.x) [DOI] [Google Scholar]
- 27.Teske PR, Golla TR, Sandoval-Castillo J, Emami-Khoyi A, van der Lingen CD, von der Heyden S, Chiazzari B, van Vuuren BJ, Beheregaray LB. 2018. Mitochondrial DNA is unsuitable to test for isolation by distance. Sci. Rep. 8, 1–9. ( 10.1038/s41598-018-25138-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 ( 10.1371/journal.pone.0003376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foitzik S, Haberl M, Gadau J, Heinze J. 1997. Mating frequency of Leptothorax nylanderi ant queens determined by microsatellite analysis. Insectes Soc. 44, 219–227. ( 10.1007/s000400050043) [DOI] [Google Scholar]
- 30.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140. ( 10.1111/mec.12354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinforma. Oxf. Engl. 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moody ME, Mueller LD, Soltis DE. 1993. Genetic variation and random drift in autotetraploid populations. Genetics 134, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy OJ. 2016. Population genetics of autopolyploids under a mixed mating model and the estimation of selfing rate. Mol. Ecol. Resour. 16, 103–117. ( 10.1111/1755-0998.12431) [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Fu Y-X. 2015. Exploring population size changes using SNP frequency spectra. Nat. Genet. 47, 555–559. ( 10.1038/ng.3254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maisano DP, Corrigan S, Hale M, Li C, Veuille M, Planes S, Naylor G, Mona S. 2016. Population genomics of C. melanopterus using target gene capture data: demographic inferences and conservation perspectives. Sci. Rep. 6, 33753 ( 10.1038/srep33753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genuer R, Poggi J-M, Tuleau-Malot C. 2015. VSURF: an R package for variable selection using random forests. R J. 7, 9–33. ( 10.32614/RJ-2015-018) [DOI] [Google Scholar]
- 38.Kursa MB, Rudnicki WR. 2010. Feature selection with the Boruta package. J. Stat. Softw. 36, 1–13. ( 10.18637/jss.v036.i11) [DOI] [Google Scholar]
- 39.Paris JR, Stevens JR, Catchen JM. 2017. Lost in parameter space: a road map for stacks. Methods Ecol. Evol. 8, 1360–1373. ( 10.1111/2041-210X.12775) [DOI] [Google Scholar]
- 40.Seifert B. 1995. Two new Central European subspecies of Leptothorax nylanderi (Förster, 1850) and Leptothorax sordidulus Müller, 1932 (Hymenoptera: Formicidae). Abhandlungen und Berichte Des Naturkundemuseums Görlitz 68, 1–18. [Google Scholar]
- 41.Romiguier J, et al. 2014. Population genomics of eusocial insects: the costs of a vertebrate-like effective population size. J. Evol. Biol. 27, 593–603. ( 10.1111/jeb.12331) [DOI] [PubMed] [Google Scholar]
- 42.Lapierre M, Lambert A, Achaz G. 2017. Accuracy of demographic inferences from the site frequency spectrum: the case of the Yoruba population. Genetics 206, 439–449. ( 10.1534/genetics.116.192708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez W, Mazet O, Grusea S, Boitard S, Chikhi L. 2018. The IICR and the non-stationary structured coalescent: demographic inference with arbitrary changes in population structure. bioRxiv 341750 ( 10.1101/341750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laporte M, et al. 2016. RAD sequencing reveals within-generation polygenic selection in response to anthropogenic organic and metal contamination in North Atlantic eels. Mol. Ecol. 25, 219–237. ( 10.1111/mec.13466) [DOI] [PubMed] [Google Scholar]
- 45.Morandin C, et al. 2016. Comparative transcriptomics reveals the conserved building blocks involved in parallel evolution of diverse phenotypic traits in ants. Genome Biol. 17, 43 ( 10.1186/s13059-016-0902-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toth AL, Robinson GE. 2007. Evo-devo and the evolution of social behavior. Trends Genet. 23, 334–341. ( 10.1016/j.tig.2007.05.001) [DOI] [PubMed] [Google Scholar]
- 47.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinforma. Oxf. Engl. 21, 3674–3676. ( 10.1093/bioinformatics/bti610) [DOI] [PubMed] [Google Scholar]
- 48.Munshi-South J, Kharchenko K. 2010. Rapid, pervasive genetic differentiation of urban white-footed mouse (Peromyscus leucopus) populations in New York City. Mol. Ecol. 19, 4242–4254. ( 10.1111/j.1365-294X.2010.04816.x) [DOI] [PubMed] [Google Scholar]
- 49.Munshi-South J, Zolnik CP, Harris SE. 2016. Population genomics of the Anthropocene: urbanization is negatively associated with genome-wide variation in white-footed mouse populations. Evol. Appl. 9, 546–564. ( 10.1111/eva.12357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertelsmeier C, Ollier S, Liebhold A, Keller L. 2017. Recent human history governs global ant invasion dynamics. Nat. Ecol. Evol. 1, 0184 ( 10.1038/s41559-017-0184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capinha C, Essl F, Seebens H, Moser D, Pereira HM. 2015. The dispersal of alien species redefines biogeography in the Anthropocene. Science 348, 1248–1251. ( 10.1126/science.aaa8913) [DOI] [PubMed] [Google Scholar]
- 52.Vepsäläinen K, Ikonen H, Koivula MJ. 2008. The structure of ant assemblages in an urban area of Helsinki, southern Finland. Ann. Zool. Fenn. 45, 109–128. ( 10.5735/086.045.0203) [DOI] [Google Scholar]
- 53.Betts MG, Fahrig L, Hadley AS, Halstead KE, Bowman J, Robinson WD, Wiens JA, Lindenmayer DB. 2014. A species-centered approach for uncovering generalities in organism responses to habitat loss and fragmentation. Ecography 37, 517–527. ( 10.1111/ecog.00740) [DOI] [Google Scholar]
- 54.Haila Y. 2002. A conceptual genealogy of fragmentation research: from island biogeography to landscape ecology. Ecol. Appl. 12, 321–334. ( 10.2307/3060944) [DOI] [Google Scholar]
- 55.Karlik J, Epps MJ, Dunn RR, Penick CA. 2016. Life inside an acorn: how microclimate and microbes influence nest organization in Temnothorax ants. Ethology 122, 790–797. ( 10.1111/eth.12525) [DOI] [Google Scholar]
- 56.Scharf I, Modlmeier AP, Fries S, Tirard C, Foitzik S. 2012. Characterizing the collective personality of ant societies: aggressive colonies do not abandon their home. PLoS ONE 7, e33314 ( 10.1371/journal.pone.0033314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacquier L, Doums C, Péronnet R, Four-Chaboussant A, Tirard C, Molet M. In preparation. Deleterious impacts of a trace metal on forest colonies of the ant Temnothorax nylanderi are mitigated in urban colonies.
- 58.Chick LD, Strickler SA, Perez A, Martin RA, Diamond SE. 2019. Urban heat islands advance the timing of reproduction in a social insect. J. Therm. Biol. 80, 119–125. ( 10.1016/j.jtherbio.2019.01.004) [DOI] [PubMed] [Google Scholar]
- 59.Penick CA, Savage AM, Dunn RR. 2015. Stable isotopes reveal links between human food inputs and urban ant diets. Proc. R. Soc. B 282, 20142608 ( 10.1098/rspb.2014.2608) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Home-made code to reproduce the analysis of population structure (PCA, Fst and bootstrap confidence interval, NMDS) and the filtered vcf with one SNP per locus are deposited in Dryad and they are accessible with this temporary review link: https://datadryad.org/stash/share/9y3WVtfMO1YPoszoIvaoHlOGFiaaW7U6EPqJIUvq3Zs.