Abstract
Sarcomere assembly and maintenance are essential physiological processes required for cardiac and skeletal muscle function and organism mobility. Over decades of research, components of the sarcomere and factors involved in the formation and maintenance of this contractile unit have been identified. Although we have a general understanding of sarcomere assembly and maintenance, much less is known about the development of the thin filaments and associated factors within the sarcomere. In the last decade, advancements in medical intervention and genome sequencing have uncovered patients with novel mutations in sarcomere thin filaments. Pairing this sequencing with reverse genetics and the ability to generate patient avatars in model organisms has begun to deepen our understanding of sarcomere thin filament development. In this review, we provide a summary of recent findings regarding sarcomere assembly, maintenance, and disease with respect to thin filaments, building on the previous knowledge in the field. We highlight debated and unknown areas within these processes to clearly define open research questions.
Keywords: sarcomere assembly, sarcomere maintenance, sarcomere thin filaments, thin filament assembly, thin filament maintenance, thin filament turnover, actin turnover, chaperone, sarcomere, myopathy
1. Introduction
Striated muscle requires the coordination of hundreds of proteins not only for cellular function but also for assembly of the contractile sarcomere units within the myofibril.
While the assembly and maintenance of myosin thick filaments and titin within the sarcomere have been the spotlight of many studies in muscle development and myopathies, the formation and maintenance of the thin filaments have been largely under-characterized despite the suite of myopathies originating from components of thin filament development. The purpose of this review is to present the discoveries of thin filament assembly and maintenance in the last decade. We will combine these findings with previous knowledge of thin filament development to highlight a model for thin filament assembly/maintenance and how this relates to muscle disease. Many factors discussed in this review have alternative names listed in Table 1. Sarcomere thin filament assembly and maintenance is highly conserved across striated muscle with a few isoform and tissue-specific exceptions. We discuss these cardiac and skeletal sarcomere thin filament differences in the appropriate sections below.
Table 1.
Alternative names for sarcomere thin filament factors.
| Common Name | Alternative Name(s) | Common Name | Alternative Name(s) |
|---|---|---|---|
| ZASP | Cypher, Oracle | HSPB7 | Cardiovascular HSP |
| actinin | α-actinin | Muscle LIM Protein (MLP) | Cardiac LIM protein (CLP) |
| NRAP | N-RAP | T-CAP | TCAP, telethonin |
| Filamin C | γ-filamin, ABP-L, sarcomere filamin | Calpain 1 | μ-calpain, mu-calpain, calpain I |
| myopodin | SYNPO2 | Calpain 2 | m-calpain, calpain II |
| Calsarcin | FATZ, myozenin | Calpain 3 | p94 |
| Calcineurin | Protein phosphatase 2B (PP2B) | MuRF1 | TRIM63 |
| KLHL41 | Krp1, sarcosin | MuRF2 | TRIM55 |
| Titin | Connectin | MuRF3 | TRIM54 |
| CapZ | β-actinin, CapZβ, CapZβ1 | Atrogin-1 | MAFbx1, Fbxo32 |
| Myotilin | TTID | Hsc70 | HSPA8 |
| TRiC | CCT, chaperonin | HspB8 | Hsp22 |
| Prefoldin | GimC | Cofilin2 | m-cofilin, ADF |
| Hsp25/27 | Hspβ1, Hsp25, Hsp27 | Wdr-1 | AIP-1 |
| αβ-crystallin | CRYAB, Hsp20 |
2. Thin Filament Assembly in the Sarcomere
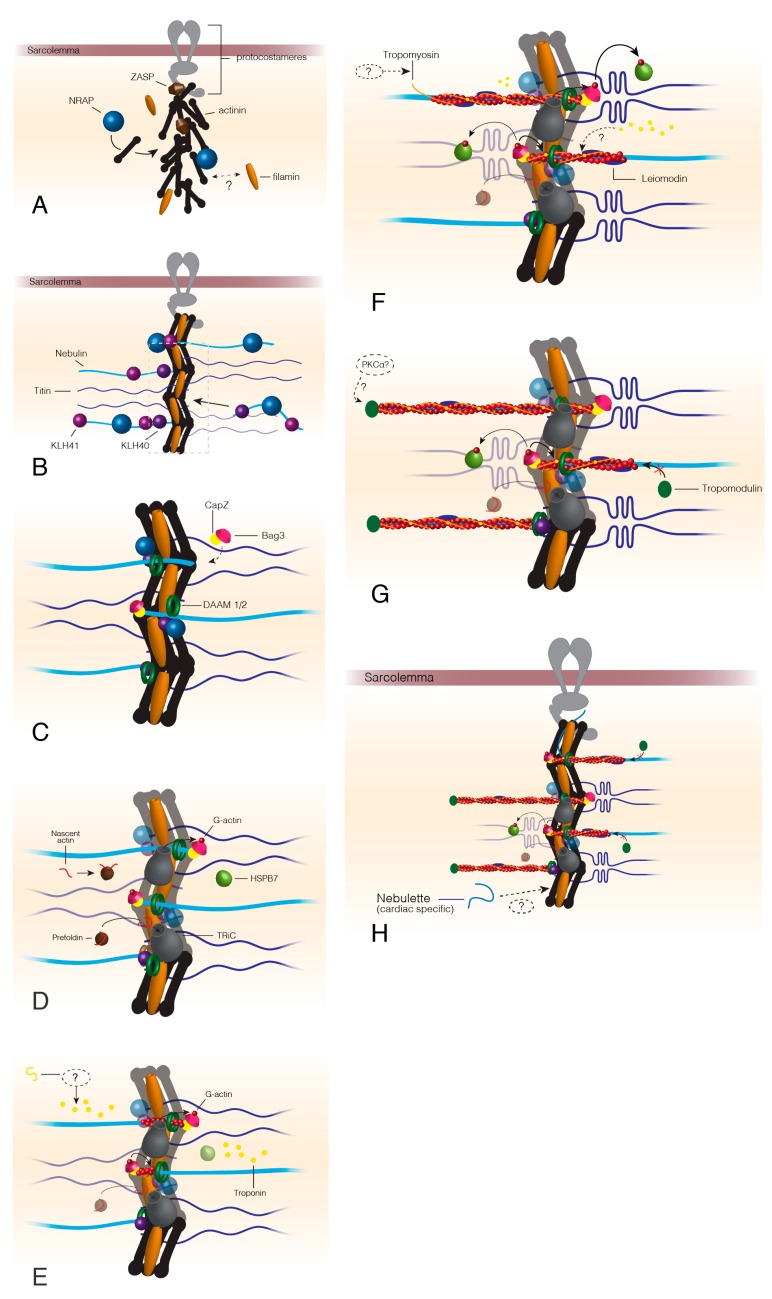
The site of thin filament formation begins with the development of the Z-disc at the protocostameres (premature costameres or sarcomere-to-membrane attachment) and muscle cell membrane (Figure 1). Protocostameres recruit ZASP, a member of the Alp/Enigma protein family, to the sarcolemma, followed by the ZASP-dependent localization of α-actinin [1,2,3,4]. α-actinin is organized into the characteristic Z-disc lattice by N-RAP, which remains in the sarcomere to act as a scaffold for thin filament assembly within the Z-disc [5,6,7]. Sarcomere filamin localizes early to the developing Z-disc but the reason for this localization is not clear and a filamin chaperone remains to be identified [8]. Filamin acts as a second anchor for thin filament attachment to the Z-disc and aids in linking thin filaments of adjacent sarcomeres together [9]. Although myopodin (SYNPO2) is expressed before α-actinin, a small amount of in vitro work demonstrates myopodin colocalizes with filamin C and α-actin during myofibril maturation suggesting myopodin may require both sarcomere proteins to be properly recruited to the sarcomere [10].
Figure 1.
Thin filament assembly beginning at the formation of the Z-disc. The formation of the Z-disc begins once ZASP is recruited to the sarcolemma via proteins of the protocostameres (A). ZASP localization draws actinin, the actinin chaperone, NRAP, and filamin C to the protocostameres (A). Actinin and filamin C organization allows for nebulin/nebulette and titin incorporation into the premature Z-disc (B). KLHL40 and KLHL41 factors stabilize nebulin as it is incorporated into the Z-disc and prevent nebulin aggregation (B). Bag3 localizes CapZ to the Z-disc at approximately the same time as DAAM1/2 appears in the Z-disc (C). Actin chaperones prefoldin and TRiC localize to the Z-disc after DAAM1/2 and Bag3. Prefoldin targets nascent actin to TRiC for folding into its final conformation. TRiC transfers folded G-actin to Bag3, which provides actin monomers for filament formation by formin proteins (D). HSPB7 arrives at the developing thin filaments around this time to regulate the speed of actin polymerization. Troponin localizes to the I-band by an unknown mechanism (E), before tropomyosin, which is also incorporated by an unknown method (F). Leiomodin is recruited to polymerizing actin thin filaments (F) and functions to both stabilize the growing filament but also prevent tropomodulin capping that stops actin polymerization (G). Once thin filaments reach their mature length, leiomodin no longer competes with tropomodulin and the thin filaments are capped by tropomodulin possibly following phosphorylation by PKCα (G,H). Dashed lines indicate unknown or hypothesized mechanisms/proteins.
Calsarcin localization coincides with ZASP recruitment to the protocostameres [11]. Calsarcin binds α-actinin, filamin C early in sarcomere formation followed by T-cap at the time of its integration into the Z-disc [12]. Calsarcin is important for regulating calcineurin-mediated signaling by sequestering calcineurin to the Z-disc [13].
Once the nascent Z-disc has formed, the protein giant, nebulin, is incorporated into the Z-line of skeletal muscle [14,15,16]. Low levels of nebulin have been detected in the sarcomere thin filaments of cardiomyocytes [17] but another member of the nebulin family, nebulette, is predominantly expressed in cardiac tissue [18,19,20,21]. For a concise comparison of nebulin family members in striated muscle, see [22]. Nebulette and nebulin are homologous in function and have similar protein structure but far more studies have been conducted on dissecting nebulin and its role in skeletal sarcomere assembly and maintenance, which is discussed in this review [22]. While no chaperones have been identified for nebulin folding, NRAP, KLHL41 (skeletal muscle-specific factor), and KLHL40 bind to nebulin and prevent the giant from aggregating or degrading [23,24,25,26]. The binding of these chaperones may result in the localization of nebulin to the Z-disc. Nebulin acts as a thin filament scaffold, providing structural stability to the sarcomere thin filaments during contraction. Thin filaments can form without nebulin but quickly degrade once sarcomere contractions begin [27,28,29,30]. There are two competing models for nebulin function in sarcomere assembly: The first suggesting that nebulin simply acts as a molecular ruler and determines thin filament length [28,29,31,32], while the second model states that nebulin is only required for thin filament assembly and stability and does not dictate thin filament length [33,34]. This latter model proposes that there are two segments of the actin thin filaments: nebulin-bound and nebulin-free [34]. The nebulin-free segment is regulated and stabilized by other thin filament proteins such as tropomodulin that caps the end of the thin filaments. Thin filament length varies across fiber and tissue type, possibly to accommodate the different force generation demands of each muscle type. While we cannot review nebulette to the same extent, some studies have shown that nebulin and nebulette properties and sarcomere physiology are homologous, suggesting that nebulette may also utilize the same chaperones and proteolysis components [22].
Based on immunostaining of titin as a landmark of myofibril assembly (occurs simultaneously with nebulin), myotilin integrates into the Z-disc at the stage of titin incorporation [35]. Myotilin binds filamin C, F-actin, and α-actinin and acts as an additional anchor/force absorber at the Z-disc [36,37]. Whether myotilin is necessary for sarcomere assembly is debated in the literature [35,38]. CapZ localizes to the Z-disc, with aid from Bag3, before actin is recruited for thin filament polymerization [39,40]. CapZ will bind the C-terminal end of nebulin and helps direct and regulate actin polymerization [41,42]. CapZ is also the site of chaperone and sarcomere maintenance factor localization, acting as a focal point for mechanosensory pathways [40,43,44].
Factors DAAM 1 and 2 appear at the Z-disc before actin filament assembly and recruit the critical factor, profilin, for actin polymerization [45,46,47]. DAAM1/2 mouse knockout models display disrupted cardiac sarcomere assembly and intercalated disc structures [48] and DAAM Drosophila mutants demonstrate disorganized sarcomeres with reduced thin filament numbers [49]. Since DAAM1/2 are members of the formin family of proteins, DAAM1/2 possess formin homology domains 1 and 2 (FH1 and FH2, respectively). The FH2 domains of a formin are arranged in a ring-like shape around the growing barbed ends of the actin filament [50,51]. Competitive binding between free profilin and actin-bound profilin to the FH1 domain may regulate the speed of actin thin filament formation [52,53]. It is debated in the literature whether an additional formin, FHOD3, is necessary for sarcomere thin filament assembly [54,55,56] or maintenance alone [45,57,58]. Unlike cytoskeletal actin polymerization, which can be initiated by the Arp2/3 complex in the dendritic model of cell migration [59], recent work found thin filament assembly in muscle sarcomeres does not require Arp2/3 but rather involves FHOD3 [60]. The thin filament maintenance aspect of formins will be discussed in a later section.
It is unclear if the co-chaperone, Bag3, arrives just prior to or simultaneously with the actin chaperones TRiC and prefoldin [40,61]. Prefoldin targets nascent actin to TRiC, which is localized within the Z-disc [61]. TRiC folds and transfers G-actin (actin monomer) to Bag3 that is bound to CapZ. Together, bag3, CapZ, and formin stabilize and begin actin polymerization along the nebulin scaffold [54,56,62,63]. HSPB7 regulates thin filament length by sequestering G-actin from the bag3-CapZ-formin-nebulin complex. In mice, mutants of HSPB7 are embryonic lethal with actin filament aggregates and longer than normal thin filaments [64].
The following steps of thin filament assembly occur simultaneously with actin polymerization. Troponin T localizes to the developing thin filament and is required for the recruitment of tropomyosin [65,66]. Troponin T is part of a transcriptional feedback pathway as abnormal expression of troponin T (overexpression or no expression) results in the down-regulation of troponin T, C, I, actin and tropomyosin [67,68,69]. Troponin T may serve as a checkpoint for proper sarcomere assembly with respect to the I-band region. No chaperones have been identified for the troponin subunits or tropomyosin.
Tropomyosin incorporation recruits leiomodin (lmod) to the thin filament and is required for the addition of tropomodulin [45,70]. Leiomodin binds to the growing ends of the actin filament and helps stabilize G-actin polymerization while inhibiting tropomodulin binding to the ends of the thin filaments [45,71]. Tropomodulin binding or “capping” prevents further actin polymerization and is critical for regulation of thin filament length. The length of thin filaments is dependent on the temporal tradeoff of leiomodin and tropomodulin. Higher concentrations of leiomodin at the thin filament during assembly ensure filament growth with lmod mutants displaying short thin filaments that result in dysfunctional contraction and force generation. As the assembly is completed, leiomodin concentrations must decrease to allow for tropomodulin binding at the ends of the thin filaments [72]. The mechanism behind the tradeoff between leiomodin and tropomodulin is not well established but may include post-translational modifications by protein kinase, PKCα, or targeted protein turnover by calpains or the ubiquitin-proteasome system [73,74].
3. Sarcomere Thin Filament Maintenance
3.1. Thin Filament Maintenance and Turnover
Sarcomere maintenance can be subdivided into two categories of factors: those that refold/reassemble the sarcomere structure and those factors that remove nonfunctional proteins from the cell. Factors of the former are likely chaperones and structural proteins such as those discussed in the previous section. Many factors involved in the assembly also act as maintenance factors, complicating the identification of activities specific to maintenance. Without conditional knockout studies, many factors are only classified as assembly factors since development does not proceed past sarcomere formation in their absence. Maintenance factors of the second category come from the suite of systems that remove sarcomere proteins that can no longer maintain functional integrity. These two categories of thin filament quality control must work together to properly maintain sarcomere thin filaments throughout the life of the organism.
There are two main processes for thin filament maintenance: 1) autophagy and 2) single protein degradation. Single protein degradation happens through the Ubiquitin-Proteasome System (UPS), following isolation from the myofiber by calpains in most cases. The UPS is one of the major quality control pathways for thin filament maintenance and turnover with many proteins destined for degradation by the proteasome [75]. This section will focus on factors that monitor and target thin filament and thin filament associated proteins for turnover (e.g., Bag3).
3.2. Calpain Mediated Breakdown of Z-Disc and Thin Filaments
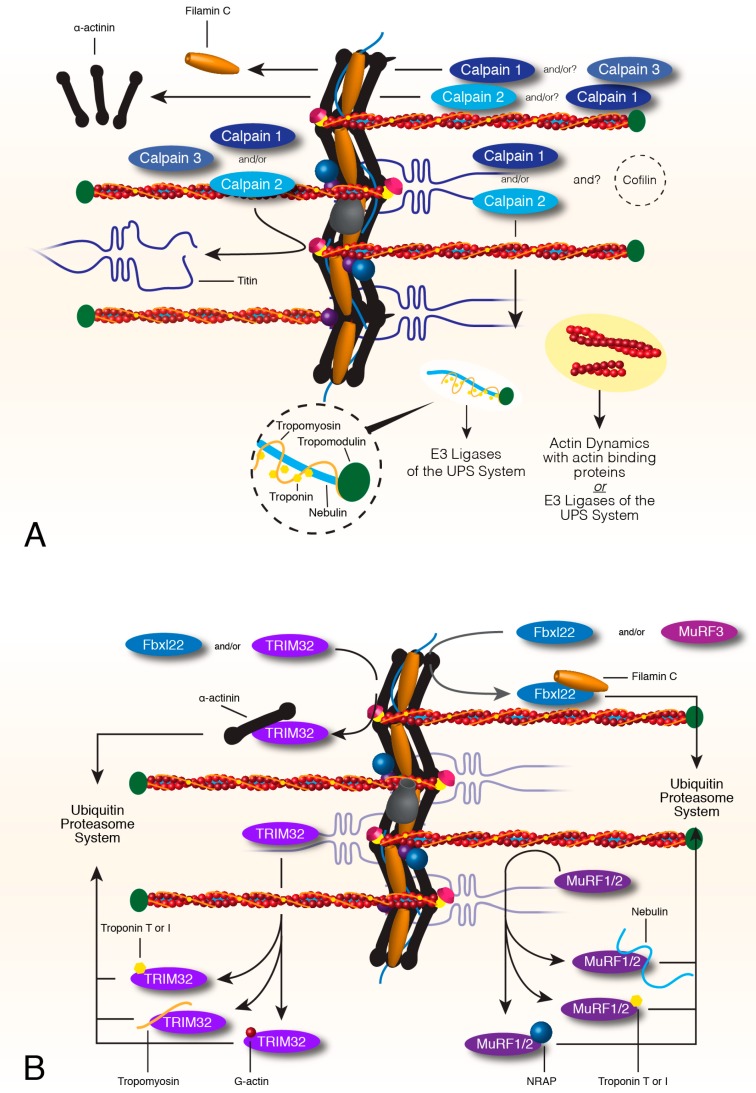
Within the last decade, the calpain protease family has emerged as an important class of factors involved in intracellular protein turnover [76]. Unlike other proteases that degrade their substrates, calpains recognize their substrates, but do not degrade them [76]. Although not well understood, calpains can target and release individual or protein complexes from the sarcomere (Figure 2A) [76]. The function of calpains appears to be the first step in a pathway required for sarcomere protein turnover as members of the ubiquitin-proteasome system (UPS) are unable to degrade some sarcomere proteins in the absence of calpains—this could be due to the complexity of the sarcomere and that the UPS is unable to segregate their client proteins [76,77,78]. Among the calpain family, only three have been identified in muscle development and health: calpain 1, 2, and 3 [77]. The calpains share functional redundancy, targeting and segregating many of the same sarcomere proteins, although calpain 1 and 2 are ubiquitously expressed and calpain 3 is a skeletal muscle-specific homolog [79].
Figure 2.
Calpain and Ubiquitin Proteasome Turnover of sarcomere thin filaments and associated factors. Calpains 1, 2 and 3 remove or release proteins from the complex contractile network of the sarcomere. A combination of calpains can release filamin C, actinin, titin, and the sarcomere thin filament, although actin is not degraded by calpains (A). Targets of the calpain system that are removed from the sarcomere become target proteins of the UPS system. MuRF1 and 2 can target proteins of the thin filament such as nebulin, troponin T and I and the nebulin chaperone, NRAP, for proteasome degradation (B). MuRF3 or Fbxl22, the recognition factor of the SCF complex, target filamin C for degradation. TRIM32 can also target proteins of the thin filament such as troponin T and I, tropomyosin and actin, for degradation. TRIM32 or Fbxl22 can also target actinin for ubiquitin-dependent degradation (B). All targets of the E3 ligases are directed to E2 and E1 ligases for ubiquitination and eventually degradation by the proteasome.
Calpain 1 and 2 target the thin filament protein complex, cleaving tropomyosin, tropomodulin, troponin T and I, and nebulin [73,80,81,82,83,84]. Calpain 1 and 2 do not degrade actin, suggesting that another factor involved in F-actin maintenance may also be required during thin filament turnover [84]. Calpain 1 and/or calpain 2 will release α-actinin while calpain 1 and/or calpain 3 cleave filamin C from the Z-disc [78,80,85,86]. All three calpains will free titin at the Z-disc, with most studies focused on calpain 3 and its localization to titin throughout muscle development [84,87,88,89].
Calpain activity can be modified by substrate phosphorylation. Phosphorylation of troponin I by cyclic AMP-dependent protein kinase (PKA) increases the resistance of troponin I to calpain 1 proteolysis [82]. However, troponin I phosphorylation by protein kinase C (PKC) significantly increases troponin I cleavage by calpain 1, while calpain 2-targeted degradation of troponin I was not altered by phosphorylation. Calpain 1-mediated cleavage of filamin C can be inhibited by phosphorylation using PKCα [78].
Calpain proteolysis is the first step in the turnover of several sarcomere proteins for protein quality control systems but calpain cleavage is not the endpoint for these proteins. Damaged proteins released from the myofiber are targeted and degraded by either the UPS or autophagy pathways.
3.3. Turnover of Thin Filament Components via E3 Ligases and the Ubiquitin Proteasome System
The UPS is dependent on three ubiquitin ligases, E1, E2, and E3, which recognize and deliver poly-ubiquitinated target proteins to the proteasome for degradation. E3 ubiquitin ligases (MuRF(s)) recognize damaged client proteins and transport them to E1 and E2 ligases for ubiquitination and subsequent degradation by the proteasome. For a recent detailed review of the UPS in muscle, see [75].
MuRF1 or MuRF2 recognizes nebulin, troponin T and I, and NRAP for UPS-mediated degradation (Figure 2B) [90,91,92]. Troponin T and I, tropomyosin, G-actin, myotilin, and α-actinin are all recognized and targeted for degradation by TRIM32 [93,94,95]. Despite the discovery of MuRF3 at the beginning of the century, filamin C and FHL2 are the only documented targets of MuRF3 [96,97]. A fourth muscle-specific ring finger protein has been discovered in the vertebrate lineage, excluding avian and placental mammals, but no targets of this MuRF4 have been identified yet [98].
Although MuRF factors work alone or in combination with another MuRF to target sarcomere proteins to the proteasome, other E3-ligases are composed of several proteins. The SCF (SKP1-CUL1-F-box protein) protein complex is composed of four proteins that recognize and target sarcomere components for degradation via the UPS system [99]. Fbxl22 is a cardiac-specific interchangeable recognition component of the SCF-E3 ligase complex that targets filamin C and α-actinin (Figure 2B) [99,100]. Fbxl22 can be replaced by atrogin-1, which localizes to the Z-disc and targets calcineurin for degradation, identifying a UPS therapeutic target for hypertrophy signaling [100,101,102]. Mutants of atrogin-1 show decreased troponin I degradation, suggesting troponin I might be an SCF-atrogin-1 target.
The co-chaperone bag3 and chaperone Hsc70 regulate UPS-mediated degradation of CapZ and indirectly F-actin stability [40,103]. Bag3 directs CapZ localization to the ends of F-actin anchored within the Z-disc. Bag3 association with CapZ recruits Hsc70 and binding of Hsc70 to the CapZβ1 subunit induces a conformational change in CapZ that stabilizes F-actin [42,104,105]. Without bag3, CapZ does not localize properly nor can it form stable bonds with F-actin, leading to degradation via the ubiquitin-proteasome system.
The 26S proteasome is the final destination for proteins targeted by the ubiquitin-proteasome system [75,106,107]. Although no calpain or E3 ligase that recognizes troponin C has been identified to date, troponin C is degraded by the 26S proteasome [80,106]. KLHL40 remains in the sarcomere after assembly to help stabilize nebulin and eventually leiomodin 3 [26]. KLHL40 protects nebulin and leiomodin 3 from targeted degradation by the ubiquitin-proteasome pathway.
3.4. Autophagy-Dependent Mechanisms for Thin Filament Maintenance
Autophagy is the second major degradation mechanism and differs from the UPS in that autophagy targets insoluble proteins, protein aggregates and lasting proteins [108]. The autophagy-lysosomal pathway (ALP) shares many similarities to the UPS system in that ALP uses E1, E2, and E3-like factors to expand an isolation membrane around the target organelle or insoluble proteins [109]; for a detailed review of this pathway, see [77,110]. Autophagy, like the UPS, is required for the maintenance of striated muscle due to the necessity of protein turnover and clearing of damaged sarcomere proteins to prevent protein toxicity [111,112,113].
Bag3, which was discussed in the UPS section, is also a member of an autophagy-dependent pathway called chaperone-assisted selective autophagy (CASA) [114,115]. Bag3 and SYNPO2 colocalize to the Z-disc and bind α-actinin and filamin C. Bag3 and SYNPO2 monitor filamin C integrity and recruit the CASA complex upon filamin C unfolding. The CASA complex is composed of Hsp70, Hsp27, HspB8, and the chaperone-associated ubiquitin ligase, CHIP [113,116,117]. Filamin C and SYNPO2 are removed from the sarcomere, ubiquitinated and targeted to the lysosome [116,118]. SYNPO2 is consumed in this pathway and would require continuous replacement as damaged/unfolded filamin C is turned over [115].
The autophagy-lysosomal pathway can also be triggered by the aggregation of actin protein. Actin aggregates also recruit and contain proteasomes (UPS) but the proteasomes are unable to degrade the actin aggregate, possibly due to the size or insolubility of the aggregate [116,119].
3.5. Dynamic Actin Turnover
Actin is the major component of the thin filament, interacting with all the dynamic proteins of sarcomere contraction. Therefore, it should be no surprise that actin must be consistently monitored and replaced due to normal wear and tear. This section describes actin protein turnover from CapZ’s weak dissociation to the re-polymerization of actin in the thin filament.
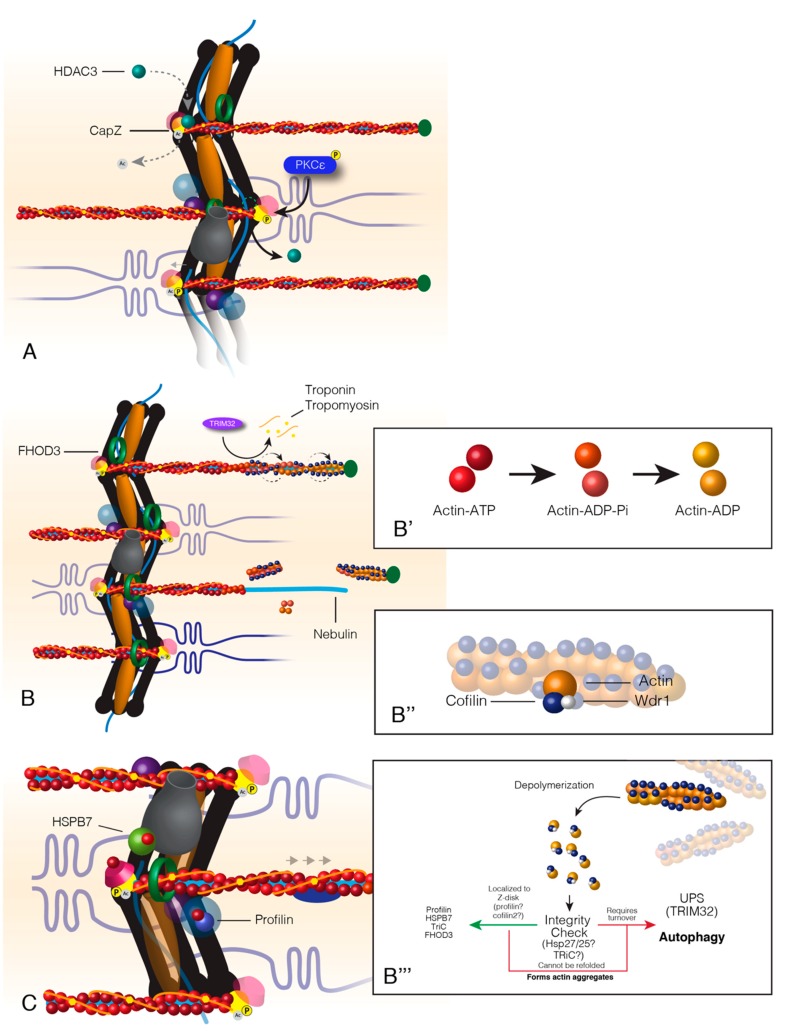
The binding between CapZ and actin is weakened by the phosphorylation of CapZ by PKCε, which causes HDAC3 to disassociate from CapZ (Figure 3A) [44,120]. HDAC3 disassociation allows for the acetylation of CapZ that then releases actin and allows for polymerization of the thin filament [41,44]. These posttranslational modifications of CapZ are reversible upon completion of thin filament turnover.
Figure 3.
Turnover of actin within sarcomere thin filaments. Actin turnover begins with the relaxation of CapZ-thin filament binding at the Z-disc (A). This relaxation occurs from the competition between HDAC3 and PKCε. HDAC3 acetylates CapZ, which promotes a tight bond between CapZ and actin. PKCε phosphorylates CapZ that causes HDAC3 to disassociate from the Z-disc and CapZ to loosely bind the thin filaments. Following CapZ relaxation, FHOD3 encompasses the thin filaments, and actin-binding proteins associate with actin at the distal end of the thin filament (B). Actin at the distal end of the thin filaments is “aged” by the associated state of ATP, ADP+Pi or ADP (B’). Actin-binding proteins, Cofilin and Wdr1, bind “aged” actin, twist and break it away from the filament (B&B’’). Cofilin breaks filamentous actin into monomeric actin that is escorted back to the Z-disc where it is recycled back into the thin filaments (B’’’). Hypothetically, actin should pass through an “integrity check” as most sarcomere proteins do. The outcomes of this check would result in recycling back into the thin filament or targeted degradation (B’’’). Healthy actin monomers (G-actin) are reincorporated into the thin filament by Z-disc localized factors profilin, formin, and HSPB7; a process that maintains the oldest actin within the sarcomere A-band or the distal portion of the thin filament (C).
Cofilin2 is the only ADF (actin-depolymerizing factor)/cofilin protein family member present in muscle [121]. Cofilin2 is not required for sarcomere assembly as knockout mouse models develop muscle normally but quickly lose sarcomere integrity shortly after birth [122,123]. The cofilin co-factor Wdr1 (AIP-1) is required for cofilin-dependent actin depolymerization and disassembly of the thin filament (Figure 3B) [124]. Cofilin2 contains a region of surface residues that are hypothesized to act as a sensor of the nucleotide state of actin [125]. Cofilin2 preferentially binds actin-ADP at the pointed ends of the actin thin filaments by localization to the M-band of the sarcomere (Figure 3B, B’ & B’’). ADF factors regulate actin disassembly by changing the twist of actin filaments allowing for the severing of F-actin [122]. F-actin is further depolymerized into G-actin monomers to recycle actin and prevent aggregation of F-actin (Figure 3B’’’,C) [122,126,127,128,129].
Major sarcomere structural proteins (e.g., myosin, actinin) must be correctly folded and maintain structural integrity to be recycled back into the sarcomere during protein turnover. Protein recycling is possible if chaperones can correctly refold their client proteins but weakened or damaged targets that cannot reach their final conformation prevent reuse. In the event of a damaged/misfolded client protein, chaperones aid in the targeting of client proteins for degradation. Although no studies have identified an integrity check for actin recycling, it can be hypothesized that actin must also be correctly folded for re-incorporation into the thin filament (Figure 3B’’’,C). Unfolded/misfolded actin may recruit actin-binding partners due to exposed target residues. TRiC and/or Hsp27/25 are likely candidates for chaperones involved in a hypothetical actin integrity check due to their actin folding activity and localization to F-actin during muscle injury, respectively [61,130,131,132,133]. G-actin monomers recruit several binding partners, which supports a tight regulation of cytoplasmic concentrations of G-actin [61,125,134,135]. G-actin, when left “unattended” will polymerize into unstable/unregulated F-actin on its own [61,134,136,137].
After being severed from F-actin by cofilin2, ADP is exchanged for ATP in G-actin before being reincorporated into a developing thin filament [135,138]. While not definitively understood, profilin may facilitate this process by binding and forcing a conformational change in actin that favors the release of ADP [138,139,140]. Borrowing from the dendritic model for actin recycling at the leading edge of motile cells [59], profilin binds ADP-G-actin, undergoes nucleotide exchange to ATP-G-actin and is transferred from profilin to thymosin β4 (Tβ4). Tβ4 localizes G-actin to the barbed end of an actin thin filament at the Z-disc where G-actin is transferred to a separate pool of Z-disc localized profilin [141,142]. It is suggested that Tβ4 acts to create a separate pool of ATP-G-actin that both regulates thin filament polymerization and prevents uncontrolled F-actin synthesis and mouse Tβ4 mutants have short sarcomere thin filaments [59,141,142]. However, the use of Tβ4 to transfer G-actin from pointed to barbed ends of actin filaments seems inefficient since profilin is required at both ends for actin recycling.
A second model for actin recycling relies on reusing factors that were involved in sarcomere assembly. DAAM1/2, a formin protein, is localized to the Z-disc, where it recruits profilin and synthesizes F-actin during sarcomere assembly [48,51,55,143]. FHOD3, the debated formin discussed in thin filament assembly, is required for sarcomere maintenance by regulating actin dynamics and a lack of formin proteins increases the cytoplasmic concentration of G-actin [51,143,144]. FHOD3 likely accepts G-actin from profilin, but it is debated if this transfer occurs at the Z-disc or along the thin filament within the A-band [47,52,53,144,145,146].
Growing thin filaments will extend out from the Z-disc and are stabilized by leiomodin [71,147]. PKCα regulates thin filament length via phosphorylation of tropomodulin, which associate with the ends of actin filaments and stop polymerization [74]. How tropomyosin and troponin re-associate with the refreshed sarcomeric actin thin filament is unknown.
4. Diseases of Components of Striated Muscle Thin Filaments
This section will focus on diseases that arise solely from mutations that affect thin filament structural proteins or the factors that are required for proper assembly and maintenance. As some factors/pathways are shared with the development and health of other tissues (e.g., autophagy), the mutations discussed in this section will be specific to sarcomeres.
4.1. Mutations That Interfere with Proper Thin Filament Assembly
Mutations that disturb the proper assembly of sarcomere thin filaments can be further subdivided into several categories but in this review, we simplify the mutations to those that inhibit sarcomere formation and those mutations that develop weakened or improper thin filaments. Mutations that completely inhibit sarcomere formation are typically embryonic lethal due to the lack of a contractile heart and do not allow for a full pathophysiological study of these fatal mutations. The second category, which we will focus on in this review, encompasses those mutations that result in an initial weakly functional protein or misregulation of sarcomeric thin filament formation. Myopathies that manifest later in life from mutations in structural components should not be confused with maintenance or turnover myopathies as structural proteins fall under the umbrella of thin filament formation. Maintenance and turnover factors and their resulting myopathies will be discussed in the next section.
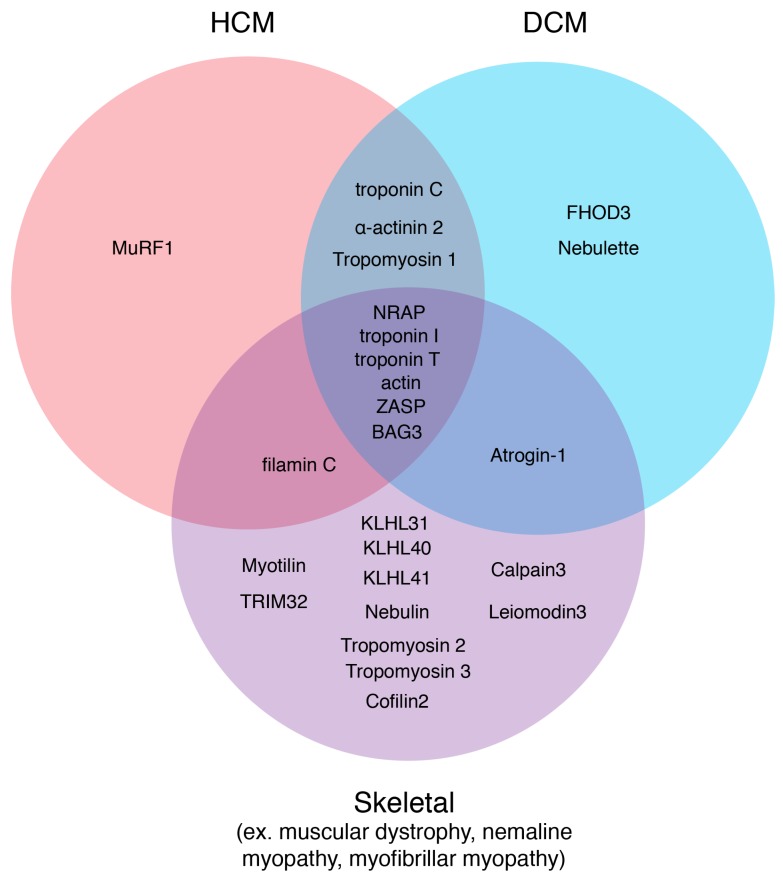
Weakly functional proteins are largely structural proteins of the sarcomere (e.g., actin, troponin T or I, tropomyosin, etc.) that do not maintain integrity over a period of time or for which the system can no longer compensate [148]. That time period can range such that myopathy presentations occur perinatally, pediatrically or with late onset. Improper functional leiomodin or tropomodulin result in irregular thin filament lengths that are either too short or too long; both inhibit efficient muscle contraction [149]. Mutations in many sarcomere structural proteins result in dilated cardiomyopathy (DCM) that can present early in childhood but many develop as late-onset DCM (Figure 4) [148]. Mutations in ZASP result in myopathies including muscular dystrophy, myofibrillar myopathy, DCM or HCM that manifest within and after the fourth decade [150,151,152]. NRAP mutations manifest as DCM in young adults [153], NRAP overexpression results in right ventricle cardiomyopathy in mice [154] and nemaline myopathy in zebrafish [24]. KLHL40 mutations have a perinatal presentation with severe paralysis to complete “lock-in” diagnoses due to a lack of nebulin and leiomodin stability [26]. Only a subset of thin filament proteins can carry DCM, HCM or nemaline-causing mutations that manifest at a variety of ages with a number of severities, suggesting that multiple disease mechanisms exist within muscle and possibly coordinate through these select factors (Figure 4) [148]. The mechanisms behind the age of onset are unknown but would be particularly powerful for therapeutic intervention.
Figure 4.
Sarcomere thin filament disease candidates. Using the online database, Online Mendelian Inheritance in Man, diseases originating from sarcomere thin filament factors were sorted into three disease states: HCM, DCM, and Skeletal. Factors that could carry mutations that could result in several different myopathies are depicted in the overlapping regions.
4.2. Mutations that Interfere with Thin Filament Maintenance and Turnover
Maintenance and turnover of thin filaments refer specifically to those proteins/factors that were not required for the initial assembly of the sarcomeric thin filaments but are necessary to maintain thin filaments throughout the life of the organism. This category is composed mainly of accessory factors to the thin filaments and in rare cases, includes factors that are also necessary for thin filament formation (e.g., Kelch factors).
KLHL31 maintains muscle integrity by maintaining functional filamin C protein levels in the cell through ubiquitination [155]. KLHL31 knockout mice develop postnatal myopathies due to large FLNC aggregates that reduce Z-disc stability. TRIM32 mutations lead to variations of limb-girdle muscular dystrophy 2H and myofibrillar myopathy with accumulations of client protein aggregates (ex. myotilin) that interfere with normal muscle function [95]. Atrogin-1 mutations result in the misregulation of calcineurin and the autophagy pathway allowing for toxic protein accumulation within the cardiomyocytes that presents as pediatric DCM [101].
Bag3 is a unique factor that is involved in both the proper assembly and maintenance of the sarcomere [40,156]. Mutations in bag3 can result in embryonic lethality due to improper sarcomere assembly [157,158] or child-onset skeletal myopathies, DCM, or HCM (Figure 4) [159,160].
5. Summary
Although the mechanisms and development of sarcomere assembly have been, and are currently, explored, the focus of these investigations is primarily myosin, titin, and actinin. Only in the last decade has the importance of assembly and protein turnover studies of thin filament proteins been revealed due to a growing population with devastating mutations in these factors. Understanding the mechanisms behind sarcomere thin filament formation and maintenance is vital to creating a powerful and effective treatment for such myopathies.
Author Contributions
Conceptualization, K.P. and J.F.D.; resources, J.F.D.; data curation, K.P.; writing—original draft preparation, K.P.; writing—review and editing, K.P. and J.F.D.; visualization, K.P.; supervision, J.F.D.; funding acquisition, J.F.D. All authors have read and agree to the published version of the manuscript.
Funding
This research was funded by the Heart and Stroke Foundation, grant number G-18-0020424 and the APC was funded by G-18-0020424.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Katzemich A., Liao K.A., Czerniecki S., Schöck F. Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly. PLoS Genet. 2013;9:e1003342. doi: 10.1371/journal.pgen.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Q., Ruiz-Lozano P., Martone M.E., Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to α-actinin-2 and protein kinase C. J. Biol. Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- 3.Passier R., Richardson J.A., Olson E.N. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech. Dev. 2000;92:277–284. doi: 10.1016/S0925-4773(99)00330-5. [DOI] [PubMed] [Google Scholar]
- 4.Jani K., Schöck F. Zasp is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 2007;179:1583–1597. doi: 10.1083/jcb.200707045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll S.L., Herrera A.H., Horowits R. Targeting and functional role of N-RAP, a nebulin-related LIM protein, during myofibril assembly in cultured chick cardiomyocytes. J. Cell Sci. 2001;114:4229–4238. doi: 10.1242/jcs.114.23.4229. [DOI] [PubMed] [Google Scholar]
- 6.Carroll S., Lu S., Herrera A.H., Horowits R. N-RAP scaffolds I-Z-I assembly during myofibrillogenesis in cultured chick cardiomyocytes. J. Cell Sci. 2004;117:105–114. doi: 10.1242/jcs.00847. [DOI] [PubMed] [Google Scholar]
- 7.Manisastry S.M., Zaal K.J.M., Horowits R. Myofibril assembly visualized by imaging N-RAP, alpha-actinin, and actin in living cardiomyocytes. Exp. Cell Res. 2009;315:2126–2139. doi: 10.1016/j.yexcr.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Der Ven P.F.M., Obermann W.M.J., Lemke B., Gautel M., Weber K., Fürst D.O. Characterization of muscle filamin isoforms suggests a possible role of γ-Filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskelet. 2000;45:149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 9.González-Morales N., Holenka T.K., Schöck F. Filamin actin-binding and titin-binding fulfill distinct functions in Z-disc cohesion. PLoS Genet. 2017;13:e1006880. doi: 10.1371/journal.pgen.1006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linnemann A., Vakeel P., Bezerra E., Orfanos Z., Djinović-Carugo K., Van Der Ven P.F.M., Kirfel G., Fürst D.O. Myopodin is an F-actin bundling protein with multiple independent actin-binding regions. J. Muscle Res. Cell Motil. 2013;34:61–69. doi: 10.1007/s10974-012-9334-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J.M., Sanger J.W. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskelet. 2005;61:34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner G., Pallavicini A., Comelli A., Salamon M., Bortoletto G., Ievolella C., Trevisan S., Kojić S., Vecchia F.D., Laveder P., et al. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 2000;275:41234–41242. doi: 10.1074/jbc.M007493200. [DOI] [PubMed] [Google Scholar]
- 13.Frey N., Richardson J.A., Olson E.N. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. USA. 2000;97:14632–14637. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu M., Gregorio C.C., Pappas C.T. Nebulin, a multi-functional giant. J. Exp. Biol. 2016;219:146–152. doi: 10.1242/jeb.126383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moncman C.L., Wang K. Assembly of nebulin into the sarcomeres of avian skeletal muscle. Cell Motil. Cytoskelet. 1996;34:167–184. doi: 10.1002/(SICI)1097-0169(1996)34:3<167::AID-CM1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Labeit S., Kolmerer B. The complete primary structure of human nebulinand its correlation to muscle structure. J. Mol. Biol. 1995;248:308–315. doi: 10.1016/S0022-2836(95)80052-2. [DOI] [PubMed] [Google Scholar]
- 17.Kazmierski S.T., Antin P.B., Witt C.C., Huebner N., McElhinny A.S., Labeit S., Gregorio C.C. The complete mouse nebulin gene sequence and the identification of cardiac nebulin. J. Mol. Biol. 2003;328:835–846. doi: 10.1016/S0022-2836(03)00348-6. [DOI] [PubMed] [Google Scholar]
- 18.Millevoi S., Trombitas K., Kolmerer B., Kostin S., Schaper J., Pelin K., Granzier H., Labeit S. Characterization of nebulette and nebulin and emerging concepts of their roles for vertebrate Z-discs. J. Mol. Biol. 1998;282:111–123. doi: 10.1006/jmbi.1998.1999. [DOI] [PubMed] [Google Scholar]
- 19.Moncman C.L., Wang K. Targeted disruption of nebulette protein expression alters cardiac myofibril assembly and function. Exp. Cell Res. 2002;273:204–218. doi: 10.1006/excr.2001.5423. [DOI] [PubMed] [Google Scholar]
- 20.Esham M., Bryan K., Milnes J., Holmes W.B., Moncman C.L. Expression of nebulette during early cardiac development. Cell Motil. Cytoskelet. 2007;64:258–273. doi: 10.1002/cm.20180. [DOI] [PubMed] [Google Scholar]
- 21.Moncman C.L., Wang K. Nebulette: A 107 kD nebulin-like protein in cardiac muscle. Cell Motil. Cytoskelet. 1995;32:205–225. doi: 10.1002/cm.970320305. [DOI] [PubMed] [Google Scholar]
- 22.Bang M.L., Chen J. Roles of nebulin family members in the heart. Circ. J. 2015;79:2081–2087. doi: 10.1253/circj.CJ-15-0854. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg C.C., Connelly P.S., Daniels M.P., Horowits R. Krp1 (Sarcosin) promotes lateral fusion of myofibril assembly intermediates in cultured mouse cardiomyocytes. Exp. Cell Res. 2008;314:1177–1191. doi: 10.1016/j.yexcr.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jirka C., Pak J.H., Grosgogeat C.A., Marchetii M.M., Gupta V.A. Dysregulation of NRAP degradation by KLHL41 contributes to pathophysiology in nemaline myopathy. Hum. Mol. Genet. 2019;28:2549–2560. doi: 10.1093/hmg/ddz078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Martinez A., Cenik B.K., Bezprozvannaya S., Chen B., Bassel-Duby R., Liu N., Olson E.N. KLHL41 stabilizes skeletal muscle sarcomeres by nonproteolytic ubiquitination. Elife. 2017;6:e26439. doi: 10.7554/eLife.26439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg A., O’Rourke J., Long C., Doering J., Ravenscroft G., Bezprozvannaya S., Nelson B.R., Beetz N., Li L., Chen S., et al. KLHL40 deficiency destabilizes thin filament proteins and promotes Nemaline myopathy. J. Clin. Investig. 2014;124:3529–3539. doi: 10.1172/JCI74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottenheijm C.A.C., Hooijman P., DeChene E.T., Stienen G.J., Beggs A.H., Granzier H. Altered myofilament function depresses force generation in patients with nebulin-based nemaline myopathy (NEM2) J. Struct. Biol. 2010;170:334–343. doi: 10.1016/j.jsb.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witt C.C., Burkart C., Labeit D., McNabb M., Wu Y., Granzier H., Labeit S. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tskhovrebova L., Trinick J. Titin and Nebulin in thick and thin filament length regulation. Subcell. Biochem. 2017:285–318. doi: 10.1007/978-3-319-49674-0_10. [DOI] [PubMed] [Google Scholar]
- 30.Bang M.L., Li X., Littlefield R., Bremner S., Thor A., Knowlton K.U., Lieber R.L., Chen J. Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J. Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pappas C.T., Krieg P.A., Gregorio C.C. Nebulin regulates actin filament lengths by a stabilization mechanism. J. Cell Biol. 2010;189:859–870. doi: 10.1083/jcb.201001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labeit S., Gibson T., Lakey A., Leonard K., Zeviani M., Knight P., Wardale J., Trinick J. Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett. 1991;282:313–316. doi: 10.1016/0014-5793(91)80503-U. [DOI] [PubMed] [Google Scholar]
- 33.Castillo A., Nowak R., Littlefield K.P., Fowler V.M., Littlefield R.S. A nebulin ruler does not dictate thin filament lengths. Biophys. J. 2009;96:1856–1865. doi: 10.1016/j.bpj.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gokhin D.S., Fowler V.M. A two-segment model for thin filament architecture in skeletal muscle. Nat. Rev. Mol. Cell Biol. 2013;14:113. doi: 10.1038/nrm3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmikangas P., van der Ven P.F.M., Lalowski M., Taivainen A., Zhao F., Suila H., Schröder R., Lappalainen P., Fürst D.O., Carpén O. Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum. Mol. Genet. 2003;12:189–203. doi: 10.1093/hmg/ddg020. [DOI] [PubMed] [Google Scholar]
- 36.Sanger J.W., Wang J., Holloway B., Du A., Sanger J.M. Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell Motil. Cytoskelet. 2009;66:556–566. doi: 10.1002/cm.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmikangas P., Mykkänen O.M., Grönholm M., Heiska L., Kere J., Carpén O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- 38.Moza M., Mologni L., Trokovic R., Faulkner G., Partanen J., Carpen O. Targeted Deletion of the Muscular Dystrophy Gene myotilin Does Not Perturb Muscle Structure or Function in Mice. Mol. Cell. Biol. 2007;27:244–252. doi: 10.1128/MCB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer D.A., Waddle J.A., Cooper J.A. Localization of CapZ during myofibrillogenesis in cultured chicken muscle. Cell Motil. Cytoskelet. 1993;25:317–335. doi: 10.1002/cm.970250403. [DOI] [PubMed] [Google Scholar]
- 40.Hishiya A., Kitazawa T., Takayama S. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ. Res. 2010;107:1220–1231. doi: 10.1161/CIRCRESAHA.110.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita A., Maeda K., Maéda Y. Crystal structure of CapZ: Structural basis for actin filament barbed end capping. EMBO J. 2003;22:1529–1538. doi: 10.1093/emboj/cdg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer D.A., Hug C., Cooper J.A. Inhibition of CapZ during myofibrillogenesis alters assembly of actin filaments. J. Cell Biol. 1995;128:61–70. doi: 10.1083/jcb.128.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y.H., Warren C.M., Li J., McKinsey T.A., Russell B. Myofibril growth during cardiac hypertrophy is regulated through dual phosphorylation and acetylation of the actin capping protein CapZ. Cell. Signal. 2016;28:1015–1024. doi: 10.1016/j.cellsig.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y.H., Swanson E.R., Li J., Mkrtschjan M.A., Russell B. Cyclic mechanical strain of myocytes modifies CapZβ1 post translationally via PKCε. J. Muscle Res. Cell Motil. 2015;36:329–337. doi: 10.1007/s10974-015-9420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanger J.W., Wang J., Fan Y., White J., Mi-Mi L., Dube D.K., Sanger J.M., Pruyne D. Assembly and Maintenance of Myofibrils in Striated Muscle. In: Jockusch B., editor. The Actin Cytoskeleton. Vol. 235. Springer; Cham, Switzerland: 2016. Handbook of Experimental Pharmacology. [DOI] [PubMed] [Google Scholar]
- 46.David Pruyne L.M.M. Loss of Sarcomere-associated Formins Disrupts Z-line Organization, but does not Prevent Thin Filament Assembly in Caenorhabditis elegans Muscle. J. Cytol. Histol. 2015;6:318. doi: 10.4172/2157-7099.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aspenström P. Formin-binding proteins: Modulators of formin-dependent actin polymerization. Biochim. Biophys. Acta -Mol. Cell Res. 2010;1803:174–182. doi: 10.1016/j.bbamcr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Ajima R., Bisson J.A., Helt J.C., Nakaya M.A., Habas R., Tessarollo L., He X., Morrisey E.E., Yamaguchi T.P., Cohen E.D. DAAM1 and DAAM2 are co-required for myocardial maturation and sarcomere assembly. Dev. Biol. 2015;408:126–139. doi: 10.1016/j.ydbio.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molnár I., Migh E., Szikora S., Kalmár T., Végh A.G., Molnár I., Migh E., Szikora S., Kalmár T., Végh A.G., et al. DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila. PLoS Genet. 2014;10:e1004166. doi: 10.1371/journal.pgen.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/S0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 51.Paul A.S., Pollard T.D. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskelet. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul A., Pollard T. The Role of the FH1 Domain and Profilin in Formin-Mediated Actin-Filament Elongation and Nucleation. Curr. Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vavylonis D., Kovar D.R., O’Shaughnessy B., Pollard T.D. Model of formin-associated actin filament elongation. Mol. Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taniguchi K., Takeya R., Suetsugu S., Kan-o M., Narusawa M., Shiose A., Tominaga R., Sumimoto H. Mammalian formin Fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 2009;284:29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goode B.L., Eck M.J. Mechanism and Function of Formins in the Control of Actin Assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 56.Fujimoto N., Kan-O M., Ushijima T., Kage Y., Tominaga R., Sumimoto H., Takeya R. Transgenic expression of the formin protein fhod3 selectively in the embryonic heart: Role of actin-binding activity of fhod3 and its sarcomeric localization during myofibrillogenesis. PLoS ONE. 2016;11:e0148472. doi: 10.1371/journal.pone.0148472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iskratsch T., Lange S., Dwyer J., Kho A.L., Dos Remedios C., Ehler E. Formin follows function: A muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 2010;191:1159–1172. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iskratsch T., Reijntjes S., Dwyer J., Toselli P., Dégano I.R., Dominguez I., Ehler E. Two distinct phosphorylation events govern the function of muscle FHOD3. Cell. Mol. Life Sci. 2013;70:893–908. doi: 10.1007/s00018-012-1154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 60.Fenix A.M., Neininger A.C., Taneja N., Hyde K., Visetsouk M.R., Garde R.J., Liu B., Nixon B.R., Manalo A.E., Becker J.R., et al. Muscle-specific stress fibers give rise to sarcomeres in cardiomyocytes. Elife. 2018;7:e42144. doi: 10.7554/eLife.42144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berger J., Berger S., Li M., Jacoby A.S., Arner A., Bavi N., Stewart A.G., Currie P.D. In Vivo Function of the Chaperonin TRiC in α-Actin Folding during Sarcomere Assembly. Cell Rep. 2018;22:313–322. doi: 10.1016/j.celrep.2017.12.069. [DOI] [PubMed] [Google Scholar]
- 62.Lutsch G., Vetter R., Offhauss U., Wieske M., Gröne H.J., Klemenz R., Schimke I., Stahl J., Benndorf R. Abundance and location of the small heat shock proteins HSP25 and αB- crystallin in rat and human heart. Circulation. 1997;96:3466–3476. doi: 10.1161/01.CIR.96.10.3466. [DOI] [PubMed] [Google Scholar]
- 63.Brown D.D., Christine K.S., Showell C., Conlon F.L. Small heat shock protein Hsp27 is required for proper heart tube formation. Genesis. 2007;45:667–678. doi: 10.1002/dvg.20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu T., Mu Y., Bogomolovas J., Fang X., Veevers J., Nowak R.B., Pappas C.T., Gregorio C.C., Evans S.M., Fowler V.M., et al. HSPB7 is indispensable for heart development by modulating actin filament assembly. Proc. Natl. Acad. Sci. USA. 2017;114:11956–11961. doi: 10.1073/pnas.1713763114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferrante M.I., Kiff R.M., Goulding D.A., Stemple D.L. Troponin T is essential for sarcomere assembly in zebrafish skeletal muscle. J. Cell Sci. 2011;124:565–577. doi: 10.1242/jcs.071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang W., Zhang R., Xu X. Myofibrillogenesis in the developing zebrafish heart: A functional study of tnnt2. Dev. Biol. 2009;331:237–249. doi: 10.1016/j.ydbio.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marco-Ferreres R., Arredondo J.J., Fraile B., Cervera M. Overexpression of troponin T in Drosophila muscles causes a decrease in the levels of thin-filament proteins. Biochem. J. 2005;386:145–152. doi: 10.1042/BJ20041240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sehnert A.J., Huq A., Weinstein B.M., Walker C., Fishman M., Stainier D.Y.R. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002;31:106. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 69.Hinkle A., Goranson A., Butters C.A., Tobacman L.S. Roles for the troponin tail domain in thin filament assembly and regulation: A deletional study of cardiac troponin T. J. Biol. Chem. 1999;274:7157–7164. doi: 10.1074/jbc.274.11.7157. [DOI] [PubMed] [Google Scholar]
- 70.Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szatmári D., Bugyi B., Ujfalusi Z., Grama L., Dudás R., Nyitrai M. Cardiac leiomodin2 binds to the sides of actin filaments and regulates the ATPase activity of myosin. PLoS ONE. 2017;12:e0186288. doi: 10.1371/journal.pone.0186288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pappas C.T., Farman G.P., Mayfield R.M., Konhilas J.P., Gregorio C.C. Cardiac-specific knockout of Lmod2 results in a severe reduction in myofilament force production and rapid cardiac failure. J. Mol. Cell. Cardiol. 2018;122:88–97. doi: 10.1016/j.yjmcc.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gokhin D.S., Tierney M.T., Sui Z., Sacco A., Fowler V.M. Calpain-mediated proteolysis of tropomodulin isoforms leads to thin filament elongation in dystrophic skeletal muscle. Mol. Biol. Cell. 2014;25:852–865. doi: 10.1091/mbc.e13-10-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bliss K.T., Tsukada T., Novak S.M., Dorovkov M.V., Shah S.P., Nworu C., Kostyukova A.S., Gregorio C.C. Phosphorylation of tropomodulin1 contributes to the regulation of actin filament architecture in cardiac muscle. FASEB J. 2014;28:3987–3995. doi: 10.1096/fj.13-246009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carlisle C., Prill K., Pilgrim D. Chaperones and the proteasome system: Regulating the construction and demolition of striated muscle. Int. J. Mol. Sci. 2018;19:32. doi: 10.3390/ijms19010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ono Y., Sorimachi H. Calpains—An elaborate proteolytic system. Biochim. Biophys. Acta-Proteins Proteom. 2012;1824:224–236. doi: 10.1016/j.bbapap.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Portbury A.L., Willis M.S., Patterson C. Tearin’ up my heart: Proteolysis in the cardiac sarcomere. J. Biol. Chem. 2011;286:9929–9934. doi: 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reimann L., Wiese H., Leber Y., Schwable A.N., Fricke A.L., Rohland A., Knapp B., Peikert C.D., Drepper F., Van Der Ven P.F.M., et al. Myofibrillar Z-discs are a protein phosphorylation hot spot with protein kinase C (PKCα) modulating protein dynamics. Mol. Cell. Proteom. 2017;16:346–367. doi: 10.1074/mcp.M116.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Letavernier E., Zafrani L., Perez J., Letavernier B., Haymann J.P., Baud L. The role of calpains in myocardial remodelling and heart failure. Cardiovasc. Res. 2012;96:38–45. doi: 10.1093/cvr/cvs099. [DOI] [PubMed] [Google Scholar]
- 80.Koohmaraie M. Ovine skeletal muscle multicatalytic proteinase complex (proteasome): Purification, characterization, and comparison of its effects on myofibrils with μ-calpains. J. Anim. Sci. 1992;70:3697–3708. doi: 10.2527/1992.70123697x. [DOI] [PubMed] [Google Scholar]
- 81.Dayton W.R., Goll D., Stromer M., Reville W., Zeece M., Robson R. Some properties of a Ca2+-activated protease that may be involved in myofibrillar protein turnover. Cold Spring Harb. Conf. Cell Prolif. 1975;2:551–577. [Google Scholar]
- 82.Di Lisa F., De Tullio R., Salamino F., Barbato R., Melloni E., Siliprandi N., Schiaffino S., Pontremoli S. Specific degradation of troponin T and I by μ-calpain and its modulation by substrate phosphorylation. Biochem. J. 1995;308:57–61. doi: 10.1042/bj3080057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huff-Lonergan E., Mitsuhashi T., Beekman D.D., Parrish F.C., Olson D.G., Robson R.M. Proteolysis of Specific Muscle Structural Proteins by μ-Calpain at Low pH and Temperature is Similar to Degradation in Postmortem Bovine Muscle. J. Anim. Sci. 1996;74:993–1008. doi: 10.2527/1996.745993x. [DOI] [PubMed] [Google Scholar]
- 84.Goll D.E., Thompson V.F., Taylor R.G., Ouali A. The calpain system and skeletal muscle growth. Can. J. Anim. Sci. 1998;78:503–512. doi: 10.4141/A98-081. [DOI] [Google Scholar]
- 85.Bullard B., Sainsbury G., Miller N. Digestion of proteins associated with the Z-disc by calpain. J. Muscle Res. Cell Motil. 1990;11:271–279. doi: 10.1007/BF01843580. [DOI] [PubMed] [Google Scholar]
- 86.Guyon J.R., Kudryashova E., Potts A., Dalkilic I., Brosius M.A., Thompson T.G., Beckmann J.S., Kunkel L.M., Spencer M.J. Calpain 3 cleaves filamin C and regulates its ability to interact with γ- and δ-sarcoglycans. Muscle Nerve. 2003;28:472–483. doi: 10.1002/mus.10465. [DOI] [PubMed] [Google Scholar]
- 87.Goll D.E., Thompson V.F., Taylor R.G., Christiansen J.A. Role of the calpain system in muscle growth. Biochimie. 1992;74:225–237. doi: 10.1016/0300-9084(92)90121-T. [DOI] [PubMed] [Google Scholar]
- 88.Taveau M., Bourg N., Sillon G., Roudaut C., Bartoli M., Richard I. Calpain 3 Is Activated through Autolysis within the Active Site and Lyses Sarcomeric and Sarcolemmal Components. Mol. Cell. Biol. 2003;23:9127–9135. doi: 10.1128/MCB.23.24.9127-9135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hayashi C., Ono Y., Doi N., Kitamura F., Tagami M., Mineki R., Arai T., Taguchi H., Yanagida M., Hirner S., et al. Multiple molecular interactions implicate the connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J. Biol. Chem. 2008;283:14801–14814. doi: 10.1074/jbc.M708262200. [DOI] [PubMed] [Google Scholar]
- 90.Witt S.H., Granzier H., Witt C.C., Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: Towards understanding MURF-dependent muscle ubiquitination. J. Mol. Biol. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 91.Kedar V., McDonough H., Arya R., Li H.H., Rockman H.A., Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. USA. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen S.N., Czernuszewicz G., Tan Y., Lombardi R., Jin J., Willerson J.T., Marian A.J. Human molecular genetic and functional studies identify TRIM63, encoding muscle RING finger protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 2012;111:907–919. doi: 10.1161/CIRCRESAHA.112.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cohen S., Zhai B., Gygi S.P., Goldberg A.L. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell Biol. 2012;198:575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kudryashova E., Kudryashov D., Kramerova I., Spencer M.J. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005;354:413424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 95.Panicucci C., Traverso M., Baratto S., Romeo C., Iacomino M., Gemelli C., Tagliafico A., Broda P., Zara F., Bruno C., et al. Novel TRIM32 mutation in sarcotubular myopathy. Acta Myol. 2019;38:8–12. [PMC free article] [PubMed] [Google Scholar]
- 96.Spencer J.A., Eliazer S., Ilaria R.L., Richardson J.A., Olson E.N. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J. Cell Biol. 2000;150:771–784. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fielitz J., Van Rooij E., Spencer J.A., Shelton J.M., Latif S., Van Der Nagel R., Bezprozvannaya S., De Windt L., Richardson J.A., Bassel-Duby R., et al. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc. Natl. Acad. Sci. USA. 2007;104:4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Macqueen D.J., Fuentes E.N., Valdés J.A., Molina A., Martin S.A.M. The vertebrate muscle-specific RING finger protein family includes MuRF4—A novel, conserved E3-ubiquitin ligase. FEBS Lett. 2014;588:4390–4397. doi: 10.1016/j.febslet.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 99.Zhou W., Wei W., Sun Y. Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases. Cell Res. 2013;23:599. doi: 10.1038/cr.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spaich S., Will R.D., Just S., Spaich S., Kuhn C., Frank D., Berger I.M., Wiemann S., Korn B., Koegl M., et al. F-box and leucine-rich repeat protein 22 is a cardiac-enriched f-box protein that regulates sarcomeric protein turnover and is essential for maintenance of contractile function in vivo. Circ. Res. 2012;111:1504–1516. doi: 10.1161/CIRCRESAHA.112.271007. [DOI] [PubMed] [Google Scholar]
- 101.Al-Yacoub N., Shaheen R., Awad S.M., Kunhi M., Dzimiri N., Nguyen H.C., Xiong Y., Al-Buraiki J., Al-Habeeb W., Alkuraya F.S., et al. FBXO32, encoding a member of the SCF complex, is mutated in dilated cardiomyopathy. Genome Biol. 2016;17:2. doi: 10.1186/s13059-015-0861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H.H., Kedar V., Zhang C., McDonough H., Arya R., Wang D.Z., Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 2004;114:1058–1071. doi: 10.1172/JCI200422220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takayama S., Reed J.C. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 2001;3:E237. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 104.Haus U., Trommler P., Fisher P.R., Hartmann H., Lottspeich F., Noegel A.A., Schleicher M. The heat shock cognate protein from Dictyostelium affects actin polymerization through interaction with the actin-binding protein cap32/34. EMBO J. 1993;12:3763–3771. doi: 10.1002/j.1460-2075.1993.tb06054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tardieux I., Baines I., Mossakowska M., Ward G.E. Actin-binding proteins of invasive malaria parasites and the regulation of actin polymerization by a complex of 32/34-kDa proteins associated with heat shock protein 70kDa. Mol. Biochem. Parasitol. 1998;93:295–308. doi: 10.1016/S0166-6851(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 106.Benaroudj N., Tarcsa E., Cascio P., Goldberg A.L. The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie. 2001;83:311–318. doi: 10.1016/S0300-9084(01)01244-5. [DOI] [PubMed] [Google Scholar]
- 107.Bedford L., Paine S., Sheppard P.W., Mayer R.J., Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klionsky D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 109.Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gustafsson Å.B., Gottlieb R.A. Recycle or die: The role of autophagy in cardioprotection. J. Mol. Cell. Cardiol. 2008;44:654–661. doi: 10.1016/j.yjmcc.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 112.Kostin S., Pool L., Elsässer A., Hein S., Drexler H.C.A., Arnon E., Hayakawa Y., Zimmermann R., Bauer E., Klövekorn W.P., et al. Myocytes die by multiple mechanisms in failing human hearts. Circ. Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 113.Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Fürst D.O., Saftig P., Saint R., Fleischmann B.K., et al. Chaperone-Assisted Selective Autophagy Is Essential for Muscle Maintenance. Curr. Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 114.Klimek C., Kathage B., Wördehoff J., Höhfeld J. BAG3-mediated proteostasis at a glance. J. Cell Sci. 2017;130:2781–2788. doi: 10.1242/jcs.203679. [DOI] [PubMed] [Google Scholar]
- 115.Ulbricht A., Höhfeld J. Tension-induced autophagy: May the chaperone be with you. Autophagy. 2013;9:920–922. doi: 10.4161/auto.24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ulbricht A., Eppler F.J., Tapia V.E., Van Der Ven P.F.M., Hampe N., Hersch N., Vakeel P., Stadel D., Haas A., Saftig P., et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 2013;23:430–435. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 117.Carra S., Seguin S.J., Landry J. HspB8 and Bag3: A new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008;4:237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 118.Joshi V., Amanullah A., Upadhyay A., Mishra R., Kumar A., Mishra A. A decade of boon or burden: What has the chip ever done for cellular protein quality control mechanism implicated in neurodegeneration and aging? Front. Mol. Neurosci. 2016;9:93. doi: 10.3389/fnmol.2016.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lázaro-Diéguez F., Aguado C., Mato E., Sánchez-Ruíz Y., Esteban I., Alberch J., Knecht E., Egea G. Dynamics of an F-actin aggresome generated by the actin-stabilizing toxin jasplakinolide. J. Cell Sci. 2008;121:1415–1425. doi: 10.1242/jcs.017665. [DOI] [PubMed] [Google Scholar]
- 120.Li J., Russell B. Phosphatidylinositol 4,5-bisphosphate regulates CapZβ1 and actin dynamics in response to mechanical strain. Am. J. Physiol.-Heart Circ. Physiol. 2013;305:H1614–H1623. doi: 10.1152/ajpheart.00477.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ono S., Minami N., Abe H., Obinata T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 1994;269:15280–15286. [PubMed] [Google Scholar]
- 122.Gurniak C.B., Chevessier F., Jokwitz M., Jönsson F., Perlas E., Richter H., Matern G., Boyl P.P., Chaponnier C., Fürst D., et al. Severe protein aggregate myopathy in a knockout mouse model points to an essential role of cofilin2 in sarcomeric actin exchange and muscle maintenance. Eur. J. Cell Biol. 2014;93:252–266. doi: 10.1016/j.ejcb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 123.Agrawal P.B., Joshi M., Savic T., Chen Z., Beggs A.H. Normal myofibrillar development followed by progressive sarcomeric disruption with actin accumulations in a mouse Cfl2 knockout demonstrates requirement of cofilin-2 for muscle maintenance. Hum. Mol. Genet. 2012;21:2341–2356. doi: 10.1093/hmg/dds053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan B., Wan P., Chu D., Nie J., Cao Y., Luo W., Lu S., Chen J., Yang Z. A cardiomyocyte-specific Wdr1 knockout demonstrates essential functional roles for actin disassembly during myocardial growth and maintenance in mice. Am. J. Pathol. 2014;184:1967–1980. doi: 10.1016/j.ajpath.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 125.Kremneva E., Makkonen M.H., Skwarek-Maruszewska A., Gateva G., Michelot A., Dominguez R., Lappalainen P. Cofilin-2 controls actin filament length in muscle sarcomeres. Dev. Cell. 2014;31:215–226. doi: 10.1016/j.devcel.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McGough A., Pope B., Chiu W., Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bamburg J.R. Proteins of the ADF/Cofilin Family: Essential Regulators of Actin Dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 128.Nakashima K., Sato N., Nakagaki T., Abe H., Ono S., Obinata T. Two mouse cofilin isoforms, muscle-type (MCF) and non-muscle type (NMCF), interact with F-actin with different efficiencies. J. Biochem. 2005;138:519–526. doi: 10.1093/jb/mvi152. [DOI] [PubMed] [Google Scholar]
- 129.Maciver S.K., Zot H.G., Pollard T.D. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guay J., Lambert H., Gingras-Breton G., Lavoie J.N., Huot J., Landry J. Regulation of actin filament dynamics by p38 map kinase-mediated phosphorylation of heat shock protein 27. J. Cell Sci. 1997;110:357–368. doi: 10.1242/jcs.110.3.357. [DOI] [PubMed] [Google Scholar]
- 131.Lavoie J.N., Gingras-Breton G., Tanguay R.M., Landry J. Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock. HSP27 stabilization of the microfilament organization. J. Biol. Chem. 1993;268:3420–3429. [PubMed] [Google Scholar]
- 132.Yoshida K.I., Aki T., Harada K., Shama K.M.A., Kamoda Y., Suzuki A., Ohno S. Translocation of HSP27 and MKBP in ischemic heart. Cell Struct. Funct. 1999;24:181–185. doi: 10.1247/csf.24.181. [DOI] [PubMed] [Google Scholar]
- 133.Sakamoto K., Urushidani T., Nagao T. Translocation of HSP27 to sarcomere induced by ischemic preconditioning in isolated rat hearts. Biochem. Biophys. Res. Commun. 2000;269:137–142. doi: 10.1006/bbrc.2000.2233. [DOI] [PubMed] [Google Scholar]
- 134.Morita T., Hayashi K. G-actin sequestering protein thymosin-β4 regulates the activity of myocardin-related transcription factor. Biochem. Biophys. Res. Commun. 2013;437:331–335. doi: 10.1016/j.bbrc.2013.06.069. [DOI] [PubMed] [Google Scholar]
- 135.Ono S. The role of cyclase-associated protein in regulating actin filament dynamics—more than a monomer-sequestration factor. J. Cell Sci. 2013;126:3249–3258. doi: 10.1242/jcs.128231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J. Molecular Cell Biology. 4th ed. W. H. Freeman; New York, NY, USA: 2000. [(accessed on 1 September 2019)]. Section 18.2, The Dynamics of Actin Assembly. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21594/ [Google Scholar]
- 137.Schaus T.E., Taylor E.W., Borisy G.G. Self-organization of actin filament orientation in the dendritic- nucleation/array-treadmilling model. Proc. Natl. Acad. Sci. USA. 2007;104:7086–7091. doi: 10.1073/pnas.0701943104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wen K.K., McKane M., Houtman J.C.D., Rubenstein P.A. Control of the ability of profilin to bind and facilitate nucleotide exchange from G-actin. J. Biol. Chem. 2008 doi: 10.1074/jbc.M709806200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carlier M.F. Actin: Protein structure and filament dynamics. J. Biol. Chem. 1991;266:1–4. [PubMed] [Google Scholar]
- 140.Mockrin S.C., Korn E.D. Acanthamoeba Profilin Interacts with G-Actin to Increase the Rate of Exchange of Actin-Bound Adenosine 5’-Triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- 141.Smart N., Riegler J., Turtle C.W., Lygate C.A., McAndrew D.J., Gehmlich K., Dubé K.N., Price A.N., Muthurangu V., Taylor A.M., et al. Aberrant developmental titin splicing and dysregulated sarcomere length in Thymosin β4 knockout mice. J. Mol. Cell. Cardiol. 2017;102:94–107. doi: 10.1016/j.yjmcc.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Skruber K., Read T.A., Vitriol E.A. Reconsidering an active role for G-actin in cytoskeletal regulation. J. Cell Sci. 2018;131:jcs203760. doi: 10.1242/jcs.203760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ushijima T., Fujimoto N., Matsuyama S., Kan-O M., Kiyonari H., Shioi G., Kage Y., Yamasaki S., Takeya R., Sumimoto H. The actin-organizing formin protein Fhod3 is required for postnatal development and functional maintenance of the adult heart in mice. J. Biol. Chem. 2018;293:148–162. doi: 10.1074/jbc.M117.813931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Arimura T., Takeya R., Ishikawa T., Yamano T., Matsuo A., Tatsumi T., Nomura T., Sumimoto H., Kimura A. Dilated cardiomyopathy-associated FHOD3 variant impairs the ability to induce activation of transcription factor serum response factor. Circ. J. 2013:2990–2996. doi: 10.1253/circj.CJ-13-0255. [DOI] [PubMed] [Google Scholar]
- 145.Kan-o M., Takeya R., Taniguchi K., Tanoue Y., Tominaga R., Sumimoto H. Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS ONE. 2012;7:e34765. doi: 10.1371/journal.pone.0034765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Matsuyama S., Kage Y., Fujimoto N., Ushijima T., Tsuruda T., Kitamura K., Shiose A., Asada Y., Sumimoto H., Takeya R. Interaction between cardiac myosin-binding protein C and formin Fhod3. Proc. Natl. Acad. Sci. USA. 2018;115:E4386–E4395. doi: 10.1073/pnas.1716498115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Skwarek-Maruszewska A., Boczkowska M., Zajac A.L., Kremneva E., Svitkina T., Dominguez R., Lappalainen P. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Mol. Biol. Cell. 2010;21:3352–3361. doi: 10.1091/mbc.e10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yotti R., Seidman C.E., Seidman J.G. Advances in the Genetic Basis and Pathogenesis of Sarcomere Cardiomyopathies. Annu. Rev. Genom. Hum. Genet. 2019;20:129–153. doi: 10.1146/annurev-genom-083118-015306. [DOI] [PubMed] [Google Scholar]
- 149.Tsukada T., Pappas C.T., Moroz N., Antin P.B., Kostyukova A.S., Gregorio C.C. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J. Cell Sci. 2010;123:3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zheng J., Chen S., Chen Y., Zhu M., Hong D. A novel mutation in the PDZ-like motif of ZASP causes distal ZASP-related myofibrillar myopathy. Neuropathology. 2017;37:45–51. doi: 10.1111/neup.12328. [DOI] [PubMed] [Google Scholar]
- 151.Selcen D., Engel A.G. Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 2005;57:269–276. doi: 10.1002/ana.20376. [DOI] [PubMed] [Google Scholar]
- 152.Theis J.L., Martijn Bos J., Bartleson V.B., Will M.L., Binder J., Vatta M., Towbin J.A., Gersh B.J., Ommen S.R., Ackerman M.J. Echocardiographic-determined septal morphology in Z-disc hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2006;351:896–902. doi: 10.1016/j.bbrc.2006.10.119. [DOI] [PubMed] [Google Scholar]
- 153.Truszkowska G.T., Bilińska Z.T., Muchowicz A., Pollak A., Biernacka A., Kozar-Kamińska K., Stawiński P., Gasperowicz P., Kosińska J., Zieliński T., et al. Homozygous truncating mutation in NRAP gene identified by whole exome sequencing in a patient with dilated cardiomyopathy. Sci. Rep. 2017;7:3362. doi: 10.1038/s41598-017-03189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lu S., Crawford G.L., Dore J., Anderson S.A., DesPres D., Horowits R. Cardiac-specific NRAP overexpression causes right ventricular dysfunction in mice. Exp. Cell Res. 2011;317:1226–1237. doi: 10.1016/j.yexcr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Papizan J.B., Garry G.A., Brezprozvannaya S., McAnally J.R., Bassel-Duby R., Liu N., Olson E.N. Deficiency in Kelch protein Klhl31 causes congenital myopathy in mice. J. Clin. Investig. 2017;127:3730–3740. doi: 10.1172/JCI93445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hishiya A., Salman M.N., Carra S., Kampinga H.H., Takayama S. BAG3 directly interacts with mutated alphaB-crystallin to suppress its aggregation and toxicity. PLoS ONE. 2011;6:e16828. doi: 10.1371/journal.pone.0016828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Homma S., Iwasaki M., Shelton G.D., Engvall E., Reed J.C., Takayama S. BAG3 deficiency results in fulminant myopathy and early lethality. Am. J. Pathol. 2006;169:761–773. doi: 10.2353/ajpath.2006.060250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Arimura T., Ishikawa T., Nunoda S., Kawai S., Kimura A. Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum. Mutat. 2011;32:1481–1491. doi: 10.1002/humu.21603. [DOI] [PubMed] [Google Scholar]
- 159.Myers V.D., McClung J.M., Wang J.F., Tahrir F.G., Gupta M.K., Gordon J., Kontos C.H., Khalili K., Cheung J.Y., Feldman A.M. The Multifunctional Protein BAG3: A Novel Therapeutic Target in Cardiovascular Disease. JACC Basic Transl. Sci. 2018;3:122–131. doi: 10.1016/j.jacbts.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Norton N., Li D., Rieder M.J., Siegfried J.D., Rampersaud E., Züchner S., Mangos S., Gonzalez-Quintana J., Wang L., McGee S., et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am. J. Hum. Genet. 2011;88:273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]