Abstract
Background
Although massive studies have been conducted to investigate the mechanisms of esophageal squamous cell carcinoma (ESCC) carcinogenesis, the understanding of molecular alterations during the malignant transformation of epithelial dysplasia is still lacking, especially regarding epigenetic changes.
Results
To better characterize the methylation changes during the malignant transformation of epithelial dysplasia, a whole-genome bisulfite sequencing analysis was performed on a series of tumor, dysplastic, and non-neoplastic epithelial tissue samples from esophageal squamous cell carcinoma (ESCC) patients. Promoter hypermethylation in TGF-β receptor type II (TGFBR2), an important mediator of TGF-β signaling, was identified. Further, we evaluated the methylation and expression of TGFBR2 in tumor samples through The Cancer Genome Atlas multiplatform data as well as immunohistochemistry. Moreover, treatment of ESCC cell lines with5-Aza-2′-deoxycytidine, a DNA methyltransferase inhibitor, reactivated the expression of TGFBR2. The lentiviral mediating the overexpression of TGFBR2 inhibited the proliferation of ESCC cell line by inducing cell cycle G2/M arrest. Furthermore, the overexpression of TGFBR2 inhibited the tumor growth obviously in vivo.
Conclusions
The characterization of methylation silencing of TGFBR2 in ESCC will enable us to further explore whether this epigenetic change could be considered as a predictor of malignant transformation in esophageal epithelial dysplasia and whether use of a TGFBR2 agonist may lead to a new therapeutic strategy in patients with ESCC.
Keywords: Esophageal squamous cell carcinoma, Whole genome bisulfite sequencing, TGFBR2, Methylation changes, Cancer diagnosis, Treatment
Background
Esophageal cancer (EC) is one of the common malignant tumors in China [1, 2]. The incidence of EC in China accounts for about 50% of new EC patients occurring worldwide [3, 4]. Notably, esophageal squamous cell carcinoma (ESCC) represents the predominant histological type with a 90% prevalence in China [2, 5]. The standard treatments include surgery and chemotherapy, but due to the absence of obvious symptoms in early stage of ESCC, patients are frequently diagnosed only after reaching an advanced stage [6, 7]. The 5-year overall survival rate following comprehensive treatment for advanced patients is approximately 25–30%, and these treatments are often accompanied by highly undesirable side effects. However, surgical treatment at early stages for ESCC may increase the 5-year survival rate to 70%. Therefore, there is a need for further in-depth study of disease mechanism and the development of new treatment strategies.
It is widely recognized that the carcinogenesis of ESCC is a multistep process, which progresses from dysplasia and involves multiple genetic changes [8]. Epigenetic changes including DNA methylation play a critical character in the management of gene expression patterns and are independent of mutations in the DNA sequence. Methylation of CpG islands in promoter regions frequently contributing to gene transcriptional silencing may serve as an important mechanism to inactivate tumor suppressor genes in cancer [9–11]. Moreover, promoter methylation occurs early in the development of cancer. Thus, the identification of methylation changes in tumor suppressor genes is of tremendous importance since it could contribute to early detection and new drug development for ESCC patients.
Although tumor methylomes have been extensively characterized, conventional methods, such as methylation-specific PCR (MSP), or enrichment-based approaches, such as methylated DNA immunoprecipitation sequencing (MeDIP-Seq), have been mostly used to study methylation changes in cancers including ESCC [12–16]. However, a more comprehensive landscape of the ESCC methylome is still lacking. In addition, little is known about the development of dysplasia into ESCC. Recent advances in high throughput sequencing enable mapping of DNA methylation at single-base resolution (whole genome bisulfite sequencing; WGBS), which would also help to comprehensively characterize changes in DNA methylation. As such, it is necessary to re-evaluate DNA methylation changes during the development of ESCC by comparing esophageal tumors with corresponding normal as well as dysplastic tissues.
In this study, the transforming growth factor-β (TGF-β) receptor type II gene (TGFBR2), a key mediator of TGF-β signaling which has been implicated in ESCC carcinogenesis, was identified as a putative tumor suppressor in ESCC based on WGBS of paired and unpaired ESCC tissues. We further investigated TGFBR2 methylation status and expression level both in ESCC tissues and cell lines, and determined the relationship between TGFBR2 and ESCC. This study provides significant insight into the epigenetic regulation in ESCC associated with TGFBR2 which could be a potential molecular target in the ESCC diagnosis and treatment.
Results
Methylation landscape of esophageal squamous dysplasia and ESCC
To better characterize the methylation profiles of ESCC and precursor lesions, and the relationship between them, we performed whole-genome bisulfite sequencing (WGBS) on ESCC (n = 3), dysplastic (n = 7), and non-neoplastic epithelial tissue samples (n = 6) from ESCC patients. From two of these patients, we obtained matched non-neoplastic, dysplastic, and tumor samples. On average, the alignment rate of sequencing read mapping to the reference genome was 87.8%. The average coverage of all libraries was 6.83-fold. To evaluate the bisulfite conversion rate, unmethylated lambda DNA was spiked in as the control during library construction. The average bisulfite conversion (unmethylated cytosine to uracil) rate was considerably high (99.2%). Two of the samples were of insufficient quality and therefore did not undergo further analysis.
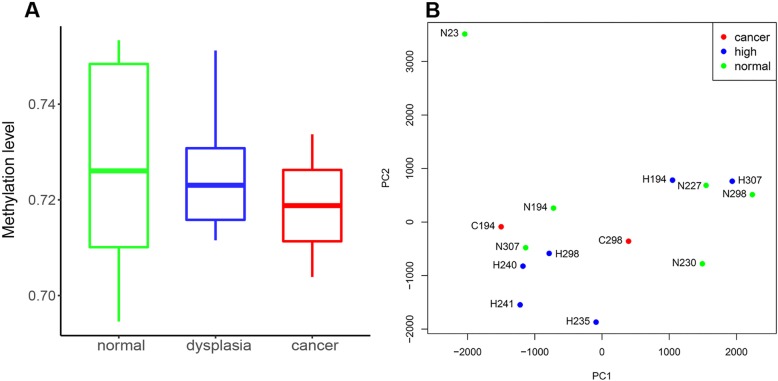
WGBS data revealed a bimodal distribution of methylation in these samples. However, the genome-wide methylation levels of non-neoplastic, dysplastic, and ESCC samples did not differ significantly. Only a trend toward a decrease in the methylation levels from non-neoplastic to cancer tissue samples emerged from these data (Fig. 1a). In addition, using the principal component analysis, we observed a relatively high degree of epigenetic heterogeneity between non-neoplastic, dysplastic, and cancer samples in each patient (Fig. 1b). These results support the previous finding, based on the genomic analysis of ESCC, that significant heterogeneity exists between matched dysplastic and ESCC samples in patients [17].
Fig. 1.
Whole genome methylation profiling of ESCC and esophageal dysplasia samples. a Genome-wide methylation level of ESCC, dysplasia, and non-neoplastic samples. b Principal component analysis of whole genome bisulfite sequencing data
Characterization of differentially methylated regions in dysplasia and ESCC
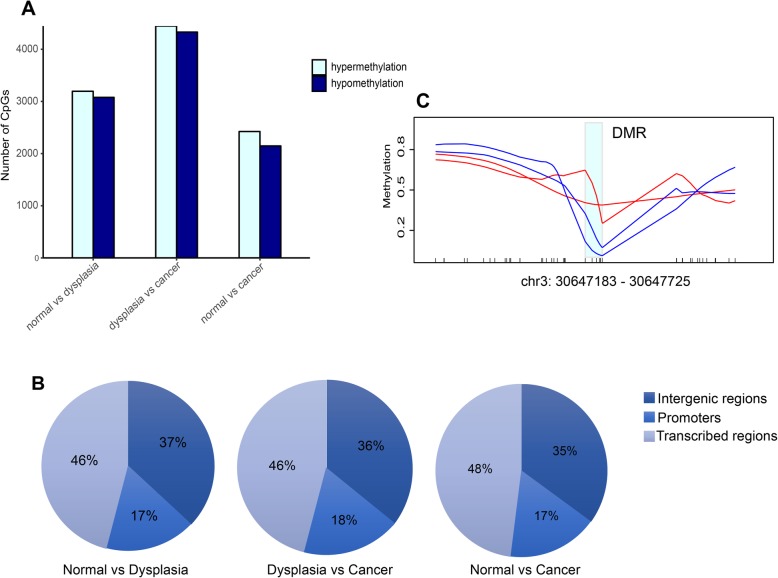
To investigate whether DNA methylation variations are associated with alterations in gene expression in the development of ESCC, we identified differentially methylated regions (DMRs) between the different stages. In total, we identified 969 differentially methylated regions between non-neoplastic and tumor samples, 1293 DMRs between non-neoplastic and dysplastic samples, and 1838 DMRs between dysplastic and tumor samples. There were no obvious differences in the number between hypermethylated and hypomethylated CpG sites in these comparisons (Fig. 2a). In addition, almost half of these DMRs were located in transcribed regions (including transcriptional start sites to transcriptional end sites) rather than in intergenic or promoter regions (Fig. 2b). In some genes, including LHFPL6 and ABL2, promoter methylation from the non-neoplastic to the dysplastic stage had decreased. This result suggested that in these genes, promoter hypomethylation likely occurred very early in ESCC development and might therefore serve as potential biomarkers for the diagnosis of esophageal dysplasia. We did not observe promoter hypermethylation in certain tumor suppressor genes such as CDKN2A [18], TFF1 [14], and CDH1 [19] as previously reported in other cohorts. However, we found promoter hypermethylation in the putative tumor suppressor gene TGFBR2 during the transition from dysplasia to ESCC. We observed similar results when comparing methylation levels in two paired samples (Fig. 2c). The TGFBR2 promoter exhibited hypermethylation not only in the transition from dysplasia to cancer, but also from normal epithelium to cancer. Notably, previous studies have reported mutations in TGFBR2 in ESCC, but at a comparatively low mutation rate [20, 21].
Fig. 2.
Differential DNA methylation in esophageal dysplasia and ESCC. a Distribution of hypermethylated and hypomethylated CpG sites between different stages. b Proportion of differentially methylated regions in transcribed regions, intergenic regions, and promoters. c Methylation difference in TGFBR2 promoter between dysplastic and tumor stages in two paired samples
TGFBR2 mRNA and protein are decreased in primary tumor samples relative to normal tissue
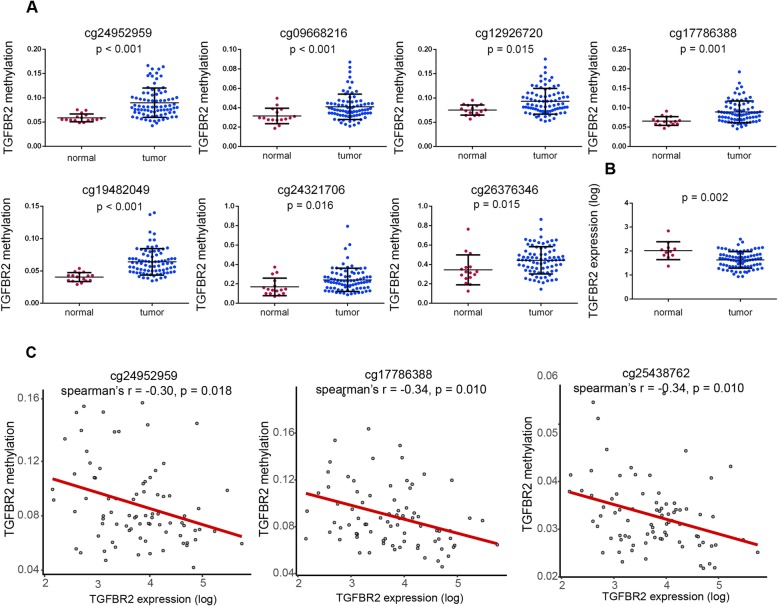
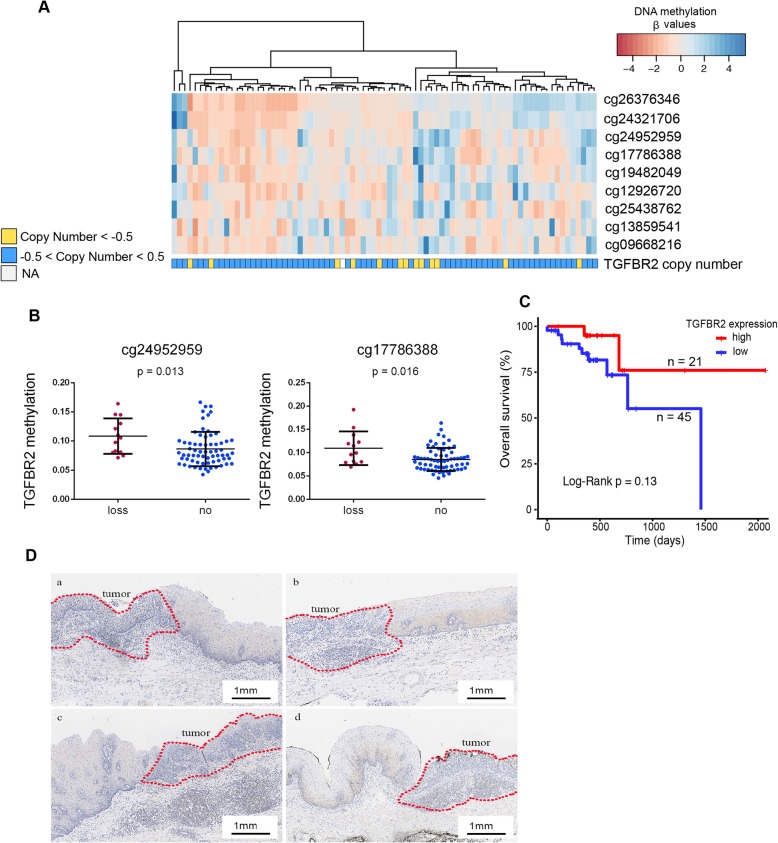
To examine the association between TGFBR2 methylation and expression levels, we explored the multiplatform TGFBR2 profiles, including methylation, RNA-seq, and copy number data for 81 ESCC samples and 16 adjacent normal tissue samples from the TCGA dataset. CpG sites of the TGFBR2 promoter were significantly hypermethylated in tumor samples compared to normal samples (Fig. 3a). Based on the RNA-seq data, TGFBR2 was downregulated in tumor relative to normal tissue samples (Fig. 3b). To investigate the influence of DNA methylation on the gene expression, we calculated the Spearman rank correlation coefficient between methylation of each TGFBR2 promoter CpG site and the expression level. All three CpG sites were significantly negatively correlated with TGFBR2 expression (Fig. 3c). We also examined the relationship between TGFBR2 copy number and methylation levels. Interestingly, samples with copy number loss had significantly higher methylation levels in two CpG sites (Fig. 4a, b), which implicated the loss of tumor suppression gene function in accord with Knudson’s two-hit theory. Finally, we examined whether TGFBR2 expression levels were associated with clinical outcome. Although not statistically significant, patients with high TGFBR2 expression had a relatively favor prognosis compared to those with low expression levels (expression threshold 63.5; Fig. 4c).
Fig. 3.
TGFBR2 is hypermethylated and downregulated in TCGA ESCC dataset. a DNA methylation comparison of TGFBR2 promoter-associated CpG sites in normal and tumor samples. b Expression levels of TGFBR2 in normal and ESCC samples. c Correlations of promoter methylation and expression for TGFBR2
Fig. 4.
TGFBR2 expression and its relationship with copy number and patients’ outcome. a, b Correlation of TGFBR2 copy number and promoter methylation. c Kaplan-Meier curves of overall survival according to TGFBR2 expression level. d IHC performed on sections from ESCC and adjacent tissues with TGFBR2 antibody
We validated these results by performing IHC on 66 samples from our own cohort. In 34 of these samples, the level of TGFBR2 protein was downregulated significantly in tumor tissues compared to dysplastic and normal tissue samples (Fig. 4d).
Promoter hypermethylation is associated with TGFBR2 transcriptional silencing in ESCC cell lines
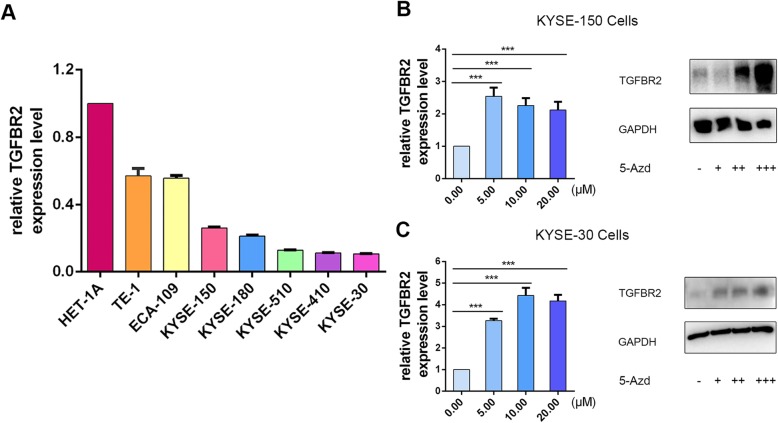
Using RT-qPCR, we obtained that TGFBR2 mRNA levels were also downregulated in several ESCC cell lines (HET-1A, TE-1, ECA-109, KYSE-150, KYSE-180, KYSE-510, KYSE-410, KYSE-30) dramatically compared to an immortalized esophageal epithelial cell line, Het-1A (Fig. 5a, Additional file 1: Figure S1; P < 0.05). To further explore the association between gene expression and the promoter methylation status of TGFBR2, we examined the expression of TGFBR2 in KYSE-150 cells exposed to the treatment with 5-Aza-2′-deoxycytidine. We treated KYSE-150 cells with different dosages of the DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine. The results of RT-qPCR illustrated that the TGFBR2 expression was significantly upregulated in cells exposed to increasing concentrations of 5-Aza-2′-deoxycytidine (Fig. 5b). Western blot analysis corroborated these results (Fig. 5b). The same results were observed in KYSE-30 cells (Fig. 5c). In summary, these data demonstrated that the methylation of promoter mediated transcriptional silencing of TGFBR2 in ESCC cell lines.
Fig. 5.
Treatment of ESCC cells in culture increases the expression of TGFBR2. a RT-qPCR to detect levels of TGFBR2 mRNA in Het-1A and ESCC cell lines using GAPDH as a control gene. b RT-qPCR and western blot analysis performed on RNA and protein isolated from KYSE-150 cells exposed to increasing concentrations of 5-Aza-2′-deoxycytidine (mean ± SD. P < 0.001). c RT-qPCR and western blot analysis performed on RNA and protein isolated from KYSE-30 cells exposed to increasing concentrations of 5-Aza-2′-deoxycytidine (mean ± SD. P < 0.001)
TGFBR2 induces ESCC cell cycle arrest but not cell apoptosis
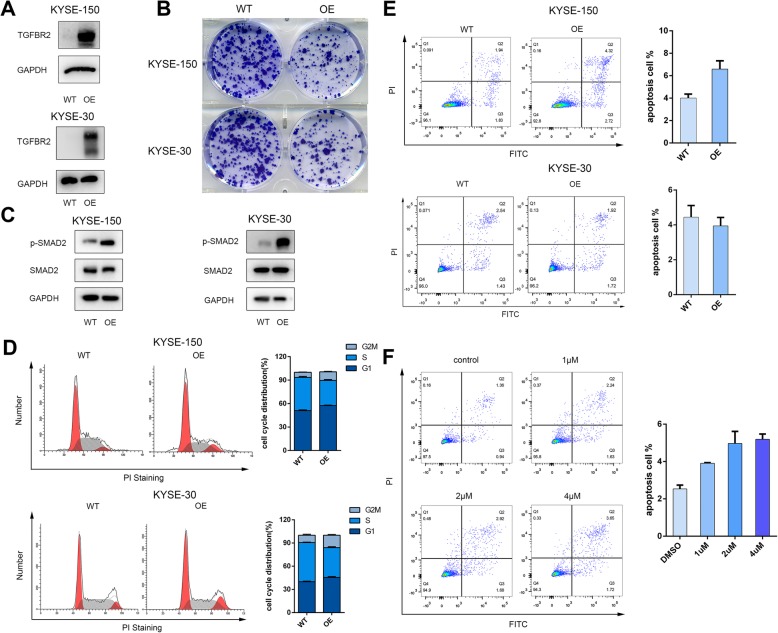
To validate whether the expression of TGFBR2 could prevent the tumor progression, we constructed the TGFBR2 overexpression tumor cells in KYSE-150 and KYSE-30 (Fig. 6a). The expression of phospho-SMAD2 was dramatically reactivated in the TGFBR2 overexpression cells which suggested the TGFβ signaling is restored (Fig. 6b). Overexpression of TGFBR2 suppressed the growth of tumor cells significantly through colony formation assays (Fig. 6c). Consistently, the TGFBR2 overexpression cells induced cell cycle G2/M arrest relative to the wild-type cells as determined through flow cytometry analysis of propidium staining (Fig. 6c). However, 5-Aza-2′-deoxycytidine treatment or TGFBR2 overexpression did not increase the apoptosis rate obviously (Fig. 6d). Collectively, these results suggested that TGFBR2 inhibits the growth of cells by inducing the cell cycle G2/M delayed, but not due to causing cell apoptosis.
Fig. 6.
TGFBR2 overexpression induces ESCC cell cycle arrest but not cell apoptosis. a Western blot analysis of wild-type (WT) cells and lentiviral mediating the overexpression of TGFBR2 (OE) cells in KYSE-150 and KYSE-30 cell lines. b The morphology of WT and OE cells in colony formation assay. c The expression level of phospho-SMAD2 and SMAD2 in WT and OE cells. d Cell cycle distribution in WT and OE cells. Graphic representation of results from cell cycle analysis in WT and OE cells. e Annexin V staining of parental WT and KO cells to detect apoptosis using flow cytometry. Graphic representation of the percentage of apoptotic cells in parental versus OE cells. f Annexin V staining of KYSE-150 cells exposed to increasing concentrations of 5-Aza-2′-deoxycytidine detected with flow cytometry. Graphic representation of the percentage of apoptotic cells with increasing 5-Aza-2′-deoxycytidine concentration
TGFBR2 suppresses ESCC growth in vivo
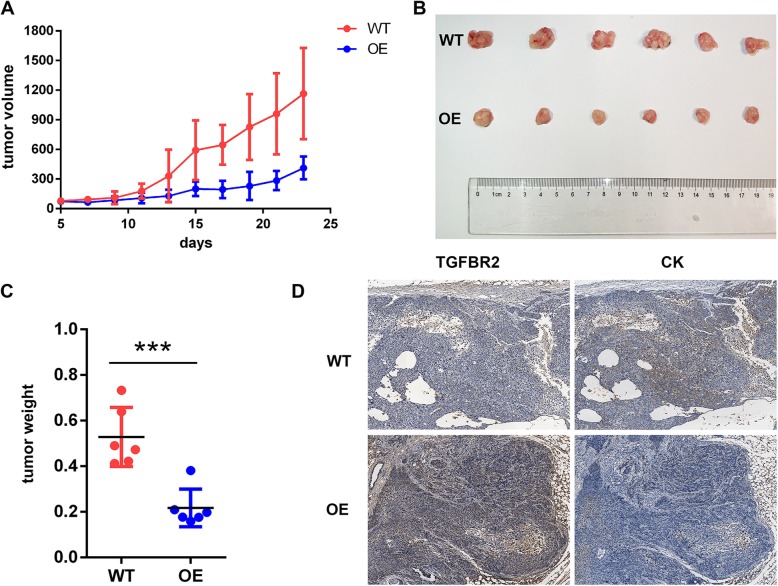
To determine whether TGFBR2 suppressed ESCC proliferation in vivo, we established a subcutaneous ESCC xenograft model in nude mice using KYSE-150-TGFBR2 and control KYSE-150-vector cells. Tumor weight and volume were decreased significantly in KYSE-150-TGFBR2 xenografts compared to KYSE150-vector xenografts (weight 0.22 ± 0.08 g and 0.53 ± 0.13 g, KYSE-150-TGFBR2 vs KYSE-150-vector; Fig. 7a–c). IHC confirmed that the level of TGFBR2 protein was upregulated and Pan-Keratin (CK) was downregulated in KYSE-150-TGFBR2 tumors when compared to the controls. There was a significantly negative correlation between TGFBR2 and CK protein which suggested that the overexpression of TGFBR2 can inhibit the proliferation of ESCC (Fig. 7d). Taken together, these results indicated that TGFBR2 expression significantly inhibited ESCC growth in vivo.
Fig. 7.
TGFBR2 overexpression inhibits tumor growth in vivo. a Tumor volume as measured in xenografts over 21 days derived from KYSE-150-vector (WT) and KYSE-150-TGFBR2 (OE) cells. b Image of subcutaneous xenografts derived from the cells indicated after growth in vivo for 3 weeks. c Tumor weight associated with WT- and OE-derived xenografts at 21 days. d IHC reveals the expression of TGFBR2 and CK in WT and OE cell xenografts
Discussion
In the present study, we analyzed DNA methylation in different stages of ESCC carcinogenesis at single-base resolution. This approach allowed us to identify the methylation profiling in each ESCC patient and the exact boundaries of DMRs on a genome-wide scale. To the best of our knowledge, this is the first time methylation changes in ESCC have been characterized compared with those occurring in both normal esophageal and dysplasia tissues. In addition to the well-described genetic heterogeneity existing between dysplasia and ESCCs, we also observed epigenetic heterogeneity in each ESCC patient.
Carcinogenesis has been recognized as a composition of altered epigenetic and genetic events. Many cancers display global hypomethylation and site-specific hypermethylation at the CpG islands [22, 23]. We observed a similar genome-wide trend toward hypomethylation in the development of normal esophageal tissue into cancer. We also found that most DMRs were located in transcribed regions of the genome. However, we did not observe previously reported gene methylation changes, such as CDKN2A [18] and TFF1 [14], in ESCC possibly due to our small sample size. Nevertheless, the hypermethylation pattern was identified to play an unequivocal character in the management of TGFBR2 expression in ESCC. We further validated our bisulfite sequencing results through analysis of the TCGA ESCC dataset. Tumor samples showed higher methylation levels compared to normal esophageal tissue samples at TGFBR2 promoter-related CpG sites. TGFBR2 methylation was inversely correlated to its expression in tumor samples. In addition, TGFBR2 promoter methylation was increased in tumors with copy number loss compared to those remaining intact at the gene site.
TGFBR2, as a member of TGF-β/Smad signal pathway, is an important tumor suppressor, which mediates TGF-β signaling and induces cell cycle arrest and apoptosis [24]. Under physiological condition, TGFBR2 is normally expressed in the epithelial cells of esophageal mucosal. However, in pathological conditions, its expression specifically disappears [25, 26]. Downregulation or missense mutation of TGFBR2 has been found in several cancers [27–29]. A previous study in vivo demonstrated that mice lacking TGFBR2 expression developed anal or genital carcinoma. These results have indicated that loss of TGFBR2 expression promotes epithelial oncogenesis. Several studies have shown that TGFBR2 inhibits cell growth, invasion, migration, and metastasis in breast and pancreatic cancer [30, 31]. TCGA consortium and others have reported the mutations of TGFBR2 in ESCC [26]. Here, we provide evidences to favor an alternative hypothesis which DNA promoter methylation is a crucial mechanism leading to suppression of TGFBR2 expression in ESCC. Furthermore, we identified specific regions methylated in the TGFBR2 gene. Our study confirmed that TGFBR2 expression was highly suppressed in ESCC cells and tumor tissues.
Hypermethylation of the CpG islands in promoter region is highly associated with silenced tumor-related genes through the reduction of mRNA transcription. Furthermore, gene expression could be rehabilitated with methylation inhibitors since DNA methylation is a reversible procedure. Thus, the regular growth regulation mode could be restored with demethylating genes before other genetic changes. In the present study, TGFBR2 expression in ESCC cell lines could be restored with the demethylating reagent 5-Aza-2′-deoxycytidine. In addition, epigenetic silencing genes are often involved in several circuit of carcinogenesis, such as apoptosis, cell cycle, and DNA repair. The imbalance between cell growth and cell death can be recognized as an early and significant event in the carcinogenic process. Our results demonstrated that the overexpression of TGFBR2 or treatment with the demethylating agent 5-Aza-2′-deoxycytidine significantly induced cell cycle arrest in ESCC cell lines. Moreover, the overexpression of TGFBR2 suppressed ESCC growth in vivo. All together, we hypothesize that TGFBR2 plays a role in suppressing ESCC tumorigenesis.
DNA methylation changes in tumor-related genes are frequent and early events during carcinogenesis [32]. Several methylation shifts occur during development from dysplasia to tumor. Methylation of specific sites may therefore be of biological and further clinical value in the early detection of ESCC, which is urgent for more favorable outcomes in the treatment of patients. Hotspots for DNA methylation are also valuable as biomarkers in the so-called liquid biopsy for cancer diagnosis and therapy since they are not only detected in resected tissues, but also in various body fluids, including peripheral blood [33–36], saliva [37–39], and urine [40–42]. In fact, methylated APC [43] and CDKN2A [44] have already been detected in the plasma of a subset of ESCC patients. The feasibility of the detection of TGFBR2 methylation in serum of ESCC patients is therefore warranted.
Conclusions
In summary, TGFBR2 is downregulated in ESCC due to DNA hypermethylation of its promoter regions. The high level of methylated CpGs in TGFBR2 in ESCC suggests that DNA methylation in TGFBR2 promoter region would contribute to absent or reduced TGFBR2 mRNA expression, and hence promote ESCC carcinogenesis. Cancer cells with treatment of DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine reversed methylation levels in the TGFBR2 promoter and induced cell cycle arrest. Characterizing the role of TGFBR2 in ESCC could pave the way to a deeper understanding of the potential mechanisms underlying disease development as well as illuminate its potential as a biomarker for early diagnosis and personalized therapeutic agent for ESCC patients.
Methods
Patient samples
Primary tissue samples were collected from patients who had undergone endoscopic surveillance and were analyzed by experienced pathologists. Individuals with ESCC were all inpatients who underwent surgical operations in Chinese PLA General Hospital between 2017 and 2018. Tissues were separated into two sections, one of which was stored at − 80 °C and the other was formalin fixed and paraffin embedded. Histological tumor characterization of resected specimens was carried out in the Department of Pathology in Chinese PLA General Hospital. All sample sections were stained in hematoxylin and eosin and were reviewed by two experienced pathologists. The pathology of the samples collected was the following: dysplasia, n = 6; and ESCC cancer, n = 3.
Whole-genome bisulfite sequencing library preparation
DNA was extracted with DNeasy Blood and Tissue Kit (Qiagen; Valencia, CA, USA), and each DNA sample was spiked with 1% unmethylated lambda DNA (Promega; Madison, WI, USA) to evaluate the bisulfite conversion efficiency. The genomic DNA (500 ng) was fragmented with Covaris M220 ultrasonicator (Covaris; Woburn, MA, USA) to an average size of 350 bp. End repair and methylated adaptor ligation was conducted with NEBNext Ultra End Repair/dA-Tailing Module, Ligation Module and NEBNext Multiplex Oligos for Illumina (Methylated Adaptor, Index Primers Set 1; New England Biolabs; Ipswich, MA, USA). DNA fragments between 400 and 500 bp were selected for library construction with Ampure XP beads (Beckman Coulter; Brea, CA, USA). Bisulfite conversion was performed on samples using the EZ DNA Methylation kit (Zymo Research; Irvine, CA, USA) with modified single-stranded DNA fragments amplified using the Kapa HiFi U+ HotStart ReadyMix (Kapa Biosystems; Wilmington, MA, USA) with primers (NEBNext Multiplex Oligos for Illumina). A final size selection was performed to enrich the library for a range between 300 and 500 bp. Constructed libraries were evaluated on the Agilent 2100 Bioanalyzer (Agilent Technologies; Santa Clara, CA, USA) and then sequenced on the Illumina HiSeq X Ten (Illumina; San Diego, CA, USA) using the 150-bp paired-end mode.
Whole-genome bisulfite sequencing data analysis
Sequencing reads were processed with the Bsmooth software package (http://rafalab.jhsph.edu/bsmooth), as described previously (PMID: 23034175). Briefly, reads were aligned to the human genome (hg19) together with the lambda phage genome using Bowtie2 v. 2.2.3. Following alignment, methylation measurements for each CpG site were obtained, and the bisulfite conversion rates were calculated based on the spiked-in unmethylated lambda phage DNA. To identify DMRs, the bsseq package in Bsmooth was utilized to smooth the data with default parameters (ns = 70, h = 1000), to characterize DMRs containing either 70 CpGs or a width of 1 kb, whichever was larger. Regions fulfilling the following criteria were deemed putative DMRs: (1) t-statistics passed the cutoff criteria of (− 3, 3); (2) containing at least three CpG sites; and (3) methylation difference of at least 10%. DMRs were then annotated with ANNOVAR software. Promoter regions of genes were defined as up to 1500 nt regions upstream of transcriptional start sites.
Analysis of TCGA data
RNA-seq level 3 data, DNA methylation array data, mean segments of copy number, and clinical data of patients were downloaded from The Cancer Genome Atlas (TCGA) portal (https://portal.gdc.cancer.gov/). We chose FPKM values to represent gene expression levels (81 ESCCs and 11 normal tissue samples). FPKM values were then transformed into TPM values (transcript per million) to compare expression between samples. The methylation levels of CpG sites were measured using the Illumina Infinium Human Methylation 450 BeadArray platform and represented as the β value (81 ESCCs and 16 normal tissue samples; β value = intensity of the methylated allele/(intensity of the methylated allele + the unmethylated allele). Differentially methylated or expressed analyses were performed using the Mann-Whitney U test. The Benjamini-Hochberg method was applied for adjusting P values to control the false discovery rate. The CpG sites with adjusted P values less than 0.05 were considered to be differentially methylated. For copy number variation analysis, a segment mean of 0.5 was defined as the cutoff for amplifications and − 0.5 for deletions. Survival analysis was conducted with the Kaplan-Meier method, and the log-rank test was performed to test difference in survival between two groups. Optimal cutoff value for TGFBR2 expression in survival analysis was determined using maximally selected rank statistics.
Cell lines and cell culture
The cell lines KYSE-150 and KYSE-30 were kindly gifted from Dr. Shimada Y (Kyoto University, Kyoto, Japan). All ESCC cell lines were cultured in RPMI1640 medium supplemented with 10% FBS. The cell line Het-1A was acquired from ATCC and cultured in BEGM™ medium prepared with the Bronchial Epithelial Cell Growth Medium Bullet Kit along with all the additives (Lonza/Clonetics Corporation, CC-3170; Hayward, CA, USA).
Construction of stable TGFBR2 expressing cell lines
To generate lentivirus, the TGFBR2 lentiviral plasmid (pLVX-IRE-Puro-TGFBR2; Wuhan Miaoling Bioscience & Technology Co., Ltd; Wuhan, China) was cotransfected with the psPAX2 and pMD2.G plasmids using Neofect™ DNA transfection reagent (1 μL/mL; Neofect; Beijing, China) for packaging in HEK-293 T cells. The viruses were harvested 72 h after transfection. KYSE-150 cells were transduced with the lentivirus. Stably infected cells were selected in puromycin (2 μg/mL; Life Technologies, Waltham, MA, USA) for 2 days and confirmed by RT-qPCR and western blot.
Quantitative real-time PCR
Total RNA was extracted from cultured cell lines with TRIzol (Thermo Fisher Scientific; Waltham, MA, USA), and cDNA was synthesized with the PrimeScript™ RT Master Mix (TaKaRa; Beijing, China). Quantitative real-time PCR was performed in triplicate using TB Green™ Premix Ex Taq™ (Tli RnaseH Plus; TaKaRa) on the ABI (7900HT) system (Applied Biosystems; Foster City, CA, USA). The expression of TGFBR2 was calculated using the 2−△△CT method. The primer sequences used are the following: TGFBR2 forward primer: 5′-GTAGCTCTGATGAGTGCAATGAC-3′; TGFBR2 reverse primer: 5′-CAGATATGGCAACTCCCAGTG-3′; GAPDH forward primer: 5′-GGAGCGAGATCCCTCCAAAAT-3′; GAPDH reverse primer: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western blot analysis
Cells were harvested and lysed in RIPA lysis buffer. Protein concentrations were evaluated by the BCA protein assay kit (PLYGEN, China) according to the manufacturer’s instructions. The protein lysates were separated on 10% SDS-PAGE and electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were overnight incubated with primary antibodies at 4 °C. The protein bands were detected and quantitated using enhanced chemiluminescence (ECL).
Colony formation assay
For colony formation assay, cells were seeded onto 6-well plates with 800 cells per well and the medium was changed every 3 days. After 10 days, cells were fixed with 4% formaldehyde for 20 min and stained with 1% crystal violet solution for 10 min.
Flow cytometry
KYSE-150 cells were incubated with different dosages of DNA methyltransferase inhibitor for 48 h before flow cytometry analysis. DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine was acquired from MedChemExpress (Monmouth Junction, NJ, USA) and fully dissolved in dimethylsufoxide (DMSO) at a concentration of 10 mM. Apoptosis was assessed using the Annexin V, 633 Apoptosis Detection Kit (Dojindo, Kumamoto, Japan). Cell cycle analysis was performed using the Cell Cycle and Apoptosis Analysis Kit (Beyotime; Jiangsu, China). Both assays were carried out and analyzed on a flow cytometer (Beckman Coulter) according to the manufacturer’s instructions. Data from the apoptosis assay were analyzed using FlowJo v10 (FlowJo, LLC). Cell cycle distributions were statistically determined by Modfit LT 3.2 software (Verity Software House; www.vsh.com; Topsham, ME).
Immunohistochemistry (IHC)
Immunohistochemistry was performed using an indirect peroxidase method. Paraffin embedded sections of esophageal tissues on slides were dewaxed fully in xylene and rehydrated thoroughly in a decreasing graded series of ethanol concentrations. Endogenous peroxidase was quenched with 3% hydrogen peroxide, and sections were blocked with 10% goat serum (ZSGB-BIO; Beijing, China) to reduce non-specific binding of antibodies. All tissues were incubated overnight with primary antibody at 4 °C. Antibody against TGFBR2 was obtained from Abcam (Shanghai, China). For detection, slides were returned to room temperature and incubated with horse radish peroxidase (HRP)-labeled goat anti-rabbit IgG (1:200, Proteintech, Wuhan, China). Diaminobenzidine (DAB, ZSGB-BIO, Beijing, China) was used as the chromogenic substrate. Slides were counterstained with hematoxylin and mounted in resin. Images were acquired through Aperio pathology scanner.
Xenografts
KYSE-150 cells and KYSE-150-TGFBR2-overexpressed cell suspensions were subcutaneously injected into female BALB/c nude mice (age 4–5 weeks; n = 6 in each group). Tumor volumes were subsequently measured every 3 days and calculated (volume = R × r2/2, R represents the longest diameter and r represents the shortest diameter). After 3 weeks, the mice were sacrificed, and tumor samples were processed for further analysis.
Supplementary information
Additional file 1: Figure S1. TGFBR2 mRNA levels in several ESCC cell lines. (A) RT-qPCR to detect levels of TGFBR2 mRNA in Het-1A and ESCC cell lines using ACTB as a control gene. (B) RT-qPCR to detect levels of TGFBR2 mRNA in Het-1A and ESCC cell lines using 18S RNA as a control gene.
Acknowledgements
Not applicable.
Abbreviations
- DAB
Diaminobenzidine
- DMRs
Differentially methylated regions
- DMSO
Dimethylsufoxide
- EC
Esophageal cancer
- ECL
Enhanced chemiluminescence
- ESCC
Esophageal squamous cell carcinoma
- HRP
Horse radish peroxidase
- IHC
Immunohistochemistry
- MeDIP-Seq
Methylated DNA immunoprecipitation sequencing
- MSP
Methylation-specific PCR
- PVDF
Polyvinylidene fluoride
- TCGA
The Cancer Genome Atlas
- TGFBR2
Transforming growth factor-β receptor type II gene
- TGF-β
Transforming growth factor-β
- WGBS
Whole genome bisulfite sequencing
Authors’ contributions
YM, SH, and XW performed the experiments, data curation, and wrote the manuscript. AG, YZ, MG, and DZ contributed to the patient specimen collection and clinical data. QZ, PW, BY, HY, YL, JS, PY, and YL contributed to the research data. ML coordinated the experimental work with human cell lines. YJ conceived and designed the study and provided financial support for the study. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Basic Research Program of China (grant 2015CB553902), National Key R&D Program of China (2018YFA0208902, 2018YFC0115204), the National Natural Science Foundation Fund (81472559, 81502023, 81772490, U1604281), Beijing Science Foundation of China (BJSFC no.7171008), and the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (grants 2016-I2M-1-001, 2017-I2M-3-004, and 2019-I2M-1-003).
Availability of data and materials
All the datasets are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The study was reviewed and approved by the Ethics Committee of Chinese PLA general hospital, and informed consent was obtained from all recruited individuals.
All the animal procedures were approved by the animal ethical committee at the Cancer Hospital, Chinese Academy of Medical Sciences.
Consent for publication
All authors have reviewed the manuscript and given consent for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yarui Ma, Siyuan He, Aiai Gao and Ying Zhang contributed equally to this work.
Contributor Information
Xiaobing Wang, Email: wangxb@cicams.ac.cn.
Mingzhou Guo, Email: mzguo@hotmail.com.
Yuchen Jiao, Email: Jiaoyuchen@cicams.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13148-020-0819-6.
References
- 1.Chen W, Zheng R, Zeng H, Zhang S. The incidence and mortality of major cancers in China, 2012. Chin J Cancer. 2016;35(1):73. doi: 10.1186/s40880-016-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14(1):33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J, He YT, Zheng RS, Zhang SW, Chen WQ. Analysis of esophageal cancer time trends in China, 1989-2008. Asian Pac J Cancer Prev. 2012;13(9):4613–4617. doi: 10.7314/APJCP.2012.13.9.4613. [DOI] [PubMed] [Google Scholar]
- 6.Sohda M, Kuwano H. Current status and future prospects for esophageal cancer treatment. Ann Thorac Cardiovasc Surg. 2017;23(1):1–11. doi: 10.5761/atcs.ra.16-00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higuchi K, Koizumi W, Tanabe S, Sasaki T, Katada C, Azuma M, et al. Current management of esophageal squamous-cell carcinoma in Japan and other countries. Gastrointest Cancer Res. 2009;3(4):153–161. [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor PR, Abnet CC, Dawsey SM. Squamous dysplasia--the precursor lesion for esophageal squamous cell carcinoma. Cancer Epidemiol Biomark Prev. 2013;22(4):540–552. doi: 10.1158/1055-9965.EPI-12-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 10.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 12.Kishino T, Niwa T, Yamashita S, Takahashi T, Nakazato H, Nakajima T, et al. Integrated analysis of DNA methylation and mutations in esophageal squamous cell carcinoma. Mol Carcinog. 2016;55(12):2077–2088. doi: 10.1002/mc.22452. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Peng H, Huang X, Zhao M, Li Z, Yin N, et al. Genome-wide profiling of DNA methylation and gene expression in esophageal squamous cell carcinoma. Oncotarget. 2016;7(4):4507–4521. doi: 10.18632/oncotarget.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima SC, Hernandez-Vargas H, Simao T, Durand G, Kruel CD, Le Calvez-Kelm F, et al. Identification of a DNA methylome signature of esophageal squamous cell carcinoma and potential epigenetic biomarkers. Epigenetics. 2011;6(10):1217–1227. doi: 10.4161/epi.6.10.17199. [DOI] [PubMed] [Google Scholar]
- 15.Taghavi N, Biramijamal F, Sotoudeh M, Khademi H, Malekzadeh R, Moaven O, et al. p16INK4a hypermethylation and p53, p16 and MDM2 protein expression in esophageal squamous cell carcinoma. BMC Cancer. 2010;10:138. doi: 10.1186/1471-2407-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo M, Ren J, House MG. Qi Y, Brock MV, Herman JG. Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res. 2006;12(15):4515–4522. doi: 10.1158/1078-0432.CCR-05-2858. [DOI] [PubMed] [Google Scholar]
- 17.Chen XX, Zhong Q, Liu Y, Yan SM, Chen ZH, Jin SZ, et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat Commun. 2017;8(1):524. doi: 10.1038/s41467-017-00650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maesawa C, Tamura G, Nishizuka S, Ogasawara S, Ishida K, Terashima M, et al. Inactivation of the CDKN2 gene by homozygous deletion and de novo methylation is associated with advanced stage esophageal squamous cell carcinoma. Cancer Res. 1996;56(17):3875–3878. [PubMed] [Google Scholar]
- 19.Ling ZQ, Li P, Ge MH, Zhao X, Hu FJ, Fang XH, et al. Hypermethylation-modulated down-regulation of CDH1 expression contributes to the progression of esophageal cancer. Int J Mol Med. 2011;27(5):625–635. doi: 10.3892/ijmm.2011.640. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, Brigham, Women’s H, Broad I et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 2016;150(5):1171–1182. doi: 10.1053/j.gastro.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300(5618):455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, Chen C, Liu LU, Zhan NA, Li B. Expression of Smad4, TGF-betaRII, and p21(waf1) in esophageal squamous cell carcinoma tissue. Oncol Lett. 2015;9(6):2847–2853. doi: 10.3892/ol.2015.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XC, Wang MY, Yang M, Dai HJ, Zhang BF, Wang W, et al. A mutational signature associated with alcohol consumption and prognostically significantly mutated driver genes in esophageal squamous cell carcinoma. Ann Oncol. 2018;29(4):938–944. doi: 10.1093/annonc/mdy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Miranda NF, van Dinther M, van den Akker BE, van Wezel T, ten Dijke P, Morreau H. Transforming growth factor beta signaling in colorectal cancer cells with microsatellite instability despite biallelic mutations in TGFBR2. Gastroenterology. 2015;148(7):1427–1437. doi: 10.1053/j.gastro.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 28.Sakai E, Nakayama M, Oshima H, Kouyama Y, Niida A, Fujii S, et al. Combined mutation of Apc, Kras, and Tgfbr2 effectively drives metastasis of intestinal cancer. Cancer Res. 2018;78(5):1334–1346. doi: 10.1158/0008-5472.CAN-17-3303. [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research N, Albert Einstein College of M, Analytical Biological S, Barretos Cancer H, Baylor College of M, Beckman Research Institute of City of H et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Zhao KN, Masci PP, Lakhani SR, Antonsson A, Simpson PT, et al. TGFbeta isoforms and receptors mRNA expression in breast tumours: prognostic value and clinical implications. BMC Cancer. 2015;15:1010. doi: 10.1186/s12885-015-1993-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung HH, Lee TL, Rennert OM, Chan WY. DNA methylation of cancer genome. Birth Defects Res C Embryo Today. 2009;87(4):335–350. doi: 10.1002/bdrc.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan Z, Yu H, Cuk K, Zhang Y, Brenner H. Whole-blood DNA methylation markers in early detection of breast cancer: a systematic literature review. Cancer Epidemiol Biomark Prev. 2019;28(3):496–505. doi: 10.1158/1055-9965.EPI-18-0378. [DOI] [PubMed] [Google Scholar]
- 34.Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin Epigenetics. 2016;8:115. doi: 10.1186/s13148-016-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shridhar K, Walia GK, Aggarwal A, Gulati S, Geetha AV, Prabhakaran D, et al. DNA methylation markers for oral pre-cancer progression: a critical review. Oral Oncol. 2016;53:1–9. doi: 10.1016/j.oraloncology.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campan M, Moffitt M, Houshdaran S, Shen H, Widschwendter M, Daxenbichler G, et al. Genome-scale screen for DNA methylation-based detection markers for ovarian cancer. PLoS One. 2011;6(12):e28141. doi: 10.1371/journal.pone.0028141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim Y, Wan Y, Vagenas D, Ovchinnikov DA, Perry CF, Davis MJ, et al. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer. 2016;16(1):749. doi: 10.1186/s12885-016-2785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markopoulou S, Nikolaidis G, Liloglou T. DNA methylation biomarkers in biological fluids for early detection of respiratory tract cancer. Clin Chem Lab Med. 2012;50(10):1723–1731. doi: 10.1515/cclm-2012-0124. [DOI] [PubMed] [Google Scholar]
- 39.Rettori MM, de Carvalho AC, Bomfim Longo AL, de Oliveira CZ, Kowalski LP, Carvalho AL, et al. Prognostic significance of TIMP3 hypermethylation in post-treatment salivary rinse from head and neck squamous cell carcinoma patients. Carcinogenesis. 2013;34(1):20–27. doi: 10.1093/carcin/bgs311. [DOI] [PubMed] [Google Scholar]
- 40.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16(23):5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 41.Pietrusinski M, Kepczynski L, Jedrzejczyk A, Borkowska E, Traczyk-Borszynska M, Constantinou M, et al. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark. 2017;18(1):47–59. doi: 10.3233/CBM-160673. [DOI] [PubMed] [Google Scholar]
- 42.Moreira-Barbosa C, Barros-Silva D, Costa-Pinheiro P, Torres-Ferreira J, Constancio V, Freitas R, et al. Comparing diagnostic and prognostic performance of two-gene promoter methylation panels in tissue biopsies and urines of prostate cancer patients. Clin Epigenetics. 2018;10(1):132. doi: 10.1186/s13148-018-0564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawakami K, Brabender J, Lord RV, Groshen S, Greenwald BD, Krasna MJ, et al. Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst. 2000;92(22):1805–1811. doi: 10.1093/jnci/92.22.1805. [DOI] [PubMed] [Google Scholar]
- 44.Hibi K, Taguchi M, Nakayama H, Takase T, Kasai Y, Ito K, et al. Molecular detection of p16 promoter methylation in the serum of patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2001;7(10):3135–3138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. TGFBR2 mRNA levels in several ESCC cell lines. (A) RT-qPCR to detect levels of TGFBR2 mRNA in Het-1A and ESCC cell lines using ACTB as a control gene. (B) RT-qPCR to detect levels of TGFBR2 mRNA in Het-1A and ESCC cell lines using 18S RNA as a control gene.
Data Availability Statement
All the datasets are available from the corresponding author upon reasonable request.