Abstract
Reaction heterogeneity, poor pH control, and catalyst decomposition in the ring-closing metathesis (RCM) of DNA–chemical conjugates lead to poor yields of the cyclized products. Herein we address these issues with a RCM reaction system that includes a novel aqueous solvent combination to enable reaction homogeneity, an acidic buffer system which masks traditionally problematic functional groups, and a decomposition-resistant catalyst which maximizes conversion to the cyclized product. Additionally, we provide a systematic study of the substrate scope of the on-DNA RCM reaction, a demonstration of its applicability to a single-substrate DNA-encoded chemical library that includes sequencing analysis, and the first successful stapling of an unprotected on-DNA [i, i+4] peptide.
Keywords: DNA-encoded chemistry, DNA-encoded libraries, homogeneous ring-closing metathesis, functional group-tolerant ring-closing metathesis, stapled peptides
Introduction
Since the conception of DNA-encoded combinatorial chemical libraries by Brenner and Lerner,1 many variations of DNA-encoded chemical screening technologies have been developed to aid drug discovery efforts.2 Advancements both in the production of large quantities of synthetic DNA oligomers3 and in high-throughput DNA-sequencing methods4 now allow for rapid and cost-effective screens of vast DNA-encoded chemical library (DECL) collections against biological targets.5 DECLs are single-pot chemical libraries consisting of compounds that each possess a covalently attached, unique DNA sequence “barcode”, which enables identification of binder compounds by DNA sequencing after multiplexed target-based screens. Compared to high-throughput compound collections and screens, DECLs are relatively inexpensive to prepare and use6 and, most importantly, offer the opportunity for deeper exploration of chemical space5 enabled by unprecedented numbers of compounds per DECL—millions to trillions compared to only hundreds of thousands per HTS chemical library.7 Additionally, DECLs have provided starting points for the development of several clinical candidates6,8 and are considered by many to have become one of the pillars of drug discovery.9 However, while large numbers are the main advantage of DECLs, the chemical diversity achievable under its umbrella is limited by the number of effective DNA-compatible reactions. Currently, only a limited set of DNA-compatible, solution-phase chemical reactions has been reported,10−18 and expanding the repertoire of chemical reactions to more effectively sample chemical space is a major goal within this area.
Within the compendium of synthetic methodologies, the ring-closing metathesis (RCM) reaction has become a mainstay for the construction of organic molecules spanning a wide range of structural diversity and complexity. Accordingly, it has been applied across medicinal, natural product, and diversity-oriented synthetic endeavors.19−22 The widespread popularity of RCM is grounded in several reasons: its broad and well-studied functional group and substrate tolerance, the commercial availability of a wide array of air-stable, tunable Ru-based catalysts,23,24 its successful application in aqueous media,25,26 and its relevance to the production of drug-like compounds27,28 as well as novel molecular frameworks for the probing of unexplored chemical space.22 DECL productions are based on combinatorial chemistry, and the RCM reaction would therefore be a significant enhancement to the technology. Stimulated by our interest in combining chemical diversity and large numbers for drug discovery, we became interested in the application of RCM to the production of DECLs. Our work was not done in a vacuum, however, and the precedents upon which we built need mentioning.
The modification of proteins using the cross-metathesis (CM) reaction29 has been demonstrated in aqueous tert-butanol, using a high excess (10,000 equiv) of MgCl2 as a Lewis acidic masking agent. Additionally, RCM-stapling of unprotected peptides has been achieved in water using the water-soluble AquaMet catalyst, also in the presence of MgCl2 (400 equiv).30 The Mg2+ ion is believed to act as a mild Lewis acid, masking coordinating functional groups typically prevalent in biomolecules.29,31 Finally Lu et al.32 reported the use of a third generation Grubbs catalyst/MgCl2/aq. tert-butanol system for the on-DNA RCM and cross metathesis (CM) reactions. While the latter study established that RCM could be achieved on DNA–chemical conjugates, the reported conditions were not adequate, in our hands, for the use of RCM in a DECL production.
The addressed limitations of the previously reported work32 include a) the insolubility of the Ru catalyst used in aqueous tert-butanol, b) the tendency of phase separation between high-salt (MgCl2) aqueous solutions and tert-butanol leading to heterogeneity, c) the absence of pH control, d) a narrowly explored substrate scope, and e) the formation of significant amounts of side products arising from catalyst decomposition, which limits the yield of the reaction. Furthermore, we demonstrate 1) the applicability of the developed conditions to a single-substrate DECL as a way to test for susceptibility to varying DNA sequences, 2) the maintenance of DNA integrity post-RCM via sequencing analysis, and 3) the first successful stapling of an unprotected on-DNA [i, i+4] peptide as an illustration of the robust functional group tolerance of the developed conditions.
Results and Discussion
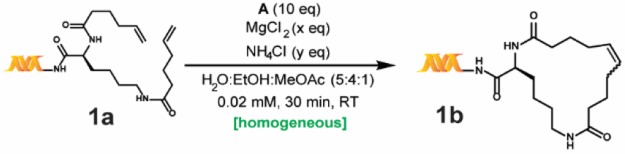
Our work relies on three key findings: 1) a solvent system that allows for homogeneous reaction conditions with much lower catalyst loading, 2) a nonclassical acidic “buffer” that not only avoids basicity but also masks coordinating functional groups—in addition to the effect of MgCl2, and 3) the use of a decomposition-resistant catalyst, B, that minimizes side reactions and maximizes conversion. Our developed reaction conditions are presented in Scheme 1, and our studies toward its development and application are discussed in the following sections.
Scheme 1. Reaction Conditions and Performance Summary of This Work versus Previously Reported Work.
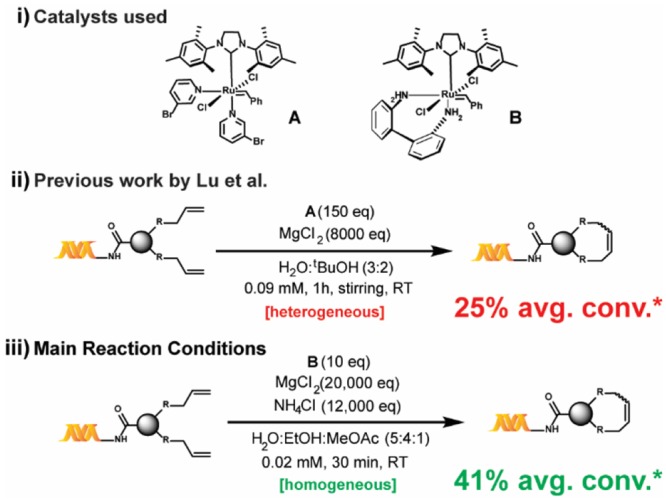
(i) Grubbs third generation catalyst A and its 2,2′-biphenyldiamine derivative B; (ii) previously reported conditions for the on-DNA RCM and CM reactions; (iii) our main conditions for the on-DNA RCM and CM reactions; *average percent conversion for the investigated substrate scope (22 substrates).
Development of a Solvent System for Reaction Homogeneity
To enhance the reproducibility of RCM on substrates with variably sized DNA tags at a range of scales, our initial work focused on developing a fully homogeneous catalyst–aqueous solvent system combination. Unfortunately, many conventional RCM catalysts (e.g., Grubbs I, Grubbs II, Hoveyda–Grubbs I, Hoveyda–Grubbs II, Grela) are extremely insoluble in most aqueous alcoholic mixtures and were ultimately found to be unproductive for on-DNA RCM under a variety of conditions. Ammonium-functionalized RCM catalysts (e.g., Aquamet)30,33 have high solubility in aqueous solutions but were also unproductive for on-DNA RCM, presumably due to DNA–ammonium interactions.34 Even the fast-initiating Grubbs III catalyst A (Scheme 1) utilized in the previously reported on-DNA RCM exhibits limited solubility in alcohols (soluble up to 1 mM in pure tBuOH). In addition, most nonalcoholic water-miscible solvents (e.g., 1,2-dimethoxyethane or 1,4-dioxane) are metal-coordinating and generally suppress metathesis. Informed by reports of conventional RCM in ethyl acetate as solvent,35 we were pleased to discover methyl acetate as a much better solvent for both catalysts A and B (∼4 mM). When a methyl acetate/ethanol/water solvent mixture was used, fully homogeneous reaction mixtures were observed, even for solutions at high ionic strengths. Due to the fixed solubility of B in methyl acetate, a reaction concentration of only 0.02 mM was possible to maintain adequate solvent percentages. While high dilution favors intramolecular metathesis, it can also stress the catalyst36—a trade-off we attempted to mitigate through the use of the more robust catalyst B. Additionally, a reaction concentration of 0.02 mM would be manageable during DECL production, which is the central motivation behind our efforts.
Development of a Brønsted Acidic “Buffer”
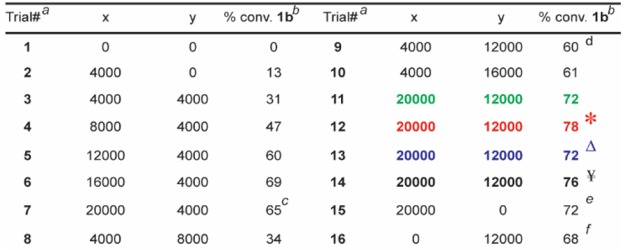
To avoid catalyst degradation at basic pH37 and to enhance the masking of coordinating functional groups, we sought to include an acidic buffering system. Despite numerous reports of RCM in the presence of aqueous buffers,38,39 we found that even modest amounts of common buffers (e.g., phosphate) or “noncoordinating” buffers40,41 (e.g., 2-(N-morpholino)ethanesulfonic acid) inhibited reactivity. Hypothesizing the suppression was due to undesired coordination of the buffer’s nonhalide conjugate bases to the Ru center, we considered whether the inclusion of large amounts of NH4Cl could induce an acidic pH without presenting RCM-suppressing buffer anions. Although not a buffer in the traditional sense, NH4Cl may block Lewis-base interactions via protonation and/or hydrogen-bonding (e.g., a 4 M aqueous solution of NH4Cl has a pH of ∼5). Indeed, as illustrated in Table 1, adding NH4Cl significantly enhances conversion (e.g., entries 2 vs 9). With a homogeneous solvent system capable of tolerating high ionic strength in hand, we optimized the proportions of NH4Cl and MgCl2 for both high conversion and quality of the LC-MS trace. Ultimately a combination of 20,000 equiv of MgCl2 and 12,000 equiv of NH4Cl for 30 min with 10 equiv of catalyst42 was found to be optimal (Table 1).
Table 1. Screen for the Optimal Equivalences of MgCl2 and NH4Clg.
All reactions were run with 1 nmol of 1a.
The percent conversions (% conv.) were determined by LC/MS after the quenching procedure, as described in the Supporting Information.
The MS trace was of higher quality relative to that of Trial 6 despite a lower conversion.
The MS trace was of higher quality relative to Trial 10.
The MS signals for those were of lower quality relative to Trial 10.
*Catalyst B instead of A. ΔCatalyst B instead of A, reaction time 5 min. ¥Catalyst B instead of A, reaction time 60 min.
Application of a Decomposition-Resistant Catalyst
Despite these encouraging results, conversions remained limited by the formation of side products. These appeared to originate in the reduction of one or both olefins, the loss of methylene from one of the olefins and/or the isomerization of the starting material to an unreactive internal olefin (see the Supporting Information). Unfortunately, attempts to suppress those undesired reactions with standard additives such as benzoquinones36 were unsuccessful (various benzoquinones were incompatible with NH4Cl and also degraded DNA). While several catalytic species have been proposed as culprits behind such side reactions,37,43,44 a compelling study by Fogg and co-workers45—showing that the addition of substoichiometric amounts of a poisoning ligand limits isomerization—proved to be key to our discovery of the adequacy of B for our purposes. Indeed, we found that the addition of 2,2′-biphenyldiamine, C, to A, as shown in Scheme 2(ii), enhances the percent conversion (see the Supporting Information). However, we were aware of another study by Fogg and co-workers46 which reported the 40% equilibrium yield of decomposition-resistant catalyst B from A in the presence of C, at RT. We therefore suspected that the in situ formation of B was responsible for the conversion enhancement observed. The substitution of B for A, as shown in Scheme 1(iii), indeed proved to significantly enhance conversion beyond what an A–C combination could achieve (Tables 2–4). Since catalyst B was not commercially available at the time of the study and given its reported synthesis requires a glovebox,46 we also studied alternative reaction conditions involving catalyst A instead (Scheme 2).
Scheme 2. Alternative Reaction Conditions using Catalyst A.
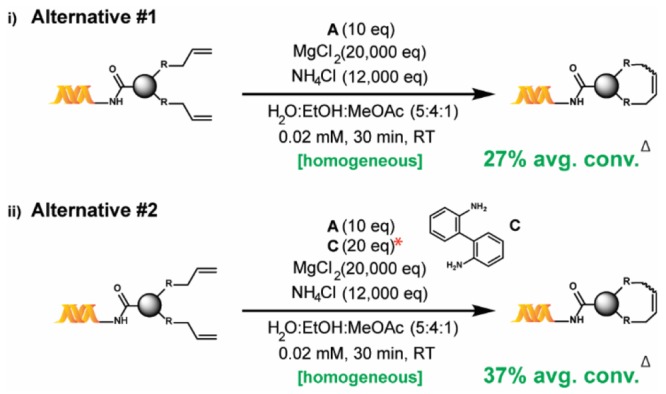
*The screening results used to determine the optimal equivalence of C are provided in the Supporting Information, Table S1. Δ Average percent conversion for the investigated substrate scope (22 substrates).
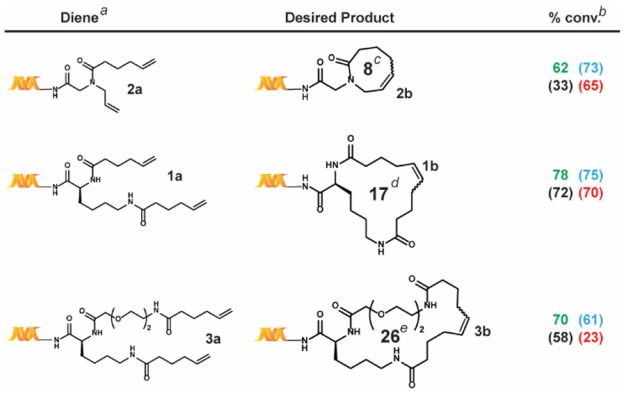
Table 2. Ring Size Scope Study.
All reactions were run with 1 nmol of each diene.
The percent conversions (% conv.) were determined by LC/MS after quenching, as described in the Supporting Information.
Ring sizes in terms of number of member atoms; reaction conditions: green = main reaction conditions, blue = alternative #2, black = alternative #1, and red = Lu et al.
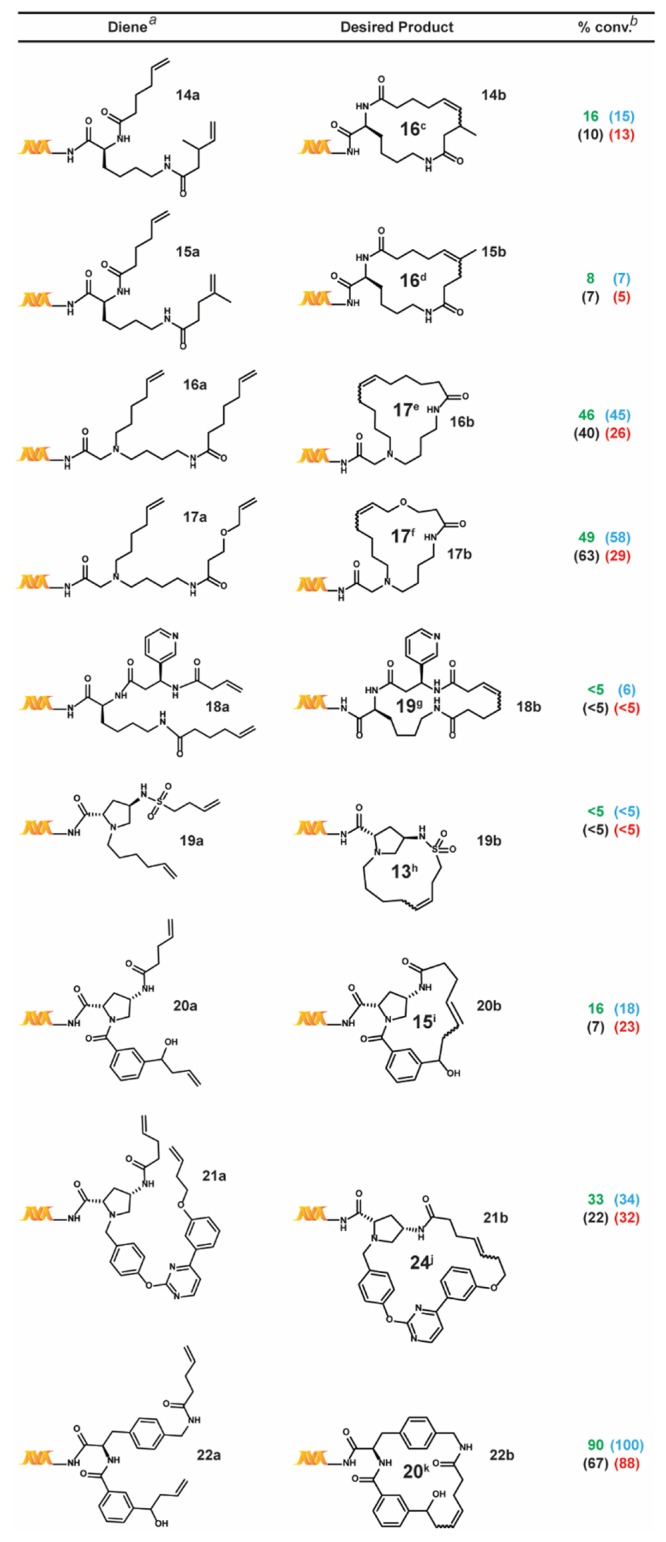
Table 4. Additional Substrate Scope Study.
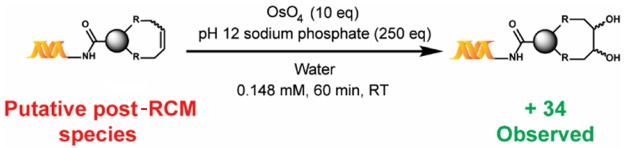
Of note, reactions within DECLs are typically performed on nano- to micromole scales, and mass spectrometry is the main method of characterization. Therefore, to further corroborate the formation of the putative cyclized product, we developed a chemical derivatization protocol through OsO4-mediated dihydroxylation, which reveals the number of olefins present (Scheme 3). Results of the dihydroxylation of the post-RCM reaction mixture of 1a were consistent with the formation of the cyclized product as the major product (see the Supporting Information).
Scheme 3. Confirmation of Ring Formation via Chemical Modification.
We next turned to applying these four conditions—the previously reported conditions from Lu et al. (Scheme 1, (ii), our main reaction conditions (Scheme 1, (iii), and the two alternative conditions (Scheme 2)—to a series of substrate scope studies (Tables 2–4). Overall, while the conditions reported by Lu et al. exhibit comparable conversions for simple substrates, significant differences were observed for those containing unfavorable coordinating functional groups.
We first investigated the ring-size scope of the reaction (Table 2). Although all four conditions generally produced similar results, the tBuOH/MgCl2 system significantly underperformed in the case of 3a. This may stem from the presence of the glycol chain within this substrate. Of note, these reactions were complete after only 30 min at room temperature, which is remarkable given it can take days and high temperatures for rings of comparable size to close in organic media.24
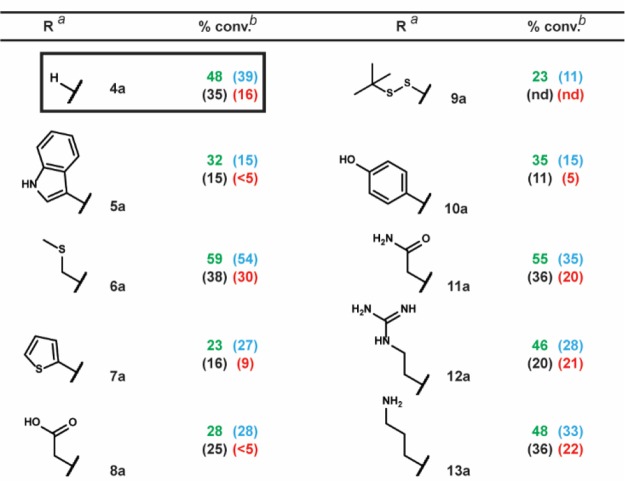
To investigate the functional group tolerance of the four reaction conditions, we synthesized a series of substrates differentiated only by substitution at a single position (Table 3). Using substrate 4a (R = H) as benchmark, we were able to demonstrate the remarkable functional group tolerance of our main reaction conditions. Indeed, functional groups such as the carboxylic acid of 8a or the amine of 13a—groups typically protected in organic-phase RCM—exhibited at least half the conversion obtained for reference compound 4a. This suggests that the incorporation of NH4Cl exerts an acidic masking effect, consistent with other reports of RCM in the presence of acidic additives.47,48 In contrast, the previously reported unbuffered MgCl2/tBuOH condition exhibited significant sensitivity to the indole of 5a, the thiopheneof 7a, the carboxylic acid of 8a, and the phenol of 10a.
Table 3. Functional Group Tolerance Study.
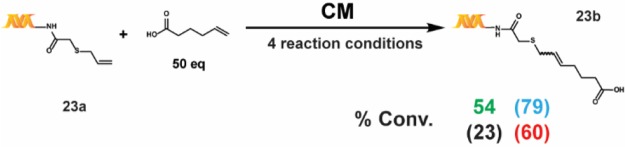
We also tested a series of on-DNA compounds designed to investigate other DECL-relevant substrate features (Table 4). While we had previously observed that internal olefins were unreactive to our RCM conditions, we sought to assess the impact of small substituents in β-methyl 14a and α-methyl 15a. When compared to the analogous unsubstituted 1a, 14a and 15a were cyclized in modest conversions, suggesting a low-tolerance for steric-encumbrance at or near the olefin. The presence of an allylic chalcogen is known to enhance RCM efficiency through a preassociation effect.49 However, comparable conversions were observed for alkyl 16a and related allyl ether 17a. The proximity of coordinating groups to the olefin may also inhibit successful cyclization. Although all four RCM conditions effected negligible cyclization on 18a and 19a—bearing a proximally located pyridyl and sulfonamide, respectively—a modest amount of cyclization was observed for 21a—exhibiting a more distally located pyrimidine. Homoallylic alcohols are a potential substrate series for on-DNA RCM, as the water-compatible allylation of aldehydes is well-known.50 Homoallyl alcohols 20a and 22a were successfully cyclized, although sterically encumbered 20a afforded lower conversion. Finally we applied these conditions to the CM of 5-hexenoic acid with the CM-favored thioether substrate5123a. All four conditions provided the CM product in moderate to good conversion (Scheme 4).
Scheme 4. On-DNA Cross Metathesis (CM).
The reaction was run with 1 nmol of 23a. A 50 mM stock solution of the 5-hexenoic acid building block was prepared in ethanol. The percent conversions (% conv.) were determined by LC/MS after the quenching procedure, as described in the Supporting Information. Reaction conditions: green = main reaction conditions, blue = alternative #2, black = alternative #1, and red = Lu et al.
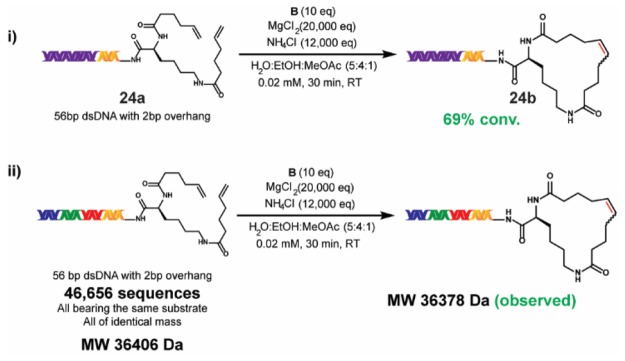
Certain on-DNA reactions exhibit sensitivity to the length of the DNA-tag, which may be due to differences in solubility and/or intermolecular DNA interactions. To test the viability of our new RCM conditions on a late-stage DECL substrate, we prepared 56-bp dsDNA substrate 24a from 17-bp dsDNA substrate 1a. Application of our main reaction conditions (Scheme 1, iii) to 24a afforded 24b at 69% conversion, which was close to the 78% conversion observed for 1a (Scheme 5). After an adapted precipitation procedure to minimize chaotropic effects (see the Supporting Information), the 56-bp dsDNA was obtained in 20% yield (quantified by Bioanalyzer electropherogram analysis). Although the 56-bp dsDNA was the dominant component within the post-RCM reaction mixture, small amounts of larger DNA segments were observed (see the Supporting Information), which may arise from intermolecular metathesis. These may require removal by HPLC during full-scale DECL synthesis.
Scheme 5. Application of the Main Reaction Conditions to a 56-bp DNA Substrate and to a Single-Substrate DECL.
(i)The reaction was run with 5 nmol of 24a. (ii) The reaction was run with 5 nmol of the single-substrate DECL.
The application of newly developed on-DNA conditions to a pilot DECL and subsequent analysis by DNA sequencing are useful tests prior to beginning a full DECL production. To investigate the potential for DNA base-related effects, we prepared a small single-substrate DECL of ∼47,000 unique 56-bp dsDNA sequences from substrate 1a. This DECL was built through a three-cycle “split-and-pool” approach, featuring three cycles of splitting into portions, performing 36 ligations of unique dsDNA tags, and pooling. All DNA tags used within a cycle were of identical molecular weight, and thus the DECL was observed as a single ensemble mass after purification by HPLC (see the Supporting Information). The application of our main reaction conditions to the DECL resulted in the observable loss of MW = 28 (loss of ethylene from RCM cyclization). Additionally, sequenced samples of the DECL before and after the RCM reaction showed no significant differences, thus ascertaining DNA integrity post-RCM (see the Supporting Information).
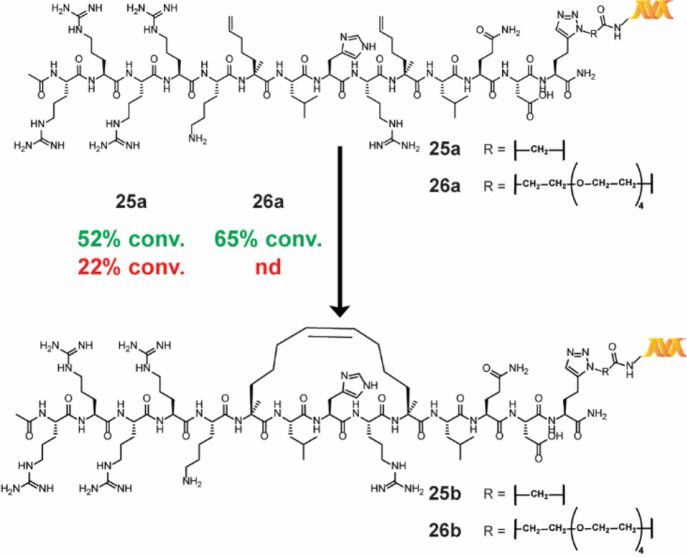
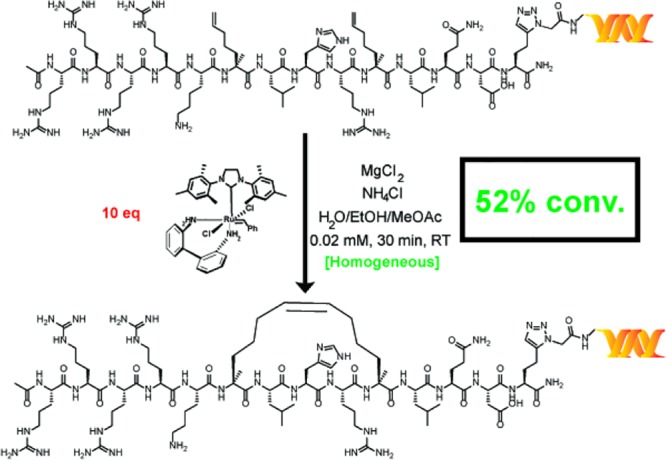
In addition to the synthesis of small-molecule macrocycles, RCM has also been used to prepare stapled peptides—linear peptides conformationally constrained through an intramolecular linkage—to simulate protein–protein interactions.52 To test the relevance of our reaction conditions for the production of on-DNA stapled peptides, we synthesized substrates 25a and 26a through copper-catalyzed azide–alkyne cycloaddition of an alkynylated [i, i+4] SRC stapled peptide precursor known to bind the coactivator region of ERα.53 The application of our main reaction conditions to both substrates provided the stapled peptide in moderate conversions while, in the case of 25a, significantly outperforming the conditions reported by Lu et al. (Scheme 6)—the comparison was not studied for 26a. To further corroborate the successful production of the stapled peptide, adapted samples of a control, peptide 26a, and stapled peptide 26b were tested within a homogeneous time-resolved fluorescence (HTRF) assay for binding to the coactivator region of ERα. Both 26a and 26b displayed a dose-dependent HTRF response, and significantly enhanced coactivator region binding was observed for stapled peptide 26b (see the Supporting Information). It is noteworthy that DECL technology is analogous to phage display libraries.54 However, our methodology is particularly useful for incorporating unnatural amino acids such as the α-methyl-α-pentenyl amino acid required for RCM stapling. Although highly specialized systems for incorporating unnatural amino acids for phage display have been reported,55 there have been no reports of making stapled peptides by phage display.
Scheme 6. RCM Stapling of an Unprotected [i, i+4] Peptide Connected to DNA via Two Different Linkers.
Each reaction was run with 1 nmol of material. The percent conversion (% conv.) was determined by LC/MS after the quenching procedure, as described in the Supporting Information. Reaction conditions: green = main reaction conditions, and red = Lu et al.
Conclusions
In summary, we have developed a RCM reaction system promoting homogeneity, minimization of side reactions, and masking of coordinating functional groups and 1) studied and contrasted its applicability to a diverse range of substrates, 2) ensured its compatibility to DNA through the successful cyclization of a single-substrate DECL, and 3) demonstrated its functional group tolerance through the production of the first on-DNA stapled peptide. We believe this work will allow for the integration of the well-documented capacity of RCM to generate chemical diversity into DECLs. Efforts to apply this work toward the production of RCM-based DECLs and their screening are ongoing within these laboratories and will be reported in due course.
Experimental Procedures
Each RCM substrate was constructed from a 17-bp dsDNA DNA headpiece (see the Supporting Information for structure) and was purified by HPLC due to RCM-inhibiting effects induced by residual chemical building block impurities. Procedural details for all four RCM methods are included in the Supporting Information. Additionally, while reaction mixtures involving 17-bp dsDNA tagged substrates were fully homogeneous, those with longer dsDNA tags (56 bp dsDNA) tended to be slightly cloudy. While this did not appear to affect the reaction conversion, a specific precipitation procedure was required to obtain suitable DNA recovery (see the Supporting Information). For application to substrates with DNA tags of alternative sizes (>56-bp) or composition, adjustments may be required to ensure substrate solubility.
Acknowledgments
We would like to acknowledge Kevin MacKenzie (Baylor College of Medicine—BCM) for NMR spectroscopic assistance, Murugesan Palaniappan (BCM) and Zhifeng Yu (BCM) for naive sequencing assistance, Kevin Riehle (BCM) for bioinformatic analysis assistance, John C. Faver (BCM) for cheminformatic analysis assistance, and the Fogg Research Group (University of Ottawa) for the generous provision of catalyst B. We would also like to acknowledge Srinivas Chamakuri (BCM) for his unwavering generosity and invaluable guidance.
Glossary
Abbreviations
- CM
cross metathesis
- DECL
DNA-encoded chemical library
- DNA
DNA
- ERα
estrogen receptor alpha
- HPLC
high performance liquid chromatography
- HTRF
homogeneous time-resolved fluorescence
- RCM
ring-closing metathesis
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscombsci.9b00199.
General procedures and reaction conditions; catalyst and substrate synthesis information; RCM-specific experimental procedures; experiments for confirmation of RCM cyclization; DNA sequencing results for verification of DNA integrity post-RCM; characterization information; postmetathesis MS deconvolution spectra (PDF)
Author Contributions
§ O.B.C.M. and N.S. contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by the Welch Foundation (Grant Q-0042), a Core Facility Support Award (RP160805) from the Cancer Prevention Research Institute of Texas (CPRIT), grant OPP1160866 from The Bill and Melinda Gates Foundation, and National Institutes of Health grant P01HD087157 from The Eunice Kennedy Shriver National Institute of Child Health and Human Development.
The authors declare no competing financial interest.
Supplementary Material
References
- Brenner S.; Lerner R. a. Encoded Combinatorial Chemistry. Proc. Natl. Acad. Sci. U. S. A. 1992, 89 (12), 5381–5383. 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favalli N.; Bassi G.; Scheuermann J.; Neri D. DNA-Encoded Chemical Libraries - Achievements and Remaining Challenges. FEBS Lett. 2018, 592 (12), 2168–2180. 10.1002/1873-3468.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S.; Church G. M. Large-Scale de Novo DNA Synthesis: Technologies and Applications. Nat. Methods 2014, 11 (5), 499–507. 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller F.; Steiner M.; Scheuermann J.; Mannocci L.; Nissen I.; Kohler M.; Beisel C.; Neri D. High-Throughput Sequencing for the Identification of Binding Molecules from DNA-Encoded Chemical Libraries. Bioorg. Med. Chem. Lett. 2010, 20 (14), 4188–4192. 10.1016/j.bmcl.2010.05.053. [DOI] [PubMed] [Google Scholar]
- Goodnow R. A.; Dumelin C. E.; Keefe A. D. DNA-Encoded Chemistry: Enabling the Deeper Sampling of Chemical Space. Nat. Rev. Drug Discovery 2017, 16 (2), 131–147. 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- Yuen L. H.; Franzini R. M. Achievements, Challenges, and Opportunities in DNA-Encoded Library Research: An Academic Point of View. ChemBioChem 2017, 18 (9), 829–836. 10.1002/cbic.201600567. [DOI] [PubMed] [Google Scholar]
- Dickson P.; Kodadek T. Chemical Composition of DNA-Encoded Libraries, Past Present and Future. Org. Biomol. Chem. 2019, 17 (19), 4676–4688. 10.1039/C9OB00581A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. A.; King B. W.; Bandyopadhyay D.; Berger S. B.; Campobasso N.; Capriotti C. A.; Cox J. A.; Dare L.; Dong X.; Finger J. N.; et al. DNA-Encoded Library Screening Identifies Benzo[b][1,4]Oxazepin-4-Ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors. J. Med. Chem. 2016, 59 (5), 2163–2178. 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]
- Neri D.; Lerner R. A. DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu. Rev. Biochem. 2018, 87 (1), 479–502. 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satz A. L.; Cai J.; Chen Y.; Goodnow R.; Gruber F.; Kowalczyk A.; Petersen A.; Naderi-Oboodi G.; Orzechowski L.; Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjugate Chem. 2015, 26 (8), 1623–1632. 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- Kölmel D. K.; Loach R. P.; Knauber T.; Flanagan M. E. Employing Photoredox Catalysis for DNA-Encoded Chemistry: Decarboxylative Alkylation of α-Amino Acids. ChemMedChem 2018, 13 (20), 2159–2165. 10.1002/cmdc.201800492. [DOI] [PubMed] [Google Scholar]
- Li J.-Y.; Huang H. Development of DNA-Compatible Suzuki-Miyaura Reaction in Aqueous Media. Bioconjugate Chem. 2018, 29 (11), 3841–3846. 10.1021/acs.bioconjchem.8b00676. [DOI] [PubMed] [Google Scholar]
- Ruff Y.; Berst F. Efficient Copper-Catalyzed Amination of DNA-Conjugated Aryl Iodides under Mild Aqueous Conditions. MedChemComm 2018, 9 (7), 1188–1193. 10.1039/C8MD00185E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Gabriele E.; Samain F.; Favalli N.; Sladojevich F.; Scheuermann J.; Neri D. Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb. Sci. 2016, 18 (8), 438–443. 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H. C.; Simmons N.; Faver J. C.; Yu Z.; Palaniappan M.; Riehle K.; Matzuk M. M. A Mild, DNA-Compatible Nitro Reduction Using B 2 (OH) 4. Org. Lett. 2019, 21 (7), 2194–2199. 10.1021/acs.orglett.9b00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Sun H.; Liu J.; Dai D.; Zhang M.; Zhou H.; Zhong W.; Lu X. Ruthenium-Promoted C-H Activation Reactions between DNA-Conjugated Acrylamide and Aromatic Acids. Org. Lett. 2018, 20 (16), 4764–4768. 10.1021/acs.orglett.8b01837. [DOI] [PubMed] [Google Scholar]
- Xu H.; Ma F.; Wang N.; Hou W.; Xiong H.; Lu F.; Li J.; Wang S.; Ma P.; Yang G.; et al. DNA-Encoded Libraries: Aryl Fluorosulfonates as Versatile Electrophiles Enabling Facile On-DNA Suzuki, Sonogashira, and Buchwald Reactions. Adv. Sci. 2019, 6 (23), 1901551. 10.1002/advs.201901551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gironda-Martínez A.; Neri D.; Samain F.; Donckele E. J. DNA-Compatible Diazo-Transfer Reaction in Aqueous Media Suitable for DNA-Encoded Chemical Library Synthesis. Org. Lett. 2019, 21 (23), 9555–9558. 10.1021/acs.orglett.9b03726. [DOI] [PubMed] [Google Scholar]
- Stanton B. Z.; Peng L. F.; Maloof N.; Nakai K.; Wang X.; Duffner J. L.; Taveras K. M.; Hyman J. M.; Lee S. W.; Koehler A. N.; et al. A Small Molecule That Binds Hedgehog and Blocks Its Signaling in Human Cells. Nat. Chem. Biol. 2009, 5 (3), 154–156. 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallinson J.; Collins I. Macrocycles in New Drug Discovery. Future Med. Chem. 2012, 4 (11), 1409–1438. 10.4155/fmc.12.93. [DOI] [PubMed] [Google Scholar]
- Nicolaou K. C.; Bulger P. G.; Sarlah D. Metathesis Reactions in Total Synthesis. Angew. Chem., Int. Ed. 2005, 44 (29), 4490–4527. 10.1002/anie.200500369. [DOI] [PubMed] [Google Scholar]
- Hung A. W.; Ramek A.; Wang Y.; Kaya T.; Wilson J. A.; Clemons P. a; Young D. W. Route to Three-Dimensional Fragments Using Diversity-Oriented Synthesis. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (17), 6799–6804. 10.1073/pnas.1015271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs R. H. Olefin Metathesis. Tetrahedron 2004, 60 (34), 7117–7140. 10.1016/j.tet.2004.05.124. [DOI] [Google Scholar]
- Fürstner A. Olefin Metathesis and Beyond. Angew. Chem., Int. Ed. 2000, 39 (17), 3012–3043. . [DOI] [PubMed] [Google Scholar]
- Tomasek J.; Schatz J. Olefin Metathesis in Aqueous Media. Green Chem. 2013, 15 (9), 2317. 10.1039/c3gc41042k. [DOI] [Google Scholar]
- Kirkland T. A.; Lynn D. M.; Grubbs R. H. Ring-Closing Metathesis in Methanol and Water. J. Org. Chem. 1998, 63 (26), 9904–9909. 10.1021/jo981678o. [DOI] [Google Scholar]
- Hughes D.; Wheeler P.; Ene D. Olefin Metathesis in Drug Discovery and Development—Examples from Recent Patent Literature. Org. Process Res. Dev. 2017, 21 (12), 1938–1962. 10.1021/acs.oprd.7b00319. [DOI] [Google Scholar]
- Higman C. S.; Lummiss J. A. M.; Fogg D. E. Olefin Metathesis at the Dawn of Implementation in Pharmaceutical and Specialty-Chemicals Manufacturing. Angew. Chem., Int. Ed. 2016, 55 (11), 3552–3565. 10.1002/anie.201506846. [DOI] [PubMed] [Google Scholar]
- Lin Y. A.; Chalker J. M.; Davis B. G. Olefin Metathesis for Site-Selective Protein Modification. ChemBioChem 2009, 10 (6), 959–969. 10.1002/cbic.200900002. [DOI] [PubMed] [Google Scholar]
- Masuda S.; Tsuda S.; Yoshiya T. Ring-Closing Metathesis of Unprotected Peptides in Water. Org. Biomol. Chem. 2018, 16 (48), 9364–9367. 10.1039/C8OB02778A. [DOI] [PubMed] [Google Scholar]
- Ahmad R.; Arakawa H.; Tajmir-Riahi H. a. A Comparative Study of DNA Complexation with Mg(II) and Ca(II) in Aqueous Solution: Major and Minor Grooves Bindings. Biophys. J. 2003, 84 (4), 2460–2466. 10.1016/S0006-3495(03)75050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Fan L.; Phelps C. B.; Davie C. P.; Donahue C. P. Ruthenium Promoted On-DNA Ring-Closing Metathesis and Cross-Metathesis. Bioconjugate Chem. 2017, 28 (6), 1625–1629. 10.1021/acs.bioconjchem.7b00292. [DOI] [PubMed] [Google Scholar]
- Hong S. H.; Grubbs R. H. Highly Active Water-Soluble Olefin Metathesis Catalyst. J. Am. Chem. Soc. 2006, 128 (11), 3508–3509. 10.1021/ja058451c. [DOI] [PubMed] [Google Scholar]
- Tancini F.; Wu Y.-L.; Schweizer W. B.; Gisselbrecht J.-P.; Boudon C.; Jarowski P. D.; Beels M. T.; Biaggio I.; Diederich F. 1,1-Dicyano-4-[4-(Diethylamino)Phenyl]Buta-1,3-Dienes: Structure-Property Relationships. Eur. J. Org. Chem. 2012, 2012 (14), 2756–2765. 10.1002/ejoc.201200111. [DOI] [Google Scholar]
- Porto-Carrero W.; Cupani A.; Veenstra M. J.; Dorbec M.; Eykens L.; Ormerod D. Volume Intensified Dilution of a Ring-closing Metathesis in Ethyl Acetate by Means of a Membrane-assisted Process in Solvent Recycle. J. Chem. Technol. Biotechnol. 2019, 94 (9), 2990–2998. 10.1002/jctb.6109. [DOI] [Google Scholar]
- Hong S. H.; Sanders D. P.; Lee C. W.; Grubbs R. H. Prevention of Undesirable Isomerization during Olefin Metathesis. J. Am. Chem. Soc. 2005, 127 (49), 17160–17161. 10.1021/ja052939w. [DOI] [PubMed] [Google Scholar]
- Bailey G. A.; Lummiss J. A. M.; Foscato M.; Occhipinti G.; McDonald R.; Jensen V. R.; Fogg D. E. Decomposition of Olefin Metathesis Catalysts by Brønsted Base: Metallacyclobutane Deprotonation as a Primary Deactivating Event. J. Am. Chem. Soc. 2017, 139 (46), 16446–16449. 10.1021/jacs.7b08578. [DOI] [PubMed] [Google Scholar]
- Mayer C.; Gillingham D. G.; Ward T. R.; Hilvert D. An Artificial Metalloenzyme for Olefin Metathesis. Chem. Commun. 2011, 47 (44), 12068. 10.1039/c1cc15005g. [DOI] [PubMed] [Google Scholar]
- Neville A.; Iniesta J.; Palomo J. Design of Heterogeneous Hoveyda-Grubbs Second-Generation Catalyst-Lipase Conjugates. Molecules 2016, 21 (12), 1680. 10.3390/molecules21121680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson W. J.; Braunschweiger K. I.; Braunschweiger W. R.; Smith J. R.; McCormick J. J.; Wasmann C. C.; Jarvis N. P.; Bell D. H.; Good N. E. Hydrogen Ion Buffers for Biological Research. Anal. Biochem. 1980, 104 (2), 300–310. 10.1016/0003-2697(80)90079-2. [DOI] [PubMed] [Google Scholar]
- Kandegedara A.; Rorabacher D. B. Noncomplexing Tertiary Amines as “better” Buffers Covering the Range of PH 3–11. Temperature Dependence of Their Acid Dissociation Constants. Anal. Chem. 1999, 71 (15), 3140–3144. 10.1021/ac9902594. [DOI] [PubMed] [Google Scholar]
- Winter G. E.; Buckley D. L.; Paulk J.; Roberts J. M.; Souza A.; Dhe-Paganon S.; Bradner J. E. Phthalimide Conjugation as a Strategy for in Vivo Target Protein Degradation. Science (Washington, DC, U. S.) 2015, 348 (6241), 1376–1381. 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. H.; Day M. W.; Grubbs R. H. Decomposition of a Key Intermediate in Ruthenium-Catalyzed Olefin Metathesis Reactions. J. Am. Chem. Soc. 2004, 126 (24), 7414–7415. 10.1021/ja0488380. [DOI] [PubMed] [Google Scholar]
- Higman C. S.; Plais L.; Fogg D. E. Isomerization during Olefin Metathesis: An Assessment of Potential Catalyst Culprits. ChemCatChem 2013, 5 (12), 3548–3551. 10.1002/cctc.201300886. [DOI] [Google Scholar]
- Higman C. S.; Lanterna A. E.; Marin M. L.; Scaiano J. C.; Fogg D. E. Catalyst Decomposition during Olefin Metathesis Yields Isomerization-Active Ruthenium Nanoparticles. ChemCatChem 2016, 8 (15), 2446–2449. 10.1002/cctc.201600738. [DOI] [Google Scholar]
- Higman C. S.; Nascimento D. L.; Ireland B. J.; Audörsch S.; Bailey G. A.; McDonald R.; Fogg D. E. Chelate-Assisted Ring-Closing Metathesis: A Strategy for Accelerating Macrocyclization at Ambient Temperatures. J. Am. Chem. Soc. 2018, 140 (5), 1604–1607. 10.1021/jacs.7b13257. [DOI] [PubMed] [Google Scholar]
- Fürstner A.; Leitner A. A Catalytic Approach to (R)-(+)-Muscopyridine with Integrated “Self-Clearance.. Angew. Chem., Int. Ed. 2003, 42 (3), 308–311. 10.1002/anie.200390103. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Dias H. V. R.; Lovely C. J. Synthesis of Fused Bicyclic Imidazoles by Ring-Closing Metathesis. Tetrahedron Lett. 2003, 44 (7), 1379–1382. 10.1016/S0040-4039(02)02864-2. [DOI] [Google Scholar]
- Lin Y. A.; Davis B. G. The Allylic Chalcogen Effect in Olefin Metathesis. Beilstein J. Org. Chem. 2010, 6, 1219–1228. 10.3762/bjoc.6.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.; Li Y.; Wang F.; Zhou W.; Wang J.-X. Indium-Mediated Synthesis of Homoallyl Alcohols in the Aqueous Phase. Synth. Commun. 2011, 41 (11), 1664–1671. 10.1080/00397911.2010.492072. [DOI] [Google Scholar]
- Lin Y. A.; Chalker J. M.; Floyd N.; Bernardes G. J. L.; Davis B. G. Allyl Sulfides Are Privileged Substrates in Aqueous Cross-Metathesis: Application to Site-Selective Protein Modification. J. Am. Chem. Soc. 2008, 130 (30), 9642–9643. 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]
- Walensky L. D.; Bird G. H. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J. Med. Chem. 2014, 57 (15), 6275–6288. 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speltz T. E.; Danes J. M.; Stender J. D.; Frasor J.; Moore T. W. A Cell-Permeable Stapled Peptide Inhibitor of the Estrogen Receptor/Coactivator Interaction. ACS Chem. Biol. 2018, 13 (3), 676–684. 10.1021/acschembio.7b01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe J. W.; Kay B. K. Filamentous Phage Display in the New Millennium. Chem. Rev. 2005, 105 (11), 4056–4072. 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- Tian F.; Tsao M.-L.; Schultz P. G. A Phage Display System with Unnatural Amino Acids. J. Am. Chem. Soc. 2004, 126 (49), 15962–15963. 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.