Abstract
Parasites often infect genetically diverse host populations, and the evolutionary trajectories of parasite populations may be shaped by levels of host heterogeneity. Mixed genotype host populations, compared to homogeneous host populations, can reduce parasite prevalence and potentially reduce rates of parasite adaptation due to trade-offs associated with adapting to specific host genotypes. Here, we used experimental evolution to select for increased virulence in populations of the bacterial parasite Serratia marcescens exposed to either heterogeneous or homogeneous populations of Caenorhabditis elegans. We found that parasites exposed to heterogeneous host populations evolved significantly less virulence than parasites exposed to homogeneous host populations over several hundred bacterial generations. Thus, host heterogeneity impeded parasite adaptation to host populations. While we detected trade-offs in virulence evolution, parasite adaptation to two specific host genotypes also resulted in modestly increased virulence against the reciprocal host genotypes. These results suggest that parasite adaptation to heterogeneous host populations may be impeded by both trade-offs and a reduction in the efficacy of selection as different host genotypes exert different selective pressures on a parasite population.
Keywords: Caenorhabditis elegans, Serratia marcescens, virulence, parasite evolution, monoculture, host heterogeneity
1. Introduction
Hosts and parasites are ubiquitous in nature. A long-standing goal in evolutionary biology is to understand the reciprocal selective pressures exerted by host and parasite interactions [1]. Theoretical and empirical studies point to multiple factors that can determine the rate and magnitude of parasite adaptation to hosts. These factors include host genetic heterogeneity [2,3], host spatial structure [4,5], competition [6,7], and migration and gene flow [8,9]. Of particular interest is how host genotypes influence the evolutionary trajectory of parasites populations as they adapt to host populations.
Historically, host heterogeneity has been overlooked in theoretical models of infection dynamics [10,11], yet host heterogeneity is both biologically relevant and a potential source of selection driving parasite evolution. Host homogeneity is generally rare in natural populations, even in many asexual hosts [12,13]. Theoretical models of host heterogeneity predict that specialization on similar host genotypes results in reduced transmission between dissimilar genotypes, which leads to lower parasite prevalence [3,14]. Due to this trade-off, parasite prevalence tends to be mitigated compared with homogeneous populations, known as the monoculture effect [15]. Evidence for the monoculture effect has been found in agriculture systems [16–19] and natural populations [20–27], in which prevalence differs between homogeneous and heterogeneous populations.
Heterogeneous populations may impede parasite adaptation and thus limit virulence. In some cases, host genetic diversity can even prevent parasite adaptation altogether [3]. Host diversity reduces the average rate at which parasites successfully infect hosts [28], thereby limiting specialization on a single host genotype. Here, we asked whether heterogeneity per se is sufficient to alter parasite evolution by examining virulence in populations with different ratios of host genotypes. Further, if homogeneity leads to greater virulence, is there a cost of adapting to one specific genotype when parasitizing novel hosts, resulting in a fitness loss?
We used experimental evolution to select for virulence while passaging parasites through either genetically homogeneous or heterogeneous host populations. We predicted that heterogeneous host populations would impede virulence evolution and that parasites evolved in homogeneous host populations would evolve greater virulence by specializing on a single host genotype. Further, we expected to see a cost of specialization when infecting a new host genotype. To test these predictions, we evolved a clonal bacterial parasite, Serratia marcescens (Sm2170), in two genotypes of the host Caenorhabditis elegans. The C. elegans genotypes used were CB4856 and ewIR 68 [29]. The two strains have genetically diverse backgrounds but identical portions of chromosome V, where many innate immune loci reside. CB4856 and ewIR 68 were chosen to minimize trade-offs of specialization as a means to better isolate heterogeneity as a variable. For our experimental treatments, we varied the ratio of the host genotypes in each host population. We then compared the mortality rates of the evolved parasites from each treatment to the ancestral parasites by infecting each host genotype separately.
2. Methods
(a). Experimental evolution
Using experimental evolution, we imposed selection for increased virulence on S. marcescens Sm2170 parasites exposed to either homogeneous or heterogeneous host populations. Hosts were the C. elegans strains ewIR 68 and CB4586. S. marcescens infection occurs upon feeding. Some live bacterial cells survive ingestion [30] and infect the host [31]. We measured virulence as infection-induced host mortality rate, and imposed selection for increased virulence by passaging Sm2170 only from hosts that died after 24 hpi (see electronic supplementary material, figure S1 for detailed experimental design). Thus, parasite genotypes that facilitated rapid killing were favoured. We passaged Sm2170 populations through five different host treatments and a control in which parasites were passaged in the absence of hosts (in vitro, 0–0) (electronic supplementary material, figure S2).
For each passage of experimental evolution, we plated 1000 worms on a Serratia selection plate (electronic supplementary material, figure S1) and allowed the worms to consume Sm2170 for 24 h [32,33]. We then isolated 30 dead worms from the Sm2170 lawn. Dead worms were identified by a lack of movement in response to provocation with a platinum wire [34]. Then, we extracted Sm2170 from the hosts, cultured them in standard laboratory conditions (28°C shaker overnight) and inoculated an unseeded nematode growth media (US Biological, Salem, MA, USA) plate to grow colony-forming units (CFUs) for 48 h at room temperature. From these plates, we randomly picked 40 CFUs per Sm2170 population, to inoculate the next passage. New naive (non-evolved) hosts (from homozygous host lines kept at −80°C) were then placed on the evolved bacteria and the process was repeated. For our in vitro control (0–0), 40 CFUs of Sm2170 were picked from the bacterial lawn. This treatment served as our control for passage conditions. The selection experiment concluded at the end of 10 passages (totalling hundreds of bacterial generations). At the end of each passage, a subset of the evolved bacteria was stored at −80°C.
(b). Mortality assays
Mortality assays were used to determine virulence at the beginning and end of the experiment. Bacteria from passage 10 were used to infect homogeneous groups of either host genotype, and mortality rates were compared to the ancestral bacteria. The steps outlined in the creation of the Serratia selection plates were identical to those of the mortality assays (see electronic supplementary material, figures S1 and S2).
We placed 200 worms from one genotype on a mortality assay plate (electronic supplementary material, figure S2, step 1). After 48 h at 20°C, the number of dead worms on each plate was counted (electronic supplementary material, figure S2, step 2). Mortality rates were calculated as the proportion of dead worms divided by the number plated. When performing mortality assays, each replicate population had 3–6 technical replicates for a total of 360 mortality assay plates (electronic supplementary material, figure S2, step 3). Ancestral mortality assays were performed both at the outset of the experiment and again when performing evolved Sm2170 mortality assays at the end of the experiment (electronic supplementary material, figures S2 and S3).
(c). Statistical analysis
To assess the mean changes in mortality rate between ancestral and evolved populations, we used JMP Pro 14 (SAS, Cary, NC, USA) to perform a generalized linear model (GLM) with a link logit function and normal distribution. Factors in the model include treatment (e.g. homogeneous, heterogeneous, in vitro), host genotypes in mortality assays (ewIR 68 or CB4856) and the interaction. We did not detect overdispersion using a Pearson test. Post-analysis Tukey contrast tests were used to determine the significance of pair-wise comparisons. We report our values as χ2 statistics and corresponding p-values. Multiple comparisons were corrected for using a Bonferroni correction of p < 0.025 (p < a/k, where a = 0.05, k = 2 comparisons: host genotype and parasite treatment).
3. Results
The ancestral populations of Sm2170 bacteria tested at the beginning of experimental evolution produced a mean mortality rate of 49.51% (s.e.m. ± 0.03) in host strain ewIR 68 and 64.32% (s.e.m. ± 0.04) in host strain CB4856 [35]. As predicted, we found that selection for virulence resulted in an increase in mortality when experimental populations were assayed concurrently with the ancestral population. Parasites evolved in both homogeneous host populations were significantly more virulent than the in vitro controls (CB4856: χ2 = 29.13, p < 0.0001; ewIR 68: χ2 = 14.68, p = 0.0001, figure 1; electronic supplementary material, tables S2 and S3). Parasites evolved in CB4856 hosts had a 29% increase in mortality rate in CB4856 populations compared to the ancestor, while parasites evolved in ewIR 68 had a 19% increase in mortality rate in ewIR 68 populations compared to the ancestor.
Figure 1.
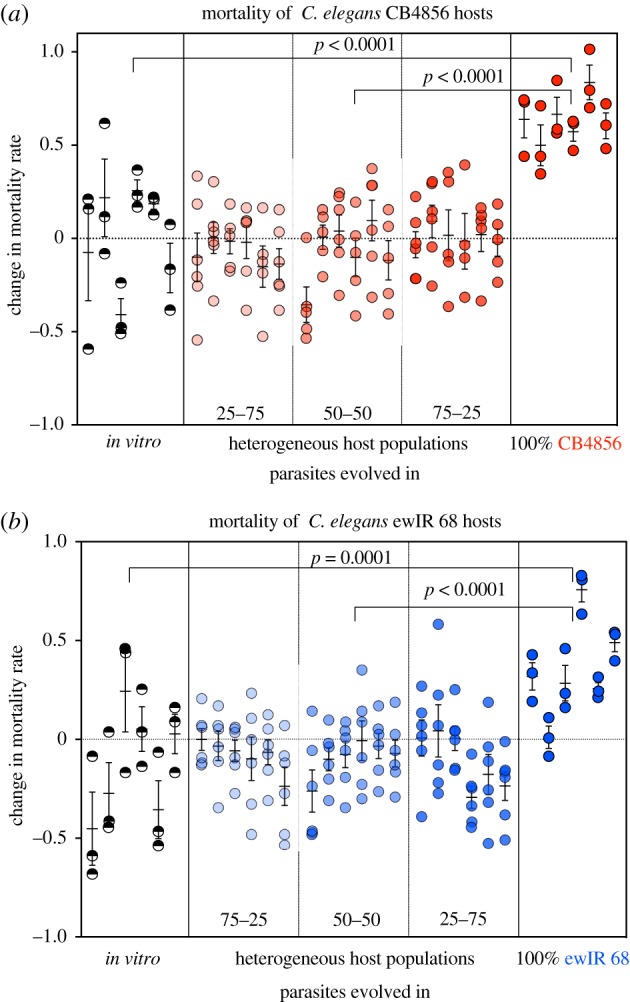
(a,b) Mean change in host mortality rate relative to the ancestral parasites in C. elegans host strains CB4856 (a) and ewIR 68 (b). All experimental populations shared a common ancestor, and thus, any change from the ancestral data is indicative of relative virulence. Parasites were evolved in heterogeneous host populations, homogeneous host populations or in vitro (no hosts), and then tested for changes in virulence. The heterogeneous populations, from left to right, are 75–25, 50–50 and 25–75. Circles represent the mean change within each technical replicate (18–36 each). Bars represent ± s.e.m. (Online version in colour.)
There were no significant differences in mortality induced by parasites in either host between any of the pairs evolved on 75–25, 50–50 or 25–75 (figure 1). When tested in CB4856 hosts, parasites evolved in heterogeneous host populations did not differ significantly in mortality rate from the in vitro parasites (χ2 = 0.0023, p = 0.96, figure 1a; electronic supplementary material, table S2), indicating little to no adaptation to the CB4856 host genotype. Further, the same parasites exhibited no significant increase in mortality rate compared to in vitro parasites when tested in ewIR 68 hosts (χ2 = 0.00002, p = 0.99, figure 1b; electronic supplementary material, table S3). Parasites evolved in homogeneous ewIR 68 populations caused greater virulence in ewIR 68 than parasites evolved in heterogeneous populations (χ2 = 18.37, p < 0.0001, figure 1b; electronic supplementary material, table S3). Additionally, parasites evolved in homogeneous CB4856 populations caused greater virulence in CB4856 hosts than parasites evolved in heterogeneous populations (χ2 = 52.99, p < 0.0001, figure 1a; electronic supplementary material, table S2). Overall, these results demonstrate that host heterogeneity impedes parasite adaptation relative to host homogeneity.
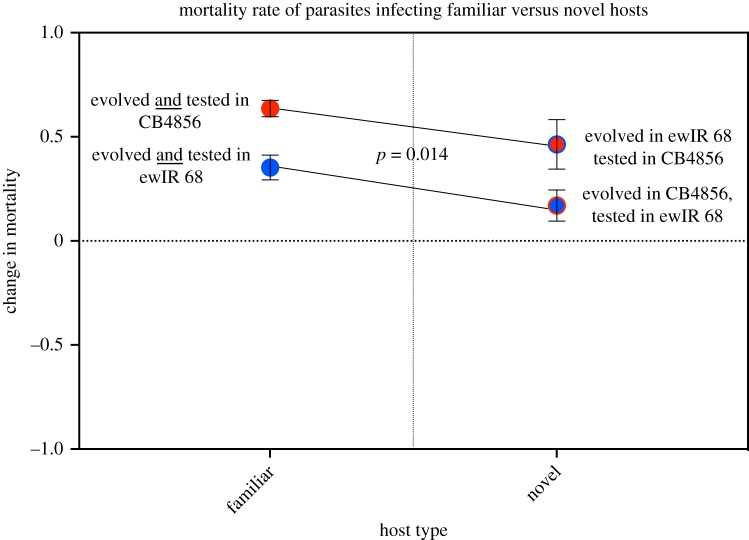
Next, we determined if parasites that were evolved on homogeneous hosts and then exposed to a novel host exhibited reduced virulence, and thus lowered fitness, as predicted by trade-off theory. In both cross-infections, there was an increase in mortality rate relative to the ancestral strain and relative to the in vitro controls (figure 2; electronic supplementary material, table S1). Further, cross-infections were significantly different from one another (χ2 = 6.04, p = 0.014, figure 2; electronic supplementary material, table S1), indicating that although parasites caused high mortality in novel hosts, they did not increase to the same extent as parasites in familiar hosts. Despite this difference, the result overall is in accordance with the previous finding: that heterogeneous host populations limit the evolution of parasite virulence and indicate a trade-off imposed by host heterogeneity.
Figure 2.
Each dot represents the treatment's average change in mortality rate of all populations and replicates relative to ancestral parasites. All experimental populations shared a common ancestor, and thus, any change from the ancestral data is indicative of relative virulence. The x-axis shows the type of host infected: either familiar to the parasite or novel. The p-value is based on a post-GLM Tukey contrast test between all familiar hosts (left panel) and all novel hosts (right panel) (χ2 = 6.04, p = 0.01). In both cases, although all treatments had an increased mortality rate relative to the ancestor, novel hosts had a lower mortality rate than do the familiar hosts. Bars around mean represent s.e.m. (Online version in colour.)
4. Discussion
In our selection regime, higher host mortality equates to higher virulence, and thus greater parasite fitness. Our results show that parasites selected in homogeneous host populations evolved substantial increases in virulence when infecting those same hosts (for both ewIR 68 and CB4856) when compared with in vitro controls (figure 1). However, parasites that were selected in mixed genotype host populations and then tested on homogeneous host genotypes exhibited limited increases in virulence (figure 1), despite strong selection favouring increased virulence. We found no differences in the mortality rates of hosts infected by parasites evolved with any mixed host population on either host—neither comparing between each mixed treatment nor compared with the in vitro control. Thus, exposure to heterogeneous host populations impeded virulence evolution relative to exposure to homogeneous hosts. Further, parasites evolved in homogeneous populations and then used to cross-infect the other (novel) host genotype exhibited smaller increases from the ancestral virulence than when infecting their familiar host (figure 2). Therefore, we observed trade-offs in virulence due to specialization on the parasites' familiar host genotype.
Trade-offs in parasite virulence due to specialization on a particular host genotype are often invoked as a reason that heterogeneous host populations may impede parasite adaptation. Here, we found that heterogeneous host populations impeded the evolution of parasite virulence and we found evidence of trade-offs in parasite virulence (figure 1). However, the evolved trade-offs in parasite virulence that we observed are not sufficient to explain the limited virulence evolution in parasites evolved with heterogeneous host populations. Despite parasite specialization (greater virulence) on familiar homogeneous hosts, parasites evolved in homogeneous host populations still exhibited increased virulence against novel hosts relative to the in vitro control parasites (figure 2). Therefore, any potential cost of a trade-off should have been mitigated in the heterogeneous host populations, as adaptation to either host genotype still resulted in increased virulence against the other host genotype. Yet, we still observed a limited response to selection for increased virulence in parasites evolving in heterogeneous host populations (figure 1).
One possibility for the lack of a substantial trade-off cost (i.e. a decline in parasite fitness) may be that the C. elegans genotypes used, CB4856 and ewIR 68, share an identical region of chromosome V, which harbours loci associated with innate immunity [36]. It is likely that parasites evolved in either genotype were under strong selection to evolve in response to that particular region of the genome. Despite the genetic similarity of the strains at many innate immune system loci, heterogeneous populations still impeded parasite adaptation relative to homogeneous populations. While it is plausible that trade-offs in virulence slowed parasite adaptation in the heterogeneous host populations to some degree, trade-offs alone are insufficient to explain the lack of increase in virulence exhibited by heterogeneous-selected parasites when infecting CB4856 hosts (figure 1). We hypothesize that this lack of response to selection was likely driven by a reduction in the efficacy of selection in the heterogeneous host populations relative to the homogeneous hosts. Selection imposed by different host genotypes can act on different groups of loci in the parasite genome [37]. As a result, the efficacy of selection on a particular set of loci in the parasites may be reduced in the heterogeneous hosts as parasites encounter different host genotypes with each infection [38]. Although a portion of our host genomes were identical, the diverse genetic backgrounds of the CB4856 and ewIR 68 strains may have imposed fluctuating selection on the parasite populations, resulting in limited parasite adaptation within heterogeneous host populations. Another possibility is that specialization on a single host, as opposed to a generalist strategy, may lead to a stronger strength of selection over time. Thus, our results at passage 10 may be the result of stronger selection in homogeneous populations as specialization increases [39].
Heterogeneous host populations are shown to be common in nature [40–44], and our results demonstrate that heterogeneity can alter the trajectory of parasite evolution. Importantly, parasites are capable of adapting to heterogeneous host populations [45]. Nonetheless, our results indicate that parasite adaptation can be impeded by heterogeneous relative to homogeneous host populations. While we observed little cost to host specialization in our experiment, trade-offs are likely to impede rates of parasite adaptation in heterogeneous host populations [46]. We anticipate that changes in the efficacy of selection imposed by heterogeneous host populations may also contribute to reduce rates of parasite adaptation. Therefore, we believe it is critical to understand the implications of host heterogeneity for disease evolution. The ability to manage parasite virulence in both human infectious diseases, agriculture and in the conservation of wildlife has long been a goal of research on parasite evolution [47]. Our results indicate that increasing host heterogeneity may not only be useful for decreasing disease prevalence and spread, but also for hindering parasite adaptation and virulence evolution.
Supplementary Material
Data accessibility
The datasets supporting this article are available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.3bk3j9kdw [35].
Authors' contributions
P.S.W., J.d.R. and L.M. conceived of the experiment. P.S.W., L.M. and A.K.G. designed the experiment. P.S.W., A.C., R.P. and A.M. carried out the experimental evolution. P.S.W., A.M. and M.P. performed and counted mortality assays, P.S.W., L.M. and J.d.R. carried out data analysis. P.S.W. wrote the manuscript. L.M. and J.d.R. critically revised the manuscript; A.C., R.P., A.M., M.P. and A.K.G. all provided manuscript edits and suggestions. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by National Science Foundation (grant no. DEB-1750553).
References
- 1.Ebert D, Bull JJ. 2008. The evolution and expression of virulence. In Evolution in health and disease (eds Stearns SC, Koella JC), pp. 153–168. Oxford, UK: Oxford University Press; ( 10.1093/acprof:oso/9780199207466.003.0012) [DOI] [Google Scholar]
- 2.Regoes RR, Nowak MA, Bonhoeffer S. 2000. Evolution of virulence in a heterogeneous host population. Evolution (NY) . 54, 64–71. ( 10.1111/j.0014-3820.2000.tb00008.x) [DOI] [PubMed] [Google Scholar]
- 3.Morley D, Broniewski JM, Westra ER, Buckling A, van Houte S.. 2017. Host diversity limits the evolution of parasite local adaptation. Mol. Ecol. 26, 1756–1763. ( 10.1111/mec.13917) [DOI] [PubMed] [Google Scholar]
- 4.Haraguchi Y, Sasaki A. 2000. The evolution of parasite virulence and transmission rate in a spatially structured population. J. Theor. Biol. 203, 85–96. ( 10.1006/jtbi.1999.1065) [DOI] [PubMed] [Google Scholar]
- 5.Boots M, Mealor M. 2007. Local interactions select for lowering pathogen infectivity. Science 315, 1284–1286. ( 10.1126/science.1137126) [DOI] [PubMed] [Google Scholar]
- 6.De Roode JC, Culleton R, Cheesman SJ, Carter R, Read AF. 2004. Host heterogeneity is a determinant of competitive exclusion or coexistence in genetically diverse malaria infections. Proc. R. Soc. B 271, 1073–1080. ( 10.1098/rspb.2004.2695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mideo N, Alizon S, Day T. 2008. Linking within- and between-host dynamics in the evolutionary epidemiology of infectious diseases. Trends Ecol. Evol. 23, 511–517. ( 10.1016/j.tree.2008.05.009) [DOI] [PubMed] [Google Scholar]
- 8.Lively CM. 1996. Host–parasite coevolution and sex. Bioscience 46, 107–114. ( 10.2307/1312813) [DOI] [Google Scholar]
- 9.Lion S, Gandon S. 2015. Evolution of spatially structured host–parasite interactions. J. Evol. Biol. 28, 10–28. ( 10.1111/jeb.12551) [DOI] [PubMed] [Google Scholar]
- 10.May RM, Anderson RM. 1983. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. Lond. B 219, 281–313. ( 10.1098/rspb.1983.0075) [DOI] [PubMed] [Google Scholar]
- 11.Bremermann HJ, Pickering J. 1983. A game-theoretical model of parasite virulence. J. Theor. Biol. 100, 411–426. ( 10.1016/0022-5193(83)90438-1) [DOI] [PubMed] [Google Scholar]
- 12.Fontcuberta García-Cuenca A, Dumas Z, Schwander T. 2016. Extreme genetic diversity in asexual grass thrips populations. J. Evol. Biol. 29, 887–899. ( 10.1111/jeb.12843) [DOI] [PubMed] [Google Scholar]
- 13.Dybdahl MF, Lively CM. 1995. Diverse, endemic and polyphyletic clones in mixed populations of a freshwater snail (Potamopyrgus antipodarum). J. Evol. Biol. 8, 385–398. ( 10.1046/j.1420-9101.1995.8030385.x) [DOI] [Google Scholar]
- 14.Kaltz O, Shykoff JA. 1998. Local adaptation in host–parasite systems. Heredity (Edinb). 81, 361–370. ( 10.1046/j.1365-2540.1998.00435.x) [DOI] [Google Scholar]
- 15.Ekroth AKE, Rafaluk-Mohr C, King KC. 2019. Host genetic diversity limits parasite success beyond agricultural systems: a meta-analysis. Proc. R. Soc. B 286, 20191811 ( 10.1098/rspb.2019.1811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett KA, Mundt CC. 1999. Epidemiology in mixed host populations. Phytopathology 89, 984–990. ( 10.1094/PHYTO.1999.89.11.984) [DOI] [PubMed] [Google Scholar]
- 17.Elton CS. 1958. The ecology of invasions by animals and plants. New York, NY: John Wiley. [Google Scholar]
- 18.Zhu Y, et al. 2000. Genetic diversity and disease control in rice. Nature 406, 718–722. ( 10.1038/35021046) [DOI] [PubMed] [Google Scholar]
- 19.Pilet F, Chacón G, Forbes GA, Andrivon D. 2006. Protection of susceptible potato cultivars against late blight in mixtures increases with decreasing disease pressure. Phytopathology 96, 777–783. ( 10.1094/PHYTO-96-0777) [DOI] [PubMed] [Google Scholar]
- 20.Schmid B. 1994. Effects of genetic diversity in experimental stands of Solidago altissima—evidence for the potential role of pathogens as selective agents in plant populations. J. Ecol. 82, 165 ( 10.2307/2261395) [DOI] [Google Scholar]
- 21.van Houte S, et al. 2016. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. ( 10.1038/nature17436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baer B, Schmid-Hempel P. 1999. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature 397, 151–154. ( 10.1038/16451) [DOI] [Google Scholar]
- 23.Baer B, Schmid-Hempel P. 2001. Unexpected consequences of polyandry for parasitism and fitness in the bumblebee, Bombus terrestris. Evolution (NY) 55, 1639–1643. ( 10.1111/j.0014-3820.2001.tb00683.x) [DOI] [PubMed] [Google Scholar]
- 24.Ganz HH, Ebert D. 2010. Benefits of host genetic diversity for resistance to infection depend on parasite diversity. Ecology 91, 1263–1268. ( 10.1890/09-1243.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altermatt F, Ebert D. 2008. Genetic diversity of Daphnia magna populations enhances resistance to parasites. Ecol. Lett. 11, 918–928. ( 10.1111/j.1461-0248.2008.01203.x) [DOI] [PubMed] [Google Scholar]
- 26.Campbell G, Noble LR, Rollinson D, Southgate VR, Webster JP, Jones CS. 2010. Low genetic diversity in a snail intermediate host (Biomphalaria pfeifferi Krass, 1848) and schistosomiasis transmission in the Senegal River Basin. Mol. Ecol. 19, 241–256. ( 10.1111/j.1365-294X.2009.04463.x) [DOI] [PubMed] [Google Scholar]
- 27.Pearman PB, Garner TWJ. 2005. Susceptibility of Italian agile frog populations to an emerging strain of Ranavirus parallels population genetic diversity. Ecol. Lett. 8, 401–408. ( 10.1111/j.1461-0248.2005.00735.x) [DOI] [Google Scholar]
- 28.Gandon S, Nuismer SL. 2009. Interactions between genetic drift, gene flow, and selection mosaics drive parasite local adaptation. Am. Nat. 173, 212–224. ( 10.1086/593706) [DOI] [PubMed] [Google Scholar]
- 29.Doroszuk A, Snoek LB, Fradin E, Riksen J, Kammenga J. 2009. A genome-wide library of CB4856/N2 introgression lines of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 37, e110 ( 10.1093/nar/gkp528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avery L, You Y. 2012. C. elegans feeding. (21 May 2012). WormBook (ed. The C. elegans Research Community). See http://www.wormbook.org. ( 10.1895/wormbook.1.150.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulenburg H, Kurz CL, Ewbank JJ. 2004. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198, 36–58. ( 10.1111/j.0105-2896.2004.0125.x) [DOI] [PubMed] [Google Scholar]
- 32.Morran LT, Parmenter MD, Phillips PC. 2009. Mutation load and rapid adaptation favour outcrossing over self-fertilization (supplementary information). Nature 462, 350–352. ( 10.1038/nature08496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penley MJ, Morran LT. 2018. Assessment of Caenorhabditis elegans competitive fitness in the presence of a bacterial parasite. Bio-Protocol 8, 1–14. ( 10.21769/BioProtoc.2971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amrit FRG, Ratnappan R, Keith SA, Ghazi A. 2014. The C. elegans lifespan assay toolkit. Methods 68, 465–475. ( 10.1016/j.ymeth.2014.04.002) [DOI] [PubMed] [Google Scholar]
- 35.White PS, Choi A, Menezes A, Pandey R, Penley MJ, Gibson AK, de Roode JC, Morran LT. 2019. Data from: Host heterogeneity mitigates virulence evolution Dryad Digital Repository. ( 10.5061/dryad.3bk3j9kdw) [DOI] [PMC free article] [PubMed]
- 36.Glater EE, Rockman MV, Bargmann CI. 2014. Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens. G3 4, 265–276. ( 10.1534/g3.113.008649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croll D, McDonald BA. 2017. The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems. Mol. Ecol. 26, 2027–2040. ( 10.1111/mec.13870) [DOI] [PubMed] [Google Scholar]
- 38.Bell G. 2010. Fluctuating selection: the perpetual renewal of adaptation in variable environments. Phil. Trans. R. Soc. B 365, 87–97. ( 10.1098/rstb.2009.0150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawecki TJ. 1998. Red queen meets Santa Rosalia: arms races and the evolution of host specialization in organisms with parasitic lifestyles. Am. Nat. 152, 635–651. ( 10.1086/286195) [DOI] [PubMed] [Google Scholar]
- 40.van Baalen M, Beekman M.. 2006. The costs and benefits of genetic heterogeneity in resistance against parasites in social insects. Am. Nat. 167, 568–577. ( 10.1086/501169) [DOI] [PubMed] [Google Scholar]
- 41.Lively CM. 2010. The effect of host genetic diversity on disease spread. Am. Nat. 175, 1–4. ( 10.1086/652430) [DOI] [PubMed] [Google Scholar]
- 42.Berngruber TW, Lion S, Gandon S. 2015. Spatial structure, transmission modes and the evolution of viral exploitation strategies. PLoS Pathog. 11, e1004810 ( 10.1371/journal.ppat.1004810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubinak JL, Potts WK. 2013. Host resistance influences patterns of experimental viral adaptation and virulence evolution. Virulence 4, 410–418. ( 10.4161/viru.24724) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan J, Mundt CC, Hoffer ME, McDonald BA. 2002. Local adaptation and effect of host genotype on the rate of pathogen evolution: an experimental test in a plant pathosystem. J. Evol. Biol. 15, 634–647. ( 10.1046/j.1420-9101.2002.00428.x) [DOI] [Google Scholar]
- 45.Koskella B, Lively CM. 2007. Advice of the rose: experimental coevolution of a trematode parasite and its snail host. Evolution (NY) 61, 152–159. ( 10.1111/j.1558-5646.2007.00012.x) [DOI] [PubMed] [Google Scholar]
- 46.Gibson AK, Baffoe-Bonnie HS, Penley MJ, Lin J, Owens R, Khalid A, Morran LT. 2019. The evolution of parasite host range in genetically diverse host populations. bioRxiv ( 10.1101/653675). [DOI] [PMC free article] [PubMed]
- 47.Dieckmann U, Metz JAJ, Sabelis MW, Sigmund K (eds). 2002. Adaptive dynamics of infectious disease. In Pursuit of virulence management. Cambridge, UK: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- White PS, Choi A, Menezes A, Pandey R, Penley MJ, Gibson AK, de Roode JC, Morran LT. 2019. Data from: Host heterogeneity mitigates virulence evolution Dryad Digital Repository. ( 10.5061/dryad.3bk3j9kdw) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The datasets supporting this article are available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.3bk3j9kdw [35].