Abstract
The mechanisms of action of the complex including entomopathogenic nematodes of the genera Steinernema and Heterorhabditis and their mutualistic partners, i.e., bacteria Xenorhabdus and Photorhabdus, have been well explained, and the nematodes have been commercialized as biological control agents against many soil insect pests. However, little is known regarding the nature of the relationships between these bacteria and the gut microbiota of infected insects. In the present study, 900 bacterial isolates that were obtained from the midgut samples of Melolontha melolontha larvae were screened for their antagonistic activity against the selected species of the genera Xenorhabdus and Photorhabdus. Twelve strains exhibited significant antibacterial activity in the applied tests. They were identified based on 16S rRNA and rpoB, rpoD, or recA gene sequences as Pseudomonas chlororaphis, Citrobacter murliniae, Acinetobacter calcoaceticus, Chryseobacterium lathyri, Chryseobacterium sp., Serratia liquefaciens, and Serratia sp. The culture filtrate of the isolate P. chlororaphis MMC3 L3 04 exerted the strongest inhibitory effect on the tested bacteria. The results of the preliminary study that are presented here, which focused on interactions between the insect gut microbiota and mutualistic bacteria of entomopathogenic nematodes, show that bacteria inhabiting the gut of insects might play a key role in insect resistance to entomopathogenic nematode pressure.
Keywords: Melolontha melolontha, midgut microbiota, entomopathogenic nematodes, bacterial interactions, Xenorhabdus, Photorhabdus
1. Introduction
Coleoptera is the largest order of insects, representing over 400,000 of known species. During evolution, representatives of this order have colonized diverse ecological niches and climate zones. The most abundant (over 30,000 species) family of these insects is Scarabaeidae, with characteristic lamellate club antennae in imagines [1]. The evolutionary success of scarabs lies in the activity of the gut microbiota of their larvae, which allows for them to feed on a wide range of low energy foods, grass roots, and organic matter, with inconsiderable competition from other insects. Hence, scarab larvae exploit a variety of niches, which range from rotting organic matter and dead tree trunks to freshly growing roots. Using these resources, they have easily become pests in agriculture and forestry. Of the approximately 150 scarab species that were recorded in Central Europe, damage is predominantly caused by only four native species: Melolontha melolontha, Melolontha hippocastani, Amphimallon solstitialis, and Phyllopertha horticola [2]. These pests are difficult to control due to the cryptic position of larvae in the soil and the usually nocturnal activity of adults [3].
Entomopathogenic nematodes (EPN) of the genera Steinernema and Heterorhabditis (Nematoda: Rhabditida) are effective biocontrol agents against soil-dwelling stages of many insect pests [4,5,6]. They are safe for vertebrates, plants, and numerous invertebrates [7,8]. In recent years, significant progress in applying these bioagents to reduce the populations of Scarabaeidae pests was observed [9,10,11,12]. Entomopathogenic nematodes are symbiotically associated with entomopathogenic bacteria (EPB) Xenorhabdus spp. and Photorhabdus spp. EPB are motile, gram-negative, non-spore-forming, facultatively anaerobic rods of the family Enterobacteriaceae. They live in the intestinal lumen of infective juveniles (the only free-living stage of EPN) and in the body cavity of the infected insects [13]. Infective juveniles migrate to the hemocoel and release bacterial symbionts, which multiply quickly causing a lethal bacteremia within 24–48 h, after entering the body of an insect through the orifices of the respiratory and digestive systems [14,15]. However, findings regarding Steinernema carpocapsae indicate that some of the bacteria (Xenorhabdus nematophila) transported by nematodes may be released in the insect gut as early as several hours after the entrance of the parasites into the gastrointestinal canal [12]. It has also been found that, during early infection, Photorhabdus bacteria specifically proliferate in the midgut, where they release toxins and a metalloprotease that destroy the midgut epithelium [16]. A growing bacterial population provides an optimal environment for the rapid development of the nematodes. Compounds that are secreted by these bacteria, such as lytic enzymes and substances with antimicrobial properties, give EPB a competitive advantage, which facilitates their rapid invasion of an attractive environment, despite the initial presence of the species-rich indigenous microbiota of the insect gut [17,18].
Entomopathogenic nematodes with their mutualistic bacteria and infected insects have become the subject of numerous research studies, which have already provided considerable data, e.g., on the mechanisms of pathogenesis, insect immunity, and the ecological aspects of the biological complex [19]. It can be expected that the interactions between the mutualistic bacteria of EPN and the gut-associated bacteria of the infected insects represent a highly important component of the complicated nematode-bacterium-insect complex, but they have not been investigated in detail. Previous studies in this area primarily focused on the antibacterial activities of EPB. To date, there have been no studies on antagonistic mechanisms that would work in the opposite direction, i.e., growth inhibition of the nematode bacterial symbionts by bacteria of the insect gut microbiota. This type of interaction is to be expected, based on the fact that the body of the insect harbors two groups of bacteria with conflicting interests when a nematode larva has released EPB. The optimal habitat for the insect gut microbiota is the digestive system of a living host, while developing EPB lead to a rapid death of the insect. It is possible that some of the gut bacteria are capable of producing substances that have an antagonistic action against EPB, thus protecting both the intestinal microbiota and the entire body of the insect. This type of bacterial activity might substantially reduce the effectiveness of biopesticides containing entomopathogenic nematodes. The isolation and identification of insect gut bacteria, which exhibit antimicrobial activity against Xenorhabdus spp. or Photorhabdus spp., would allow for a much better understanding of the mechanism of EPN infection of insects and the bacterial interactions that occur during this process.
The objective of this study was to isolate and identify the bacteria colonizing the midgut of the common cockchafer M. melolontha, which exhibit antibacterial activity against selected species of the genera Xenorhabdus and Photorhabdus.
2. Results
2.1. Isolation of Bacteria from the Midgut of M. melolontha Larvae
Sixty samples of the midgut of the second and third instar M. melolontha larvae (L2 and L3) were used in the study. The guts were sampled from six groups of larvae. The first group comprised the specimens that had been freshly collected in the natural habitat. The other five groups of larvae were subjected to initial 12-day exposure to entomopathogenic nematodes and ten live larvae from each group were selected for further analyses. This procedure aimed at increasing the probability of acquisition of bacterial isolates with the ability to inhibit the growth of bacteria colonizing the entomopathogenic nematodes. The nematode pressure can be regarded as strong, since the incubation of M. melolontha in the presence of EPN resulted in the death of 42.5% of larvae in the presence of Heterorhabditis megidis, as well as 55%, 52.5%, 25%, and 35% of larvae that were exposed to Steinernema arenarium, Steinernema bicornutum, Steinernema carpocapsae, and Steinernema silvaticum, respectively.
In total, 900 bacterial strains were isolated from the gut samples (15 strains from each gut). The isolation and growth inhibition assays were carried out in aerobic conditions in the case of half of the obtained strains (i.e., 450) and in microaerobic conditions in the case of the other half since the aerobic conditions in the midgut of scarab larvae may vary [20].
2.2. Screening of Midgut Bacteria with Antagonistic Activity Against EPB
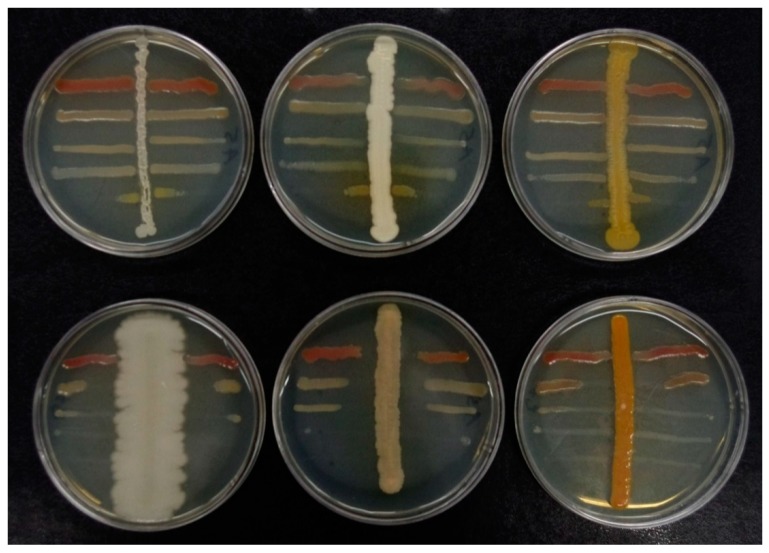
In the first stage of the investigations, the antibacterial activities of the isolates were analyzed while using cross-streak tests (Figure 1). Thirty-eight isolates inhibited the growth of the selected EPB species, i.e., Photorhabdus temperata, Xenorhabdus kozodoii, Xenorhabdus bovienii, Xenorhabdus nematophila, and Xenorhabdus budapestensis (Table 1). Twenty-three and fifteen strains with this ability were isolated in the aerobic and microaerobic conditions, respectively. The greatest number of positive isolates (12) was obtained from the midgut of larvae that were exposed to H. megidis. Some isolates completely inhibited the growth of the symbiotic bacteria of nematodes over the entire surface of the Petri dishes in the cross-streak tests. In most cases, the lowest susceptibility to growth inhibition characterized P. temperata (Table 1).
Figure 1.
Interactions between M. melolontha midgut isolates and nematode bacteria in cross-streak test. Horizontal streaks—five entomopathogenic bacteria (EPB) strains, vertical streak—midgut isolate. Top row—no inhibition effect; bottom row—isolates with high antibacterial activity against EPB.
Table 1.
Antagonistic activities of selected M. melolontha midgut isolates against five entomopathogenic bacteria (EPB) strains shown by cross-streak tests.
| Strain ID * | Size of Inhibition Zones (mm) Left Inhibition Zone|Right Inhibition Zone |
||||
|---|---|---|---|---|---|
| P. temperata | X. kozodoii | X. bovienii | X. nematophila | X. budapestensis | |
| MT1 L2 01 | 0|0 | 12|13 | 20|18 | no gr. **| no gr. | no gr.|no gr. |
| MT1 L2 02 | 1|1 | 1|1 | no gr.|no gr. | no gr.|no gr. | 7|4 |
| MT3 L2 02 | 2|4 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MT3 L2 05 | 12|7 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MT3 L2 06 | 6|6 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MT3 L2 08 | 10|12 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MT3 L2 10 | 12|11 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MT3 L2 13 | 7|5 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MT4 L2 11 | 0|0 | 0|0 | 30|22 | no gr.|24 | 18|no gr. |
| MTA1 L2 01 | 1|1 | no gr.|no gr. | no gr.|12 | 6|6 | 30|20 |
| MTA3 L2 13 | 5|5 | 25|25 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTA5 L3 10 | 3|5 | 7|5 | no gr.|no gr. | no gr.|no gr. | 20|25 |
| MMB2 L3 03 | 0|0 | 0|1 | 11|12 | 4|0 | 3|3 |
| MMB2 L3 04 | 0|0 | 0|0 | 7|6 | 0|0 | 1| 0 |
| MMB2 L3 07 | 0|0 | 0|0 | 12|12 | 9|10 | 10|9 |
| MMB4 L3 10 | 0|0 | 0|0 | 11|12 | 1|0 | 0|0 |
| MTB1 L3 08 | 0|0 | 15|14 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTB1 L3 09 | 0|0 | 15|20 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTB1 L3 12 | 0|0 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTB5 L3 11 | 0|0 | 7|10 | no gr.|no gr. | no gr.|no gr. | 17|22 |
| MMC3 L3 03 | 0|0 | 11|12 | 14|16 | 14|13 | 12|13 |
| MMC3 L3 04 | 0|0 | 11|13 | 15|17 | 15|17 | 15|14 |
| MMC3 L3 07 | 0|0 | 13|14 | 16|18 | 17|19 | 17|17 |
| MMC3 L3 12 | 0|0 | 12|14 | 15|17 | 15|16 | 15|14 |
| MTC1 L3 03 | 0|0 | 10|10 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTS3 L3 15 | 8|7 | 25|25 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MMH3 L2 04 | 2|2 | 3|2 | 17|18 | 17|16 | 13|14 |
| MMH3 L2 05 | 2|1 | 3|2 | 14|16 | 12|15 | 12|10 |
| MMH3 L2 06 | 2|1 | 4|3 | 16|17 | 16|15 | 11|13 |
| MMH4 L2 06 | 1|1 | 2|3 | 17|15 | 10|13 | 9 |12 |
| MMH4 L2 07 | 1|1 | 2|2 | 14|13 | 12|10 | 12|11 |
| MMH5 L2 04 | 0|1 | 2|3 | 2|3 | 1|1 | 7|5 |
| MMH5 L2 08 | 0|0 | 0|0 | 4|4 | 5|5 | 3|3 |
| MTH3 L2 08 | 0|0 | 16|14 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTH3 L2 09 | 0|0 | 10|9 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTH3 L2 10 | 0|0 | 10|10 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTH3 L2 14 | 0|0 | 14|15 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
| MTH3 L2 15 | 0|0 | 10|10 | no gr.|no gr. | no gr.|no gr. | no gr.|no gr. |
* Strain ID provides information about the conditions of the isolation and the most important data on the M. melolontha larva used for isolation thereof. The second letter in strain ID: M—isolation in microaerobic conditions; T—isolation in aerobic conditions. The third letter in strain ID: information about the initial exposure of the M. melolontha larva to: A—S. arenarium; B—S. bicornutum; C—S. carpocapsae; S—S. silvaticum; H—H. megidis. L2 or L3: developmental stage of the M. melolontha larva. ** no gr.—no growth. Complete inhibition of bacterial strain growth.
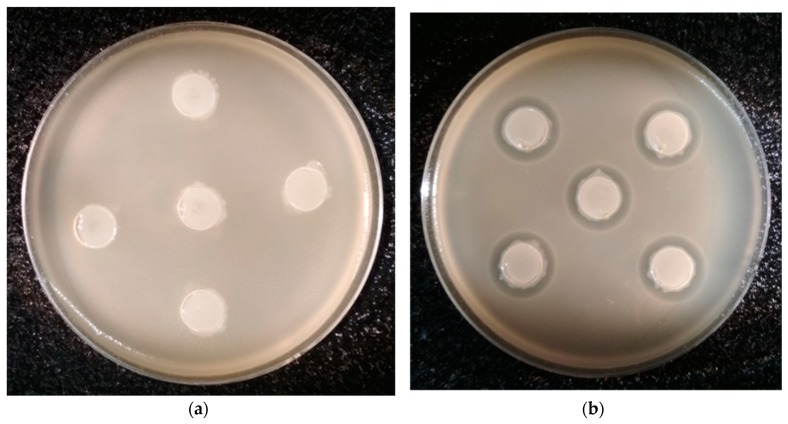
Next, the selected isolates were subjected to modified agar well diffusion tests to confirm their antibacterial activity. Twelve isolates were shown to have the ability to inhibit the growth of the symbiotic bacteria of nematodes. The highest antibacterial activity was detected for the isolate MMC3 L3 04 (Figure 2 and Table 2).
Figure 2.
Interactions between M. melolontha midgut isolates and nematode bacteria in a modified agar well diffusion test. (a) Isolate incapable of inhibition of the growth of EPB. (b) MMC3 L3 04 isolate showing antagonistic activity against X. kozodoii.
Table 2.
Antagonistic activities of selected M. melolontha midgut isolates against five EPB strains that were shown by modified agar well diffusion tests.
| Strain ID | Diameter of the Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| P. temperata | X. kozodoii | X. bovienii | X. nematophila | X. budapestensis | |
| MT3 L2 05 | 0.00 | 0.00 | 14.00 ± 0.41 | 15.85 ± 0.41 | 15.35 ± 0.58 |
| MT3 L2 08 | 14.10 ± 0.46 | 16.95 ± 0.64 | 0.00 | 17.05 ± 0.55 | 17.45 ± 0.55 |
| MT3 L2 10 | 15.00 ± 0.33 | 18.55 ± 0.64 | 0.00 | 18.05 ± 0.64 | 17.05 ± 0.50 |
| MT3 L2 13 | 0.00 | 0.00 | 14.00 ± 0.58 | 16.05 ± 0.28 | 14.95 ± 0.44 |
| MTA1 L2 01 | 13.9 ± 0.66 | 0.00 | 15.05 ± 0.59 | 14.45 ± 0.39 | 0.00 |
| MTA3 L2 13 | 0.00 | 0.00 | 0.00 | 0.00 | 19.20 ± 0.48 |
| MTB1 L3 12 | 0.00 | 17.10 ± 0.57 | 0.00 | 0.00 | 18.05 ± 0.28 |
| MTC1 L3 03 | 0.00 | 0.00 | 0.00 | 0.00 | 19.25 ± 0.63 |
| MMC3 L3 04 | 16.00 ± 0.41 | 18.95 ± 0.60 | 16.10 ± 0.39 | 16.15 ± 0.34 | 19.90 ± 0.70 |
| MMH3 L2 04 | 0.00 | 16.05 ± 0.44 | 15.40 ± 0.32 | 17.05 ± 0.37 | 17.45 ± 0.39 |
| MMH4 L2 06 | 0.00 | 15.50 ± 0.52 | 0.00 | 17.00 ± 0.58 | 16.45 ± 0.55 |
| MTS3 L3 15 | 0.00 | 17.95 ± 0.68 | 14.05 ± 0.37 | 18.45 ± 0.44 | 16.10 ± 0.57 |
All data are expressed as mean ± SD, n = 10.
2.3. Identification of Selected Midgut Isolates from M. melolontha Larvae
Subsequently, isolates that inhibit the growth of the Xenorhabdus and Photorhabdus bacteria in both the cross-streak tests and the modified agar well diffusion tests were identified with the use of molecular methods. Preliminary identification was based on 16S rRNA gene sequence analysis. Nearly full-length 16S rDNA sequences of all tested isolates were determined while using a pair of universal primers 27F and 1492R. Additionally, the rpoD, rpoB or recA gene sequences were analyzed while using genus specific primers (Table 3).
Table 3.
Oligonucleotides used in the study.
| Primer | Sequence | Target Gene | Target Bacterial Genus | PCR Cycling | Product Length | Reference |
|---|---|---|---|---|---|---|
| 27F 1492R |
5′-AGAGTTTGATCCTGGCTCAG-3′ 5′-GGTTACCTTGTTACGACTT-3′ |
16S rDNA | All tested | 3 min 95 °C, 30 × (30 s 94 °C, 45 s 55 °C, 90 s 72 °C), 5 min 72 °C | 1500 bp | [21] |
| PsEG30F PsEG790R |
5′- ATYGAA ATCGCCAARCG-3′ 5′-CGGTTGATKT CCTTGA-3′ |
rpoD | Pseudomonas | 3 min 95 °C, 30 × (60 s 94 °C, 45 s 55 °C, 50 s 72 °C), 5 min 72 °C | 750 bp | [22] |
| Vic3 Vic2 |
5′-GGCGAAATGGCWGAGAACCA-3′ 5′-GAGTCTTCGAAGTTGTAACC-3′ |
rpoB | Citrobacter | 4 min 94 °C, 30 × (30 s 94 °C, 30 s 50 °C, 45 s 72 °C), 5 min 72 °C | 410 bp | [23] |
| Ac696F Ac1093R |
5′-TAYCGYAAAGAYTTGAAAGAAG-3′ 5′-CMACACCYTTGTTMCCRTGA-3′ |
rpoB | Acinetobacter | 3 min 95 °C, 30 × (60 s 94 °C, 52 s 45 °C, 60 s 72 °C), 5 min 72 °C | 370 bp | [24] |
| 359f 359r |
5′-TTATCGCTCAGGCGAACTCCAAC-3′ 5′-TGCTGGATTCGCCTTTGCTACG-3′ |
rpoB | Serratia | 3 min 95 °C, 30 × (50 s 94 °C, 40 s 52 °C, 60s 72 °C), 5 min 72 °C | 530 bp | [25] |
| ESchr-rpoF ESchr-rpoR |
5′GGTGAAGTAGTTTCTATCGAAAGA-3′ 5′-ATGTTTGGTCCTTCCGGAGTT-3′ |
rpoB | Chryseobacterium | 3 min 95 °C, 30 × (35 s 95 °C, 35 s 52 °C, 50 s 72 °C), 5 min 72 °C | 790 bp | This work |
| recAF recAR |
5′-TCSGGYAARACCACSCTGAC-3′ 5′-RTACCAGGCRCCGGACTTCT-3′ |
recA | Pseudomonas | 4 min 94 °C, 30 × (30 s 94 °C, 30 s 55 °C, 40 s 72 °C), 5 min 72 °C | 600 bp | [26] |
Based on the 16S rDNA and protein-coding housekeeping gene sequences, six isolates were identified as Pseudomonas chlororaphis and single isolates represented Citrobacter murliniae, Acinetobacter calcoaceticus, Chryseobacterium lathyri, Serratia liquefaciens, Serratia sp. and Chryseobacterium sp. (Table 4). The molecular identification of the isolates was supported by phylogenetic analysis, whose results are shown in supplementary data (Supplementary Materials Figures S1–S6).
Table 4.
Molecular identification of bacterial strains with antimicrobial activity against EPB isolated from the midgut of M. melolontha larvae.
| Strain ID | Identification Result/Gene Accession Numbers | Strain with the Highest Similarity to the Isolate in the Gene Bank Based on the Gene Sequence/Gene Accession Number/% Nucleotide Identity | ||
|---|---|---|---|---|
| 16S rDNA | rpoD/rpoB | recA | ||
|
MT3
L2 05 |
Pseudomonas chlororaphis 16S rDNA—MM421924 rpoD—MN445046 |
P. chlororaphis subsp. piscum JF3835T (FJ168539)—99.8% | * P. chlororaphis subsp. piscum ZJU60 (CP027656)—98.2%; P. chlororaphis subsp. aurefaciens NCIMB 9265 (AB039555)—98.2% |
n.d. |
|
MT3
L2 08 |
Pseudomonas chlororaphis 16S rDNA—MM421925 rpoD—MN445047 |
P. chlororaphis subsp. piscum JF3835T (FJ168539)—99.8% | * P. chlororaphis subsp. piscum ZJU60 (CP027656)—98.2%; P. chlororaphis subsp. aurefaciens NCIMB 9265 (AB039555)—98.2% |
n.d. |
|
MT3
L2 10 |
Pseudomonas chlororaphis 16S rDNA—MM421926 rpoD—MN445048 recA—MN477250 |
P. chlororaphis subsp. piscum JF3835T (FJ168539)—99.8% | * P. chlororaphis subsp. piscum ZJU60 (CP027656)—98.0%; P. chlororaphis subsp. aurefaciens NCIMB 9265 (AB039555)—98.0% |
P. chlororaphis subsp. piscum JF3835T (FJ168540)—98.4% |
|
MT3
L2 13 |
Pseudomonas chlororaphis 16S rDNA—MM421923 rpoD—MN445049 |
P. chlororaphis subsp. piscum JF3835T (FJ168539)—99.9% | * P. chlororaphis subsp. piscum ZJU60 (CP027656)—98.0%; P. chlororaphis subsp. aurefaciens NCIMB 9265 (AB039555)—98.0% |
n.d. |
|
MTA1
L2 01 |
Citrobacter murlinae 16S rDNA—MM421947 rpoB—MN445055 |
C. murliniae CIP 104556T (KY178281)—99.3% | ** C. murliniae CIP 104556T (KM516007)—99.5% | n.d. |
|
MMC3
L3 04 |
Pseudomonas chlororaphis 16S rDNA—MM421927 rpoD—MN445050 recA—MN477251 |
P. chlororaphis subsp. piscum JF3835T (FJ168539)—99.8% | * P. chlororaphis subsp. piscum ZJU60 (CP027656)—98.2%; P. chlororaphis subsp. aurefaciens NCIMB 9265 (AB039555)—98.2% |
P. chlororaphis subsp. piscum JF3835T (FJ168540)—98.6% |
|
MTB1
L3 12 |
Serratia liquefaciens 16S rDNA—MM422010 rpoB—MN445052 |
Serratia quinivorans DSM4597T (AJ233435)—99.4%; S. liquefaciens CIP 103238T (NR 042062)—98.9% |
** S. liquefaciens LMG7884T (JX425335)—99.4% S. quinivorans LMG7887T (JX425309)—97.7% |
n.d. |
|
MTA3
L2 13 |
Acinetobacter calcoaceticus 16S rDNA—MN429316 rpoB—MN445056 |
A. calcoaceticus DSM 30006T (AJ633632)—99.8% | ** A. calcoaceticus CIP 81.8T (DQ207474)—97.9%; A. calcoaceticus CA16 (NZ_CP020000)—98.7% |
n.d. |
| MTS3 L3 15 |
Pseudomonas chlororaphis 16S rDNA—MM421928 rpoD—MN445051 recA—MN477252 |
P. chlororaphis subsp. piscum JF3835T (FJ168539)—99.7% | * P. chlororaphis subsp. piscum ZJU60 (CP027656)—98.4%; P. chlororaphis subsp. aurefaciens NCIMB 9265—98.4% |
P. chlororaphis subsp. piscum JF3835T (FJ168540)—98.6% |
|
MTC1
L3 03 |
Serratia sp. 16S rDNA—MM422009 rpoB—MN445053 |
Serratia grimesii DSM 30063T (AJ233430)—99.3%; Serratia proteamaculans DSM 4543T (AJ233434)—99.2% S. liquefaciens CIP 103238 (AJ306725)—99.1% |
** S. liquefaciens LMG7884T (JX425335)—98.8% S. quinivorans LMG7887T (JX425309)—98.3% |
n.d. |
|
MMH3
L2 04 |
Chryseobacterium lathyri 16S rDNA—MM429317 rpoB—MN445063 |
C. lathyri RBA2-6T (DQ673674)—99.8% | ** C. lathyri KCTCT 22544T (NZ_QNFY01000004)—99.1% | n.d. |
|
MMH4
L2 06 |
Chryseobacterium sp. 16S rDNA—MN429318 rpoB—MN445064 |
C. nakagawai NCTC 13529T (NZLR134386)—98.7% | ** C. joostei DSM 16927T (CP033926)—93.6%; C. nakagawai NCTC 13529TT (LR134386)—91.0% |
n.d. |
n.d.—not determined, * rpoD gene sequences analysis; ** rpoB gene sequences analysis.
2.4. Detailed Evaluation of Antibacterial Activity of Selected Gut Isolates from M. melolontha Larvae
Finally, the antibacterial activity of the isolates that were identified in the study was compared while using Maximum Inhibitory Dilution (MID) tests. In this stage of the study, seven strains, including five strains that represent P. chlororaphis and two from the genus Chryseobacterium, were found to be able to inhibit the growth of the symbiotic bacteria of nematodes (Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7).
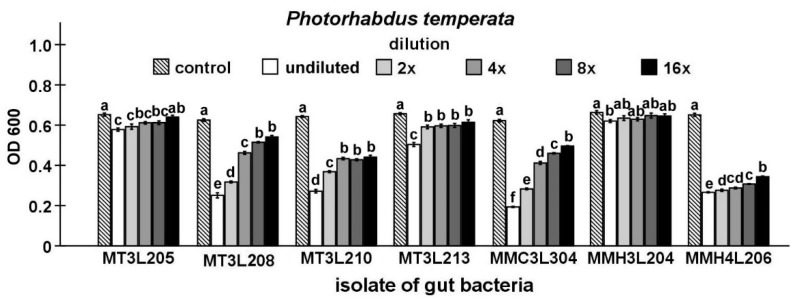
Figure 3.
Antagonistic activities of different dilutions of culture supernatants from selected M. melolontha midgut isolates against P. temperata shown by Maximum Inhibitory Dilution (MID) tests. The same letters mean no significant differences between the activities of supernatants from the same midgut isolate.
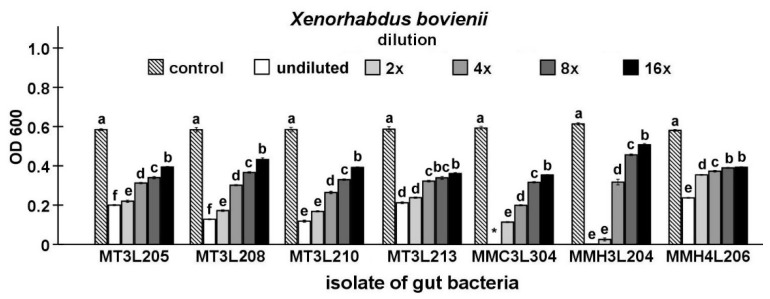
Figure 4.
Antagonistic activities of different dilutions of culture supernatants from selected M. melolontha midgut isolates against X. bovienii shown by MID tests. The same letters mean no significant differences between the activities of supernatants from the same midgut isolate. *—no bacterial growth.
Figure 5.
Antagonistic activities of different dilutions of culture supernatants from selected M. melolontha midgut isolates against X. budapestensis shown by MID tests. The same letters mean no significant differences between the activities of supernatants from the same midgut isolate.
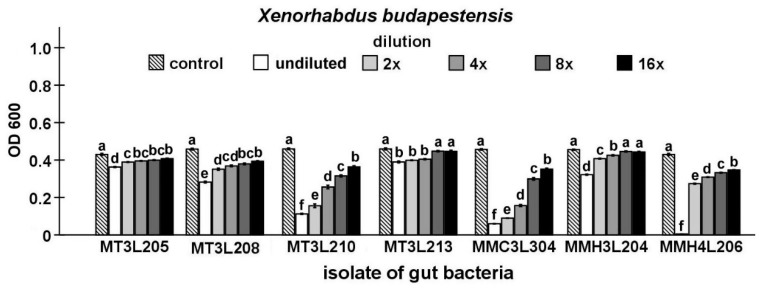
Figure 6.
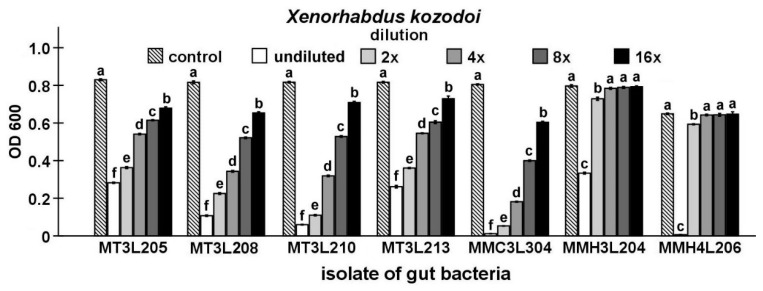
Antagonistic activities of different dilutions of culture supernatants from selected M. melolontha midgut isolates against X. kozodoi shown by MID tests. The same letters mean no significant differences between the activities of supernatants from the same midgut isolate.
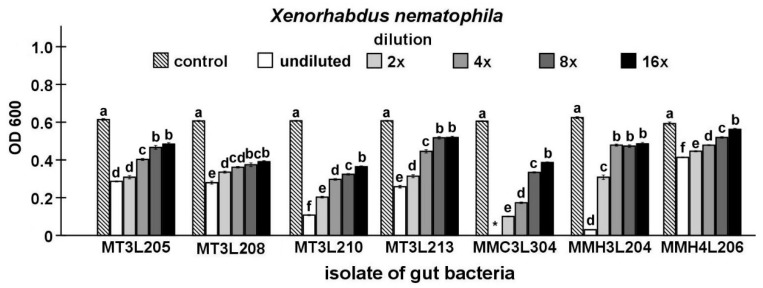
Figure 7.
Antagonistic activities of different dilutions of culture supernatants from selected M. melolontha midgut isolates against X. nematophila shown by MID tests. The same letters mean no significant differences between the activities of supernatants from the same midgut isolate. *—no bacterial growth.
The undiluted supernatant of the P. chlororaphis MMC3L304 isolate caused the strongest inhibition of P. temperata growth (t-test t18 = 58.86, p < 0.001). In the undiluted supernatant groups, the supernatant of the C. lathyri MMH3L204 isolate exerted the lowest inhibitory effect on P. temperata growth (t-test t18 = 3.68, p < 0.01). The supernatant dilution had a statistically significant influence on P. temperata growth (ANOVA, F5,54 ≥ 9.40, p < 0.001). The supernatant of the Chryseobacterium sp. MMH4L206 isolate (t-test t18 = 34.72, p < 0.001) exerted the highest inhibitory effect on P. temperata growth in the 16-fold diluted supernatant group. This dilution of the P. chlororaphis MT3L205 and C. lathyri MMH3L204 supernatants had no significant influence on P. temperata growth (Figure 3).
The undiluted supernatant of the P. chlororaphis MMC3L304 isolate caused the complete inhibition of X. bovienii growth (t-test, t18 = 111.31, p < 0.001). In the undiluted supernatant groups, the supernatant of the P. chlororaphis MT3L213 isolate exerted the lowest inhibitory effect on X. bovienii growth (t-test t18 = 27.29, p < 0.001). The supernatant dilution had a statistically significant influence on X. bovienii growth (ANOVA, F5,54 ≥ 343.52, p < 0.001). The supernatant of the P. chlororaphis MT3L205 isolate (t-test t18 = 36.66, p < 0.001) exerted the highest inhibitory effect on X. bovienii growth, while the lowest inhibitory effect was exhibited by the C. lathyri MMH3L204 isolate supernatant (t-test t18 = 10.55, p < 0.001) in the 16-fold diluted supernatant group (Figure 4).
The undiluted supernatant of the P. chlororaphis MMC3L304 isolate caused the strongest inhibition of X. budapestensis growth (t-test, t18 = 100.31, p < 0.001). In the undiluted supernatant groups, the supernatant of the P. chlororaphis MT3L213 isolate exerted the lowest inhibitory effect on X. budapestensis growth (t-test t18 = 9.42, p < 0.001). The supernatant dilution had a statistically significant influence on X. budapestensis growth (ANOVA, F5,54 ≥ 43.29, p < 0.001). The supernatant of the P. chlororaphis MMC3L304 isolate (t-test t18 = 16.14, p < 0.001) exerted the highest inhibitory effect on X. budapestensis growth in the 16-fold diluted supernatant group. There was no significant effect of the 8- and 16-fold P. chlororaphis MT3L213 and C. lathyri MMH3L204 supernatant dilutions on X. budapestensis growth (Figure 5).
The undiluted supernatant of the P. chlororaphis MMC3L304 isolate caused the strongest inhibition of X. kozodoi growth (t-test t18 = 222.50, p < 0.001). In the undiluted supernatant groups, the supernatant of the C. lathyri MMH3L204 isolate exerted the lowest inhibitory effect on X. kozodoi growth (t-test t18 = 54.79, p < 0.001). The supernatant dilution had a statistically significant influence on X. kozodoi growth (ANOVA, F5,54 ≥ 968.90, p < 0.001). The supernatant of the P. chlororaphis MMC3L304 isolate (t-test t18 = 31.49, p < 0.001) exerted the highest inhibitory effect on X. kozodoi growth in the 16-fold diluted supernatant group. There was no significant effect of the 4-, 8-, and 16-fold C. lathyri MMH3L204 and Chryseobacterium sp. MMH4L206 supernatant dilutions on X. kozodoi growth (Figure 6).
The undiluted supernatant of the P. chlororaphis MMC3L304 isolate caused the complete inhibition of X. nematophila growth (t-test, t18 = 260.45, p < 0.001). In the undiluted supernatant groups, the supernatant of the Chryseobacterium sp. MMH4L206 isolate exerted the lowest inhibitory effect on X. nematophila growth (t-test t18 = 19.58, p < 0.001). The supernatant dilution had a statistically significant influence on X. nematophila growth (ANOVA, F5,54 ≥ 204.68, p < 0.001). The supernatant of the P. chlororaphis MT3L210 isolate (t-test t18 = 55.19, p < 0.001) exerted the highest inhibitory effect on X. nematophila growth, while the lowest inhibitory effect was exhibited by the Chryseobacterium sp. MMH4L206 isolate supernatant (t-test t18 = 2.98, p < 0.05) in the 16-fold diluted supernatant group (Figure 7).
As shown above, one of the isolates, i.e., P. chlororaphis MMC3 L3 04, exhibited the strongest antagonistic properties against the tested EPB. This strain was isolated under microaerobic conditions from the midgut of a larva that was exposed to S. carpocapsae infective juveniles. The culture filtrate of this bacterium significantly inhibited the growth of all Xenorhabdus species, with the strongest effect on X. bovienii and X. nematophila, which was evidenced by the complete inhibition of bacterial growth by the undiluted filtrate (Figure 4 and Figure 7).
3. Discussion
In the past years, considerable new information regarding the interactions between mutualistic bacteria of entomopathogenic nematodes and gut-associated bacteria of insects has been provided; however, the findings had a one-sided and limited character. To date, there has only been a unidirectional relationship imposed by EPB through the production of a number of substances that inhibit the growth of insect gut bacteria. It is known that the bacteriocins produced by Xenorhabdus and Photorhabdus bacteria exhibit strong growth-inhibitory activity against other EPB or bacteria from the host. For example, xenorhabdicin that is produced by X. nematophila inhibits the growth of other species of Xenorhabdus as well as P. luminescens and bacteria of the genus Proteus [27]. Similarly, Photorhabdus sp. produces photorhabdicin and lumicin-bacteriocins, which are active against other Photorhabdus strains and Escherichia coli [28,29]. In addition to bacteriocins, EPB generate an array of antimicrobial secondary metabolites, many of which have a wider scope of activity than bacteriocins [30,31]. As bacteria proliferate in the insect’s body, their antibiotic activity steadily grows to reach a maximum between days 3 and 5 of infection, i.e., when the host is already dead [32,33].
The method of selection that was applied in the study helped to isolate strains with antagonistic effects against Xenorhabdus and Photorhabdus bacteria in the material containing an extremely species-rich insect midgut microbiota. The cross-streak tests used in the initial stage are relatively quick to perform and allow for preliminary analysis of a large number of isolates. The next two stages facilitated the analysis of the antibacterial properties of the isolates with greater precision. Evidently, only some of the strains that were selected by the cross-streak tests had their activity confirmed by the other two tests. This might have been related to the different types of bacterial growth in each test. In the cross-streak tests, the bacteria grew on the surface, whereas, in our modification of the agar well diffusion tests, they were suspended in agar medium. We used a modified version of the agar well diffusion method, as we found that it yielded substantially larger growth inhibition zones in the case of the tested strains, which facilitated the identification of isolates with antibacterial properties. The modification consisted in introduction of LB agar medium inoculated with the tested bacterial isolates into the wells instead of the culture filtrate of these isolates (Materials and Methods 4.6.2).
In the last test, i.e., MID, contrary to the previous ones, there was no direct interaction between the investigated bacterial groups, as bacterium-free culture filtrates were used. This might have contributed to the detection of the antagonistic activity against Xenorhabdus and Photorhabdus in only seven isolates at this stage. Another explanation of such results might also be the loss of antibacterial activity during storage: the cross-streak tests were carried out immediately after the isolation of the bacteria from larval midguts, whereas the subsequent tests were performed while using freeze-stored bacteria.
As shown in Table 1, most of the isolates displaying antagonistic properties against EPB (23 of the 38 isolates that were obtained in the first stage of the study) were isolated in aerobic conditions. However, the P. chlororaphis MMC3 L3 04 strain, which exhibited the highest antibacterial activity in most assays, was isolated in microaerobic conditions. It is noteworthy that the vast majority of the selected strains with antagonistic properties, including all strains identified taxonomically, exhibited the ability to grow in aerobic conditions (data not shown), although some of them were isolated in microaerobic conditions. Therefore, it can be assumed that higher efficiency of screening of the bacteria from the midgut of M. melolontha larvae in terms of their antibacterial activity against Xenorhabdus and Photorhabdus species can be achieved in aerobic conditions.
Comparative analyses of 16S rRNA gene sequences are useful for the classification of cultured microorganisms, especially given the established taxonomic thresholds [34]. However, it is known that the sequencing of the 16S rRNA gene is only not sufficient for the identification of most bacteria at the species level [35,36]. We analyzed the sequences of 16S rRNA genes and those coding for proteins with conserved functions i.e., rpoB, rpoD, and/or recA, which efficiently supplemented the 16S rRNA gene-based identification of the bacteria to reliably identify the isolates at the species level [22,23,24,25,26]. As shown in the present study, the 12 analyzed bacterial isolates represented five genera, namely, Pseudomonas, Citrobacter, Serratia, Acinetobacter, and Chryseobacterium (Table 4, Figures S1–S6). Half of the isolates with strong antagonistic activity against EPB were identified as P. chlororaphis. Four of them were isolated from the midgut of the same M. melolontha larva, while the other two were isolated from other specimens collected from different locations at different times (Table 1). The P. chlororaphis strains were isolated from the larvae in different developmental phases, i.e., four isolates were obtained from a stage L2 larva and the other two were isolated from two L3 larvae. In most cases, the P. chlororaphis strains were characterized by a stronger ability to inhibit the growth of the symbiotic bacteria of nematodes than the other isolates. These results suggest that P. chlororaphis bacteria may be an important factor inhibiting the growth of bacteria from the genera Xenorhabdus and Photorhabdus also in in vivo conditions, i.e., in the organism of M. melolontha.
The presence of the Pseudomonas sp. bacteria has been repeatedly detected in samples that were collected from Scarabaeidae larvae, e.g., in the midgut of M. hippocastani L3 larvae [37], in the hindgut of Holotrichia parallela L3 larvae [38], or in the midgut of 3rd instar Protaetia brevitarsis larvae, where these bacteria were the most dominant genera [39]. Pseudomonas sp. bacteria have also been isolated in studies on their insecticidal activity [40]. To date, the ability of P. chlororaphis to inhibit the growth of Photorhabdus or Xenorhabdus bacteria has not been shown, but its antibacterial activity against other species, e.g., Clavibacter michiganensis [41], Bacillus subtilis, and Salmonella enteritidis [42], or Staphylococcus aureus [43], has been reported.
Most of the bacteria that represent the other genera identified in the present study were previously isolated from the digestive system of insects from the genus Melolontha. For example, Serratia sp., Acinetobacter sp., and Citrobacter sp. were isolated from the midgut of M. hippocastani L3 larvae [37], while a Serratia marcescens strain producing a highly active bacteriocin-like substance and Acinetobacter sp. have been isolated from M. melolontha [40,44]. In turn, Chryseobacterium sp. bacteria have been isolated from the gut of another member of the Scarabaeidae family, i.e., Protaetia brevitarsis seulensis [40]. Furthermore, the antibacterial activities of bacteria from the genera Chryseobacterium [45] and Citrobacter have been described before [46,47]; however, their interactions with the symbiotic bacteria of entomopathogenic nematode, have not been reported so far, as in the case of Pseudomonas or Serratia.
Importantly, 10 of the 12 selected isolates, i.e., representatives of the genera Acinetobacter, Citrobacter, Pseudomonas, and Serratia, belong to γ-proteobacteria, i.e., a class that comprises a number of human and animal pathogens, including many insect species. Various studies have reported the production of many compounds that inhibit the development of fungi, insects, and nematodes (e.g., phenazine-type antibiotics, hydrogen cyanide, chitinases, and proteases) by P. chlororaphis [48,49]. It has been evidenced that P. chlororaphis injected directly into the hemocoel caused the high mortality of Galleria mellonella (Lepidoptera: Pyralidae) larvae [50]. Oral and injectable toxicity to Manduca sexta (Lepidoptera: Sphingidae) and Drosophila melanogaster (Diptera: Drosophiladae) larvae have both also been described [51]. As shown by Schellenberger et al. [52], P. chlororaphis isolated from soil produces insecticidal protein, which is effective in Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae) larvae. In turn, a S. liquefaciens strain that was isolated from M. melolontha larvae exerted a pathogenic effect on the larvae of Dendroctonus micans (Coleoptera: Curculionidae), Thaumetopoea pityocampa (Lepidoptera: Thaumetopoeidae), and Lymantria dispar (Lepidoptera: Erebidae) [53]. Similarly, Chryseobacterium sp. was shown to exhibit high pathogenicity to D. melanogaster when injected into the hemocoel [54].
Based on the literature data and the results of the present study, it can be concluded that the bacterial strains that are present in the M. melolontha larvae gut can potentially exert both adverse and beneficial effects on the health status of their hosts and on their survival under pressure from entomopathogenic nematodes. Consideration of the potential interactions between insect gut bacteria and symbiotic Xenorhabdus and Photorhabdus bacteria of entomopathogenic nematodes should address the question of whether these two groups of bacteria have a chance of mutual contact in natural conditions, i.e., in the insect organism. The natural environment for the development of Xenorhabdus spp. and Photorhabdus spp. is the hemocoel, where entomopathogenic nematodes, rather than the insect gut, release the bacteria. However, nematodes sometimes release their symbiotic bacteria still in the intestine, as mentioned earlier [12,16]. In such a case, it is possible that the bacteria carried by the nematode and those colonizing the insect can compete with each other in a direct manner. This suggests that such antagonism might be the insect’s first line of defense, being triggered before the immune response, which is initiated later when the bacteria enter the hemolymph. Importantly, a nematode must perforate the insect′s gut when penetrating from the alimentary canal to the hemocoel. While considering the size of bacterial cells, it might be assumed that the damaged gut becomes a gateway through which a certain amount of intestinal bacteria may enter the hemocoel. This seems to create good conditions for interactions between the two groups of bacteria at an early stage of infection. Furthermore, EPB, which find their way into the hemocoel proliferate at a fast rate, as this space provides optimal conditions for their growth, and the natural barriers that separate gut-associated bacteria from EPB, are quickly removed by bacterial enzymes that hydrolyze insect tissues, including the gut wall.
These preliminary investigations only provide a fragmentary description of the interactions between bacteria inhabiting the gut of scarab larvae and entomopathogenic nematode bacterial symbionts. Comprehensive research in this field was conducted with the use of different culture media, diverse screening methods for the determination of antibacterial activity, or a wider spectrum of insect and nematode species would certainly help to fully elucidate the mechanisms of bacterial interspecies competition and the insect defense against infection by entomopathogenic nematodes. Furthermore, a better understanding of the scarab larva-associated microbiota is necessary for elucidating the effect of natural microbiota on host resistance to pathogens. The identification of specific bacteria that protect insects from entomopathogenic nematodes might have great importance for manipulating insect fitness and susceptibility to pathogens, thus opening avenues for increasing the efficacy of pest management programs.
4. Materials and Methods
4.1. Entomopathogenic Nematodes
The investigations were carried out on five species of EPN that were isolated from the natural environment in Poland and identified with molecular methods. The nematodes were propagated in Galleria mellonella larvae. The infective larvae were collected after the nematodes have developed in the insect, while using the modified “white trap” method [55]. Straight genetic lines were derived from the isolates. Infective juveniles of EPN were kept in a sterile aqueous solution at 8 °C. The larvae were kept under these conditions for no longer than 15 days. The larvae of G. mellonella were grown on a natural diet. The experiments were conducted while using caterpillars weighing 180–200 mg.
4.2. Entomopathogenic Nematodes Symbiotic Bacteria
Five species of EPN bacterial symbionts were isolated from nematodes and used in the research (in the brackets names of the source nematodes): X. bovienii (S. silvaticum), X. nematophila (S. carpocapsae), X. kozodoii (S. arenarium), X. budapestensis (S. bicornutum), and P. temperata (H. megidis). The bacteria were stored frozen at −85 °C in lysogeny broth (LB) medium that was supplemented with 20% glycerol. Each time after thawing, the bacteria were tested on the NBTA, and blue or green colonies were used in subsequent analyses, since all of the experiments were exclusively based on EPB in the primary form.
4.3. Collection of M. melolontha Larvae from Their Natural Environment
M. melolontha L2 and L3 larvae were collected in forest and agricultural areas of the Lublin region (Eastern Poland). The collected grubs were put separately in plastic boxes with soil to prevent the insects from harmful biting. In the laboratory, the species and the grub stage of development were identified. Insects in good condition were selected for experiments and for the control group.
4.4. Exposure of M. melolontha to Selected EPN Species
The larvae of M. melolontha were placed separately in 150-mL plastic cups containing soil (−50 kPa water potential) from the larvae harvesting field. A scarab larva was placed at the bottom of each cup prior to addition of the soil. Infective juvenile stages of EPN were introduced in doses of 1000 IJs per insect. The cups were kept in incubators at 20 °C for 12 days.
4.5. Isolation of Bacteria from M. melolontha Midgut
All of the scarab larvae were surface sterilized with 70% alcohol, washed twice in sterile distilled water, and allowed to dry for 1 min. The digestive tract was dissected; then, the midgut was isolated and homogenized in 1 mL of 0.5% NaCl while using a glass tissue grinder. Serial 10-fold dilutions were spread on duplicated plates of LB agar and then incubated at 20 °C in aerobic or microaerobic (6% oxygen) conditions for 2–4 days. Single colonies were picked, purified by subculturing on plates, and then transferred to agar slants for further tests.
4.6. Antimicrobial Activity Assays
4.6.1. Cross-Streak Tests
The nematode bacteria and the midgut isolates were grown separately in liquid LB medium for two days at 20 °C to prepare the inocula for the cross-streak tests. Each midgut isolate was then subinoculated as a middle line on a plate with LB agar medium. Each of all isolated Xenorhabdus or Photorhabdus strains was seeded in perpendicular lines on both sides of the midgut isolate line, and the plates were incubated at 20 °C for 72 h in aerobic or microaerophilic conditions. The antibacterial potential of gut isolates was measured (in mm) as an EFB inhibition zone.
4.6.2. Modified Agar Well Diffusion Tests
The nematode bacteria and the midgut isolates were separately cultured in liquid LB medium for two days at 20 °C in the preparation of the inocula for the agar well diffusion assay. Next, warm (43 °C) LB medium containing 2% of agar placed in five conical flasks was separately inoculated with 2% of two-day old culture of nematode bacterial strains and then spilled (20 mL) onto disposable plastic Petri dishes with a 9-cm diameter. An appropriate number of wells (12 mm diameter) were cut out in the solidified agar medium. Each well was then filled with 260 μL of the warm LB medium with 2.0% agar inoculated with 2% of two-day old culture of a respective isolate strain. Two plates with one standard strain were intended for the study of its interaction with each midgut isolate. The plates were incubated at 20 °C for three days in the dark. The diameters (in mm) of the zones of inhibition of the nematode strain growth around the wells were determined.
4.6.3. Maximum Inhibitory Dilution (MID) Tests
The isolates selected in previous tests were cultured in 100 mL Erlenmeyer flasks with 20 mL of LB medium. After 48 h of incubation at 20 °C, the cultures were centrifuged at 16,600× g for 15 min. The supernatants were filtered through sterile 0.22-μm nylon filters. Subsequently, series of two-fold dilutions of cell-free supernatants were prepared. At the same time, two-day-old cultures of EPB were suspended in sterile LB medium to a density of 0.5 McFarland standard. Afterwards, 100 μL of diluted supernatants and 100 μL of EPB suspensions were dispensed in a 96-well sterile microtiter plate and then incubated at 20 °C for 72 h. Afterwards, the growth of bacteria was measured spectrophotometrically at 600 nm (OD600) while using the Synergy HT Microplate Reader (Bio-Tek Instruments, Winooksi, VT, USA). All of the experiments were performed in two replicates, with five independent groups in each replication.
4.7. Molecular Identification of the Bacterial Isolates
Total genomic DNA was extracted while using a Genomic Mini AX Bacteria Spin Kit (A&A Biotechnology, Gdynia, Poland and then stored at −20 °C. All of the PCR amplifications were carried out with PCR mix RAPID (A&A Biotechnology, Gdynia, Poland), according to the manufacturer’s recommendations. The primer sequences and PCR conditions used are listed in Table 3. The amplified PCR products were purified with Clean-Up purification columns (A&A Biotechnology, Gdynia, Poland) and then sequenced in Genomed S.A. (Warsaw, Poland). Preliminarily, the 16S rRNA gene sequences were compared with the EzBioCloude database. All of the obtained sequences were analyzed while using BLAST available on the NCBI website. Multiple sequence alignment matrices of the individual gene sequences were created using ClustalW included in the MEGA 6.06 software [56]. The sequence identity values were calculated while using BioEdit 7.0.5 software.
The 16S rRNA gene sequence similarity threshold value of 98.7% between the isolate and species type strain was used as an indicator that an isolate can be a member of a given species [34]. The identification of the isolates at the species level was considered to be final when the searching results that were based on 16S rRNA gene sequences were concordant with those based on the rpoB, rpoD, or recA gene sequences.
4.8. Phylogenetic Analysis
Phylogenetic analyses of gene sequences were performed to confirm the identification results. The 16S rRNA, rpoB, rpoD, and recA gene sequences of isolates that were obtained in this study were compared to GenBank nucleotide sequences using BLAST (NCBI). Multiple sequence alignments were created using ClustalW at the default configuration and manually checked. The evolutionary distances were computed using the Tamura–Nei algorithm and the phylogenetic trees were generated using the neighbor-joining method in MEGA 6.06. All of the positions containing gaps and missing data were eliminated. Bootstrapping with 1000 replicates of the data was conducted to determine the statistical support for the branches.
All of the gene sequences that were obtained in this study were deposited in the GeneBank database under the accession numbers given in Table 4 and depicted in the phylogenetic trees (Supplementary Materials Figures S1–S6).
4.9. Statistical Analysis
The data were pooled before statistical analysis since there were no statistically significant differences between the results of the replicates of all experiments. The test results were subjected to one-way ANOVA and Tukey′s post-hoc tests (supernatant dilutions). The t-student test was used for pairwise comparisons between bacteria species. The normality of the data distribution was determined while using the Shapiro–Wilk test and the homogeneity of variance was assessed by the Levene test. The occurrence of statistically significant differences in these experiments was based on the overlap of 95% confidence intervals. Differences among means were considered to be significant at p < 0.05. All of the statistical analyses were performed while using the IBM SPSS Statistics 24 software package.
Acknowledgments
The authors gratefully acknowledge use of the services and facilities of the Center for Interdisciplinary Research of The John Paul II Catholic University of Lublin, Lublin, Poland, co-funded by the European Union from the European Regional Development Fund in the frame of the Operational Programme Development of Eastern Poland 2007-2013 (POPW.01.03.00-06-003/09-00).
Abbreviations
| ANOVA | Analysis of variance |
| BLAST | Basic local alignment search tool |
| EPB | Entomopathogenic bacteria |
| EPN | Entomopathogenic nematodes |
| IJs | Infective juveniles |
| LB | Lysogeny broth (or Luria broth) |
| MID | Maximum inhibitory dilution |
| NBTA | Nutrient bromothymol blue agar |
| NCBI | National Center for Biotechnology Information |
| PCR | Polymerase chain reaction |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/2/580/s1, Figure S1: Neighbor joining tree based on 16S rRNA gene sequences showing the phylogenetic relationships of the studied M. melolontha midgut isolates (bolded) exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes., Figure S2 Neighbor joining tree based on rpoD gene sequences showing the phylogenetic relationships of the studied M. melolontha midgut Pseudomonas isolates (bolded) exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes., Figure S3: Neighbor joining tree based on rpoB gene sequences showing the phylogenetic relationships of the studied M. melolontha midgut Serratia and Citrobacter isolates (bolded) exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes., Figure S4: Neighbor joining tree based on rpoB gene sequences showing the phylogenetic relationships of the studied M. melolontha midgut Acinetobacer isolate (bolded) exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes., Figure S5: Neighbor joining tree based on rpoB gene sequences showing the phylogenetic relationships of the studied M. melolontha midgut Chryseobacterium isolates (bolded) exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes., Figure S6: Neighbor joining tree based on recA gene sequences showing the phylogenetic relationships of the studied M. melolontha midgut Pseudomonas isolates (bolded) exhibiting antagonistic activity against bacterial symbionts of entomopathogenic nematodes.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “Conceptualization, M.S. and A.W.; methodology, M.S., A.W., M.P., W.K., M.L. and E.S.; software, W.K., M.L. and E.S.; validation, M.S., W.K. and A.W.; formal analysis, M.P. and M.S.; investigation, M.S., A.W., M.P., W.K., M.L. and E.S.; resources, M.L.; data curation, M.S.; writing—original draft preparation, M.S., M.P., W.K., M.L. and E.S.; writing—review and editing, M.S., A.W., M.P., W.K., M.L. and E.S.; visualization, M.S., A.W., M.L. and E.S.; supervision, M.S. and A.W.; project administration, M.L.; funding acquisition, M.S. and A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the National Science Centre (Poland), grant number 2016/21/B/NZ9/01865.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jameson M.L., Ratcliffe B.C. Introduction. Series Scarabaeiformia Crowson 1960 (Lamellicornia), Superfamily Scarabaeoidea Latreille 1802. In: Arnett R.H., Thomas M.C., Skelley P.E., Frank J.H., editors. American Beetles. Polyphaga: Scarabaeoidea through Curculionoidea. Volume 2. CRC Press; New York, NY, USA: 2002. pp. 1–5. [Google Scholar]
- 2.Zimmermann G. Use of the fungus, Beauveria brongniartii for the control of European cockchafers, Melolontha spp. In: Jackson T.A., Glare T.R., editors. Europe. In Use of Pathogens in Scarab Pest Management. Andover, Hampshire; Intercept, UK: 1992. pp. 199–208. [Google Scholar]
- 3.Jackson T.A., Klein M.G. Scarabs as pests: A continuing problem. Coleopt. Soc. Monogr. 2006;5:102–119. doi: 10.1649/0010-065X(2006)60[102:SAPACP]2.0.CO;2. [DOI] [Google Scholar]
- 4.Ansari M.A., Shah F.A., Tirry L., Moens M. Field trials against Hoplia philanthus (Coleoptera: Scarabaeidae) with a combination of an entomopathogenic nematode and the fungus Metarhizium anisopliae CLO 53. Biol. Control. 2006;39:453–459. doi: 10.1016/j.biocontrol.2006.07.004. [DOI] [Google Scholar]
- 5.Kaya H.K., Aguillera M.M., Alumai A., Choo H.Y., de la Torre M., Fodor A., Ganguly S., Hazir S., Lakatos T., Pye A., et al. Status of entomopathogenic nematodes and their symbiotic bacteria from selected countries or regions of the world. Biol. Control. 2006;38:134–155. doi: 10.1016/j.biocontrol.2005.11.004. [DOI] [Google Scholar]
- 6.Lacey L., Georgis R. Entomopathogenic nematodes for control of insect pests above and below ground with comments on commercial production. J. Nematol. 2012;44:218–225. [PMC free article] [PubMed] [Google Scholar]
- 7.Akhurst R.J. Safety to nontarget invertebrates of nematodes of economically important pests. In: Laird M., Lacey L.A., Davidson E.W., editors. Safety of Microbial Insecticides. CRC Press; Boca Raton, FL, USA: 1990. pp. 234–238. [Google Scholar]
- 8.Wilson M., Gaugler R. Terrestrial slug pests. In: Lacey L., Kaya H., editors. Field Manual for Application and Evaluation of Invertebrate Pathogens. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. pp. 784–804. [Google Scholar]
- 9.An R., Grewal P.S. Differences in the virulence of Heterorhabditis bacteriophora and Steinernema scarabaei to three white grub species: The relative contribution of the nematodes and their symbiotic bacteria. Biol. Control. 2007;43:310–316. doi: 10.1016/j.biocontrol.2007.07.004. [DOI] [Google Scholar]
- 10.Beliën T. Entomopathogenic nematodes as biocontrol agents of insect pests in orchards. CAB Rev. 2018;13:1–11. doi: 10.1079/PAVSNNR201813058. [DOI] [Google Scholar]
- 11.Marianelli L., Paoli F., Torrini G., Mazza G., Benvenuti C., Binazzi F., Sabbatini Peverieri G., Bosio G., Venanzio D., Giacometto E., et al. Entomopathogenic nematodes as potential biological control agents of Popillia japonica (Coleoptera, Scarabaeidae) in Piedmont Region (Italy) J. Appl. Entomol. 2017;142:311–318. doi: 10.1111/jen.12470. [DOI] [Google Scholar]
- 12.Polavarapu S., Koppenhöfer A.M., Barry J.D., Holdcraft R.J., Fuzy E.M. Entomopathogenic nematodes and neonicotinoids for remedial control of oriental beetle, Anomala orientalis (Coleoptera: Scarabaeidae), in highbush blueberry. Crop Prot. 2007;26:1266–1271. doi: 10.1016/j.cropro.2006.10.026. [DOI] [Google Scholar]
- 13.Boemare N.E. Biology, Taxonomy and Systematics of Photorhabdus and Xenorhabdus. In: Gaugler R., editor. Entomopathogenic Nematology. CABI Publishing; New York, NY, USA: 2002. pp. 35–56. [DOI] [Google Scholar]
- 14.Gaugler R. Ecological consideration in the biological control of soil-Habitating insect with entomopathogenic nematode. Agr. Ecosyst. Environ. 1988;24:351–360. doi: 10.1016/0167-8809(88)90078-3. [DOI] [Google Scholar]
- 15.Gaugler R., Kaya H.K. Entomophathogenic Nematodes in Biological Control. CRC Press; Boca Raton, FL, USA: 1990. [Google Scholar]
- 16.Adams B.J., Fodor A., Koppenhofer H.S., Stackebrandt E., Stock S.P., Klein M.G. Reprint of “Biodiversity and systematics of nematode-Bacterium entomopathogens”. Biol. Control. 2006;38:4–21. doi: 10.1016/S1049-9644(06)00126-5. [DOI] [Google Scholar]
- 17.Akhurst R.J. Antibiotic activity of Xenohabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J. Gen. Microbiol. 1982;128:3061–3065. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- 18.Boemare N.E., Givaudan A., Brehelin M., Laumond C. Symbiosis and pathogenicity of nematode-Bacterium complexes. Symbiosys. 1997;22:21–45. [Google Scholar]
- 19.Nguyen K.B., Hunt D.J. Entomopathogenic Nematodes: Systematics, Phylogeny and Bacterial Symbionts. Brill; Leiden, The Netherlands: 2007. [Google Scholar]
- 20.Huang S.-W., Zhang H.-Y., Marshall S., Jackson E.A. The scarab gut: A potential bioreactor for bio-Fuel production. Insect Sci. 2010;17:175–183. doi: 10.1111/j.1744-7917.2010.01320.x. [DOI] [Google Scholar]
- 21.Miller C.S., Handley K.M., Wrighton K.C., Frischkorn K.R., Thomas B.C., Banfield J.F. Short-Read assembly of full-Length 16S amplicons reveals bacterial diversity in subsurface sediments. PLoS ONE. 2013;8:e56018. doi: 10.1371/journal.pone.0056018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajwar A., Sahgal M. Phylogenetic relationships of fluorescent pseudomonas deduced from the sequence analysis of 16S rRNA, Pseudomonas-Specific and rpoD genes. 3 Biotech. 2016;6:80. doi: 10.1007/s13205-016-0386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delétoile A., Decré D., Courant S., Passet V., Audo J., Grimont P., Arlet G., Brisse S. Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J. Clin. Microbiol. 2009;47:300–310. doi: 10.1128/JCM.01916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khosravi A.D., Sadeghi P., Shahraki A.H., Heidarieh P., Sheikhi N. Molecular methods for identification of Acinetobacter species by partial sequencing of the rpoB and 16S rRNA Genes. J. Clin. Diagn. Res. 2015;9:DC09–DC13. doi: 10.7860/JCDR/2015/13867.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehouse C.A., Baldwin C., Wasieloski L., Kondig J., Scherer J. Molecular identification of the biowarfare simulant Serratia marcescens from a 50-Year-Old munition buried at Fort Detrick, Maryland. Mil. Med. 2007;172:860–863. doi: 10.7205/MILMED.172.8.860. [DOI] [PubMed] [Google Scholar]
- 26.Hilario E., Buckley T.R., Young J.M. Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie Van Leeuwenhoek. 2004;86:51–64. doi: 10.1023/B:ANTO.0000024910.57117.16. [DOI] [PubMed] [Google Scholar]
- 27.Thaler J.O., Baghdiguian S., Boemare N. Purification and characterization of xenorhabdicin, a phage tail-Like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl. Environ. Microb. 1995;61:2049–2052. doi: 10.1128/AEM.61.5.2049-2052.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ffrench-Constant R., Waterfield N., Daborn P., Joyce S., Bennett H., Au C., Dowling A., Boundy S., Reynolds S., Clarke D. Photorhabdus: Towards a functional genomic analysis of a symbiont and pathogen. Fems Microbiol. Rev. 2003;26:433–456. doi: 10.1111/j.1574-6976.2003.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 29.Sharma S., Waterfield N., Bowen D., Rocheleau T., Holland L., James R., Ffrench-Constant R. The lumicins: Novel bacteriocins from Photorhabdus luminescens with similarity to the uropathogenic-Specific protein (USP) from uropathogenic Escherichia coli. Fems Microbiol. Lett. 2002;214:241–249. doi: 10.1111/j.1574-6968.2002.tb11354.x. [DOI] [PubMed] [Google Scholar]
- 30.Hu K., Webster J.M. Antibiotic production in relation to bacterial growth and nematode development in Photorhabdus heterorhabditis infected Galleria mellonella larvae. Fems Microbiol. Lett. 2000;189:219–223. doi: 10.1016/S0378-1097(00)00288-3. [DOI] [PubMed] [Google Scholar]
- 31.White J.F., Torres M.S. Defensive Mutualism in Microbial Symbiosis. CRC Press; Boca Raton, FL, USA: 2009. [Google Scholar]
- 32.Isaacson P.J., Webster J.M. Antimicrobial activity of Xenorhabus sp RIO (Enterobacteriaceae), symbiont of the entomopathogenic nematode, Steinernema riobrave (Rhabditida: Steinernematidae) J. Invertebr. Pathol. 2002;79:146–153. doi: 10.1016/S0022-2011(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 33.Walsh K.T., Webster J.M. Interaction of microbial populations in Steinernema (Steinernematidae, Nematoda) infected Galleria mellonella larvae. J. Invertebr. Pathol. 2003;83:118–126. doi: 10.1016/S0022-2011(03)00079-X. [DOI] [PubMed] [Google Scholar]
- 34.Stackebrandt E., Ebers J. Taxonomic parameters revised: Tarnished gold standards. Microbiol. Today. 2006;33:152–155. [Google Scholar]
- 35.Rossi-Tamisier M., Benamar S., Raoult D., Fournier P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-Associated bacterial species. Int. J. Syst. Evol. Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 36.Yarza P., Yilmaz P., Pruesse E., Glöckner F.O., Ludwig W., Schleifer K.-H., Whitman W.B., Euzéb J., Amann R., Rosselló-Móra R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 37.Arias-Cordero E., Ping L., Reichwald K., Delb H., Platzer M., Boland W. Comparative evaluation of the gut microbiota associated with the below- and above-Ground life stages (larvae and beetles) of the forest cockchafer, Melolontha hippocastani. PLoS ONE. 2012;7:e51557. doi: 10.1371/journal.pone.0051557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S., Sheng P., Zhang H. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae. Int. J. Mol. Sci. 2012;13:2563–2577. doi: 10.3390/ijms13032563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian X.-Y., Song F.-P., Zhang J., Liu R.-M., Zhang X.-P., Duan J.-Y., Shu C.-L. Diversity of gut bacteria in larval Protaetia brevitarsis (Coleoptera: Scarabaedia) fed on corn stalk. Acta Entomol. Sin. 2017;60:632–641. [Google Scholar]
- 40.Deng P., Wang X., Baird S.M., Lu S.E. Complete genome of Pseudomonas chlororaphis strain UFB2, a soil bacterium with antibacterial activity against bacterial canker pathogen of tomato. Stand. Genom. Sci. 2015;10:e117. doi: 10.1186/s40793-015-0106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aichour S.M., Nicklin J.L., Haichour N., Zerroug M.M. Antimicrobial activity of potato rhizospheric Pseudomonas chlororaphis subsp. aureofaciens from Sétif Algeria. Ann. Res. Rev. Biol. 2016;11:1–7. doi: 10.9734/ARRB/2016/31651. [DOI] [Google Scholar]
- 42.Veselova A., Klein S.H., Bass I.A., Lipasova V.A., Metlitskaya A.Z., Ovadis M.I., Chernin L.S., Khmel I.A. Quorum sensing systems of regulation, synthesis of phenazine antibiotics, and antifungal activity in rhizospheric bacterium Pseudomonas chlororaphis 449. Russ. J. Genet. 2008;44:1400–1408. doi: 10.1134/S102279540812003X. [DOI] [PubMed] [Google Scholar]
- 43.Urgas S., Sezen K., Kati H., Demirbag Z. Purification and characterization of an antibacterial substance produced by pest-Originated Serratia marcescens Mm3. Turk. J. Biol. 2014;38:177–184. doi: 10.3906/biy-1305-46]. [DOI] [Google Scholar]
- 44.Sezen K., Demir Ŷ., Demirbağ Z. Identification and pathogenicity of entomopathogenic bacteria from common cockchafer, Melolontha melolontha (Coleoptera: Scarabaeidae) N. Z. J. Crop. Hort. 2007;35:79–85. doi: 10.1080/01140670709510171. [DOI] [Google Scholar]
- 45.Lee J., Hwang S., Cho S. Immune tolerance to an intestine-adapted bacteria, Chryseobacterium sp., injected into the hemocoel of Protaetia brevitarsis seulensis. Sci. Rep. UK. 2016;6:31722. doi: 10.1038/srep31722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shanks R.M.Q., Dashiff A., Alster J.S., Kadouri D.E. Isolation and identification of a bacteriocin with antibacterial and antibiofilm activity from Citrobacter freundii. Arch. Microbiol. 2012;194:575–587. doi: 10.1007/s00203-012-0793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandal S.M., Sharma S., Pinnaka A.K., Kumari A., Korpole S. Isolation and characterization of diverse antimicrobial lipopeptides produced by Citrobacter and Enterobacter. BMC Microbiol. 2013;13:e152. doi: 10.1186/1471-2180-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nam H.S., Anderson A.J., Kim Y.C. Biocontrol efficacy of formulated Pseudomonas chlororaphis O6 against plant diseases and root-Knot nematodes. Plant Pathol. J. 2018;34:241–249. doi: 10.5423/PPJ.NT.12.2017.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson J.A., Staley J., Challender M., Heuton J. Safety of Pseudomonas chlororaphis as a gene source for genetically modified crops. Transgenic Res. 2018;27:103–113. doi: 10.1007/s11248-018-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flury P., Vesga P., Pechy-Tarr M., Aellen N., Dennert F., Hofer N., Kupferschmied K.P., Kupferschmied P., Melta Z., Ma Z., et al. Antimicrobial and insecticidal: Cyclic lipopeptides and hydrogen cyanide produced by plant-Beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 2017;8:100. doi: 10.3389/fmicb.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rangel L.I., Henkels M.D., Shaffer B.T., Walker F.L., Davis II E.W., Stockwell V.O., Bruck D., Taylor B.J., Loper J.E. Characterization of toxin complex gene clusters and insect toxicity of bacteria representing four subgroups of Pseudomonas fluorescens. PLoS ONE. 2016;11:e0161120. doi: 10.1371/journal.pone.0161120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schellenberger U., Oral J., Rosen B.A., Wei J.Z., Xie W., McDonald M.J., Cerf D.C., Diehn S.H., Crane V.C., Sandhal G.A., et al. A selective insecticidal protein from Pseudomonas for controlling corn rootworms. Science. 2016;354:634–637. doi: 10.1126/science.aaf6056. [DOI] [PubMed] [Google Scholar]
- 53.Ertürk Ö., Yaman M. Potential of five non-Spore-Forming bacteria, originated from the European cockchafer, Melolontha melolontha (Linnaeus, 1758) (Coleoptera: Scarabaeidae), on three economic insect pests. Egypt. J. Biol. Pest Control. 2019;29:59. doi: 10.1186/s41938-019-0160-6. [DOI] [Google Scholar]
- 54.Aryal S.K., Carter-House D., Stajich J.E., Dillman A.R. Microbial associates of the southern mole cricket (Scapteriscus borellii) are highly pathogenic. J. Invertebr. Pathol. 2017;150:54–62. doi: 10.1016/j.jip.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 55.White G. A method for obtaining infective nematode larvae from culture. Science. 1927;66:302–303. doi: 10.1126/science.66.1709.302-a. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.