Abstract
The Th17 immune response plays a key role in autoimmune diseases such as multiple sclerosis (MS) and inflammatory bowel disease (IBD). Expression of Th17-related genes in inflamed tissues has been reported in autoimmune diseases. However, values are frequently obtained using invasive methods. We aimed to identify biomarkers of MS in an accessible sample, such as blood, by quantifying the relative expression of 91 Th17-related genes in CD4+ T lymphocytes from patients with MS during a relapse or during a remitting phase. We also compared our findings with those of healthy controls. After confirmation in a validation cohort, expression of SMAD7 and S1PR1 mRNAs was decreased in remitting disease (–2.3-fold and –1.3-fold, respectively) and relapsing disease (–2.2-fold and –1.3-fold, respectively). No differential expression was observed for other SMAD7-related genes, namely, SMAD2, SMAD3, and SMAD4. Under-regulation of SMAD7 and S1PR1 was also observed in another autoimmune disease, Crohn’s disease (CD) (−4.6-fold, -1.6-fold, respectively), suggesting the presence of common markers for autoimmune diseases. In addition, expression of TNF, SMAD2, SMAD3, and SMAD4 were also decreased in CD (–2.2-fold, –1.4-fold, –1.6-fold, and –1.6-fold, respectively). Our study suggests that expression of SMAD7 and S1PR1 mRNA in blood samples are markers for MS and CD, and TNF, SMAD2, SMAD3, and SMAD4 for CD. These genes could prove useful as markers of autoimmune diseases, thus obviating the need for invasive methods.
Keywords: TGF-β signaling, Th17 response, multiple sclerosis, inflammatory bowel disease
1. Introduction
Human T helper 17 (Th17) cells, together with Th1 cells, play a role in the immunopathogenesis of several human inflammatory diseases, such as multiple sclerosis (MS), Crohn’s disease (CD), rheumatoid arthritis, and psoriasis [1,2,3]. Th17 cells are considered proinflammatory cells and produce cytokines including IL17-A, IL17-F, TNF, IL-22, IL-6, and IL-21 [4]. The TGF-β pathway is a major regulator of T-cell differentiation that governs the Th1/Th17 decision in autoimmune inflammation of the central nervous system (CNS). TGF-β–mediated Th17 differentiation is transmitted via SMAD-dependent or non-SMAD pathways [5,6]. SMAD7 negatively regulates phosphorylation of the SMAD2/SMAD3 complex, which is necessary for TGF-β signaling [7,8]. Thus, TGF-β, along with other proinflammatory cytokines, such as IL-1β, IL-6, and IL-23 are inducers of human Th17 differentiation [9]. However, TGF-β is considered an anti-inflammatory factor, and the role of TGF-β in the differentiation of Th17 cells remains unclear. In this sense, various roles have been ascribed to subsets of Th17, such as the very pathogenic Th1-like Th17 cells expressing interferon (IFN) [10] and the anti-inflammatory regulatory Th17 cells [11]. A low TGF-β level supports the generation of inflammatory Th17 cells, while a high level increases the generation of regulatory Th17 cells [12]. Furthermore, it has been suggested that this regulation could be driven by the cytokine CCL2 [13].
MS is an autoimmune disease that causes inflammation and neurodegeneration in the CNS. During the course of the disease, patients usually experience acute exacerbations of inflammation (relapses) and periods of stable disease (remission). Th17 cells promote blood-brain barrier disruption, thus altering traffic and inducing chronic inflammation, which in turn leads to the degradation of myelin sheaths and axonal injury [14]. Th17 cells and their pro-inflammatory cytokines are involved in most of the autoimmune disorders affecting the CNS [12,15].
IL-17A is over-expressed in brain lesions in MS patients and in experimental autoimmune encephalomyelitis (EAE), a murine model of MS [16]. Preferential recruitment of pathogenic Th17 expressing IFN and IL-17 through the blood-brain barrier has been shown in EAE [17]. In addition, S1PR1, a regulator of lymphocyte egress from lymphoid organs into systemic circulation has also been associated with EAE due to Th17 activation via IL-6 [18].
Identifying markers of the Th17 response in MS is difficult because the damaged area is not easily accessible. However, compared with healthy donors, MS patients were found to have a higher proportion of Th17 cells among CD4+ T cells and higher serum IL-17 and IL-23 levels in peripheral blood [19]. Our objective was to compare the differential expression of a set of Th17-related genes in CD4+ T lymphocytes between MS patients during relapse and remission and healthy donors. We also aimed to validate the results in CD.
2. Results
2.1. Patients Characteristics
One hundred subjects were included in the study and distributed in four groups: Remittent recurrent multiple sclerosis (RRMS) during a relapse (n = 43), RRMS during a remitting phase (n = 21), healthy donors (n = 20), and Crohn’s disease (CD) during a relapse (n = 16). The patients’ characteristics are shown in Table 1. The main differences between the groups were the higher proportion of men, longer time from diagnosis to sample collection, and the absence of treatment-naïve patients in the CD group.
Table 1.
Patients characteristics.
| Characteristics | RRMS Rel (n = 43) | RRMS Rem (n = 21) | HD (n = 20) | CD (n =16) |
|---|---|---|---|---|
| Median age, years (IQR, range) |
35 (14; 22–53) |
40 (14.5; 25–52) |
29.5 (17.5; 25–61) |
28.5 (21.3; 18–69) |
| Gender Women n, (%) Men n, (%) |
38 (88.4%) 5 (11.6%) |
19 (90.5%) 2 (9.5%) |
16 (80%) 4 (20%) |
7 (43.8%) 9 (56.2%) |
| Months from diagnosisto sample collection, median (IQR, range) |
9 (144; 0–36) |
12 (70.5; 0–216) |
192 (231; 1–456) |
|
| Type of MS/CIS MS n, (%) CIS n, (%) |
33 (76.7%) 10 (23.3%) |
13 (61.9%) 8 (38.1%) |
||
| Naïve Yes n, (%) No n, (%) |
24 (55.8%) 19 (44.2%) |
16 (76.2%) 5 (23.8%) |
0 16 (100%) |
|
| Type of treatment in non-naïve (n) | ||||
| Interferon beta-1a sc | 4 | 2 | ||
| Glatitamer Acetate | 5 | 1 | ||
| Interferon beta-1b | 2 | 1 | ||
| Azathioprine | 1 | 1 | ||
| Interferon beta-1b im | 5 | |||
| Methylprednisolone | 1 | |||
| Dimethyl fumarate | 1 | 1 | ||
| Adalimumab | 5 | |||
| Adalimumab + azathioprine | 2 | |||
| Infliximab | 1 | |||
| Vedolizumab | 4 | |||
| Certolizumab + Methylprednisolone | 1 | |||
| Azathioprine + Prednisone | 1 | |||
| Prednisone | 1 |
RRMS, remitting-recurrent multiple sclerosis; rel, relapsing; rem, remitting; HD, healthy donor; CD, Crohn’s disease; MS, multiple sclerosis; CIS, clinically isolated syndrome; mAb, monoclonal antibody; sc, subcutaneous; im, intramuscular.
2.2. StellARrays
Among 95 Th17-related genes, we found that the only statistically significant differences were observed for expression of SMAD7, which decreased by –4.12-fold in RRMS patients during a relapse compared with healthy donors (p = 0.023) (Table 2) (see also Table A1 in Appendix A). We decided to test the top three genes. CSF3 was ruled out because it had very low expression and the melting curve showed the amplification of multiple fragments. Thus, SMAD7, TNF, and S1PR1 were selected for further analysis.
Table 2.
Top ten* differentially expressed genes in healthy donors versus RRMS patients with respect to Th17-related genes included in the Human T helper 17 (Th17) 96 StellARray qPCR Array.
| Rank | Gene Name | p-Value | GPR Fold Change |
|---|---|---|---|
| 1 | SMAD7 | 0.023116 | −4.121045 |
| 2 | TNF | 0.097984 | −1.975738 |
| 3 | CSF3 | 0.115770 | 3.099492 |
| 4 | S1PR1 | 0.169676 | −1.489735 |
| 5 | CEBPD | 0.173658 | 2.447022 |
| 6 | IL18R1 | 0.181638 | 1.741877 |
| 7 | MMP9 | 0.183453 | 2.454003 |
| 8 | ICAM1 | 0.204162 | −1.397425 |
| 9 | IL10 | 0.217187 | −1.478354 |
| 10 | MAP3K14 | 0.227840 | −1.232434 |
* Excluded genes used for normalization; GPR, Global Pattern Recognition.
2.3. Gene Expression of Th17-Selected Genes in MS
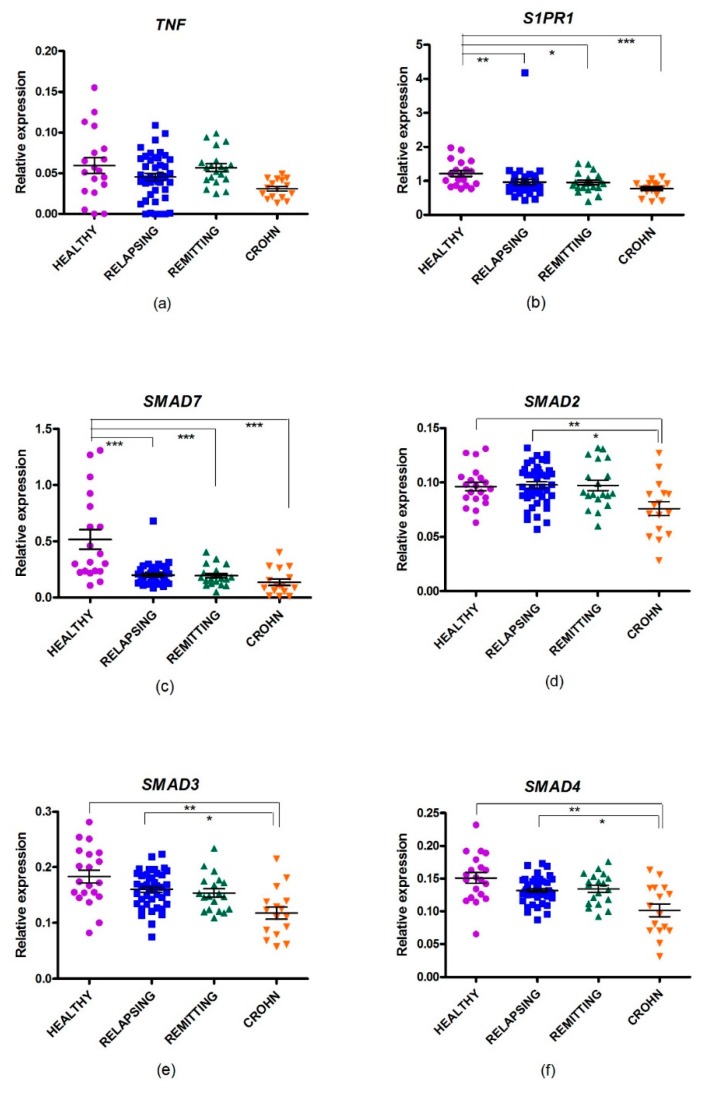
Gene expression of the previously selected genes was analyzed in the four groups of individuals (Figure 1, Table 3). Comparison of healthy donors (HD) with the RRMS relapse group showed that mRNA expression of SMAD7 was decreased in the last group (–2.17-fold, PFDR = 0.001), thus confirming the differential expression of SMAD7 between the groups, as previously observed in the StellARrays approach (Figure 1). In addition, the trend toward differential expression of S1PR1 observed in the StellARrays was confirmed, albeit now with a statistically significant result. S1PR1 was expressed –1.31-fold in RRMS patients during a relapse compared with HD (PFDR = 0.010).
Figure 1.
Relative expression of selected genes in multiple sclerosis patients in a relapsing or a remitting phase, in relapsing Crohn’s disease, and in healthy donors. (a) TNF; (b) S1PR1; (c) SMAD7; (d) SMAD2; (e) SMAD3; and (f) SMAD4. * p < 0.05; ** p < 0.01; *** p < 0.001.
Table 3.
Statistical analysis and ratios of differential gene expression between groups.
| Comparison |
SMAD7 FC (PFDR) |
S1PR1 FC (PFDR) |
TNF FC (PFDR) |
SMAD2 FC (PFDR) |
SMAD3 FC (PFDR) |
SMAD4 FC (PFDR) |
|---|---|---|---|---|---|---|
| HD vs. RRMS rem | −2.29 (0.001) |
−1.28 (0.041) |
−1.19 (0.405) |
−1.28 (0.870) |
−1.17 (0.720) |
−1.12 (0.155) |
| HD vs. RRMS rel | −2.17 (0.001) |
−1.31 (0.010) |
−1.34 (0.123) |
−1.05 (0.372) |
−1.18 (0.130) |
−1.17 (0.110) |
| HD vs.CD rel | −4.65 (0.001) |
−1.59 (0.001) |
−2.25 (0.001) |
−1.40 (0.001) |
−1.64 (0.001) |
−1.60 (< 0.001) |
| RRMS rem vs. RRMS rel | 1.05 (1.000) |
−1.02 (1.000) |
−1.13 (0.690) |
1.02 (1.000) |
1.01 (1.000) |
−1.04 (1.738) |
| RRMS rem vs. CD rel | −2.02 (0.101) |
−1.24 (0.158) |
−1.89 (0.003) |
−1.3 (0.008) |
−1.40 (0.023) |
−1.43 (0.004) |
| RRMS rel vs. CD rel | −2.14 (0.046) |
−1.22 (0.137) |
−1.67 (0.006) |
−1.33 (0.006) |
−1.38 (0.016) |
−1.40 (0.006) |
HD, healthy donor; RRMS, remitting-recurrent multiple sclerosis; CD, Crohn’s disease; rel, relapsing; rem, remitting; PFDR, P value false discovery range.
Comparison of HD with RRMS remitting patients showed that SMAD7 (–2.29-fold, PFDR = 0.001) and S1PR1 (–1.28-fold, PFDR = 0.041) were decreased in RRMS. However, there were no differences in the expression of the three genes analyzed when remitting RRMS patients were compared with relapsing RRMS patients
2.4. Gene Expression of SMAD Genes in MS
We then wanted to assess whether other members of the SMAD pathway were differentially expressed. SMAD7 inhibits phosphorylation of SMAD2 and SMAD3, thus blocking TGF-β signaling. SMAD4 binds to phosphorylated SMAD2/3, and the resulting complex drives gene expression in the nucleus [7]. Changes in expression of SMAD2, SMAD3, and SMAD4 were analyzed in RRMS during remission and relapse compared with HD (Figure 1, Table 1). No changes were detected in RRMS patients during either relapse or remission. However, a non–statistically significant trend for weak under-expression of SMAD4 was observed in RRMS during relapse compared with HD.
2.5. Changes in Gene Expression in CD
All of the genes analyzed were evaluated in relapsing CD patients (Figure 1, Table 3). A population of CD patients during a relapse was not included due to the absence of differential gene expression observed between MS patients in a relapse or in remission. The expression of all of them was decreased when compared with the HD group. SMAD7 was the most repressed gene (−4.65-fold, PFDR = 0.001), followed by S1PR1 (−1.59-fold, PFDR = 0.001), TNF (−2.25-fold, PFDR = 0.001), SMAD3 (−1.64-fold, PFDR = 0.001), SMAD4 (−1.60-fold, PFDR < 0.001), and SMAD2 (−1.40-fold, PFDR = 0.001).
Comparison between MS and CD patients showed that expression of SMAD7, TNF, SMAD2, SMAD3, and SMAD4 was lower in CD patients (Table 3).
3. Discussion
The dysregulation of the balance between pro-inflammatory Th17 cells and anti-inflammatory regulatory T (Tregs) cells has an important yet elusive role in many autoimmune diseases. This balance depends on several factors, many of which are still to be elucidated. One such factor is TGF-β, which participates in induction of Th17 and Treg cells. This differential role seems to be tissue-dependent, and TGF-β expression seems to be level-dependent [20]. Measurement of Th17-related genes in local lesions could be of immense interest for identification of new biomarkers in autoimmune diseases. However, in MS, the inaccessibility of the lesions requires an alternative approach. Even in diseases such as CD, with easy, yet invasive, access to biopsies through endoscopies, accessing samples for monitoring the outcome of these parameters is subject to ethical requirements.
In this study, circulating CD4+ T lymphocytes were screened for changes in expression of Th17-related genes. The genes selected were then tested in CD patients to assess whether these changes were common to other autoimmune diseases and could serve as potential biomarkers. This approach has been used successfully in human systemic lupus erythematosus, which is also an autoimmune disease, and in CD4+ T cells differentiating to Th17 in the presence of an inhibitor of Th17 response [21,22].
In our study, S1PR1 and SMAD7 mRNA levels were downregulated peripherally in CD4+ T lymphocytes of RRMS patients, both during relapse and in stable disease. These genes were also downregulated in the acute phase of CD.
Sphingosine-1 phosphate (S1P) signaling has an essential role in the regulation of lymphocyte egress from lymphoid nodules to peripheral blood [23]. Lymphocyte egress is dependent upon an S1P gradient between the lymphoid tissue (lower concentration) and plasma (higher concentration). Inhibition of this signaling by an S1P1R agonist, such as fingolimod, has been effective in MS. Fingolimod reduces circulating T lymphocytes (including autoreactive ones), protects from neuroinflammation by blocking the effect of S1PR1 expression in astrocytes [24], and helps to regulate the blood-brain barrier [25]. In rats, a positron emission tomography imaging study showed how S1PR1 is upregulated in the lumbar spinal cord of EAE rats and associated with glial cell activation and immune cell infiltration [26]. It has been suggested that fingolimod might exert its action via this dual central-peripheral mechanism [27] and also through modulation of the Treg/Th17 cell balance by regulation of the Akt-mTOR and MAPK/ERK pathways [28]. Consistent with our results, in systemic lupus erythematosus, S1PR1 is expressed less in peripheral blood mononuclear cells (PBMCs) of patients than in healthy controls [21,29,30].
As previously mentioned, SMAD7 negatively regulates phosphorylation of the SMAD2/SMAD3 complex [7,8]. This inhibition increases expression of IL2, a negative regulator of Th17 differentiation, thus leading to inflammation [31]. Unfortunately, the effect of the decrease of SMAD7 gene expression on SMAD2/SMAD3 phosphorylation could not be studied in our patients because no protein extract was collected from CD4+T cells. In agreement with the lower SMAD7 levels in CD4+ T cells from the peripheral blood of MS patients observed in the present work, Zhang et al. found values for the microRNA miR-181, a SMAD7 inhibitor, to be increased in the same cells of MS patients, thus suggesting an increase in TGF-β-mediated Th17 differentiation [32]. In the same way, Meoli et al. showed that SMAD7 was downregulated in the CD4+ T lymphocytes of RRMS patients and that TGF-β regulates overexpression of SMAD7 [33]. Other studies based on PBMCs of MS patients underpin our findings for downregulation of SMAD7 [34,35]. However, contradictory results were reported by other authors, who found that SMAD7 was upregulated in the CD4+ T lymphocytes of patients with RRMS during a relapse compared with remitting patients or healthy donors [36]. Our results fully agree with those obtained by Meoli et al. and Zhang et al. and support the downregulation of SMAD7 in the CD4+ T cells of MS patients.
The opposite is observed in inflamed tissue. SMAD7 is overexpressed in brain lesions in EAE mice, in human MS [37], and in the intestine of CD patients [38]. Furthermore, in patients with inflammatory bowel disease and high concentrations of SMAD7 in the intestine, an antisense RNA has proven successful against SMAD7 [39,40,41]. The high S1PR1 and SMAD7 levels reported in inflammatory lesions in MS and other autoimmune diseases and our finding that those levels were suppressed in peripheral blood, points to enrichment of highly expressed SMAD7 cells at inflammation sites and, in parallel, enrichment of poorly expressed SMAD7 CD4+ T cells in peripheral blood. In addition, the recently described association of the intestinal Smad7 expression with multiple sclerosis in a murine model suggests a relevant role of SMAD7 in these autoimmune diseases [42].
SMAD7 expression is induced by TNF inhibiting TGF-β signaling [43]. In this regard, we found a trend toward downregulation of TNF in the CD4+ T cells of MS patients and that this was more pronounced in CD patients. The trend was statistically significant when CD patients were compared with MS patients and was consistent with SMAD7 expression.
SMAD7 was the only SMAD gene downregulated in MS. This finding is in agreement with those of a previous study, in which SMAD2, SMAD3, SMAD4, and SMAD7 were measured in methylprednisolone-treated RRMS patients [44]. However, in CD, we found downregulation of SMAD2, SMAD3, and SMAD4, in addition to SMAD7, probably owing to more pronounced downregulation of SMAD transcription in CD than in MS, although this hypothesis requires further investigation.
The consequences of SMAD regulation are not altogether clear, because TGF-β has a dual role in inflammation by promoting anti-inflammatory Tregs, as well as pro-inflammatory Th17 cells [45]. In fact, all T helper subsets have shown this dual pathogenic and protective role.
Finally, we investigated the expression of Th17 genes. It is known that IL17F levels are higher in RRMS patients than in healthy donors and that these levels can discriminate between different phenotypes of MS [46]. However, in our cohorts, no differences were found for IL17A or IL17F, probably owing to the low expression level observed in our samples. Recently, IL22, which also codes for a classical Th17 cytokine, has drawn attention. It is not co-expressed with IL17A in the CD4+ T lymphocytes of MS patients [47], and IL22 mRNA is upregulated in circulating cells of relapsing MS patients compared with remitting patients and healthy donors. Our results for the Th17 gene panel support both findings, albeit without statistical significance. IL22 mRNA was expressed two-fold higher in RRMS patients during a relapse than in healthy donors. However, this gene was not selected for further analysis and confirmation because of its high p-value, suggesting that Th populations other than Th17, such as Th22, could play a role in MS. Consequently, they cannot be ruled out.
The apparent inverse correlation of SMAD7 and S1PR1 expression in peripheral CD4+ T cells and at sites of inflammation of patients with immune-mediated diseases could help to obtain information about inflamed sites using a simple blood extraction, a non-invasive method, instead a biopsy. As a limitation of the study, CD patients were more heterogeneous than MS patients in terms of concomitant medication with putative influence in gene expression. In addition, a group with non-active CD was not included because of no differences were observed between relapsing or remitting MS patients.
A regulatory effect of CCL2 in TGF-β regulation and in the balance between Th1/Th17 and Treg in EAE mice has been suggested [13]. However, in our human cohort no differential expression was recorded for CCL2 or its receptor CCR2. Given that the regulatory effect was observed after administering a very low dose of CCL2, it could be interesting to analyze CCL2 expression in larger cohorts.
In summary, we found that expression of SMAD7 and S1PR1 in CD4+ T cells in peripheral blood were biomarkers of MS and CD. In addition, TNF, SMAD2, SMAD3, and SMAD4 were downregulated in CD.
4. Materials and Methods
4.1. Patients
Samples from patients were provided by the Neurology and Gastroenterology Departments of Hospital General Universitario Gregorio Marañón, Madrid, Spain. Samples were processed immediately upon reception. The inclusion criteria were as follows: a diagnosis of MS or clinically isolated syndrome either during relapse or remission; Crohn’s disease during an acute phase. MS disease activity was defined as any new symptoms or worsening of pre-existing neurologic symptoms lasting more than 24 h after a period of 30 days of improvement or stability in the absence of infection or fever. This study followed the Declaration of Helsinki and was approved by the Gregorio Marañón Hospital ethics committee. All patients signed a written informed consent. The demographic and clinical data collected included sex, age, type of multiple sclerosis, and treatment status (naïve or active treatment).
4.2. Isolation and Culture of CD4+ T cells
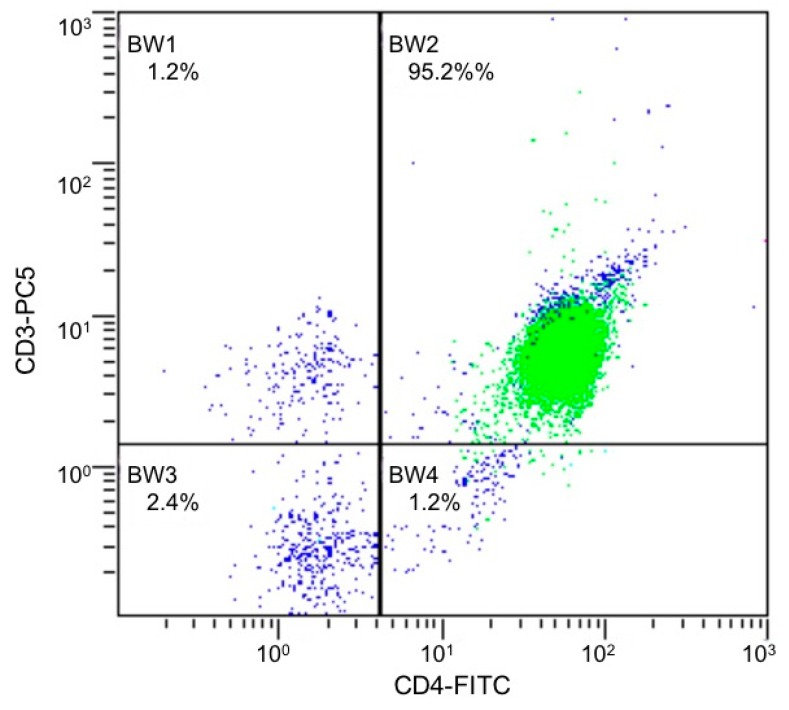
CD4 T-cells were negatively selected from fresh PBMCs as described by de Andrés et al. [44]. Purity of CD4+ T cells was >95%, as measured by flow cytometry (Figure 2).
Figure 2.
Flow cytometry graph of CD4+ T cells isolated from PBMCs from a representative patient. CD4+/CD3+ represent 95.2%. FICT, fluorescein isothiocyanate; PC5, phycoerythrin-cyanine5. In green, gate of lymphocytes; in blue, total cells.
4.3. Isolation of RNA and Synthesis of cDNA
RNA was isolated from CD4+ T cells, measured and integrity verified as described [44]. When necessary, RNA was concentrated using Concentrator 5301 (Eppendorf, Hamburg, Germany). Good quality samples (RNA integrity number > 8 were selected. cDNA was generated from 1000 ng (for StellARrays) or 500 ng (for qRT-PCR) of total RNA as described [44].
4.4. StellARrays Procedure
cDNAs from 6 RRMS patients during a relapse and 6 healthy donors were generated as described elsewhere. Samples were pooled in pairs. Each pair was used to measure the expression of Th17-related genes using a single Human T helper 17 (Th17) 96 StellARray qPCR Array (Bar Harbor Biotechnology, Inc., Trenton, ME, USA). Real-time qRT-PCR was performed using FastStar Universal SYBR Green Master (Roche Applied Science, Penzberg, Germany) in a StepOnePlus system (ThermoFisher Scientific, Waltham, MA, USA) following the StellARrayTM qPCR instructions. Data were analyzed using the Global Pattern Recognition (GPR) algorithm, and the genes used for normalizations were HS18, NFATC2, CEBPB, STAT4, IL17RA, TRAF3IP2, GATA3, STAT3, TRAF6, and NFKB1 [48].
4.5. Real Time qRT-PCR
Changes in the selected genes were quantified by qRT-PCR in 20 RRMS patients in remission, 19 healthy donors (HD), 42 RRMS patients during a relapse, and in 16 Crohn’s disease patients during a relapse. Real-time PCR was performed in triplicate using 2 μL/well of a dilution performed for 1/10 of each cDNA (0.04 μM) for SMAD7, TNF, S1PR1, SMAD2, SMAD3, SMAD4, GAPDH, and HPRT1 (SMAD7 forward, 5′-ACC CGA TGG ATT TTC TCA A-3′; SMAD7 reverse, 5′-AGG GGC CAG ATA ATT CGT TC’; TNF forward, 5′-CAG CCT CTT CTC CTT CCT GAT-3′; TNF reverse, 5′-GCC AGA GGG CTG ATT AGA GA-3′; S1PR1 forward, 5′-AAC TTC GCC CTG CTT GAG-3; S1PR1 reverse, 5′-TCC AGG CTT TTT GTG TAG CTT-3′; SMAD2 forward, AAA GGG TGG GGA GCA GAA TA; SMAD2 reverse, GAA GTT CAA TCC AGC AAG GAG T; SMAD3 forward, CCA TCC CCG AAA ACA CTA AC; SMAD3 reverse, TCC ATC TTC ACT CAG GTA GCC; SMAD4 forward, CCT GTT CAC AAT GAG CTT GC; SMAD4 reverse, GCA ATG GAA CAC CAA TAC TCA G; GAPDH forward, 5′-AGC CAC ATC GCT CAG ACA C-3′, GAPDH reverse, 5′-GCC CAA TAC GAC CAA ATC C-3′, HPRT1 forward, 5′-GAC CAG TCA ACA GGG GAC AT-3′, HPRT1 reverse, 5′-GTG TCA ATT ATA TCT TCC ACA ATC AAG-3′), 1× SYBR Green PCR Master Mix (Roche Applied Science, Penzberg, Germany) as described [44]. GAPDH and HPRT1 and were used as normalization genes [49]. The results were analyzed using the Relative Quantification app in the Thermo Fisher cloud (Applied Biosystems, Foster City, CA, USA). Relative expression values were represented on graphs using GraphPrism 5.1. The t test was applied for comparisons between groups, and the false discovery rate (FDR) was used to correct multiple testing with a confidence level of 95% and maximum Ct of 35. Efficiency was calculated for each primer pair probe and used for correction.
5. Conclusions
Transcription of SMAD7 and S1PR1 is decreased in the peripheral blood CD4+ T lymphocytes of RRMS patients during acute relapses and in remitting phases, and in CD patients compared with healthy donors. These genes could prove useful as markers of autoimmune diseases, thus obviating the need for invasive methods.
Acknowledgments
We are particularly grateful to the study patients for their participation.
Abbreviations
| MS | Multiple sclerosis |
| IBD | Inflammatory bowel disease |
| CD | Crohn’s disease |
| Th17 | Human T helper 17 |
| CNS | Central nervous system |
| IFN | Interferon |
| EAE | Experimental autoimmune encephalomyelitis |
| RRMS | Remittent recurrent multiple sclerosis |
| HD | Healthy donors |
| CIS | Clinically isolated syndrome |
| mAb | Monoclonal antibody |
| Tregs | Anti-inflammatory regulatory T |
| S1PPBMCs | Sphingosine-1 phosphate Peripheral blood mononuclear cells |
| GPR | Global Pattern Recognition |
| FDR | False discovery rate |
Appendix A
Table A1.
Complete differential expression data of genes included in the Human T helper 17 (Th17) 96 StellARray qPCR Array in healthy donors versus RRMS patients.
| Rank | Gene Name | p-Value | GPR Fold Change |
|---|---|---|---|
| 1 | SMAD7 | 0.023116 | −4.121045 |
| 2 | TNF | 0.097984 | −1.975738 |
| 3 | CSF3 | 0.115770 | 3.099492 |
| 4 | S1PR1 | 0.169676 | −1.489735 |
| 5 | CEBPD | 0.173658 | 2.447022 |
| 6 | IL18R1 | 0.181638 | 1.741877 |
| 7 | MMP9 | 0.183453 | 2.454003 |
| 8 | ICAM1 | 0.204162 | −1.397425 |
| 9 | IL10 | 0.217187 | −1.478354 |
| 10 | MAP3K14 | 0.227840 | −1.232434 |
| 12 | IL15 | 0.230006 | 1.914874 |
| 13 | IL12RB2 | 0.230664 | 1.408320 |
| 14 | IL17RC | 0.230917 | −2.236766 |
| 15 | IL22 | 0.236646 | 2.149112 |
| 16 | CCR2 | 0.236910 | 1.298827 |
| 17 | PTGES2 | 0.238409 | 1.286509 |
| 19 | CLEC7A | 0.260678 | 1.849452 |
| 21 | CCR5 | 0.280039 | 1.463742 |
| 22 | IL27 | 0.280070 | 2.076678 |
| 23 | PRKCQ | 0.280210 | −1.203029 |
| 24 | PTGS2 | 0.287147 | −1.292593 |
| 25 | CCL5 | 0.288848 | 1.868512 |
| 30 | STAT5A | 0.302796 | 1.180038 |
| 31 | TGFB1 | 0.303116 | −1.169076 |
| 32 | IL6R | 0.305101 | −1.075965 |
| 33 | S100A9 | 0.310143 | 2.053510 |
| 36 | ITGAL | 0.312514 | −1.163330 |
| 37 | IL1B | 0.317977 | −1.351223 |
| 38 | IL1R1 | 0.323904 | −1.318258 |
| 39 | CD28 | 0.326297 | −1.154887 |
| 40 | S100A8 | 0.326948 | 2.071241 |
| 42 | IL17RE | 0.333320 | 2.006149 |
| 43 | IFNG | 0.337986 | 1.213963 |
| 44 | IL17C | 0.342972 | 1.415405 |
| 45 | FAS | 0.348147 | 1.289762 |
| 46 | CD4 | 0.354280 | −1.157741 |
| 47 | IL18 | 0.368094 | −1.327924 |
| 48 | IL12RB1 | 0.374193 | −1.151614 |
| 49 | MAF | 0.380837 | −1.055353 |
| 50 | LIF | 0.380977 | −3.1015170 |
| 51 | RORC | 0.385640 | 1.290307 |
| 52 | JAK2 | 0.388477 | 1.082868 |
| 53 | IL17D | 0.389396 | −1.094554 |
| 54 | IL23A | 0.392817 | −1.192429 |
| 55 | CXCL2 | 0.400369 | −1.428309 |
| 56 | FOXP3 | 0.409157 | 1.087963 |
| 57 | ICOS | 0.422018 | −1.134658 |
| 58 | IL17RB | 0.430393 | 1.039733 |
| 59 | CSF2 | 0.433523 | 1.478325 |
| 60 | IL23R | 0.444508 | 1.133804 |
| 61 | EOMES | 0.445174 | 1.038471 |
| 62 | CCR4 | 0.446667 | 1.160488 |
| 63 | CCL20 | 0.448723 | −2.306566 |
| 64 | CCR6 | 0.468722 | 1.107297 |
| 65 | SYK | 0.471252 | 1.434643 |
| 66 | ITGAX | 0.482659 | 1.562096 |
| 67 | CD8A | 0.482678 | −1.206777 |
| 68 | SOCS3 | 0.484580 | −1.111256 |
| 69 | LCN2 | 0.502556 | 1.358341 |
| 70 | IL21 | 0.515282 | 1.572404 |
| 71 | IL2 | 0.532927 | 1.719894 |
| 72 | ITGAM | 0.546400 | 1.047556 |
| 73 | FASLG | 0.546997 | 1.224576 |
| 74 | HLX | 0.558206 | 1.118766 |
| 75 | CXCL3 | 0.563145 | 1.062506 |
| 76 | MUC5AC | 0.567032 | 2.128834 |
| 77 | IL13 | 0.568119 | 1.404698 |
| 78 | IL4 | 0.594178 | 1.031688 |
| 79 | CARD9 | 0.599793 | 1.587336 |
| 80 | TBX21 | 0.615106 | 1.006864 |
| 81 | CXCL10 | 0.620610 | 1.085097 |
| 82 | DEFB4 | 0.673370 | −1.204941 |
| 83 | IL17RD | 0.711636 | −2.334528 |
| 84 | GZMB | 0.713332 | 1.303447 |
| 85 | CCL11 | 0.755471 | −1.509432 |
| 87 | MMP3 | NS | −1.328030 |
| 88 | MMP13 | NS | −1.605307 |
| 89 | CCL13 | NS | 1.166980 |
| 90 | CXCL12 | NS | 1.726135 |
| 91 | IL17F | NS | 1.173435 |
| 92 | IL6 | NS | −1.988051 |
| 93 | IL12B | NS | −1.152891 |
| 94 | CRP | NS | −1.287846 |
| 95 | IL17A | NS | −1.526223 |
| 96 | IL25 | NS | 1.342714 |
Genes used for normalization and excluded: HS18, NFATC2, CEBPB, STAT4, IL17RA, TRAF3IP2, GATA3, STAT3, TRAF6, NFKB1, and HS genomic.
Author Contributions
J.M.G.-D. and L.A.L.F. conceived and designed the research. M.I.G., J.A.-Z., A.L.R., and L.A.L.F. performed the experiments; J.A.-Z., M.I.G., A.L.R., I.M.-J., M.L.M.-G., B.L.-C., M.L.M.-B., S.S.-M., M.S.-S., and J.M.G.-D. participate in data acquisition and curation; J.A.-Z., A.L.R., J.M.G.-D., and L.A.L.-F. wrote the original draft. J.M.G.-D. and L.A.L.-F. obtained funds. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Consejería de Educación y Deporte de la Comunidad de Madrid (grant numbers PEJ16/MED/AI-1260 and PEJD-2018-PRE/BMD-8665), and by the Gregorio Marañón Health Research Institute (grant number PRE-2018-2), Fundación GENZYME. The study was cofunded by ERDF Funds (FEDER) from the European Commission, “A way of making Europe”.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Yadav S.K., Mindur J.E., Ito K., Dhib-Jalbut S. Advances in the immunopathogenesis of multiple sclerosis. Curr. Opin. Neurol. 2015;28:206–219. doi: 10.1097/WCO.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 2.Raza A., Yousaf W., Giannella R., Shata M.T. Th17 cells: Interactions with predisposing factors in the immunopathogenesis of inflammatory bowel disease. Expert Rev. Clin. Immunol. 2012;8:161–168. doi: 10.1586/eci.11.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azizi G., Jadidi-Niaragh F., Mirshafiey A. Th17 Cells in Immunopathogenesis and treatment of rheumatoid arthritis. Int. J. Rheum. Dis. 2013;16:243–253. doi: 10.1111/1756-185X.12132. [DOI] [PubMed] [Google Scholar]
- 4.Kuwabara T., Ishikawa F., Kondo M., Kakiuchi T. The Role of IL-17 and Related Cytokines in Inflammatory Autoimmune Diseases. Mediat. Inflamm. 2017;2017:1–11. doi: 10.1155/2017/3908061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., Weaver C.T. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 6.Moustakas A., Heldin C.-H. Non-Smad TGF- signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 7.Budi E.H., Duan D., Derynck R. Transforming Growth Factor-β Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol. 2017;27:658–672. doi: 10.1016/j.tcb.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Akhurst R.J., Hata A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockinger B., Veldhoen M. Differentiation and function of Th17 T cells. Curr. Opin. Immunol. 2007;19:281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Boniface K., Blumenschein W.M., Brovont-Porth K., McGeachy M.J., Basham B., Desai B., Pierce R., McClanahan T.K., Sadekova S., de Waal Malefyt R. Human Th17 cells comprise heterogeneous subsets including IFN-gamma-producing cells with distinct properties from the Th1 lineage. J. Immunol. 2010;185:679–687. doi: 10.4049/jimmunol.1000366. [DOI] [PubMed] [Google Scholar]
- 11.Peters A., Lee Y., Kuchroo V.K. The many faces of Th17 cells. Curr. Opin. Immunol. 2011;23:702–706. doi: 10.1016/j.coi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahmasebinia F., Pourgholaminejad A. The role of Th17 cells in auto-inflammatory neurological disorders. Prog. Neuro-Psychopharmacology Biol. Psychiatry. 2017;79:408–416. doi: 10.1016/j.pnpbp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Kaushansky N., Bakos E., Becker-Herman S., Shachar I., Ben-Nun A. Circulating Picomolar Levels of CCL2 Downregulate Ongoing Chronic Experimental Autoimmune Encephalomyelitis by Induction of Regulatory Mechanisms. J. Immunol. 2019;203:1857–1866. doi: 10.4049/jimmunol.1900424. [DOI] [PubMed] [Google Scholar]
- 14.Kebir H., Kreymborg K., Ifergan I., Dodelet-Devillers A., Cayrol R., Bernard M., Giuliani F., Arbour N., Becher B., Prat A. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 2007;13:1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waisman A., Hauptmann J., Regen T. The role of IL-17 in CNS diseases. Acta Neuropathol. 2015;129:625–637. doi: 10.1007/s00401-015-1402-7. [DOI] [PubMed] [Google Scholar]
- 16.Lock C., Hermans G., Pedotti R., Brendolan A., Schadt E., Garren H., Langer-Gould A., Strober S., Cannella B., Allard J., et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- 17.Kebir H., Ifergan I., Alvarez J.I., Bernard M., Poirier J., Arbour N., Duquette P., Prat A. Preferential recruitment of interferon-γ-expressing T H 17 cells in multiple sclerosis. Ann. Neurol. 2009;66:390–402. doi: 10.1002/ana.21748. [DOI] [PubMed] [Google Scholar]
- 18.Garris C.S., Wu L., Acharya S., Arac A., Blaho V.A., Huang Y., Moon B.S., Axtell R.C., Ho P.P., Steinberg G.K., et al. Defective sphingosine 1-phosphate receptor 1 (S1P1) phosphorylation exacerbates TH17-mediated autoimmune neuroinflammation. Nat. Immunol. 2013;14:1166–1172. doi: 10.1038/ni.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y.-F., Zhang S.-X., Ma X.-W., Xue Y.-L., Gao C., Li X.-Y. Levels of peripheral Th17 cells and serum Th17-related cytokines in patients with multiple sclerosis: A meta-analysis. Mult. Scler. Relat. Disord. 2017;18:20–25. doi: 10.1016/j.msard.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 20.You P., Chen N., Su L., Peng T., Chen G., Liu Y. Local level of TGF-β1 determines the effectiveness of dexamethasone through regulating the balance of Treg/Th17 cells in TNBS-induced mouse colitis. Exp. Ther. Med. 2018;15:3639–3649. doi: 10.3892/etm.2018.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan H.-F., Leng R.-X., Feng C.-C., Li X.-P., Chen G.-M., Li B.-Z., Xu W.-D., Zheng S.G., Ye D.-Q. Expression profiles of Th17 pathway related genes in human systemic lupus erythematosus. Mol. Biol. Rep. 2013;40:391–399. doi: 10.1007/s11033-012-2073-2. [DOI] [PubMed] [Google Scholar]
- 22.Gras R., Relloso M., García M.I., de la Mata F.J., Gómez R., López-Fernández L.A., Muñoz-Fernández M.A. The inhibition of Th17 immune response in vitro and in vivo by the carbosilane dendrimer 2G-NN16. Biomaterials. 2012;33:4002–4009. doi: 10.1016/j.biomaterials.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Healy L.M., Antel J.P. Sphingosine-1-Phosphate Receptors in the Central Nervous and Immune Systems. Curr. Drug Targets. 2016;17:1841–1850. doi: 10.2174/1389450116666151001112710. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann F.S., Hofereiter J., Rübsamen H., Melms J., Schwarz S., Faber H., Weber P., Pütz B., Loleit V., Weber F., et al. Fingolimod induces neuroprotective factors in human astrocytes. J. Neuroinflammation. 2015;12:184. doi: 10.1186/s12974-015-0393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagida K., Liu C.H., Faraco G., Galvani S., Smith H.K., Burg N., Anrather J., Sanchez T., Iadecola C., Hla T. Size-selective opening of the blood–brain barrier by targeting endothelial sphingosine 1–phosphate receptor 1. Proc. Natl. Acad. Sci. USA. 2017;114:4531–4536. doi: 10.1073/pnas.1618659114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Jin H., Yue X., Luo Z., Liu C., Rosenberg A.J., Tu Z. PET Imaging Study of S1PR1 Expression in a Rat Model of Multiple Sclerosis. Mol. Imaging Biol. 2016;18:724–732. doi: 10.1007/s11307-016-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y., Shi D., Cao K., Wu F., Zhu X., Wen S., You Q., Zhang K., Liu L., Zhou H. Fingolimod targets cerebral endothelial activation to block leukocyte recruitment in the central nervous system. J. Leukoc. Biol. 2018;103:107–118. doi: 10.1002/JLB.3A0717-287R. [DOI] [PubMed] [Google Scholar]
- 28.Hou H., Cao R., Quan M., Sun Y., Sun H., Zhang J., Li B., Guo L., Song X. Rapamycin and fingolimod modulate Treg/Th17 cells in experimental autoimmune encephalomyelitis by regulating the Akt-mTOR and MAPK/ERK pathways. J. Neuroimmunol. 2018;324:26–34. doi: 10.1016/j.jneuroim.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Xin Q., Li J., Dang J., Bian X., Shan S., Yuan J., Qian Y., Liu Z., Liu G., Yuan Q., et al. miR-155 Deficiency Ameliorates Autoimmune Inflammation of Systemic Lupus Erythematosus by Targeting S1pr1 in Fas lpr/lpr Mice. J. Immunol. 2015;194:5437–5445. doi: 10.4049/jimmunol.1403028. [DOI] [PubMed] [Google Scholar]
- 30.Shah K., Lee W.-W., Lee S.-H., Kim S.H., Kang S.W., Craft J., Kang I. Dysregulated balance of Th17 and Th1 cells in systemic lupus erythematosus. Arthritis Res. Ther. 2010;12:R53. doi: 10.1186/ar2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cejas P.J., Walsh M.C., Pearce E.L., Han D., Harms G.M., Artis D., Turka L.A., Choi Y. TRAF6 inhibits Th17 differentiation and TGF-beta-mediated suppression of IL-2. Blood. 2010;115:4750–4757. doi: 10.1182/blood-2009-09-242768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Xue Z., Liu Y., Liu H., Guo X., Li Y., Yang H., Zhang L., Da Y., Yao Z., et al. MicroRNA-181c promotes Th17 cell differentiation and mediates experimental autoimmune encephalomyelitis. Brain. Behav. Immun. 2018;70:305–314. doi: 10.1016/j.bbi.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Meoli E.M., Oh U., Grant C.W., Jacobson S. TGF-β signaling is altered in the peripheral blood of subjects with multiple sclerosis. J. Neuroimmunol. 2011;230:164–168. doi: 10.1016/j.jneuroim.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Achiron A., Gurevich M., Friedman N., Kaminski N., Mandel M. Blood transcriptional signatures of multiple sclerosis: Unique gene expression of disease activity. Ann. Neurol. 2004;55:410–417. doi: 10.1002/ana.20008. [DOI] [PubMed] [Google Scholar]
- 35.Bomprezzi R., Ringnér M., Kim S., Bittner M.L., Khan J., Chen Y., Elkahloun A., Yu A., Bielekova B., Meltzer P.S., et al. Gene expression profile in multiple sclerosis patients and healthy controls: Identifying pathways relevant to disease. Hum. Mol. Genet. 2003;12:2191–2199. doi: 10.1093/hmg/ddg221. [DOI] [PubMed] [Google Scholar]
- 36.Kleiter I., Song J., Lukas D., Hasan M., Neumann B., Croxford A.L., Pedré X., Hövelmeyer N., Yogev N., Mildner A., et al. Smad7 in T cells drives T helper 1 responses in multiple sclerosis and experimental autoimmune encephalomyelitis. Brain. 2010;133:1067–1081. doi: 10.1093/brain/awq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghorbani S., Talebi F., Chan W.F., Masoumi F., Vojgani M., Power C., Noorbakhsh F. MicroRNA-181 Variants Regulate T Cell Phenotype in the Context of Autoimmune Neuroinflammation. Front. Immunol. 2017;8:758. doi: 10.3389/fimmu.2017.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monteleone G., Del Vecchio Blanco G., Monteleone I., Fina D., Caruso R., Gioia V., Ballerini S., Federici G., Bernardini S., Pallone F., et al. Post-transcriptional Regulation of Smad7 in the Gut of Patients With Inflammatory Bowel Disease. Gastroenterology. 2005;129:1420–1429. doi: 10.1053/j.gastro.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Monteleone G., Neurath M.F., Ardizzone S., Di Sabatino A., Fantini M.C., Castiglione F., Scribano M.L., Armuzzi A., Caprioli F., Sturniolo G.C., et al. Mongersen, an Oral SMAD7 Antisense Oligonucleotide, and Crohn’s Disease. N. Engl. J. Med. 2015;372:1104–1113. doi: 10.1056/NEJMoa1407250. [DOI] [PubMed] [Google Scholar]
- 40.Ardizzone S., Bevivino G., Monteleone G. Mongersen, an oral Smad7 antisense oligonucleotide, in patients with active Crohn’s disease. Ther. Adv. Gastroenterol. 2016;9:527–532. doi: 10.1177/1756283X16636781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feagan B.G., Sands B.E., Rossiter G., Li X., Usiskin K., Zhan X., Colombel J.-F. Effects of Mongersen (GED-0301) on Endoscopic and Clinical Outcomes in Patients with Active Crohn’s Disease. Gastroenterology. 2018;154:61–64. doi: 10.1053/j.gastro.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 42.Haupeltshofer S., Leichsenring T., Berg S., Pedreiturria X., Joachim S.C., Tischoff I., Otte J.-M., Bopp T., Fantini M.C., Esser C., et al. Smad7 in intestinal CD4+ T cells determines autoimmunity in a spontaneous model of multiple sclerosis. Proc. Natl. Acad. Sci. USA. 2019;116:25860–25869. doi: 10.1073/pnas.1905955116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bitzer M., von Gersdorff G., Liang D., Dominguez-Rosales A., Beg A.A., Rojkind M., Böttinger E.P. A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- 44.De Andres C., García M.I., Goicoechea H., Martínez-Ginés M.L., García-Domínguez J.M., Martín M.L., Romero-Delgado F., Benguría A., Sanjurjo M., López-Fernández L.A. Genes differentially expressed by methylprednisolone in vivo in CD4 T lymphocytes from multiple sclerosis patients: Potential biomarkers. Pharmacogenomics J. 2018;18:98–105. doi: 10.1038/tpj.2016.71. [DOI] [PubMed] [Google Scholar]
- 45.Raphael I., Nalawade S., Eagar T.N., Forsthuber T.G. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine. 2014 doi: 10.1016/j.cyto.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arellano G., Acuña E., Reyes L.I., Ottum P.A., De Sarno P., Villarroel L., Ciampi E., Uribe-San Martín R., Cárcamo C., Naves R. Th1 and Th17 Cells and Associated Cytokines Discriminate among Clinically Isolated Syndrome and Multiple Sclerosis Phenotypes. Front. Immunol. 2017;8:753. doi: 10.3389/fimmu.2017.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muls N., Nasr Z., Dang H.A., Sindic C., van Pesch V. IL-22, GM-CSF and IL-17 in peripheral CD4+ T cell subpopulations during multiple sclerosis relapses and remission. Impact of corticosteroid therapy. PLoS ONE. 2017;12:e0173780. doi: 10.1371/journal.pone.0173780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akilesh S., Shaffer D.J., Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res. 2003;13:1719–1727. doi: 10.1101/gr.533003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickles D., Chen H.P., Li M.M., Khankhanian P., Madireddy L., Caillier S.J., Santaniello A., Cree B.A.C., Pelletier D., Hauser S.L., et al. Blood RNA profiling in a large cohort of multiple sclerosis patients and healthy controls. Hum. Mol. Genet. 2013;22:4194–4205. doi: 10.1093/hmg/ddt267. [DOI] [PMC free article] [PubMed] [Google Scholar]