Abstract
AIM
To observe the effect of inhibiting long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) on diabetic neurodegeneration.
METHODS
Thirty-six 8-week-old C57BL/6 mice were randomly divided into normal control, diabetic control, diabetic scrambled small interfering RNAs (siRNAs) and diabetic MALAT1-siRNA groups. After diabetic induction with streptozocin intraperitoneally-injection, the diabetic MALAT1-siRNA group was intravitreally injected with 1 µL 20 µmol/L MALAT1 siRNA, and the diabetic scrambled siRNA group was injected with the same amount of scrambled siRNA. Electroretinography was performed to examine photoreceptor functions 16wk after diabetes induction. MALAT1 expression was detected via real time polymerase chain reaction. Cone morphological changes were examined using immunofluorescence. Rod morphological changes were examined by determining outer nuclear layer (ONL) thickness.
RESULTS
The upregulation of retinal MALAT1 expression was detected in the diabetic control mice, while MALAT1 expression in the diabetic MALAT1-siRNA mice was decreased by 91.48% compared to diabetic control mice. The diabetic MALAT1-siRNA and diabetic control mice showed lower a-wave and b-wave amplitudes than did the normal control mice in scotopic and photopic electroretinogram, while the diabetic MALAT1-siRNA mice showed higher amplitudes than diabetic control mice. Morphological examination revealed that ONL thickness in the diabetic MALAT1-siRNA and diabetic control mice was lower than normal control mice. However, ONL thickness was greater in the diabetic MALAT1-siRNA mice than diabetic control mice. Moreover, the diabetic control mice performed a sparser cone cell arrangement and shorter outer segment morphology than diabetic MALAT1-siRNA mice.
CONCLUSION
Inhibiting retinal MALAT1 results in mitigative effects on the retinal photoreceptors, thus alleviating diabetic neurodegeneration.
Keywords: long non-coding RNA MALAT1, small interfering RNA, diabetic retinopathy, neurodegeneration
INTRODUCTION
The International Diabetes Federation predicts that the number of diabetes mellitus (DM) patients will add up to 642 million in 2040[1]. Diabetic retinopathy (DR) is a serious complication of diabetes and the leading cause of blindness in the global working population. DR has been recognized as a microvascular and neurodegenerative disorder. Excessive oxidative stress, activation of protein kinase C, and production of glycation ends products under a high glucose environment in the retina damage retinal cells and tissues, including photoreceptors and other neurons[2]–[3]. Photoreceptor degeneration is the main cause of color vision abnormalities and contrast sensitivity abnormalities in patients with diabetes before DR; moreover, this degeneration is an important early pathological change in DR[4]. Verma et al[5] employed spectral-domain optical coherence tomography (SD-OCT) to measure the thickness of the retinal photoreceptor layer in patients with diabetes but no signs of DR. In this study, the photoreceptor layer was thinner in patients than in controls. Therefore, exploring protection against retinal neurodegeneration under diabetic conditions to preserve the visual function of patients as much as possible is of great significance.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved, folded, non-coding RNA that is primarily expressed in the nucleus. MALAT1 was originally identified in tumor tissue specimens with upregulated expression. MALAT1 is involved in the regulation of tumor cell proliferation, invasion, metabolism and migration[6]–[7]. In addition, according to Michalik et al[8], MALAT1 regulates the biological functions of vascular endothelial cells. Silencing of MALAT1 expression inhibits cell proliferation and reduces retinal neovascularization. However, the role of MALAT1 in neurodegenerative diseases remains unclear. In the present study, small interfering RNAs (siRNAs) was used to downregulate retinal MALAT1 to examine and compare electroretinogram (ERG) amplitude, outer nuclear layer (ONL) thickness, cone density and morphological changes in diabetic mice model to determine changes in photoreceptor cell function and morphology as well as to investigate the effect of inhibiting retinal long non-coding (lncRNA) MALAT1 on diabetic retinal neurodegeneration.
MATERIALS AND METHODS
Ethical Approval
This study was conformed to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines and was accorded with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All mice were anesthetized and sacrificed using acceptable methods by a skilled technician. The protocol was approved by the Nanchang University Committee on the Ethics of Animal Experiments.
Reagents and Instruments
The following materials were utilized in the present study: streptozotocin (STZ) and RNA extraction kit (Sigma-Aldrich, Germany), protein content determination kit (Bio-Rad, Hercules, CA, USA), rabbit anti-mouse S-opsin primary antibody (Millipore, Billerica, MA, USA), goat anti-rabbit fluorescent secondary antibody (Vector Inc., Burlingame, CA, USA), citrate buffer (Beijing Tiangen Biotech), electronic analytical balance (Shanghai Jingke Balance Factory), compound tropicamide eye drops (Santen Pharmaceutical, Osaka, Japan), recombinant bovine basic fibroblast growth factor ophthalmic gel (Zhuhai Essex Pharmaceutical), electroretinography system (Diagnosys, Westford, MA, USA), StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and inverted fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Experimental Animals and Groups
Thirty-six healthy eight weeks old male C57BL/6 mice with no ocular diseases (weighing 20-30 g and clean grade) were purchased from the Department of Animal Science of Nanchang University [license No.SYXK(JX)2015-0001]. All mice were kept at temperature of 18°C-23°C, humidity of 40%-60% with a cycle of 14h light and 10h dark. Except fasting for diabetes establishment, food and water available at all times. The mice were randomly divided into four groups: normal control group, diabetic control group, diabetic scrambled siRNA group and diabetic MALAT1 siRNA group. A diabetes model induced by STZ was employed in the latter three groups using a previously established method[9]. Briefly, mice were intraperitoneally injected with STZ for 5 consecutive days after an 8h fast. STZ was freshly dissolved in 0.01 mol/L citrate buffer with pH 4.5 before injection. Mice in these three groups were intraperitoneally injected with 50 mg/kg STZ, while mice in the normal control group were given the same volume of citrate buffer. On the 7th day after first injection, blood samples were collected from the tail vein of normally fed mice to measure blood glucose concentration. Effective induction of the diabetes was defined as glucose levels greater than 300 mg/dL. Same blood glucose measurement was operated at week 16 after first STZ injection.
Synthesis of MALAT1 siRNA
Effective siRNA sequences were designed on the report of PubMed and submitted to Shanghai GenePharma corporation to synthesize MALAT1-targeting siRNA using the following sequences: 5′-GAUUGAAGCUAGCAAUCAAAG-3′ as the sense strand and 5′-UUGAUUGCUAGCUUCAAUCUU-3′ as the antisense strand. Nontargeting siRNA was synthesized as a negative control. siRNA was dissolved in diethyl pyrocarbonate (DEPC)-treated normal saline to prepare a 20 µmol/L solution.
Intravitreal Injection of siRNA
Compound of ketamine (100 mg/kg) and xylazine (10 mg/kg) was injected intraperitoneally to anesthetize mice. The periocular region was disinfected with iodophor after subjecting mice to intraperitoneal anesthesia. The needle of a Hamilton micro syringe was inserted toward the posterior pole from 1 mm outside the limbus under microscope, and the test article was slowly injected once the needle tip was visible in the pupil. Both eyes of the diabetic MALAT1 siRNA group was intravitreally injected with a mixture of 1 µL 20 µmol/L MALAT1 siRNA and 1 µL transfection reagent, while both eyes of the diabetic scrambled siRNA group were intravitreally injected with a mixture of 1 µL 20 µmol/L scrambled siRNA and 1 µL transfection reagent. The repeated injection were carried out in the following 2, 4, 6, 8wk.
Electroretinogram
ERG was performed 16wk after first STZ injection. Mice were treated with compound tropicamide eye drops for pupil dilation and were placed in a dark room overnight, followed by anesthesia as mentioned above. Then, mice were placed on a heated plate. A reference electrode and ground electrode were inserted into the palate and tail, respectively. Platinum corneal electrodes was placed on the cornea of both eyes and lubricated with recombinant bovine basic fibroblast growth factor ophthalmic gel. The above procedures were completed under a dim red-light lamp in a dark room. Scotopic ERG responses were recorded under luminous intensities of 0.0004, 0.04, 4, 400 and 2000 cd·s/m2. Next, the light was turned on, and mice adapted for 10min. Photopic ERG responses were recorded under luminous intensities of 2000 cd·s/m2.
Real-time Polymerase Chain Reaction
Real-time polymerase chain reaction (PCR) was conducted 16wk after first STZ injection. Retinal tissues were collected from euthanized mice. RNA Purification Kit was used to extract total RNA. Reverse transcription was operated using a Takara reverse transcription kit to acquire total DNA. MALAT1 and TNF-α target genes and the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) internal reference gene were amplified. The amplification conditions were as follows: predenaturation at 94°C for 60s, 36 cycles of denaturation at 94°C for 10s, and annealing/extension at 56°C for MALAT1 and 61°C for TNF-α and GAPDH for 30s. The gene copy number was calculated. The ratio of the copy number of MALAT1 and TNF-α to the copy number of the GAPDH was used as the relative expression level of MALAT1 and TNF-α. The primers used in real-time PCR were as follows: mouse MALAT1 sense 5′-CATGGCGGAATTGCTGGTA-3′, antisense 5′-CGTGCCAACAGCATAGCAGTA-3′; mouse TNF-α sense 5′-GCCTCTTCTCATTCCTGCTTG-3′, antisense 5′-CTGATGAGAGGGAGGCCATT-3′; mouse GAPDH sense 5′-TGCCCCCATGTTTGTGATG-3′, antisense 5′-TGTGGTCATGAGCCCTTCC-3′.
Measurement of Retinal Outer Nuclear Layer Thickness
Sixteen weeks after the establishment of diabetes model, eyeballs were removed from euthanized mice and directly fixed in 4% paraformaldehyde for 1h at room temperature. Next, the cornea and lens were removed and fixed in 4% paraformaldehyde for an additional 15min at room temperature. The subsequent steps were performed following a routine hematoxylin-eosin staining procedure, and sections with thickness of 6 µm were mounted. The sections were observed under an inverted microscope (Olympus Corporation, Tokyo, Japan), and ONL thickness was measured at 0.48, 0.96, 1.44, and 1.92 mm above and below the optic nerve head using Image-Pro Plus 6.0 (Meyer Instruments Inc., Houston, TX, USA).
Immunofluorescence
Sixteen weeks after diabetes induction, the mice eyeballs were removed from euthanized mice and fixed in 4% paraformaldehyde for 1h at room temperature. The cornea and lens were then removed and fixed in 4% paraformaldehyde for an additional 15min at room temperature. After washing in phosphate buffer saline (PBS) and dehydrating in gradient alcohol, the eyeballs were cleared in xylene I and xylene II, immersed in paraffin, embedded, sectioned (6 µm thickness), and baked. The paraffin sections were deparaffinized in xylene I and xylene II, rinsed in anhydrous alcohol, and placed in gradient alcohol. Antigen retrieval was performed in 0.01 mol/L citric acid buffer at 98°C for 20min. After cooling down, the sections were rinsed in PBS 3 times, incubated with goat serum in a moisture box for 1h, followed by incubation with an anti-s opsin primary antibody (1:1000) at 4°C overnight. The slides were incubated with secondary antibody (1:2000) at room temperature for 1h in the following day. The slides were mounted and were observed with a fluorescence microscope.
Statistical Analysis
Statistical analysis was conducted using SPSS 22 (IBM Corp., Armonk, NY). The ERG results are expressed as mean±SEM and other results are expressed as mean±SD. The comparison between groups was performed using one-way analysis of variance (ANOVA) with least-significant difference (LSD) as post hoc analysis to counteract the problem of multiple comparisons. P<0.05 was considered significant.
RESULTS
Diabetes Model Preparation, Body Mass, and Blood Glucose Levels
Twenty-seven male C57BL/6 mice were used to prepare the diabetes model for the 3 diabetic groups (the diabetic control, diabetic scrambled siRNA and MALAT1 siRNA groups). Seven days after first intraperitoneal injection of STZ, mice had blood glucose levels higher than 300 mg/dL. The success rate of diabetes modeling was 100%. Body mass and blood glucose levels were measured at day 7 and week 16 after first STZ injection. No statistically significant differences in body mass were found among the four groups following initial model establishment (P>0.05). However, body mass was greater in the normal group than in the diabetic control, diabetic scrambled siRNA and MALAT1 siRNA groups by 41.99%, 45.23%, and 34.73%, respectively, at week 16 after model establishment (P<0.05). The three diabetic mouse groups were thinner and had higher water and food intake as well as urine output than did the normal control group. At 7d and week 16 after first STZ injection, the blood glucose concentration in the normal control mice were 46.26% and 49.72% lower than those in the diabetic control mice, 46.79% and 52.08% lower than those in the diabetic scrambled siRNA group, and 47.26% and 50.89% lower than those in the diabetic MALAT1 siRNA group, respectively. These differences were significant (P<0.05). However, no differences were detected in blood glucose concentration among the diabetic control mice, diabetic scrambled siRNA mice and diabetic MALAT1 siRNA mice at 0 and 16wk (Table 1).
Table 1. Body mass and blood glucose levels of the four groups.
| Groups | Body mass (g) |
Blood glucose (mg/dL) |
||
| Day 7 after first STZ injection | Week 16 after model establishment | Day 7 after first STZ injection | Week 16 after model establishment | |
| Normal control | 23.23±1.87 | 35.61±3.49 | 182.25±26.61 | 186.20±29.36 |
| Diabetic control | 23.02±2.08 | 25.08±3.96a | 339.16±63.37a | 370.34±50.21a |
| Diabetic scrambled siRNA | 22.98±2.31 | 24.52±3.68a | 342.52±60.37a | 388.54±51.46a |
| Diabetic MALAT1 siRNA | 23.12±2.24 | 26.43±3.01a | 345.59±55.98a | 379.17±63.20a |
STZ: Streptozotocin. aP<0.05 vs normal control group, n=9.
mean±SD
Retinal Expression of MALAT1 and Downstream Target TNF-α
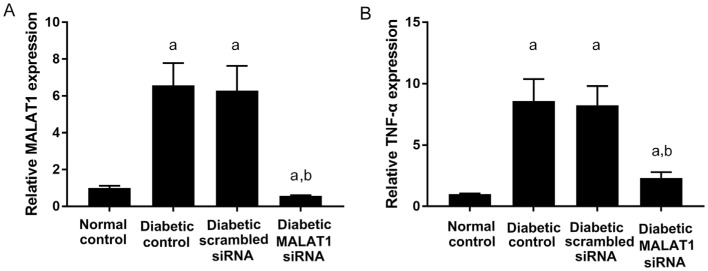
RT-PCR was employed to detect the expression of MALAT1 and its downstream target TNF-α[10] in mouse retinas. According to analyses, MALAT1 was upregulated under diabetic conditions. However, retinal MALAT1 expression was downregulated in mice after treatment with MALAT1 siRNA. Retinal MALAT1 expression was approximately 43.98% lower than that of the normal control group, and it was only 8.52% and 8.90% of the level of the diabetic control and diabetic scrambled siRNA groups, respectively. The differences were significant (P<0.05; Figure 1A). The expression of the downstream target TNF-α was also decreased in the diabetic MALAT1 siRNA group. Retinal TNF-α expression was approximately 26.89% and 28.03% of the level of the diabetic control and diabetic scrambled siRNA groups, respectively. The differences were significant (P<0.05; Figure 1B).
Figure 1. Retinal MALAT1 and TNF-α expression in diabetic mice.
A: MALAT1 expression in the neural retina layer; B: TNF-α expression in the neural retina layer. aP<0.05 vs normal control group; bP<0.05 vs diabetic control group, n=6.
Morphological and Functional Changes in Rod Cells
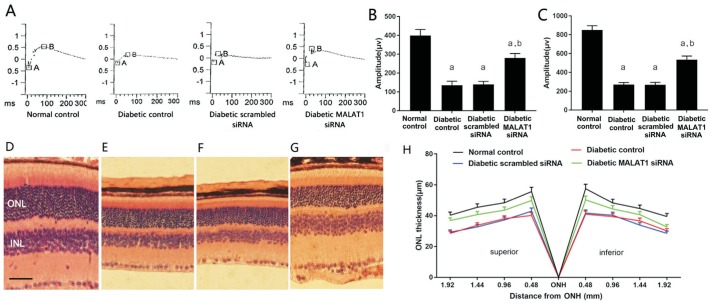
Scotopic ERG stimulates mouse retinas with gradient luminous intensity in a dark environment and reflects the function of rod cells[8]. The ERGs of the four groups are shown in Figure 2A-2C. The diabetic control, diabetic scrambled siRNA and diabetic MALAT1 siRNA groups showed significantly lower a-wave and b-wave amplitudes in scotopic ERG than did the normal control group (P<0.05). However, the MALAT1 siRNA group exhibited significantly higher a-wave and b-wave amplitudes in scotopic ERG than did the diabetic control and diabetic scrambled siRNA groups (P<0.05).
Figure 2. Scotopic ERG and ONL thickness changes in diabetic mice.
A: Representative graph of scotopic ERG; B: Statistical analysis of a-wave amplitudes in scotopic ERG (n=6, mean±SEM); C: Statistical analysis of b-wave amplitudes in scotopic ERG (n=6, mean±SEM); D: Retinal ONL of normal control mice; E: Diabetic control mice; F: Diabetic scrambled siRNA treated mice; G: Diabetic MALAT1 siRNA treated mice; H: Statistical analysis of ONL thickness (n=6, mean±SD). Scale bar: 40 µm. aP<0.05 vs normal control group, bP<0.05 vs diabetic control group.
The retinal ONL mainly comprises photoreceptor nuclei. Because 98% of photoreceptor cells are rod cells, the thickness of the ONL basically reflects changes in the number of rod cells[4],[11]. In the present study, retinal ONL thickness was measured at 0.48, 0.96, 1.44, and 1.92 mm above and below the optic nerve head. The ONL thickness of the diabetic MALAT1 siRNA, diabetic scrambled siRNA and diabetic control groups was decreased by 16.05%, 36.67% and 35.38%, respectively, compared to that of the normal control group. However, the ONL of the diabetic MALAT1 siRNA group was 28.91% and 27.65% thicker than that of the diabetic control and diabetic scrambled siRNA groups, respectively. These differences were significant (P<0.05; Figure 2D-2H).
Morphological and Functional Changes in Cone Cells
Photopic ERG was used to measure the function of cone photoreceptor[9]. Photopic ERG a-wave and b-wave amplitudes in the diabetic control, diabetic scrambled siRNA and diabetic MALAT1 siRNA mice were significantly lesser than those in the normal control mice (P<0.05). By contrast, photopic ERG a-wave and b-wave amplitudes in the diabetic MALAT1 siRNA group were greater than those in the diabetic control and diabetic scrambled siRNA mice (Figure 3A-3C). Morphological changes using s-opsin for labelling were examined with immunofluorescence[9],[12]. Compared with the normal control mice, the diabetic control, diabetic scrambled siRNA and diabetic MALAT1 siRNA mice manifested lower cone photoreceptor density with a sparse arrangement. However, changes in the diabetic control and diabetic scrambled siRNA treated mice were more obvious with dwarfish outer segments than those in the diabetic MALAT1 siRNA treated mice (Figure 3D-3G).
Figure 3. Photopic ERG and cone morphological changes in diabetic mice.
A: Representative graph of photopic ERG; B: Statistical analysis of a-wave amplitudes in photopic ERG (n=6, mean±SEM); C: Statistical analysis of b-wave amplitudes in photopic ERG (n=6, mean±SEM), aP<0.05 vs normal control group, bP<0.05 vs diabetic control group; D-G: Morphological changes in the retinal cone photoreceptors of normal control mice (D), diabetic control mice (E), diabetic scrambled siRNA mice (F), diabetic MALAT1 siRNA mice (G) detected by immunofluorescence (fluorescence microscope ×400).
DISCUSSION
DR is the most frequent ocular complication of diabetes and one of the major blinding diseases in people over 50 years old. DR was initially considered a diabetic microangiopathy. Retinal neuronal cell death is accumulated in animal models of diabetes, and abnormal glial fibrillary acidic protein (GFAP) expression in astrocytes and Müller cells as well as microglial activation occurs in diabetic animal models. Based on these findings, DR is also considered a chronic neurodegenerative disease. Photoreceptor cells, including cones and rods, are important retinal neurons involved in the primary visual pathway. As mentioned earlier, photoreceptor degeneration is an important early pathological change in DR. According to current research, the possible mechanisms of neurodegeneration in DR include oxidative stress[13], increased glycation end products[14], and activation of the renin angiotensin system[15].
lncRNA is widely present in organisms. As a transcript greater than 200 bp, non-coding RNA can regulate gene expression at the epigenetic, transcriptional and posttranscriptional levels, thereby affecting cell proliferation, cell apoptosis, cell activity, immune response and oxidative stress. MALAT1 is a 7 kb, highly conserved, folded non-coding RNA that is primarily expressed in the nucleus. In recent years, MALAT1 has been involved in the pathogenesis of diabetic ocular lesions. According to Yan et al[16], the expression of MALAT1 is upregulated in RF/6A cells under high glucose conditions as well as in the aqueous humor and proliferative fibrous membrane specimens of patients with DR. Downregulation of MALAT1 attenuates the deterioration of retinal function and alleviates retinal vascular damages caused by diabetes. Moreover, downregulation of MALAT1 reduces the expression of angiogenic factors, such as vascular endothelial growth factor (VEGF), thereby improving the ocular microvascular dysfunction caused by diabetes[17]. In addition, MALAT1 plays a role in neurologically related lesions. Based on a study by Bernard et al[18], MALAT1 regulates gene expression related to nuclear and synaptic functions as well as synaptogenesis in neurons. Kryger et al[19] found that MALAT1 is upregulated in the cerebellum, hippocampus and brainstem of alcoholics. However, reports on MALAT1 in diabetic retinal neurodegeneration are scarce. According to Yao et al[20], MALAT1 expression is increased in the retina of rats and mice following optic nerve transection, and MALAT1 knockdown inhibits Müller cell activation. MALAT1 dysregulation may also affect retinal ganglion cell survival and alter the development of glaucomatous neurodegeneration.
siRNAs are RNA fragments composed of 19-21 bp that can degrade the RNA of specific genes, thereby inhibiting the expression of these genes. In the present study, MALAT1 expression was downregulated in the retina of mice treated with MALAT1 siRNA. The effect of downregulation of MALAT1 on the function and morphology of photoreceptors was evaluated with various indicators. Flash ERG uses flashes of different light intensities to stimulate photoreceptor cell hyperpolarization, that can be detected noninvasively through a platinum electrode placed on the cornea[4]. Scotopic and photopic ERGs are frequently used to reflect the functional status of rods and cones, respectively. According to previous studies, diabetic rats have abnormal ERGs at an early stage[21]. A gradually decreasing ERG amplitude has been observed with disease progression in patients with DR as indicated by flash ERG examination[22]. In the present study, a-wave and b-wave amplitudes in scotopic and photopic ERG in the diabetic MALAT1 siRNA treated mice were lesser than those in the normal control mice but greater than those in the diabetic control and diabetic scrambled siRNA treated mice. These results suggested that downregulation of MALATI by siRNA alleviated functional impairments in photoreceptor cells in diabetic mice.
Retinal photoreceptor cells are divided into rod cells and cone cells according to the morphology of their outer segments. Rod cells account for more than 90% of photoreceptor cells. Therefore, the ONL of the retina mainly comprises the nuclei of rod cells, and its thickness reflects cell death and morphological changes in rod cells. Animal studies have confirmed that the thickness of ONL in diabetic rats is progressively reduced with the progression of the disease course[23]. The present study also found that the ONL thickness of the three diabetic mouse groups was less than that of the normal control group 16wk after model establishment, suggesting that the retinal rod cells of the diabetic mouse model in this study were continuously dying. However, the ONL thickness of the MALAT1 siRNA treated mice was greater than that of the diabetic control and diabetic scrambled siRNA treated mice at each time point. Since s-opsin responds to high intensity light and color in cones and is mainly distributed on the outer segment, we used s-opsin to label the cone outer segments for immunofluorescence to observe changes in cone cell structure. As mentioned above, the diabetic control and diabetic scrambled siRNA groups had a sparser cone arrangement and shorter outer segment morphology than did the diabetic MALAT1 siRNA group. These results suggested less damage to rods and cones in the MALAT1 siRNA-treated group despite the persistent death of photoreceptor cells under diabetic conditions, indicating that downregulation of MALAT1 has a protective effect on photoreceptor cells.
In summary, compared with regular diabetic mice, diabetic mice that received local injections of MALAT1 siRNA to inhibit MALAT1 expression in the retina showed reduced functional and morphological lesions of retinal photoreceptor cells. These results suggested that MALAT1 interference may become a novel orientation for the clinical application of intervention and therapy on diabetic neurodegeneration.
Acknowledgments
Foundations: Supported by National Natural Science Foundation of China (No.81960158, No.81760176); Key Scientific research Program of Jiangxi Education Department (No.GJJ190003).
Conflicts of Interest: Zhang YL, None; Hu HY, None; You ZP, None; Li BY, None; Shi K, None.
REFERENCES
- 1.International Diabetes Federation. IDF diabetes atlas. South Africa: International Diabetes Federation, Cape Town; 2015. [Google Scholar]
- 2.Geraldes P, Hiraoka-Yamamoto J, Matsumoto M, Clermont A, Leitges M, Marette A, Aiello LP, Kern TS, King GL. Activation of PKC-delta and SHP-1 by hyperglycemia causes vascular cell apoptosis and diabetic retinopathy. Nat Med. 2009;15(11):1298–1306. doi: 10.1038/nm.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobanovskaya N, Jürgenson M, Aonurm-Helm A, Zharkovsky A. Alterations in the polysialylated neural cell adhesion molecule and retinal ganglion cell density in mice with diabetic retinopathy. Int J Ophthalmol. 2018;11(10):1608–1615. doi: 10.18240/ijo.2018.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan SJ. Retina. 4th ed. Singapore: Elsevier; 2010. [Google Scholar]
- 5.Verma A, Rani PK, Raman R, Pal SS, Laxmi G, Gupta M, Sahu C, Vaitheeswaran K, Sharma T. Is neuronal dysfunction an early sign of diabetic retinopathy? Microperimetry and Spectral Domain Optical Coherence Tomography (SD-OCT) Study in individuals with diabetes, but no diabetic retinopathy. Eye. 2009;23(9):1824–1830. doi: 10.1038/eye.2009.184. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 8.Michalik KM, You XT, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 9.You ZP, Zhang YL, Shi K, Shi L, Zhang YZ, Zhou Y, Wang CY. Suppression of diabetic retinopathy with GLUT1 siRNA. Sci Rep. 2017;7(1):7437. doi: 10.1038/s41598-017-07942-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D, Cheng YG, Huang X, Zhong MW, Liu SZ, Hu SY. Downregulation of lncRNA MALAT1 contributes to renal functional improvement after duodenal-jejunal bypass in a diabetic rat model. J Physiol Biochem. 2018;74(3):431–439. doi: 10.1007/s13105-018-0636-y. [DOI] [PubMed] [Google Scholar]
- 11.Wang JJ, Xu XL, Elliott MH, Zhu ML, Le YZ. Müller cell-derived VEGF is essential for diabetes-induced retinal inflammation and vascular leakage. Diabetes. 2010;59(9):2297–2305. doi: 10.2337/db09-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27(3):513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 13.Wong TY, Simó R, Mitchell P. Fenofibrate-a potential systemic treatment for diabetic retinopathy? Am J Ophthalmol. 2012;154(1):6–12. doi: 10.1016/j.ajo.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Curtis TM, Stitt AW. Advanced glycation end products and diabetic retinopathy. Curr Med Chem. 2013;20(26):3234–3240. doi: 10.2174/09298673113209990025. [DOI] [PubMed] [Google Scholar]
- 15.Sjølie AK, Dodson P, Hobbs FR. Does renin-angiotensin system blockade have a role in preventing diabetic retinopathy? A clinical review. Int J Clin Pract. 2011;65(2):148–153. doi: 10.1111/j.1742-1241.2010.02552.x. [DOI] [PubMed] [Google Scholar]
- 16.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55(2):941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 17.Campochiaro PA. Ocular neovascularization. J Mol Med. 2013;91(3):311–321. doi: 10.1007/s00109-013-0993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan ZY, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kryger R, Fan L, Wilce PA, Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol. 2012;46(7):629–634. doi: 10.1016/j.alcohol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Yao J, Wang XQ, Li YJ, Shan K, Yang H, Wang YN, Yao MD, Liu C, Li XM, Shen Y, Liu JY, Cheng H, Yuan J, Zhang YY, Jiang Q, Yan B. Long non-coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol Med. 2016;8(9):1113. doi: 10.15252/emmm.201606749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Safi H, Safi S, Hafezi-Moghadam A, Ahmadieh H. Early detection of diabetic retinopathy. Surv Ophthalmol. 2018;63(5):601–608. doi: 10.1016/j.survophthal.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Jansson RW, Raeder MB, Krohn J. Photopic full-field electroretinography and optical coherence tomography in type 1 diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 2015;253(7):989–997. doi: 10.1007/s00417-015-3034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai AK, Lo AC. Animal models of diabetic retinopathy: summary and comparison. J Diabetes Res. 2013;2013:106594. doi: 10.1155/2013/106594. [DOI] [PMC free article] [PubMed] [Google Scholar]