Summary
Radial glia (RG) cells are the first neural stem cells to appear during embryonic development. Adult human glioblastomas harbor a subpopulation of RG-like cells with typical RG morphology and markers. The cells exhibit the classic and unique mitotic behavior of normal RG in a cell-autonomous manner. Single-cell RNA sequencing analyses of glioblastoma cells reveal transcriptionally dynamic clusters of RG-like cells that share the profiles of normal human fetal radial glia and that reside in quiescent and cycling states. Functional assays show a role for interleukin in triggering exit from dormancy into active cycling, suggesting a role for inflammation in tumor progression. These data are consistent with the possibility of persistence of RG into adulthood and their involvement in tumor initiation or maintenance. They also provide a putative cellular basis for the persistence of normal developmental programs in adult tumors.
Keywords: radial glia, cancer stem cells, glioblastoma, inflammation, brain tumor, scRNASeq, tumor mitosis
Highlights
-
•
Adult human glioblastomas harbor radial glia-like cells with classic mitotic behavior
-
•
Radial glia-like tumor cell clusters exist in quiescent or cycling states
-
•
Interleukin can activate dormant radial glia-like cells into the cell cycle
Tabar and colleagues report that adult brain tumors comprise neoplastic radial glia-like cells reminiscent of normal radial glia of early development. They are responsive to inflammation signals and may represent putative stem cells of origin of this lethal tumor.
Introduction
Glioblastoma (GBM) remains at the forefront of highly deadly cancers with little or no breakthrough in treatment paradigms or extension of survival over the past 15 years. Numerous studies have demonstrated that GBM tumor progression and maintenance is governed by cellular hierarchies, whereby quiescent or slow cycling stem-like cells appear to give rise to a more proliferative niche of progenitors capable of self-maintenance and tumor propagation (Lan et al., 2017, Tirosh et al., 2016). In addition, a developmental signature and different cell states (neural precursor cells, oligoprogenitor cells) have been identified within gliomas (Venteicher et al., 2017), that seem to be maintained despite the genetic heterogeneity within and across tumors (Alcantara Llaguno et al., 2015, Chen et al., 2012, Lan et al., 2017, Tirosh et al., 2016). Nonetheless, the cell type at the origin of this developmental program/hierarchy within human GBMs has not been fully elucidated.
Radial glia (RG) cells are the first neural stem cells during embryonic development. They demonstrate unique morphology, mitotic behavior, and molecular markers in the human fetus (Florio et al., 2015, Gotz and Barde, 2005, Lui et al., 2014, Pollen et al., 2015) (Figure S1A). The candidacy of RG as the putative cell of origin of GBMs or other brain tumors has been raised but not proven, in part due to the absence of markers that are highly restricted to RG cells (De Rosa et al., 2012, Mita et al., 2007, Taylor et al., 2005). Systematic characterization of RG-like cells in the adult brain will expand our understanding of the development of cortical RG cells as well as inform future investigations of the source of brain tumors.
Here, we report a series of observations of polarized mitotic cells in GBM explants and tumor dissociates that are highly typical of RG and their characteristic mitotic behavior. We systematically examine the morphology of mitotic cells and their expression of RG-specific markers. We also perform multiple timelapse video analyses that demonstrate the unique mitotic behavior associated with RG cells. RG-enriched cell dissociates form tumors in vivo. Single-cell RNA sequencing (scRNA-seq) analyses in human tumors identifies transcriptionally diverse cell clusters with characteristic RG and developmental gene expression. We also confirm that most RG clusters carry the main genomic aberrations of their parent tumor source. We further explore the role of inflammatory pathways, such as interleukin 1b (IL-1b) in driving dormant RG-like cells into the cell cycle.
Results
Identification of RG-like Cells in Human GBM
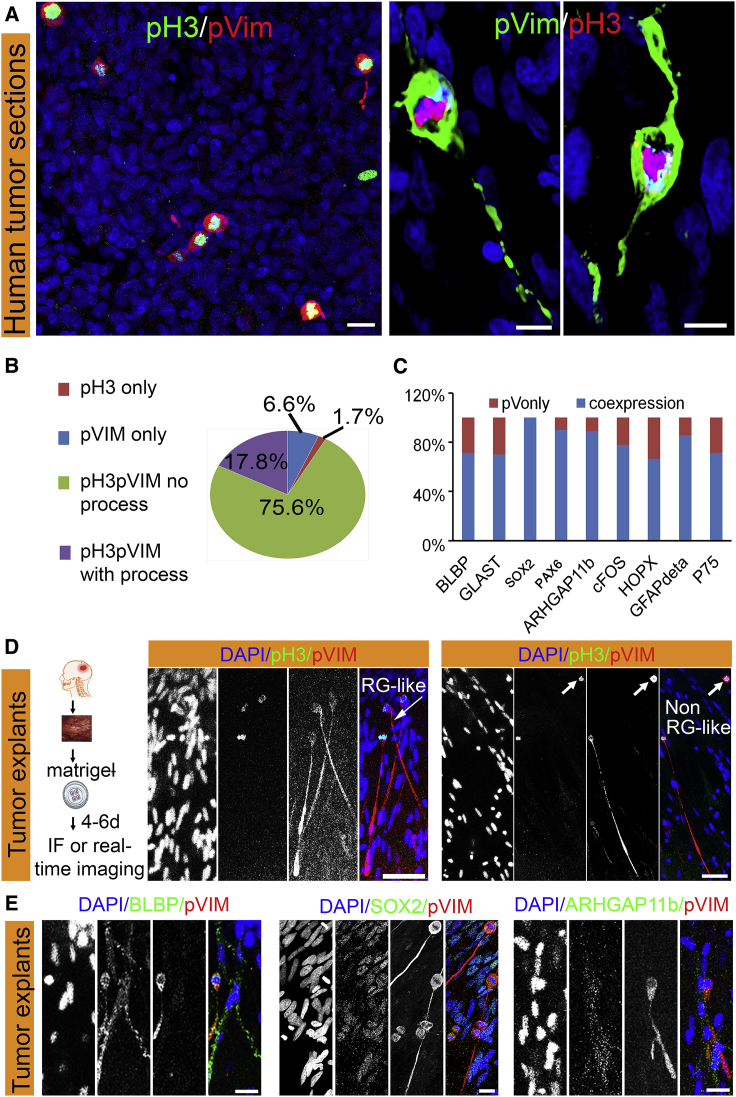
RG cells are distinguished by a unique mitotic behavior, including interkinetic nuclear migration, mitotic somal translocation (MST), and retention of a radial fiber during division, phenomena that are all distinctly uncommon in other dividing cells (Weissman et al., 2003). We evaluated sections of GBM tumors from 21 patients, for evidence of cells undergoing mitosis and exhibiting a radial fiber (Figure 1A) (Howard et al., 2008). Mitotic cells in M phase were immunostained for phospho-histone 3 (pH3+) and ranged from 0.2% to 1.2% of all cells (Table 1). A total of 17.8% of pH3+ mitotic cells had a definitive radial fiber and were immunopositive for phospho-vimentin (pVIM), which is known to be expressed in RG fibers during cell division (Kamei et al., 1998) (Figure 1B). RG cells express a range of markers that, when coupled with the appropriate developmental context and cell morphology, are characteristic of this cell state (Figure S1A). Although most known RG markers have been reported to be expressed in GBMs, the significance of these findings is unclear, nor is marker expression alone sufficient to identify a cell as RG. However, the unique mitotic behavior and retention of a radial fiber during cell division are highly pathognomonic to RG, so we based our approach on the co-existence of the division phenotype with typical RG markers. Therefore, we evaluated the same set of tumors for mitotic pVIM+ cells with a process, and co-expression of a panel of RG markers (SOX2 and FABP7, or BLBP, GLAST, PAX6, and HOPX), including the human-specific gene ARHGAP11B, as well as two human adult neural stem cell markers (P75 and GFAPδ) (van Strien et al., 2014). Most of the cells (60%–100%) were found to express one or more of these markers in addition to pVIM (Figures 1C, S1C, and S1D).
Figure 1.
RG-like Cells in Human Adult GBMs
(A) Representative confocal images of GBM sections. Cells in M phase are labeled with phospho-histone 3 (pH3) and their radial fiber-like processes are detected with pVIM.
(B) Percentage of pVIM+ and pH3+ cells in human GBMs (n = 21 tumors).
(C) Quantification of co-expression of pVim with an array of RG markers, by immunochemistry (IHC).
(D) Schematic flow of the development of 3D explants from patient GBM samples. IHC demonstrates the expression of pH3 and pVim during cell division; arrows points to mitotic cells with or without process (labeled RG-like or non-RG-like).
(E) Co-expression of RG-like markers in pVim+ cells in explants.
Scale bars, 10 and 5 μm (A), 20 and 10 μm (D and E).
Table 1.
Quantification of pVim and pH3 Cells in 21 Tumor Tissue Samples
| Patient | pVIM | pH3 | DAPI | pVIM/pH3 | %Mitotic Cells (pH3/DAPI) |
|---|---|---|---|---|---|
| GBM1 | 5 | 7 | 1,407 | 5 | 0.5 |
| GBM2 | 6 | 9 | 1,372 | 6 | 0.66 |
| GBM3 | 10 | 14 | 1,514 | 10 | 0.93 |
| GBM4 | 3 | 3 | 1,047 | 3 | 0.29 |
| GBM5 | 18 | 17 | 1,395 | 17 | 1.22 |
| GBM6 | 16 | 16 | 2,715 | 16 | 0.59 |
| GBM7 | 4 | 3 | 1,355 | 3 | 0.22 |
| GBM8 | 10 | 10 | 2,038 | 10 | 0.49 |
| GBM9 | 2 | 2 | 1,111 | 2 | 0.18 |
| GBM10 | 5 | 4 | 511 | 3 | 0.78 |
| GBM11 | 3 | 2 | 824 | 2 | 0.24 |
| GBM12 | 2 | 4 | 1,283 | 2 | 0.31 |
| GBM13 | 5 | 6 | 538 | 5 | 1.12 |
| GBM14 | 2 | 3 | 1,089 | 2 | 0.28 |
| GBM15 | 4 | 4 | 632 | 4 | 0.63 |
| GBM16 | 9 | 9 | 978 | 9 | 0.92 |
| GBM17 | 17 | 17 | 1,268 | 17 | 1.34 |
| GBM18 | 7 | 7 | 658 | 7 | 1.06 |
| GBM19 | 14 | 14 | 1,756 | 14 | 0.8 |
| GBM20 | 6 | 6 | 746 | 6 | 0.8 |
| GBM21 | 21 | 21 | 1,693 | 21 | 1.24 |
To better visualize cell morphology we performed 3D organotypic tumor cultures. These explants consist of fresh tumor tissue dissected into 1- to 2-mm pieces that are embedded in Matrigel and maintained in growth factor (GF)-free media (Figure 1D) (Shimizu et al., 2011). We analyzed whole mounts of 3D cultures obtained from five different patient tumors (Table 2). We found that 27% (122/459) of pH3+ mitotic cells maintained a radial fiber-like process and are thus RG-like, while the remaining 73% had no fiber and exhibited a classic rounded up cytoplasm during division (Figures 1D and S1B). These mitotic RG-like cells coexpressed RG markers BLBP, PAX6, SOX2, ARHGAP11B, and GLAST (Figure 1E). Interestingly, we observed a specific polarized orientation of the processes of RG-like cells within the explants (Figure S1E). In more than 90% of the cells, the short processes always faced the center of an explant and the long processes extended radially away from the center of the explant. The remaining 10% of RG-like cells (pVIM+/pH3+ and maintaining a process) had curved, horizontal, or apical (long process facing toward the center) orientations.
Table 2.
Tumor Samples Used for Examination of RG-like Cells in Explants
| Tumor | pVIM | pH3 | pVIM/pH3 Cells with Process |
|---|---|---|---|
| GBM22 | 109 | 110 | 39 |
| GBM23 | 80 | 81 | 27 |
| GBM24 | 54 | 55 | 14 |
| GBM25 | 111 | 112 | 22 |
| GBM26 | 100 | 101 | 20 |
Unique RG Mitotic Behaviors
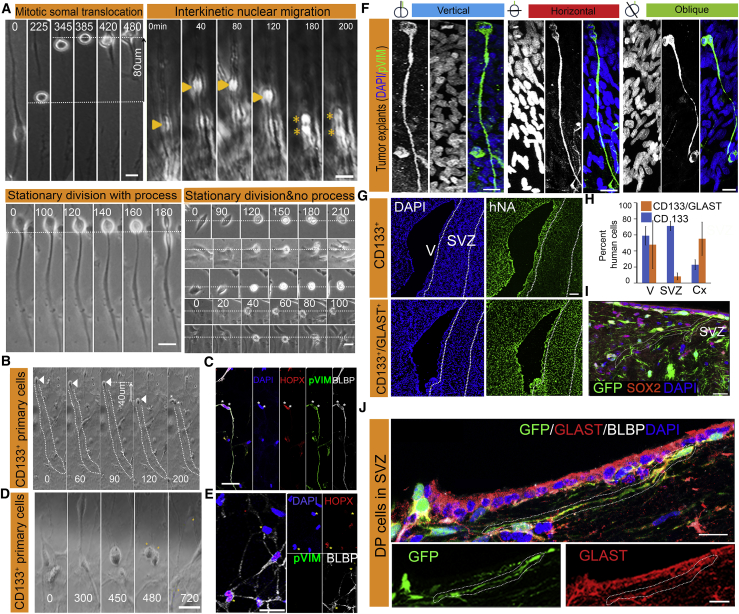
The human subventricular zone includes a dense outer zone that consists of outer RG (oRG) cells (Hansen et al., 2010). They exhibit a well-described pattern of cell division, MST, a characteristic migratory behavior, in which the soma rapidly translocates toward the cortical plate immediately prior to cytokinesis. Human oRG cells self-renew mainly by horizontal asymmetric division (LaMonica et al., 2013), whereby one daughter cell retains the basal fiber. Inheritance of the basal fiber has been hypothesized to be important for maintenance of stem cell identity. We asked whether these phenomena could be identified in the RG-like cells within tumors, and whether they were cell intrinsic or dependent on the tumor microenvironment. Therefore, we performed extensive timelapse microscopy on tumor cell cultures and multiple explants (n = 7 tumors). We observed four major mitotic patterns. In addition to MST, we observed interkinetic nuclear migration, a phenomenon observed in ventricular RG, as well as two types of stationary cell division (Figures 2A–2C; Video S1). Most cells maintaining a radial fiber-like process exhibited stationary division with process (Video S2). Half of the mitotic cells retracted their protrusion and divided in place, as is commonly observed in most cell types (stationary division without process). Out of 256 observed divisions, 57% of dividing cells maintained their processes during mitosis and 16.3% of the mitotic cells with radial fiber-like processes demonstrated unique RG mitotic behavior. Interestingly, we also observed tumor cells moving along the processes of RG-like cells, as has been described for neuronal migration during development (Figure S2A; Video S3). Furthermore, some daughter cells went through MST along a similar division trajectory as their mother cells (Figure S2B; Video S4). Having observed these behaviors both in explants and dissociated cells albeit at a lower frequency, we reasoned that cell-intrinsic mechanisms that are independent of tumor microenvironment are responsible for the establishment of cleavage angles and the control of mitotic behavior in tumor RG-like cells. To correlate this mitotic behavior with specific RG molecular markers, we performed timelapse imaging of cultures of freshly sorted CD133+ tumor cells, identified cells undergoing MST, and then fixed the samples and examined the expression of oRG-specific markers. We observed that daughter cells after MST express pVIM/HOPX/BLBP or CRYAB/HOPX/SOX2 (Figures 2C and S2C; Video S5); cells undergoing stationary division without a process were negative for the same markers (Figures 2D and 2E).
Figure 2.
Human RG-like Mitotic Behavior and Division Planes
Frames from timelapse microscopy show four major types of mitotic behavior in explants.
(A) Human oRG-specific MST pattern: the cell body moves along the process and accomplishes a fast movement over a significant distance before cytokinesis (dotted white lines); ventricular RG-specific interkinetic nuclear migration: cells move along the process up and down during mitosis; arrow heads point to the nucleus and the stars to the daughter cells; stationary divisions occur without significant movement (dotted line).
(B and C) Real-time images show a cell undergoing MST (arrowheads) in (B). Immediate fixation shows the same two daughter cells visible in the last frame (200) and identified with stars. Both cells are immunopositive for pH3, pVIM, and BLBP while only one daughter cell retains the elongated fiber process as shown in (C). Dotted contour around the RG-like fiber in (B).
(D and E) Similar approach identifies a cell without process undergoing stationary division in (D) and negative for RG markers in (E).
(F) RG cells in explants undergoing division along a horizontal plane. Schematic figures define the cleavage planes (top panel).
(G and H) Immunohistochemistry on brains injected with GBM cells expressing CD133 or CD133/GLAST (G). Strong tropism toward the subventricular zone (SVZ) in the CD133/GLAST group, quantified as percent of human cells in V (ventricle), SVZ, and Cx (cortex) in (H).
(I and J) Immunohistochemistry of tumor cells in the SVZ region shows RG-like morphology in GFP-labeled human cells lining the SVZ, and expressing SOX2 in (I) and GLAST/BLBP in (J).
Scale bars, 10 μm (A), 20 μm (C, D, E, F, and I), 100 μm (G), 20 and 10 μm (J). DP, double-positive or CD133 +/GLAST+. Time in (A)–(D) in minutes.
Cells with mitotic somal translocation (oRG-like) are highlighted in yellow and cell bodies are tracked by yellow arrow. The cells dividing in place with or without process are indicated in green and blue separately.
Daughter cell regrows the process after division.
Cell undergoing interkinetic nuclear movement is highlighted in yellow. The cell body moves along the long process toward the end of the short process then back toward the end of the long process for mitosis. The yellow arrow tracks the cell body movement. Daughter cells inherit one process and regrow another one after division. In the same field, other cells retract the cytosol/process and divide in a stationary manner (blue).
oRG-like mother and daughter cells divide via MST, a behavior suggestive of self-renewal.
Human oRG cells are self-renewed by horizontal division. Therefore, we characterized cell division in both organotypic (explants) and dissociated low-passage cell cultures. We found that 68% of RG-like mitotic cells displayed a horizontal cleavage plane in explant cultures, while 85% did so in dissociated cultures of freshly sorted CD133+ cells from surgical GBM specimens (Figures 2F, S2D, and S2E). The remaining RG-like cells exhibited a vertical or oblique plane of cleavage. Dissociated cells undergoing vertical divisions spent a significantly longer time in cytokinesis compared with cells dividing horizontally (160 and 60 min, respectively) (Figure S2F).
RG-like Tumor Cells Form Tumors In Vivo
In view of the heterogeneous nature and low prevalence of RG-like cells in GBMs, as well as the absence of pathognomonic surface markers, we attempted to empirically enrich for RG-like cells using GLAST expression, consistent with the studies using CD133/GLAST to isolate human fetal RG cells or mouse adult neural stem cells (NSCs) (Johnson et al., 2015) and with our data on markers coupled with MST behavior. Bulk qRT-PCR for an RG gene panel performed on GLAST+ cells sorted from two different GBMs and demonstrated that GLAST+ cells exhibited higher expression levels for RG markers compared with GLAST− cells, indicating that GLAST may represent a promising additional membrane marker for further studies (Figure S3A). We analyzed GLAST expression by fluorescence-activated cell sorting (FACS) in nine tumors and found it in an average of 7.37% ± 0.05% (Table 3). We also confirmed that GLAST-expressing cells are capable of propagating a tumor in vivo. We dissociated freshly obtained GBMs and sorted for CD133 and/or GLAST subsets. We injected four cell groups (CD133+, GLAST+, CD133+GLAST+, and double-negative) into the striatum of NOD/SCID gamma mice. All but the double-negative group developed brain tumors. Interestingly, the CD133+GLAST+ cells showed a significant tropism toward the subventricular zone when compared with the other groups (Figures 2G and 2H) where they exhibited typical RG-like morphology (Figure 2I) and coexpressed GLAST and BLBP (Figure 2J).
Table 3.
GLAST Expression by FACS Analysis
| Tumor | %GLAST+ |
|---|---|
| GBM39 | 11.40 |
| GBM40 | 2.90 |
| GBM29 | 0.30 |
| GBM22 | 5.50 |
| GBM23 | 13.90 |
| GBM24 | 17.10 |
| GBM25 | 6 |
| GBM26 | 2.10 |
| GBM30 | 7.10 |
Characterization of RG-like Tumor Cells via scRNA-Seq
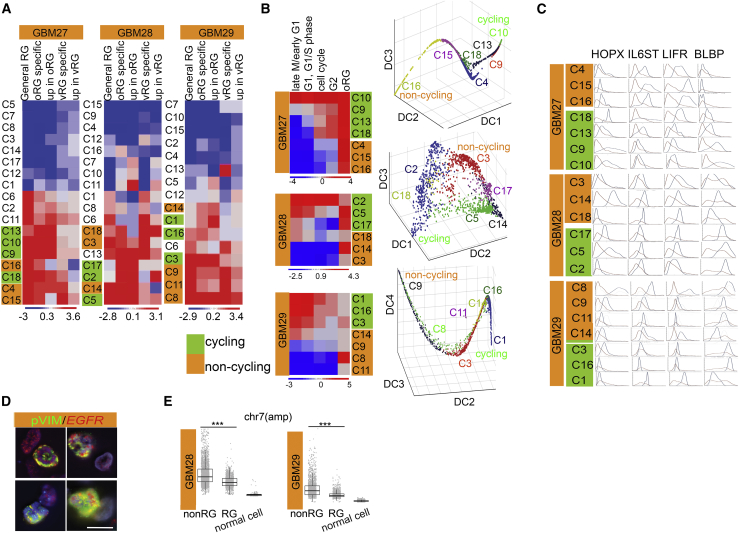
At present, RG-specific clusters have not been reported in scRNA-seq studies. We performed scRNA-seq on sorted CD133/GLAST-expressing tumor cells from three freshly procured human GBMs (GBM 27, GBM 28, and GBM 29, profiled in Table S1; available profiles of all tumors used in this article are listed in Table S2) for a total of 13,251 viable cells that passed quality controls; CD45+ cells were excluded and 12,490 cells were used in the analysis. We analyzed each tumor individually. To overcome the issue of missing transcripts, typical of scRNA-seq, and to de-noise the data we used MAGIC (Markov affinity-based Graph Imputation of Cells) (van Dijk et al., 2018). We performed unsupervised clustering of the cells using PhenoGraph, a well-established method that has previously been used to robustly infer clusters or cellular subtypes from single-cell data (Shekhar et al., 2016, Wei et al., 2017). Cluster numbers and sizes are listed in Table S3.
To identify RG-like cells, we used RG gene sets (Table S4) that were identified in studies of dissected human developing brains (Pollen et al., 2015). We then used Earth Movers' Distance (EMD) (Levine et al., 2015) to characterize and annotate clusters. For this, we computed EMD between genes belonging to the RG gene sets in each cluster compared with the rest. We detected six to seven RG-like clusters per tumor independently, confirming the presence of RG-like cells in all specimens (Figure 3A). We used the same approach to segregate the cycling and non-cycling cell compartments (Figure 3B). Cell-cycle gene sets were obtained from Kyoto Encyclopedia of Genes and Genomes pathway and gene ontology analysis (see Experimental Procedures).
Figure 3.
Single-Cell Transcriptional Analysis of GBM Cells Reveals a Population of RG-like Cells
(A) EMD heatmaps to identify RG-like clusters in the analyses of individual patient data.
(B) RG-like cells in individual patient data are segregated into non-cycling (orange) and cycling (green) based on EMD scores for cell-cycle gene sets (top panel). Diffusion maps on raw data using all genes demonstrate the distribution of RG-like cells in individual patients. Each dot represents a cell and each axis represents a diffusion component. The cells are color coded based on the clusters they belong to.
(C) Expression distribution of RG markers (HOPX, IL6ST, LIFR, and BLBP) are shown for individual tumors. Orange and blue lines represent the expression of a specific gene in the indicated cluster and its expression in the rest of the data, respectively.
(D) Immuno-FISH on sections from four patients show colocalization of pVim with the EGFR probe.
(E) CNV analysis of chromosome 7 in RG- and non-RG-like cells (n = minimum of 50 nuclei per tumor). Scale bar, 8 μm.
We then sought to study any possible relationship between cycling and non-cycling RG-like cells in each tumor. For this, we performed a diffusion component (Setty et al., 2016) analysis to get an accurate representation of the underlying structure of the data. Scatterplot of RG-like cells in individual patients along the diffusion components showed that the cycling and non-cycling cells were segregated along the data manifold with multiple intermediate states seemingly connecting them (Figure 3B). These data are highly compatible with recent literature demonstrating that NSC lineage exist on a continuum through the processes of activation and differentiation, including the presence of molecularly distinct rare intermediate stages (Dulken et al., 2017). Analysis of select individual genes demonstrated that the leukemia inhibitory factor (LIF) receptor and its coreceptor IL6ST demonstrate a trend of downregulated expression in cycling RG-like cells when compared with those in non-cycling cells, unlike HOPX and FABP, which were more heterogeneously expressed (Figure 3C). This expression pattern suggested that LIFR may be one of the membrane markers for non-cycling RG-like cells.
Genomic Aberrations in Radial Glia-like Tumor Cells
We analyzed copy number variation (CNV) in the scRNA-seq data as a means of confirming the neoplastic nature of the RG-like cells in individual GBMs. Using CD45+ immune cells as a normal cell control, CNV profiles revealed chromosomal aberrations with loss of chromosome 10 in all 3 patients, as is often reported in GBM, as well as amplifications of chromosome 7 in two tumors, as confirmed by tumor sequencing (Figure S3B; Table S1). One of the patients (GBM27) also exhibited loss of chromosome 15 (Figure S3B). The loss of a chromosome was reflected in the scRNA-seq data and was subsequently validated on matched tumor specimens using DNA-fluorescence in situ hybridization (FISH) (Figure S3C). We also carried out immuno-FISH assays for pVIM and an EGFR probe to further confirm that RG-like cells in GBM are indeed of tumor origin (Figure 3D). We further analyzed the extent of chromosomal aberration in RG clusters of each individual tumor and noted interesting variations. We found that non-cycling RG-like cells maintained a more intact chromosome 15 compared with cycling RG-like cells in GBM27 (Figure S3D). Interestingly, C18 in GBM28 (a non-cycling cluster originating from the younger 31-year-old patient) exhibits a near normal copy number of chromosomes 10 and 7 in the setting of chromosome 7 amplification and chromosome 10 loss exhibited by the remaining matched tumor cells (Figure S3E). In both GBM28 and GBM29, RG-like cells had significantly less focal chromosome 7 amplification compared with non-RG-like cells (Figure 3E). These findings largely confirm the neoplastic nature of the RG-like cells, and raise the possibility of retention of normal cells with an RG-like transcriptomal profile within the adult brain or brain tumor, but they should be interpreted cautiously in view of the small sample size.
Non-cycling Radial Glia-like Cells Are Activated by Inflammation Signaling
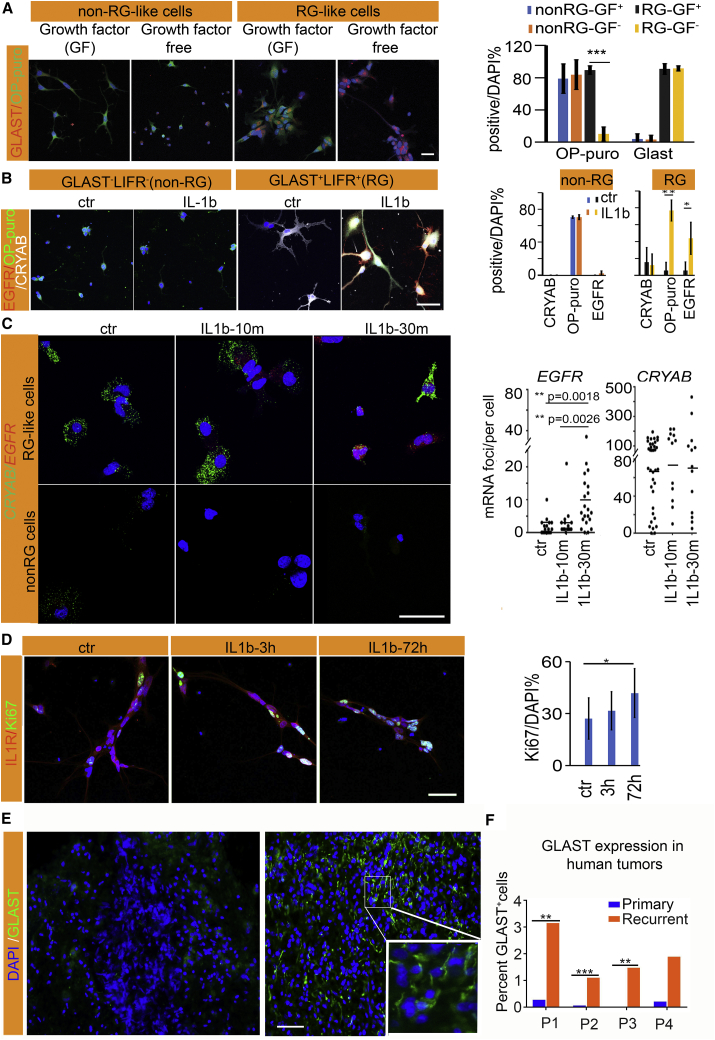
The activation of quiescent NSCs has been linked to the transcriptional upregulation of factors critical for protein synthesis machinery, such as multiple ribosomal genes, and oxidative phosphorylation (Llorens-Bobadilla et al., 2015). Several authors have also described morphological (Codega et al., 2014) and transcriptional features associated with dormancy (a deep quiescence state), such as absence of cell cycling, low protein synthesis, and EGFR gene expression (Shin et al., 2015). These factors are activated upon transition out of dormancy and progression along cell differentiation stages (Baser et al., 2017, Kokovay et al., 2010). In vitro assays showed significant differences at baseline between freshly sorted RG and non-RG-like cells. While non-RG cells maintained protein synthesis (using the O-propargyl-puromycin incorporation or Op-puro assay) in epidermal GF (EGF)- or fibroblast GF-enriched and GF-free media, the RG cells had undetectable global protein synthesis at baseline and it was upregulated in the GF media (Figure 4A). Treatment with IL-1b, but not with bone morphogenetic protein (BMP), LIF, or EGF, upregulated EGFR, the dormant cell marker (Figure S4A). Based on the observation that LIFR+ cells are significantly enriched in IL-1R1+ cells (Figure S4B), we sorted out GLAST+LIFR+ cells to test the role of IL-1 in driving dormant RG-like cells into the cell cycle. Treatment with IL-1b significantly upregulated protein synthesis (Figure 4B) and the mRNA and protein expression of EGFR (Figure 4C) in RG-like but not non-RG cells. Concomitantly, the expression of the cell-cycle marker Ki67 was also increased, indicating entry into the cell cycle (Figure 4D). Enhancement of protein synthesis occurred within 10 min of exposure to IL-1b (Figure S4C). These data suggest that the interleukin inflammation signaling pathway can activate the dormant tumor RG-like cells and trigger their entry into the cell cycle in vitro.
Figure 4.
Inflammation Regulates Cell-Cycle States of RG-like Cells
(A) Immunohistochemistry for GLAST and protein synthesis (OP-puro) in growth factor (GF) and GF-free conditions.
(B) IL-1b treatment activates RG-like cells coexpressing GLAST+LIFR+ by stimulating protein synthesis (OP-puro) and the expression of EGFR. Quantification of general protein synthesis and expression of EGFR protein in the indicated cell groups (right panel), p = 0.0008 (OP-puro), p = 0.019 (EGFR).
(C) RNA-FISH showing that IL-1b activates the transcription of EGFR in RG-like cells and while CRYAB expression is maintained.
(D) Treatment with IL-1b for 72 h significantly increases Ki67 in GLAST+LIFR+ cells, signifying entry into the cell cycle (n = 6, triplicate from two patients). Scale bars, 20 μm (A, B, and D), 50 μm (C).
(E and F) IHC for GLAST (E) in a set of matching primary (left) and recurrent (right) tumors quantified in (F). n = 4 pairs, Mann-Whitney.
Interleukin IL-1b is a potent cytokine that is a major component of the microglia secretome; its levels are increased in GBMs and it is a component of the pro-inflammatory cascade elicited by radiation. We then asked if GLAST-expressing cells might be more abundant at the time of tumor recurrence. We identified four patients with available matching tumor tissues from initial diagnosis and recurrence. GLAST expression by immunohistochemistry was heterogeneous but significantly and consistently more increased in the recurrent tissues (Figures 4E and 4F). Analysis of large datasets from cBioPortal also confirm a negative correlation between GLAST expression levels and extent of survival, as well as more increased levels in patients who have recurrence within 6 months compared with those remaining disease free (Figures S4D and S4E).
Discussion
RG cells are the earliest NSCs in the neuroectodermal lineage. In higher mammals, oRG cells contribute to the evolutionary expansion and folding of the neocortex, and to the increased diversity of neural progenitor populations (Sild and Ruthazer, 2011). It is not clear if RG cells could persist into adulthood. In this study, we demonstrate the presence of cells with RG features within adult GBMs. Initial observations showed rare but consistent RG-like morphology during mitosis as demonstrated by pH3 and pVim immunohistochemistry on a set of 21 GBM specimens. Using extensive timelapse microscopy, we demonstrate and quantify RG-specific mitotic behavior, including MST and interkinetic nuclear migration in GBM cells in 3D cultures and early passage cells. We also demonstrate that RG-like tumor cells are capable of self renewal as they give rise to generations of daughter cells with identical mitotic behavior. Combined timelapse video and immunofluorescence demonstrate co-labeling of the cells with RG-typical markers, such as GLAST, HOPX, and BLBP. Sorting for GLAST-expressing cells resulted in enrichment for RG genes, so we sorted GBM cells from three different tumors and performed scRNA-seq analysis. Unbiased clustering showed multiple cell groups within each specimen, and six to seven clusters per tumor that are enriched for fetal RG genes. Further analyses identify quiescent and active states within the RG clusters that are highly similar to those described in adult stem cell niches.
Careful analysis of the scRNA-seq data did not reveal a single pathognomonic marker or a combination thereof that would allow the prospective isolation of definitive RG-like cells from GBMs. We therefore used marker combinations to enrich for these cells. In vitro assays demonstrate a role for IL-1b in the activation of potentially dormant RG-like cells, associated with the upregulation of protein synthesis and downregulation of EGFR. Inflammation plays a critical role in tumor progression (Coussens and Werb, 2002). IL-1b is a major mediator of inflammation in the brain and is expressed in the microglia secretome (Monif et al., 2016). Our findings could suggest a role for inflammation or microglial activation in accelerating the proliferation of RG-like tumor cells. All GBMs receive radiation and chemotherapy, known to be associated with increased inflammation. We validated a potential role for GLAST protein expression in tumor recurrence, by demonstrating significant increase in its expression in matching sets of initial and recurrent tumors; in large datasets, we also found GLAST expression levels to be increased in recurrent GBMs and to correlate with poorer overall survival and early disease progression.
Estimated copy-number analyses confirmed the presence of genomic aberrations within RG clusters, matching those found in the tumors of origin. Interestingly, we found that major aberrations, such as large chromosomal losses or gains were most prevalent in the non-RG clusters, with the non-cycling RG compartment comprising genome copy numbers that are closest to the normal diploid genome. These data raise the possibility of maintenance of normal RG cells in the human adult brain.
Taken together, these data support current findings of conserved developmental hierarchies within GBM, and identify RG cells as the putative stem cells underlying this program. The paucity of the cells and the permissive expression of commonly recognized RG markers, as well as the difficulty in maintaining or expanding the cells in their RG state in vitro, pose a challenge for more advanced studies. The data also support the existence of conserved molecular dynamics of activation of quiescent NSCs within GBM, that mimic those identified in the neural stem cell niche of the developing brain. We hypothesize that RG-like tumor cells could represent the cells of origin or the cancer stem cells residing at the top of the tumor cell hierarchy, capable of transitioning into an active state and fueling tumor growth with a diversity of neural progenitors, although further data would be required to functionally validate this concept. Enhanced understanding of these cells and of the molecular factors that trigger their activation will lead to novel therapeutic targeting strategies. Hence, the role of interleukin in tumor maintenance should be further investigated. The data also hint at the possibility of persistence of normal RG cells into adulthood, when they might be vulnerable to tumorigenic transformation.
Experimental Procedures
Dissociation and FACS Sorting of Human GBM Specimens
Surgical specimens were collected from the surgical suite at Memorial Sloan Kettering Cancer Center, following diagnostic confirmation by a neuropathologist. Tissues were obtained in accordance with international review board guidelines. Single-cell suspensions were obtained based on our previously published methods (Wang et al., 2010) with minor modification to reduce processing time. In brief, tumor tissue was cut into 2-mm2 pieces and enzymatically digested with Liberase for 5–10 min at 37°C. The suspension was passed through a cell strainer. Red blood cells (RBCs) were depleted with RBC lysis buffer (eBioscience, cat no. 00-4333-57) for 3 min at 37°C. The cell suspension was then blocked with FcR (Miltenyi Biotec, cat. no. 130-059-901) for 5 min and incubated with appropriate conjugated antibodies for 10 min at room temperature. The cells were pelleted by centrifugation at 219 × g for 3 min. The approach of sorting fixed single cells was adopted from a published protocol (Thomsen et al., 2016). In brief, RBCs and CD45 hemopoietic cells in single-cell suspensions were removed by RBC lysis buffer and CD45 MACS beads with LS columns (Miltenyi Biotec, cat no. 130-045-801). Cells were then fixed with 4% paraformaldehyde (Electron Microscopy Sciences) and sorted for cell size and DAPI signal. A few hundred cells from the gate of singlet and DAPI selection were sorted to confirm single cells and absence of debris, before sorting single cells using a 120-μm nozzle for specific antibodies into an 8-strip PCR tube (Fisher) containing 5 μL of PKD buffer (QIAGEN) with 1:16 proteinase K solution (QIAGEN). Antibodies used in sorting included: APC-conjugated CD133 (AC141 and AC133, 1:500 each, Miltenyi Biotech, 130-090-854 and 130-090-826); PE-conjugated GLAST (1:1,000, Miltenyi Biotec, 130-098-804); APC-GLAST (1:800, Miltenyi Biotec, 130-098-803); PE-conjugated PAX6 (1:400, BD Bioscience, 561552); PE-conjugated LIFR (1:500, R&D Systems, FAB249P). For live cell sorting, cells positive for markers of interest but negative for DAPI were sorted on the Aria sorter and collected in N2 medium. Negative controls consisted of unstained sample and IgG control. Pilot experiments verified gating by immunostaining cells after sorting.
In Vivo Studies
Adult female NOD/SCID or male NOD/SCID gamma mice (Jackson Laboratory, Bar Harbor, Maine) were anesthetized with ketamine/xylazine (Hospira, Lake Forest, Illinois/Inc/Ben Venue Laboratories) and placed in a stereotaxic frame (Stoelting Company, Wood Dale, Illinois). Freshly sorted cells were injected into the right striatum immediately after sorting (Wang et al., 2010). Animals received 10,000 cells each from 4 groups of CD133+, GLAST+, CD133+/GLAST+, or CD133–/GLAST− cells. Animals were sacrificed upon exhibiting symptoms (n = 10). Animals were housed and cared for in accordance with the NIH guidelines for animal welfare, and all animal experiments were performed in accordance with protocols approved by our Institutional Animal Care and Use Committee.
Cell and Explant Cultures
Freshly sorted CD133+ cells were cultured in adherent culture conditions on plates coated with poly-L-ornithine hydrobromide (15 μg/mL, Sigma), mouse laminin I (4 μg/mL, R&D Systems), and fibronectin (2 μg/mL, Fisher Scientific). The adherent cells were maintained in serum-free neurobasal medium supplemented with N2, 2 mM L-glutamine, 20 ng/mL recombinant human EGF, and 10 ng/mL recombinant human fibroblast GF 2 (all from Invitrogen). About 2 mm tumor tissue explants were cultured in GF-reduced Matrigel (BD 354230) in N2 medium without supplementing any GFs. Real-time imaging was carried out 4–6 days after in vitro culture.
For activation assays, indicated cell types were cultured after sorting in the adherent culture conditions as above in GF-free N2 medium for 2 days. Cells were stimulated with 10 ng/mL final concentration of human LIF (Sigma), or 10 ng/mL final concentration of human BMP7 (R&D Systems), or 20 ng/mL final concentration of human EGF (R&D Systems), or 25 ng/mL final concentration of human IL-1b (R&D Systems) for the indicated time.
scRNA-Seq
Sorted tumor cells from human GBM specimens were washed once and resuspended in PBS containing 0.05% BSA. A dose of 21 μL of a cellular suspension at 500 cells/μL with 95% viability was loaded onto the 10X Genomics Chromium platform to generate barcoded single-cell Gel Bead-In-Emulsions. scRNA-seq libraries were prepared according to 10X Genomics specifications. The Sequence Quality Control (Azizi et al., 2018) package was used to process raw scRNA-seq reads to a transcript count matrix, including de-multiplexing, alignment, barcode and UMI error correction, and generation of a digital expression matrix. Alignment was performed to the hg38 annotation restricted to transcribed, polyadenylated RNA of length >200 nucleotides to increase mapping specificity. Viable single cells were identified based on total mRNA abundance, number of unique transcripts, and mitochondria fraction. A total of 12,367 cells from 3 patients (GBM 27, 4,570; GBM 28, 4,341; and GBM 29, 3,456 cells) with a median library size of 6,152 molecules per cell were used for downstream analysis. MAGIC and PhenoGraph were used to robustly de-noise and impute scRNA-seq data, and to perform clustering, respectively. More details in the Supplemental Methods section.
Quantification and Statistical Analysis
Immunostaining and RNA-FISH Analyses
A t test was used to determine the significance of the observations. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 are used in study. Mean and error bars with standard deviation.
CNV Analysis
Chromosomal CNVs were analyzed using scRNA-seq data as described previously (Patel et al., 2014). In brief, all genes were sorted by their genomic locations and chromosomal copy number of each gene in each cell was estimated by moving average of the relative normalized expression of the neighboring 101 genes. CD45+ immune cells in our scRNA-seq data were used as normal cell control. To estimate the copy number of each chromosome in each cell, median expression value of all genes located in the chromosome was normalized by subtracting the median and dividing by the standard deviation averaged of all CD45+ immune cells. The p values were calculated using the Wilcoxon rank-sum test.
GLAST Expression Analysis in cBioPortal Dataset
A glioblastoma multiforme (TCGA, Provisional) dataset from cBioPortal (Gao et al., 2013) was used for GLAST gene expression and patient survival analysis. A total of 528 patients with both clinical and gene expression data (mRNA expression Z scores [U133 microarray only] from cBioPortal Glioblastoma Multiforme [TCGA, Provisional]) were used for the analysis. To analyze the relationship between GLAST gene expression and patient survival, we first grouped the patients by GLAST mRNA expression Z score. The GLAST mRNA Z score distribution ranged from −4.5282 to 1.373 and the median was 0.2089. The 25% percentile was −0.4469 and the 75% percentile was 0.5707. We set up Z score = ±0.5 as the threshold to group the patient as GLAST high and low expression. The group with a GLAST Z score more than 0.5 had 143 patients and the group with a GLAST Z score less than −0.5 group had 82 patients. The overall survival in months and survival status data from those patients were used for Kaplan–Meier survival analysis using Prism 8.
Among the 528 samples, 366 samples were newly diagnosed GBM; among those patients, 152 recurred within 6 months and 23 remained disease free. We compared GLAST expression between the two groups, using a t test. PFS6 or progression-free survival at 6 months is an appropriate time point to represent disease status after recurrence, as is commonly used in clinical trials. We performed an unpaired t test to analyze the difference in GLAST expression between the two groups, and the p value was 0.04, which means that they have a statistical difference.
Mutation Profiles of Tumors
Tumors used for scRNA-seq underwent targeted sequencing by MSK-IMPACT, a hybridization capture-based next-generation sequencing assay for targeted deep sequencing of a selected cancer gene set. Full method detailed in Cheng et al. (2015).
Author Contributions
R.W. and V.T. contributed to data interpretation and manuscript writing. R.W. conceived and designed the project and performed most experiments, M.H. and D.P. supervised the scRNA-seq data analyses. R.S., X.S., A.M.L., K.F., S.H., and Y.X. contributed to the single-cell data analyses and bioinformatics; P.J.C. and M.S. contributed to live and IF imaging; P.M., I.M., M.N., and G.N. performed key technical experiments.
Acknowledgments
The work was supported by the W.M. Keck Foundation and NIH RO1 CA208405 (to V.T.) and program grant P30 CA 008748. X.S.: Scientific Research Fund of Hunan Provincial Education Department (16B237), Chinese Scholarship Council and Chan Zuckerberg Initiative DAF (183501). X.S. and M.H.: core grant to the Wellcome Sanger Institute from the Wellcome Trust (206194). K.F.: New York Stem Cell Foundation. We thank Huatai Xu for constructive discussion and critical reading, Mark Tomishima and Kiran Ramnarine; Katia Manova, Vitaly Boyko, Fujisawa Sho, Elvin Feng, Nin Fang and Mesruh Turkekul at the Molecular Cytology Core Facility; Paul Byrne, Mark Kweens, Fang Fang, Petrina Georgala, and Rui Gardner at the Flow Cytometry Core Facility at Memorial Sloan Kettering Cancer Center, and Yara Maria Studer for graphics.
Published: January 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.01.007.
Accession Numbers
Data was deposited in the GEO (GSE139448).
Supporting Citations
The following references appear in the Supplementary Methods: Amir el et al., 2013, Bakhoum et al., 2018, Coifman and Lafon, 2006, Dessimoz and Skunca, 2017, Farrell et al., 2018, Haghverdi et al., 2016, Jaitin et al., 2016, Kanehisa and Goto, 2000, Lavin et al., 2017, Takahashi et al., 2019, Vuong et al., 2018
Supplemental Information
Shaded cells highlight matching new and recurrent tumors. Data based on targeted sequencing.
Green indicates cyclingRG clusters and orange non-cycling RG like clusters.
References
- Alcantara Llaguno S.R., Wang Z., Sun D., Chen J., Xu J., Kim E., Hatanpaa K.J., Raisanen J.M., Burns D.K., Johnson J.E. Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell. 2015;28:429–440. doi: 10.1016/j.ccell.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir el A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.A., Murphy C.J., Ly P., Shah P., Sriram R.K., Watkins T.B.K., Taunk N.K. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baser A., Skabkin M., Martin-Villalba A. Neural stem cell activation and the role of protein synthesis. Brain Plast. 2017;3:27–41. doi: 10.3233/BPL-160038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li Y., Yu T.S., McKay R.M., Burns D.K., Kernie S.G., Parada L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A.R., Deleo A.M., Pastrana E., Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman R.R., Lafon S. Diffusion maps. Appl. Comput. Harmon. Anal. 2006;21:5–30. [Google Scholar]
- Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa A., Pellegatta S., Rossi M., Tunici P., Magnoni L., Speranza M.C., Malusa F., Miragliotta V., Mori E., Finocchiaro G. A radial glia gene marker, fatty acid binding protein 7 (FABP7), is involved in proliferation and invasion of glioblastoma cells. PLoS One. 2012;7:e52113. doi: 10.1371/journal.pone.0052113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessimoz C., Skunca N., editors. The Gene Ontology Handbook. Methods in Molecular Biology. Springer; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken B.W., Leeman D.S., Boulet S.C., Hebestreit K., Brunet A. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 2017;18:777–790. doi: 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J.A., Wang Y., Riesenfeld S.J., Shekhar K., Regev A., Schier A.F. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018;360 doi: 10.1126/science.aar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., Haffner C., Sykes A., Wong F.K., Peters J. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;269:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Barde Y.A. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Haghverdi L., Buttner M., Wolf F.A., Buettner F., Theis F.J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- Hansen D.V., Lui J.H., Parker P.R., Kriegstein A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Howard B.M., Zhicheng M., Filipovic R., Moore A.R., Antic S.D., Zecevic N. Radial glia cells in the developing human brain. Neuroscientist. 2008;14:459–473. doi: 10.1177/1073858407313512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin D.A., Weiner A., Yofe I., Lara-Astiaso D., Keren-Shaul H., David E., Salame T.M., Tanay A., van Oudenaarden A., Amit I. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell. 2016;167:1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- Johnson M.B., Wang P.P., Atabay K.D., Murphy E.A., Doan R.N., Hecht J.L., Walsh C.A. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y., Inagaki N., Nishizawa M., Tsutsumi O., Taketani Y., Inagaki M. Visualization of mitotic radial glial lineage cells in the developing rat brain by Cdc2 kinase-phosphorylated vimentin. Glia. 1998;23:191–199. doi: 10.1002/(sici)1098-1136(199807)23:3<191::aid-glia2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E., Goderie S., Wang Y., Lotz S., Lin G., Sun Y., Roysam B., Shen Q., Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–173. doi: 10.1016/j.stem.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonica B.E., Lui J.H., Hansen D.V., Kriegstein A.R. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat. Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X., Jorg D.J., Cavalli F.M.G., Richards L.M., Nguyen L.V., Vanner R.J., Guilhamon P., Lee L., Kushida M.M., Pellacani D. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549:227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir el A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Lui J.H., Nowakowski T.J., Pollen A.A., Javaherian A., Kriegstein A.R., Oldham M.C. Radial glia require PDGFD-PDGFRbeta signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita R., Coles J.E., Glubrecht D.D., Sung R., Sun X., Godbout R. B-FABP-expressing radial glial cells: the malignant glioma cell of origin? Neoplasia. 2007;9:734–744. doi: 10.1593/neo.07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monif M., Reid C.A., Powell K.L., Drummond K.J., O'Brien T.J., Williams D.A. Interleukin-1beta has trophic effects in microglia and its release is mediated by P2X7R pore. J. Neuroinflammation. 2016;13:173. doi: 10.1186/s12974-016-0621-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A.A., Nowakowski T.J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C.R., Shuga J., Liu S.J., Oldham M.C., Diaz A. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P., Choi K., Bendall S., Friedman N., Pe'er D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323.e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu F., Hovinga K.E., Metzner M., Soulet D., Tabar V. Organotypic explant culture of glioblastoma multiforme and subsequent single-cell suspension. Curr. Protoc. Stem Cell Biol. 2011 doi: 10.1002/9780470151808.sc0305s19. Chapter 3, Unit3 5. [DOI] [PubMed] [Google Scholar]
- Shin J., Berg D.A., Zhu Y., Shin J.Y., Song J., Bonaguidi M.A., Enikolopov G., Nauen D.W., Christian K.M., Ming G.L. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sild M., Ruthazer E.S. Radial glia: progenitor, pathway, and partner. Neuroscientist. 2011;17:288–302. doi: 10.1177/1073858410385870. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Nagata M., Gupta A., Matsushita Y., Yamaguchi T., Mizuhashi K., Maki K., Ruellas A.C., Cevidanes L.S., Kronenberg H.M. Autocrine regulation of mesenchymal progenitor cell fates orchestrates tooth eruption. Proc. Natl. Acad. Sci. U S A. 2019;116:575–580. doi: 10.1073/pnas.1810200115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.D., Poppleton H., Fuller C., Su X., Liu Y., Jensen P., Magdaleno S., Dalton J., Calabrese C., Board J. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Thomsen E.R., Mich J.K., Yao Z., Hodge R.D., Doyle A.M., Jang S., Shehata S.I., Nelson A.M., Shapovalova N.V., Levi B.P. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods. 2016;13:87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Venteicher A.S., Hebert C., Escalante L.E., Patel A.P., Yizhak K., Fisher J.M., Rodman C., Mount C., Filbin M.G. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien M.E., Sluijs J.A., Reynolds B.A., Steindler D.A., Aronica E., Hol E.M. Isolation of neural progenitor cells from the human adult subventricular zone based on expression of the cell surface marker CD271. Stem Cells Transl. Med. 2014;3:470–480. doi: 10.5966/sctm.2013-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A.S., Tirosh I., Hebert C., Yizhak K., Neftel C., Filbin M.G., Hovestadt V., Escalante L.E., Shaw M.L., Rodman C. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355 doi: 10.1126/science.aai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong N.H., Cook D.P., Forrest L.A., Carter L.E., Robineau-Charette P., Kofsky J.M., Hodgkinson K.M., Vanderhyden B.C. Single-cell RNA-sequencing reveals transcriptional dynamics of estrogen-induced dysplasia in the ovarian surface epithelium. PLoS Genet. 2018;14:e1007788. doi: 10.1371/journal.pgen.1007788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chadalavada K., Wilshire J., Kowalik U., Hovinga K.E., Geber A., Fligelman B., Leversha M., Brennan C., Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- Wei S.C., Levine J.H., Cogdill A.P., Zhao Y., Anang N.A.S., Andrews M.C., Sharma P., Wang J., Wargo J.A., Pe'er D. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 2017;170:1120–1133.e17. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman T., Noctor S.C., Clinton B.K., Honig L.S., Kriegstein A.R. Neurogenic radial glial cells in reptile, rodent and human: from mitosis to migration. Cereb. Cortex. 2003;13:550–559. doi: 10.1093/cercor/13.6.550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells with mitotic somal translocation (oRG-like) are highlighted in yellow and cell bodies are tracked by yellow arrow. The cells dividing in place with or without process are indicated in green and blue separately.
Daughter cell regrows the process after division.
Cell undergoing interkinetic nuclear movement is highlighted in yellow. The cell body moves along the long process toward the end of the short process then back toward the end of the long process for mitosis. The yellow arrow tracks the cell body movement. Daughter cells inherit one process and regrow another one after division. In the same field, other cells retract the cytosol/process and divide in a stationary manner (blue).
oRG-like mother and daughter cells divide via MST, a behavior suggestive of self-renewal.
Shaded cells highlight matching new and recurrent tumors. Data based on targeted sequencing.
Green indicates cyclingRG clusters and orange non-cycling RG like clusters.