Abstract
Background
Quality evaluation of multi-species resourced herb medicine (MSRHM) is a main problem for quality control of herb medicine. Current quality evaluation methodology lost consideration of species discrepancy. New quality evaluation strategy for MSRHM is in urgent need. Qinjiao, a representative MSRHM, originated from Gentiana macrophylla Pall., Gentiana straminea Maxim., Gentiana crassicaulis Duthie ex Burk. or Gentiana dahurica Fisch., has been used as an important herb medicine over 2000 years for expelling wind-dampness and relieving impediment pain. However, quality evaluation among species has never been revealed. The current work proposes an integrated quality evaluation strategy for MSRHM of Qinjiao, which may promote innovation of quality control of MSRHM.
Methods
In this work, 58 batches of Qinjiao covering 4 species were collected. Genetic comparative analysis based on ITS2 sequence was conducted. Metabolomics analysis based on TOF–MS and NMR spectrum were carried out. Compounds underlying species differences were identified and their discrepancies among species were investigated by ANOVA analysis and multivariate analysis.
Results
Four species of Qinjiao can be authenticated by ITS2 sequence comparation. Metabolomics analysis by TOF/MS and NMR revealed chemical discrepancies among species of Qinjiao. Maximum discrepancy was present between Gentiana crassicaulis Duthie ex Burk. and Gentiana dahurica Fisch. Chemical difference among species were tentative explored. For TOF–MS profiling, 28 constituents were tentative identified, 17 of which were further confirmed by standards. For 1H-NMR profiling, signals from 5 compounds were assigned. Contents discrepancies were investigated by ANOVA analysis. It seems that (seco)iridoids like loganic acid, gentiopicroside or swertiamarin were richer in specie of Gentiana crassicaulis Duthie ex Burk., while flavonoid (morroniside) and triterpenoids (roburic aicd, ursolic acid, oleanolic acid, β-sitosterone) were richer in specie of Gentiana dahurica Fisch. The current research demonstrates that metabolite profiling based on both UPLC/Q-TOF MS and 1H-NMR coupled with ITS2 sequence comparation can be a powerful tool for quality investigation of MSRHM of Qinjiao.
Conclusions
A comprehensive quality evaluation strategy for MSRHM was proposed by integrating UPLC-Q-TOF–MS, NMR based metabolic analysis and ITS2 sequence genetic comparation. The proposed quality evaluation strategy shall promote innovation of quality control of traditional Chinese medicine.
Keywords: Quality evaluation, Multi-species resourced herb medicine (MSRHM), Qinjiao, ITS2, Metabolomics
Background
Quality control of multi-species resourced herb medicine (MSRHM) is a main problem for quality control of herb medicine. Current quality control methodology bears disadvantages of lacking consideration of species discrepancy, ignoring the fact that discrepancies among species were inevitable and shall produce different chemical component and clinical effects. As a result, new quality investigation strategy for MSRHM is in urgent need.
Qinjiao, namely Gentianae Macrophyllae Radix, is an ancient Chinese herb medicine and has been described and recorded in several ancient Chinese Medicine monographs like Shen Nong’s Herbal Classic (Han Dynasty, Shen Nong Ben Cao Jing), Compendium of Materia Medica (Ming Dynasty, Ben Cao Gang Mu) [1] and also in Chinese pharmacopoeia. Over 2000 years, Qinjiao has been utilized to treat a wide range of diseases, including hypertension, osteoarthritis, and especially rheumatism [2]. According to China Pharmacopoeia 2015 version [3], Qinjiao consists of the dried roots of Gentiana macrophylla Pall. (G. macrophylla), Gentiana straminea Maxim. (G. straminea), Gentiana crassicaulis Duthie ex Burk. (G. crasicaulis) or Gentiana dahurica Fisch. (G. daurica). As a represented MSRHM, quality and efficacy of Qinjiao among species were inevitable variable [4, 5]. Former researches mainly focused on contents determination or chemical profiling of certain kind of Qinjiao [6, 7], or even chemical and genetic analysis of G. crasicaulis and G. macrophylla [8]. No systematic species investigation for four kinds of Qinjiao has been revealed, which may shed new light into quality control and clinical utilization of Qinjiao.
DNA barcode of internal transcribed spacer 2 (ITS2) is prevalently adopted as a universal barcode for plant, especially herbal medicinal identification [9]. ITS2 barcode has been successfully employed for species identification of Qinjiao [8, 10]. Chemical profiling combined with multivariate analysis provided systematic chemical comparison of metabolites, and can be powerful tool for species investigation of MSRHM [11]. Ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF–MS) and proton nuclear magnetic resonance spectrometer (1H-NMR) are the most frequently employed platform for metabolic profiling.
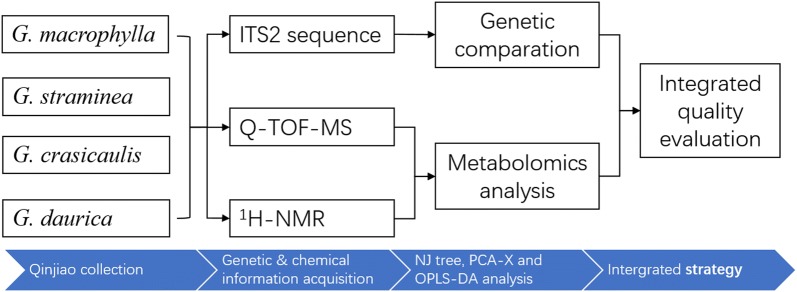
Herein, this research collected 58 batches of Qinjiao, covering four species, originated from main producing areas of China (Gansu, Shanxi, Hebei, Shaanxi, Heilongjiang, Liaoning, Nei Mongol, Yunnan, Szuchuan, Qinghai, Tibet). Genetic comparative and metabolic profiling analysis of four species of Qinjiao were carried out. First, all herbs were authenticated by morphological identification as well as DNA barcoding comparison. Then, chemical profiles were acquired by UPLC-Q-TOF–MS and 1H-NMR. Third, metabolomics based species investigation, including principal component analysis (PCA) and orthogonal partial least squares discrimination analysis (OPLS-DA), was conducted to reveal the quality discrepancy among species. Finally, an integrated quality evaluation strategy for MSRHM of Qinjiao by UPLC-Q-TOF–MS, NMR based metabolomics analysis and ITS2 sequence genetic comparation was established. The flow chart of the proposed methodology is shown in Fig. 1. The current work may facilitate quality control, utilization and species discrimination of different kinds of Qinjiao.
Fig. 1.
The flow chart of the proposed methodology
Materials and methods
Sample collection and preparation
Fifty-eight batches of rhizomes of G. macrophylla, G. straminea, G. crasicaulis and G. daurica were collected from different herbal markets or harvested from various locations of China. All samples were authenticated by Professor Jiuzhi Yuan (Shenyang Pharmaceutical University) or Professor Lihong Wu (Shanghai University of Traditional Chinese Medicine). Voucher specimens were deposited in Department of Pharmacy, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China or the Institute of Chinese Materia Medica, Shanghai University of Traditional Chinese Medicine, Shanghai, China. Detailed information of collected samples in this study is list in Table 1. The roots were gently washed and dried at 50 °C for 48 h, and then were grounded into powder and stored in glass jars in the dark at room temperature until further analysis.
Table 1.
Samples information of Qinjiao for metabolomics and ITS2 barcode analysis
| Sample ID | Origins | Species | GenBank accession nos. |
|---|---|---|---|
| QJ01 | Mongolian autonomous county of Henan, Tibetan autonomous prefecture of Huangnan, Qinghai province | G. macrophylla Pall. | MH602351 |
| QJ02 | Gonjo county, Qamdo city, Tibet | G. crassicaulis Duthie ex Burk. | MH602352 |
| QJ03 | Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. crassicaulis Duthie ex Burk. | MH602353 |
| QJ04 | Nyingchi city, Tibet autonomous region | G. crassicaulis Duthie ex Burk. | MH602354 |
| QJ05 | Ganzi Tibetan autonomous prefecture, Sichuan province | G. crassicaulis Duthie ex Burk. | MH602355 |
| QJ06 | Maqu country, Gannan Tibetan autonomous prefecture, Gansua province | G. straminea Maxim. | MH602389 |
| QJ07 | Tibetan autonomous prefecture of Golog, Qinghai province | G. straminea Maxim. | MH602390 |
| QJ08 | Mongolian autonomous county of Henan, Tibetan autonomous prefecture of Huangnan, Qinghai province | G. straminea Maxim. | MH602391 |
| QJ09 | Ebian Yi Nationality autonomous county, Minle city, Sichuan province | G. crassicaulis Duthie ex Burk. | MH602356 |
| QJ10 | Hohhot city, Inner Mongolia | G. straminea Maxim. | MH602392 |
| QJ11 | Daguan country, Zhaotong city, Yunnan province | G. crassicaulis Duthie ex Burk. | MH602357 |
| QJ13 | Yulong naxi autonomous prefecture, Lijiang city, Yunan province | G. crassicaulis Duthie ex Burk. | MH602358 |
| QJ14 | Minle country, Zhangye city, Gansu provinice | G. straminea Maxim. | MH602393 |
| QJ15 | Xinzhou city, Shanxi province | G. crassicaulis Duthie ex Burk. | MH602359 |
| QJ16 | Gonghe town, Qingyang city, Gansu province | G. macrophylla Pall. | MH602387 |
| QJ17 | Barkam city, Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. crassicaulis Duthie ex Burk. | MH602360 |
| QJ18 | Barkam city, Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. crassicaulis Duthie ex Burk. | MH602361 |
| QJ19 | Barkam city, Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. crassicaulis Duthie ex Burk. | MH602362 |
| QJ20 | Ruoergai County, Tibetan Qiang Autonomous Prefecture of Ngawau, Sichuan | G. straminea Maxim. | MH602394 |
| QJ21 | Ruoergai county, Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. straminea Maxim. | MH602395 |
| QJ22 | Huating country, Pinliang city, Gansu province | G. macrophylla Pall. | MH602388 |
| QJ23 | Tongguan country, Weinan city, Shanxi province | G. crassicaulis Duthie ex Burk. | MH602363 |
| QJ24 | Malong country, Qujing city, Yunana province | G. crassicaulis Duthie ex Burk. | MH602364 |
| QJ25 | Huichuan town, Weiyuan country, Dingxi city, Gansu province | G. crassicaulis Duthie ex Burk. | MH602365 |
| QJ26 | Ludian country, Zhaotong city, Yunnan province | G. crassicaulis Duthie ex Burk. | MH602366 |
| QJ27 | Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. dahurica Fisch. | MH602405 |
| QJ28 | Guizhou province | G. crassicaulis Duthie ex Burk. | MH602367 |
| QJ29 | Yulong naxi autonomous prefecture, Lijiang city, Yunan province | G. crassicaulis Duthie ex Burk. | MH602368 |
| QJ30 | Heishui country, Tibetan Qiang Autonomous prefecture of Ngawau, Sichuan | G. crassicaulis Duthie ex Burk. | MH602369 |
| QJ31 | Heishui country, Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. straminea Maxim. | MH602396 |
| QJ32 | Jingping country, Suzhou city, shanxi province | G. dahurica Fisch. | MH602406 |
| QJ33 | Weixi Lisu autonomous county, Diqing Tibetan autonomous Prefecture, Yunan province | G. crassicaulis Duthie ex Burk. | MH602370 |
| QJ34 | Keshiketeng Banner, Linxi County, Neimenggu province | G. straminea Maxim. | MH602397 |
| QJ35 | Nima town, Maqu country, Gannan Tibetan autonomous prefecture, Gansu province | G. straminea Maxim. | MH602398 |
| QJ36 | Tibetan Qiang autonomous prefecture of Ngawau, Sichuan | G. straminea Maxim. | MH602399 |
| QJ37 | Tibet autonomous region | G. crassicaulis Duthie ex Burk. | MH602371 |
| QJ38 | Ping’an country, Haidong city, Qinghai province | G. straminea Maxim. | MH602400 |
| QJ39 | Yunan province | G. crassicaulis Duthie ex Burk. | MH602372 |
| QJ40 | Yulong naxi autonomous prefecture, Lijiang city, Yunan province | G. crassicaulis Duthie ex Burk. | MH602373 |
| QJ41 | Dali Bai autonomous prefecture, Yunan province | G. crassicaulis Duthie ex Burk. | MH602374 |
| QJ42 | Tibet autonomous region | G. crassicaulis Duthie ex Burk. | MH602375 |
| S0 | Tu autonomous county of Huzhu, Qinghai province | G. dahurica Fisch. | MH602402 |
| S1 | Yunan province | G. crassicaulis Duthie ex Burk. | MH602377 |
| S2 | kunming city, Yunna province | G. crassicaulis Duthie ex Burk. | MH602378 |
| S3 | kunming city, Yunna province | G. crassicaulis Duthie ex Burk. | MH602379 |
| S4 | Yunan province | G. crassicaulis Duthie ex Burk. | MH602380 |
| S5 | Lijiang city, Yunan province | G. crassicaulis Duthie ex Burk. | MH602381 |
| S6 | Yunan province | G. crassicaulis Duthie ex Burk. | MH602382 |
| S7 | Tibetan autonomous county of Muli, Sichuan province | G. crassicaulis Duthie ex Burk. | MH602383 |
| S8 | Yunan province | G. crassicaulis Duthie ex Burk. | MH602376 |
| S9 | Yunan province | G. macrophylla Pall. | MH602384 |
| S10 | Unknown | G. straminea Maxim. | MH602401 |
| S11 | Gansu province | G. macrophylla Pall. | MH602385 |
| S12 | Nei Monggol autonomous region | G. dahurica Fisch. | MH602407 |
| S13 | Zhangjiakou city, Hebei province | G. dahurica Fisch. | MH602408 |
| S14 | Helongjiang province | G. dahurica Fisch. | MH602403 |
| S16 | Nei Monggol autonomous region | G. dahurica Fisch. | MH602404 |
| S18 | Gansu province | G. macrophylla Pall. | MH602386 |
QJ01–QJ42 were deposited in department of pharmacy, the first affiliated hospital of Zhengzhou University, Zhengzhou, China; S0–S18 were deposited the Institute of Chinese Materia Medica, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Chemicals and reagents
Methanol-d4 (CD3OD, 99.8%) was obtained from Cambridge Isotope Laboratories (Miami, FL, USA). Methanol of HPLC grade was purchased from Honeywell Inc. (Morristown, NJ, USA). Formic acid of LC–MS grade was bought from ROE Scientific Inc. (Newark, DE, USA). Deionized water was purified using a Milli-Q system (Millipore, Bedford, MA, USA). The chemical reference standards (CRS) of gentiopicroside, loganic acid, swertiamarin, sweroside, 6′-O-β-d-Glucosylgentiopicroside, roburic acid, morroniside, isovitexin, homoorientin, oleanolic acid, ursolic acid, β-sitosterone, citric acid, quercetin, kaempferol, and daucosterol were purchased from Chengdu Must Bio-techenology Co., Ltd. (Chengdu, China), with HPLC purity > 98%. All other chemicals and reagents were of analytical grade and commercially available.
Genetic Analysis of collected Qinjiao samples
ITS2 was prevalently adopted as a universal barcode for plant, especially herbal medicinal identification [9]. ITS2 barcode has been successfully employed for species identification of Qinjiao [8, 10]. In current research, ITS2 region was compared to verify the authentication of collected Qinjiao samples. To extract total genomic DNA from dried rhizomes (50 mg), the protocol provided by the Plant Genomic DNA Kit (Tiangen Biotech, Co., Ltd., Beijing, China) was used. Extracted DNA sample was stored at − 20 °C until use. The ITS2 sequence was amplified using a pair of primers (ITS2F: 5′-ATGCGATACTTGGTGTGAAT-3′; ITS3R: 5′-GACGCTTCTCCAGACTACAAT-3′) described previously by Chen et al. [12]. The PCR amplification was conducted as described by Luo et al. [13]. Amplification products were examined by electrophoresis in 1% (wt/vol.) agarose gels and visualized under ultraviolet light to detect successfully amplified products and the possible contamination of negative controls. After purifying, the PCR products were directly subjected to sequencing.
Sequences were edited and assembled using DNAMAN software (version 6.0) and refined manually. ITS2 resuquence were identified using DNA barcoding system for identifying herbal medicine (http://www.tcmbarcode.cn). Genetic distances were calculated using the Kimura-2-Parameter (K2P) model. All the newly obtained ITS2 sequences were uploaded to GenBank.
UPLC-Q-TOF–MS profiles analysis of the collected Qinjiao
0.5 g power (through No. 3 sieve) of each Qinjiao sample was placed into a separate 50 mL stopper conical flask followed by the addition of 20 mL methanol. The mixtures were vortexed for 1 min, sonicated (40 kHz, 500 W) for 30 min. Cooling down to ambient temperature, the lost weight was made up by methanol. After centrifugation at 10,000 rpm, 4 °C for 10 min, a 2 μL aliquot of the supernatants was injected into a UPLC-ESI-Q-TOF system (AB Sciex, Framinghan, MA, USA) for MS analysis.
Chromatography separation was achieved on an ACQUITY UPLC HSS T3 column (100 × 2.1 mm i.d., 1.8 μm) maintained at 40 °C. The mobile phase consisted of A (0.1% formic acid in water) and B (0.1% formic acid in methanol), using gradient elution: 0–1 min, 5–30% B; 1–5 min, 30–40% B; 5–6 min, 40–90% B; 6–13 min, 90–100% B; 13–21 min, 100% B; 21.01–24 min, 5% B. The flow rate was set at 0.3 mL/min and the injection volume was 2 μL.
The mass spectrometric data were collected on a SCIEX X500R QTOF mass spectrometer (AB Sciex, Framinghan, MA, USA) coupled with an electrospray ionization interface in negative ion modes (ESI-). SCIEX OS software 1.2 (AB, Milford, MA) was employed for data acquisition and procession. The following parameters settings were used: the ion spray voltage of 4000 V; turbo spray temperature (TEM) of 600 °C; declustering potential (DP) of − 80 V; collision energy (CE) of − 45 V; nebulizer gas (gas 1) of 55 psi; heater gas (gas 2) of 55 psi, CAD gas of 7 psi, and curtain gas of 35. Nitrogen was kept as the nebulizer and auxiliary gas. TOF MS and TOF MS/MS were scanned with the mass range of m/z 50–1000. Continuous recalibration was carried out every six samples. In addition, dynamic background subtraction (DBS) trigger information-dependent acquisition (IDA) was used to trigger acquisition of MS/MS information of low-level constituents. The accurate mass and composition for the precursor ions and fragment ions were analyzed using the Markerview™ software (Version 4.1, Waters Co., Milford, MA, USA) integrated with the instrument.
QC samples were prepared by combining equal aliquots from all Qinjiao samples and were injected every six specimens during the whole analysis. QC data obtained was used to assess the stability of the LC/MS platform. For all QCs, 5 characteristic features (list in Additional file 1: Table S1) were picked out to verify the stability. The results proved that variations of retention times were less than 0.2 min, drift values of m/z were less than 10 PPM, and the RSD of peak areas were all below 10% (Additional file 1: Table S1).
Raw data from Q/TOF–MS were analyzed using Markerview for peak deconvolution and peak alignment with the following parameters: initial retention time 0.5 min, final retention time 23 min, mass tolerance 10 PPM, ion intensity threshold (3000 counts) and retention time tolerance 0.1 min. The data were combined into a single matrix by aligning peaks with the same mass-retention time pair together from each data file in the data set. The ion intensities of each peak detected (2806 MS features for ESI- modes) were normalized to the sum of the peak intensities in each sample. After normalization, the data was processed according to the “80% rule”, briefly only variables with values above zero presenting in at least 80% of each group were kept for the following analysis [14].
1H-NMR profiles analysis of the collected Qinjiao
The methanol extracting supernatants (600 μL) were dried down using a centrifugal vacuum concentrator and were redissolved in 600 μL of MeOD. After mixing well, 450 μL of reconstitution wan transferred into the 5 mm NMR tubes (Norell, Landisville, NJ, USA) for NMR analysis.
NMR spectral data were obtained at 300 K on a Bruker 600-MHz AVANCE III NMR spectrometer (Bruker, Germany), equipped with a 5.0-mm BBO probe, operating at 600.13 MHz for 1H. The zg30 Bruker pulse program was used for 1D 1H NMR, with a TD of 64 k, relaxation delay of 1 s, spectral width of 20 ppm, and 256 scans. A line-broadening factor of 0.3 Hz was applied to FIDs before Fourier transformation. All NMR spectra were phased and baseline-corrected manually using TOPSPIN 3.5 (Bruker, Germany). The spectra were referenced internally to the chemical shift of H-3 signal of gentiopicroside at 7.46 ppm. Each 1H-NMR spectrum over the ranged 0.5–10.0 ppm was reduced to 238 regions of equal width (0.04 ppm) and the signal intensity in each region was integrated using AMIX (version 3.9, Bruker, Germany). The region of 4.75–5.20 ppm was removed prior to any statistical analysis in order to eliminate any residual water signal. Then data was normalized in AMIX by dividing each integrated segment by the total area of the spectrum to reduce any significant.
Statistical analysis and compound annotation
Output data from TOF–MS or NMR analysis was separately imported into SIMCA (version 14.0, Umetrics, Umeå, Sweden) for multivariate statistical analysis (MS data unit variance scaled, NMR data pareto-scaled). To provide comparative interpretations and visualization of the metabolic differences among the four species of Qinjiao, PCA and OPLS-DA were applied to the TOF–MS or NMR data set. The quality of the models was described by R2X and Q2 values. R2X shows the proportion of variance in the data explained by the models and indicates goodness of fit. The value closer to 1 indicates the goodness of fit. Q2, on the other hand, shows the proportion of variance in the data predictable by the model and indicates predictability. The results were visualized in the form of score plots, where each point represents an individual sample (to show the group clusters), and loading plots or S-plots, where each coordinate represents one mass-retention feature or 1H-NMR spectral region (to identify the variables contributing to the classification). The variable importance of projection (VIP) is the vector to summarize the total importance of the variable in explaining the model. The corresponding variables with VIP > 1.0 were chosen as potential discriminative metabolites. To justify the OPLS-DS models, analysis of variance testing of Cross-Validated predictive residuals (CV-ANOVA) were conducted. CV-ANOVA is a diagnostic tool for assessing the reliability of PLS and OPLS models. The P value produced by CV-ANOVA indicates the probability level. The common practice is to interpret a p-value lower than 0.05 as pointing to a significant model. Statistical analysis was also performed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test (SPSS, Chicago, IL, USA). A probability of P < 0.05 was considered to be statistically significant between two groups.
LC–MS Peaks were identified according to actual mass, MS/MS fragments and retention time (RT). First, the m/z value of the molecular ion of interest was searched against a self-build Qinjiao constituent Database, where data was collected from published researches [15, 16]. Then, the putative identifications were verified by comparing the MS2 fragmentations. Part of the constituents were further identified by reference standards. 1H-NMR signals were assigned by comparing the spectrum of Qinjiao with that of gentiopicroside, loganic acid, sweroside, swertiamarin or sucrose by Chenomx NMR suit (version7.6, Chenomx, Edmonton, Canada) with method described in our previous work [17].
Results
ITS2 comparation
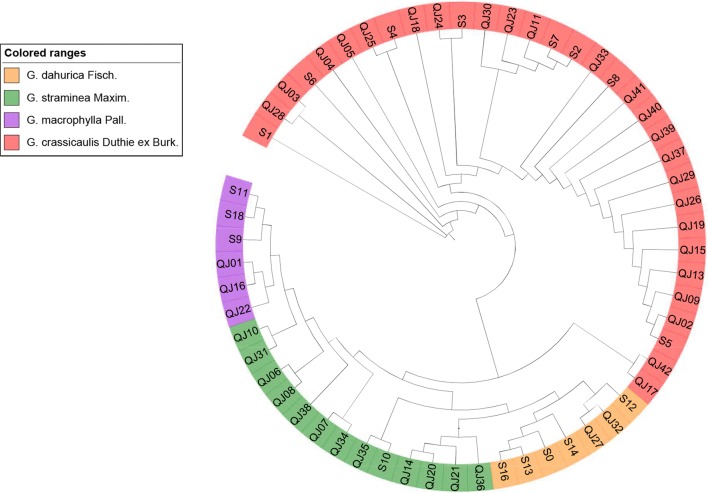
ITS2 sequences from collected Qinjiao samples were submitted to GenBank database (Accession numbers were listed in Table 1), assembled with CodonCode Aligner 3.7.1 (CodonCode Co., Dedham, MA, USA) and aligned using ClustalW. Kimura 2-Parameter (K2P) distances. GC content of base and NJ trees were calculated and constructed using the MEGA X software with the Bootstrap method (500 resampling) and K2P model [18]. The distances within or among species are separately list in Table 2. The average ITS2 region was 497 bp in length in 4 species of Qinjiao, and the G+C content ranged from 55.7% to 56.2%, with an average of 56.0% (Table 2). A neighbor-joining (NJ) tree of ITS2 barcode was formed on the basis of K2P model (Fig. 2).
Table 2.
Sequence sizes, percent G+C content and mean distance intra/inter each spices of collected Qinjiao samples
| Species | ITS2 length (bp) | G+C content (%) | Mean distance intra species | Mean distance inter species | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||
| 1 G. crassicaulis Duthie ex Burk. | 495.8 | 56.0 | 0.0274 | |||
| 2 G. dahurica Fisch. | 496.0 | 55.9 | 0.0229 | 0.0371 | ||
| 3 G. macrophylla Pall. | 498.2 | 55.7 | 0.0062 | 0.0312 | 0.0246 | |
| 4 G. straminea Maxim. | 498.8 | 56.2 | 0.0130 | 0.0288 | 0.0220 | 0.0146 |
Fig. 2.
NJ tree of G. macrophμlla, G. straminea, G. crassicaulis and G. dahurica
TOF–MS data metabolomics
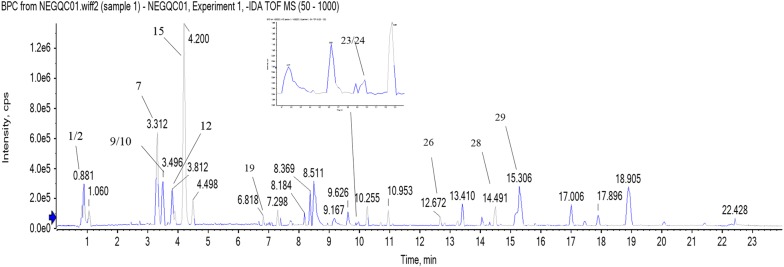
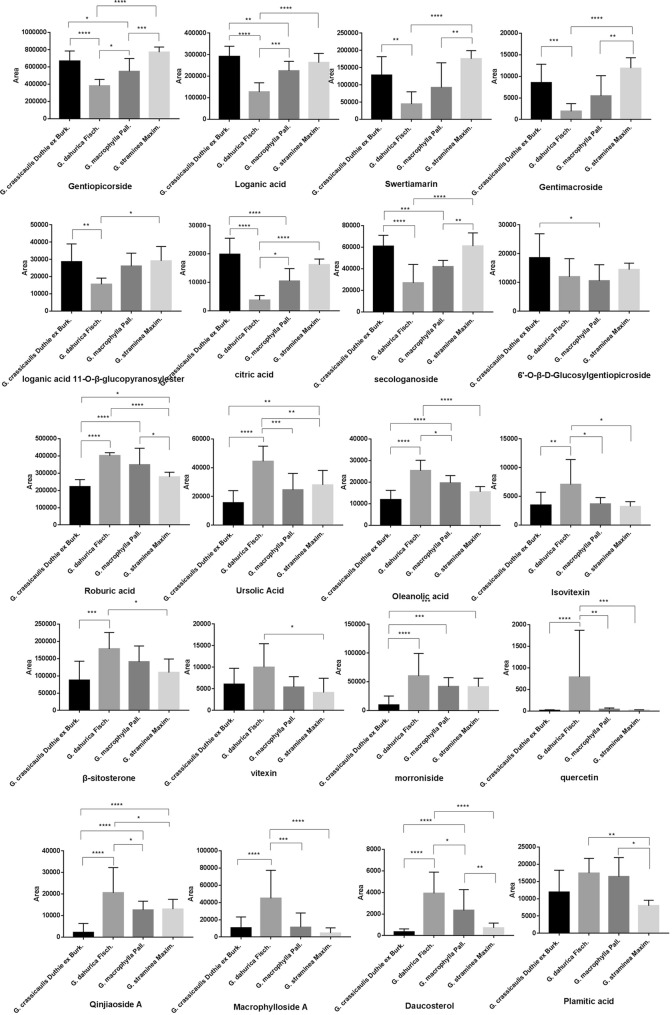
Representative base peak intensity (BPI) chromatograms of G. crassicaulis is shown in Fig. 3. By comparing actual mass, MS/MS fragments and retention time (RT) of target compounds, 28 constituents were identified, among which 17 were further identified by reference standards. The compound information is list in Table 3, with XIC chromatography shown in Additional file 2: Fig. S1. ANOVA followed by Tukey’s multiple comparison test were conducted for these 28 compounds (partly shown in Fig. 4).
Fig. 3.
Representative BPI chromatograms of G. crassicaulis, with part of the constituents identified. Note: 1. Sucrose, 2. Swertiajaposide A, 7. Loganic acid, 9. 6′-O-β-d-Glucosylgentiopicroside, 10. Qinjiaoside A, 12. Swertiamarin, 15. Gentimacroside, 19.Vitexin, 23. Ursolic Acid, 24. Oleanolic acid, 26. 2α-hydroxyl ursolic acid, 28. β-sitosterone, 29. Roburic acid
Table 3.
Characterization of chemical constituents in Qinjiao by UHPLC-QTOF–MS in ESI-mode
| No. | tR/min | Observed mass | Molecular weight | Molecular formula | MS/MS fragments ions (m/z) | Identified compound |
|---|---|---|---|---|---|---|
| 1a | 0.87 | 341.1086 | 342.1161 | C12H22O11 | 179.0562, 161.0459, 131.0351, 119.0350, 89.0241, 59.0135 | Sucrose |
| 2b | 0.88 | 387.1148 | 388.1369 | C17H24O10 | 341.1090, 195.0507,179.0562, 59.0238, 59.0136 | Swertiajaposide A |
| 3* | 1.63 | 191.0196 | 192.0270 | C6H8O7 | 111.0086, 87.0087, 67.0188, 57.0345 | Citric acid |
| 4b | 2.56 | 169.0870 | 170.0943 | C9H14O3 | 123.0806, 121.0663, 110.9975, 95.0503, 67.0559, 61.9881 | Isoboonein |
| 5b | 2.73 | 315.0719 | 316.0794 | C13H16O9 | 315.0723, 153.0181, 109.0293 | 5-(β-d-Glucopyranosyl)-2-hydroxybenzoic acid |
| 6b | 2.99 | 537.1828 | 538.1897 | C22H34O15 | 375.1297,221.0264, 213.0769, 169.0869, 113.0241, 179.0557, 89.0242, 69.0344 | Loganic acid 11-O-β-glucopyranosylester |
| 7a | 3.31 | 399.1258, | 376.1369 | C16H24O10 | 213.0766, 169.0868, 151.0761, 119.3047, 113.0240, 89.0241, 69.0342, 59.0135 | Loganic acid |
| 8a | 3.39 | 451.1446 | 406.1475 | C17H26O11 | 405.1165, 243.0872, 141.0553, 101.0242, 89.0241, 59.0135 | Morroniside |
| 9a | 3.42 | 563.1620 | 518.1635 | C22H30O14 | 221.0659, 179.0552, 161.0449, 119.0343, 89.0237 | 6′-O-β-d-Glucosylgentiopicroside |
| 10b | 4.49 | 403.1245 | 404.1319 | C17H24O11 | 151.0764, 125.0242, 113.0243, 89.0243, 59.0137 | Qinjiaoside A |
| 11b | 3.67 | 519.1726 | 520.1791 | C22H32O14 | 519.1743, 323.0995, 213.0767, 151.0762, 125.0244, 59.0142 | Swertiapunimarin;6′-O-Glucopyranosylsweroside |
| 12a | 3.73 | 419.1193 | 374.1212 | C16H22O10 | 149.0605, 141.0188, 119.0347, 89.0238 | Swertiamarin |
| 13b | 3.89 | 389.1085 | 390.1161 | C16H22O11 | 183.0660, 165.0557, 121.0655, 89.0241, 69.0343 | Secologanoside |
| 14a | 4.11 | 401.1074 | 356.1107 | C16H20O9 | 149.0604, 119.0347, 89.0239, 71.0136, 59.0133 | Gentiopicorside |
| 15b | 4.17 | 531.1529 | 532.1581 | C26H28O12 | 235.0612, 191.0714, 173.0608, 163.0767, 149.0606, 89.0243 | Gentimacroside |
| 16a | 4.4 | 403.1244 | 358.1263 | C16H22O9 | 357.1193, 195.0666, 179.0564, 125.0241, 119.0347, 151.0763, 89.0242, 81.0343, 59.0136 9 | Sweroside |
| 17a | 5.78 | 447.0936 | 448.1005 | C21H20O11 | 429.1045, 369.0630, 357.0616, 327.0508, 297.0398, 285.0404, 61.9884 | Isoorientin |
| 18a | 6.62 | 431.0983 | 432.1056 | C21H20O10 | 341.0668, 323.0563, 311.0559, 283.0608, 281.0451 | Isovitexin |
| 19b | 6.76 | 477.1032 | 432.1056 | C21H20O10 | 431.0983, 323.0766, 315.0720, 161.0238, 153.0187, 152.0048 | Vitexin |
| 20b | 7.04 | 875.2236 | 876.2324 | C40H44O22 | 875.2251, 833.2133, 739.2096, 713.1740, 577.1562, 535.1466, 315.0723, 153.0191, | Macrophylloside A |
| 21a | 7.08 | 301.0345 | 302.0426 | C15H10O7 | 193.0142, 149.0248, 121.0312 | Quercetin |
| 22a | 7.19 | 285.0397 | 286.0477 | C15H10O6 | 192.0061, 177.0194, 142.9487, 119.0135, 87.0237 | Kaempferol |
| 23a | 9.89 | 455.3531 | 456.3604 | C30H48O3 | 409.2535, 152.9962 | Ursolic acid |
| 24a | 9.92 | 455.3529 | 456.3604 | C30H48O3 | 409.2535, 152.9962 | Oleanolic acid |
| 25b | 10.73 | 255.2308 | 256.2402 | C16H32O2 | 255.2326, 237.2210 | Plamitic acid |
| 26a | 13.5 | 621.4381 | 576.4390 | C35H60O6 | 575.4681, 303.8955, 295.2278, 191.9469, 89.0241 | Daucosterol |
| 27a | 14.5 | 457.3658 | 414.3862 | C29H50O | 457.3671, 411.3623 | β-Sitosterone |
| 28a | 15.3 | 439.3581 | 440.3654 | C30H48O2 | 439.3565, 421.3469 | Roburic acid |
aIdentified by comparing with the reference standards
bPutative identifications by MS and MS2 fragmentations
Fig. 4.
Part of ANOVA test result of identified constituents by TOF–MS. *P < 0.05; **P < 0.01; ***P < 0.001;****P < 0.0001
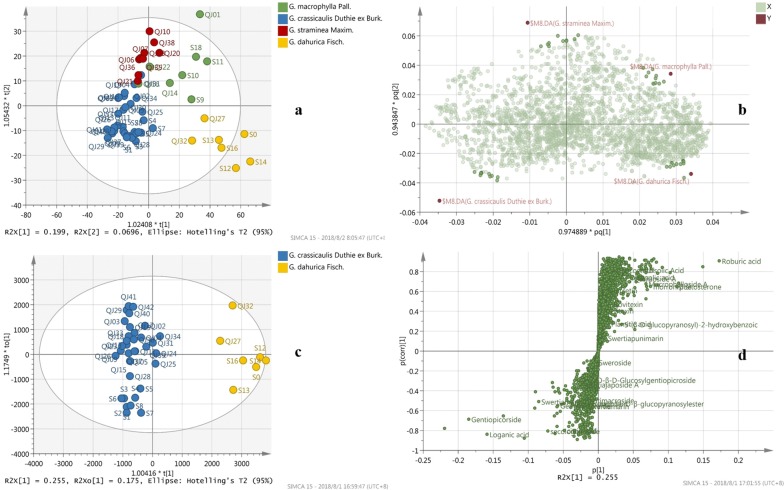
Multivariate analysis of the TOF–MS data was carried out. Initially, unsupervised PCA-X analysis were conducted among groups, showing preferably discriminative distribution (not shown, R2X = 0.661, Q2 = 0.398). Subsequently, to maximize the variation among groups and to determine the variables that contributed to this variation, supervised OPLS-DA model (Fig. 5a) was employed among four species of Qinjiao, with R2X = 0.38, R2Y = 0.666, Q2 = 0.549. The loading plot (Fig. 5b) revealed the correlations between class (species) and variables (MASS feature), where variables clustering close to each class were considered to make great contributions to the classification. Furthermore, it was noticed that G. crassicaulis and G. dahurica clustered furthest from each other in the score plot (Fig. 5a). To explore the difference, OPLS-DA analysis for these two groups was conducted, with R2Y 0.924 and Q2 0.864 (Fig. 5c). Corresponding S-plot (Fig. 5d) was analyzed, which was commonly used and effectively showed the difference between groups. Part of the variable selected from S-plot with VIP value > 1.0 and their abundances were shown in Additional file 3: Table S2. The established OPLS-DA models were further validated by CV-ANOVA test, with P valve less than 0.05 indicating significant models (Additional file 4: Table S3).
Fig. 5.
Score plot and loading plot of TOF–MS data. a Score plot of 4 species of Qinjiao; b loading plot of OPLS-DA model; c OPLS-DA score plot of G. crassicaulis and G. dahurica species; d S-plot of G. crassicaulis and G. dahurica species
1H-NMR data metabolomics
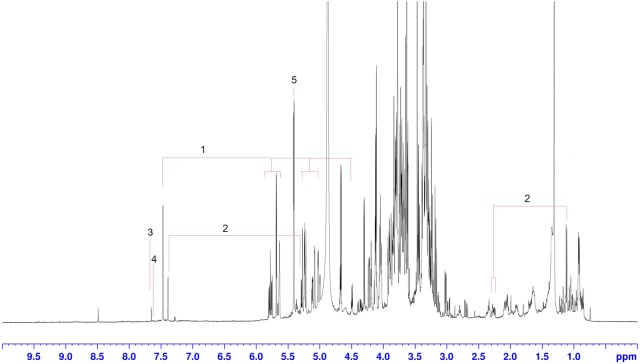
A representative 1D 1H-NMR spectrum of G. crassicaulis is shown in Fig. 6. Signal assignments were list in Table 4. Fore iridoids, gentiopiciroside, swertiamarin, loganic acid, and sweroside were identified ultimately by Chenomx NMR suit (version7.6, Chenomx, Edmonton, Canada).
Fig. 6.
Representative 1H-NMR spectrum of G. crassicaulis and signal identified. 1. Gentiopicroside, 2. Loganic acid, 3. Swertiamarin, 4. Sweroside, 5. Sucrose
Table 4.
Characteristics of 1H-NMR signals observed in Qinjiao extract
| Compound | Group | Chemical shift δ (ppm) |
|---|---|---|
| Gentiopicroside | 3-H(d) | 7.47 |
| 8-H(m) | 5.76 | |
| 1-H(d) | 5.66 | |
| 6-H(m) | 5.63 | |
| 10-CH2(dt) | 5.22 | |
| 7=CH(m) | 5.00–5.10 | |
| 1-glc-1′(d) –CH (anomeric) | 4.67 | |
| Loganic acid | 3-H(d) | 7.38 |
| 1-H(d) | 5.28 | |
| 5-H(m) | 2.24 | |
| 9-H(m) | 1.98 | |
| 8-H(d) | 1.10 | |
| 1-glc-1′(d) –CH (anomeric) | 4.66 | |
| Swertiamarin | 3-H(s) | 7.65 |
| 9-H(d) | 2.93 | |
| Sweroside | 3-H(d) | 7.60 |
| Sucrose | –CH (anomeric)(d) | 5.40 |
a Letters in parentheses indicate the peak multiplicity property of proton NMR signal
s singlet, d doublet, dd double doublet, dt double triplet, t triplet, m multiplet, br broad
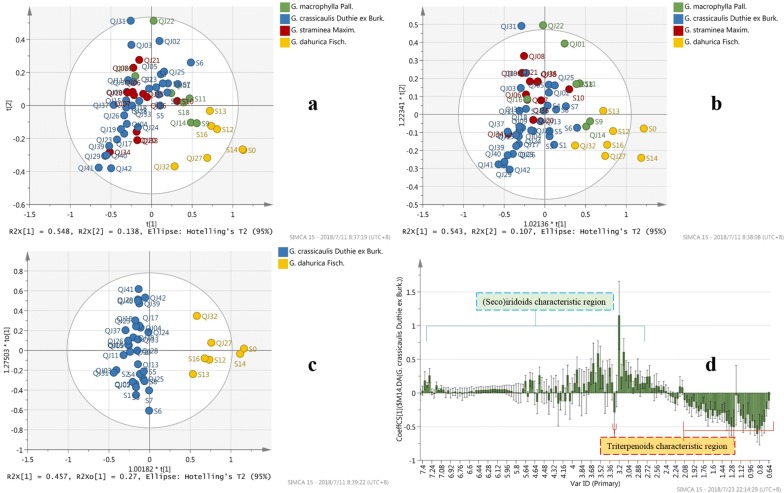
NMR data acquired was also analyzed by PCA-X model (R2X = 0.943, Q2 = 0.817) and OPLS-DA models (R2X = 0.38, R2Y = 0.666, Q2 = 0.549). As shown in Fig. 7a, b, samples from species of G. crassicaulis, G. straminea, G. macrophylla, and G. dahurica distributed regionally in the score plot, with the latter 3 species gradually deviating far away from the first one.
Fig. 7.
Score plot and loading plot of 1H-NMR data. a, b: PCA and OPLS-DA score plot of 4 species of Qinjiao; c, d: Score plot and s-plot of OPLS-DA analysis between G. crassicaulis and G. dahurica species
To explore the diversity between G. crassicaulis and G. dahurica, OPLS-DA analysis was achieved between them, with R2X 0.726, R2Y 0.884 and Q2 0.848 (Fig. 7c). Corresponding S plot (Fig. 7d) was generated. The established OPLS-DA models were further validated by CV-ANOVA test, with P valve less than 0.05 indicating significant models (Additional file 4: Table S3).
Discussion
ITS2 data analysis
Minimum distance within species was observed between G. macrophylla_and G. straminea, with K2P distance value 0.0146. While maximum K2P distance within species was present between G. crassicaulis and G. dahurica, with value 0.0371. The minimum interspecific distance of ITS2 region was higher than the maximum intraspecific distance, indicating that the ITS2 barcode performed well in the discrimination of four species of Qinjiao. The distances among species revealed were in accordance with discrepancy detected by following metabolomics analysis. It was illustrated that species of G. macrophylla, G. straminea, G. dahurica and G. crassicaulis can be clearly distinguished by the NJ tree. ITS2 analysis confirmed the potential discrepancy among species and guaranteed reliability of subsequent metabolomics-based species evaluation.
TOF–MS data analysis
The score plots (Fig. 5a) shows that samples from species of G. macrophylla, G. straminea, G. dahurica and G. crassicaulis located different areas in score plot, inferring distinctive chemical profiles of four species of Qinjiao. In general, G. crassicaulis and G. straminea were closest in the score plot, while G. macrophylla and G. dahurica were progressively far away from them. Maximum spatial distance was present between G. dahurica and G. crassicaulis. The result was consistent with discrepancy revealed by ITS2 analysis. The loading plot (Fig. 5b) revealed MASS features making great contributions to the classification, which was further confirmed by ANOVA test. Flavonoids (isovitexin, morronside, quercetin) and triterpenoids (oleanolic acid, ursolic acid, roburic acid, β-sitosterone, daucosterol), as well as certain iridoids (macrophylloside A and Qinjiaoside A) were significant higher in G. dahurica. than other 3 species; while other(seco)iridoids (swertiamarin, secologanoside, loganic acid, gentimacroside, loganic acid 11-O-β-glucopyranosylester) and citric aicd were richer in other 3 species of Qinjiao than G. daurica. No obvious distinction was detected in contents of sweroside, sucrose, swertiapunimarin, and swertiajaposide A among four species of Qinjiao.
OPLS-DA score plot (Fig. 5c) and corresponding S-plot (Fig. 5d) of G. crassicaulis and G. dahurica confirmed maximum K2P distance revealed by ITS2 analysis. It was noticed that MASS features of 401.1074_4.19 (gentiopicroside), 375.1276_3.29 (loganic aicd), 419.1193_3.81(swertiamarin), 191.0200_1.09(citric acid), 563.1601_3.50(6′-O-β-d-glucosyl-gentiopicroside), and 389.1087_3.89 (secologanic acid) were richer in G. crassicaulis, while MASS features of 439.3562_15.30 (roburic aicd), 455.3510_9.93 (ursolic Acid), 455.3505_9.99 (oleanolic acid), 457.3658_14.5(β-sitosterone) and 451.1456_3.40 (morroniside) were richer in G. dahurica. The discrepancy was consistent with ANOVA test result (Fig. 4). It seems that (seco)iridoids like loganic acid, gentiopicroside or swertiamarin were richer in specie of G. crassicaulis, while flavonoid (morroniside) and triterpenoids (roburic aicd, ursolic acid, oleanolic acid, β-sitosterone) were richer in specie of G. dahurica. The discrepancy was consistent with previous reports [19, 20], which reported higher contents of gentiopicroside, loganic acid, sertiamarin, and 6′-O-β-d-glucosyl-gentiopicroside in G. crassicaulis comparing with G. dahurica.
1H-NMR data analysis
According to the score plot (Fig. 7a, c), NMR based metabolomics validated results revealed by MS based metabolomics. The S-plot (Fig. 7d) between these two groups showed that signal abundance from 3 to 7 ppm was richer in G. crassicaulis, while which from around 0.8–2 ppm was richer in G. dahurica. Unfortunately, limited to the complexity of the 1H-NMR spectra, compounds related to the abundance difference were not directly identified. However, the difference was confirmed and can be explained by preceding MS based metabolomics. The signal intensity discrepancy present in 1H-NMR spectra coincided with abundance discrepancy MS revealed (Fig. 5d). Richer abundance of triterpenoids in G. dahurica produced richer signal intensity in characteristic region range of 0.5 to 2.4 ppm (signal from Skeleton proton) and 3.24 to 3.28 ppm(H-3) [21]. On the other side, higher concentration of (seco)iridoids produced higher signal intensity of 2.4 to 5 ppm (signal from (seco)iridoids skeleton proton) and 7.0–7.4 ppm(signal of H-3) [22]. The consistence by NMR and TOF–MS metabolomics confirmed the chemical discrepancy between G. crassicaulis and G. dahurica. The discrepancy may be helpful for distinguishing of these two species and may lead to potential pharmacodynamics discrepancy, which remains to be investigated.
ITS2, TOF–MS and NMR integration
ITS2 based gene comparison can present species distance or genetic relationship among species. Currently, ITS2 is the mostly commonly used region for the barcoding and authentication of herbal medicinal materials. In future, species authentication by ITS2 sequence (or other genetic method) may be precondition of quality evaluation of MSRHM. However, environmental implication and chemical discrepancy were not reflected. Information of quality and origin shall ultimately rely on chemical method. On the other side, TOF–MS or NMR based metabolomics analysis providing systemic chemical information among species, and can be powerful tool for quality investigations. TOF–MS bears the advantage of high sensitivity, high specificity, and thus offers maximum amount of information for quality control. However, TOF–MS based platform suffers from the disadvantage of inhomogeneous ionization propensities and time-consuming samples preparation procedures. In contrast, NMR platform possesses disadvantages of lower sensitivity and limited specificity, as well as advantages of homogenous signal response and simple sample preparations [23]. In general, a combination of ITS2 sequence comparations and TOF–MS as well as NMR based metabolomics analysis can validated each other and provided more comprehensive quality investigation for MSRHM [24].
Conclusion
In this study, a new integrated quality evaluation strategy was proposed for MSRHM of Qinjiao employing ITS2 sequence comparation, TOF–MS and NMR based metabolomics analysis. At first, gene comparison based on ITS2 sequence was conducted among 4 species of Qinijao. Then, TOF–MS and NMR based metabolic analysis were applied to investigate species discrepancy among four species of Qinjiao. It turned out that species discrepancy revealed among species were consistent by ITS2 sequencing, NMR and TOF–MS based metabolomics. Maximum species difference was noticed between G. crassicaulis and G. dahurica. Chemical difference among species based on TOF–MS and NMR were tentative explored. For TOF–MS profiling of Qinjiao, 28 constituents were tentative identified, 17 of which were further confirmed by standards. For 1H-NMR spectra of Qinjiao, signals from 5 compounds were assigned. Contents discrepancies were investigated by ANOVA analysis. It turned out that MS based metabolomics coincided with NMR based metabolomics result, and explained the intensity discrepancy in 1H-NMR spectra.
The current research demonstrates that integration of ITS2 sequence comparation and UPLC/Q-TOF MS as well as 1H-NMR based metabolomics analysis can be a powerful strategy for quality investigation of MSRHM.
Supplementary information
Additional file 1: Table S1. Drift of retention times, m/z and the RSD of peak areas of 5 selected characteristic features from QC samples during the analysis.
Additional file 2: Fig. S1. XIC chromatography of 28 identified constituents.
Additional file 3: Table S2. Part of the variable with VIP value > 1.0 from OPLS-DA analysis of G. crassicaulis and G. dahurica.
Additional file 4: TAble S3. P value of CV-ANOVA for OPLS-DA models based on MS or NMR analysis.
Acknowledgements
The authors thank the award of Part of the Qinjiao herbs of Institute of Chinese Materia Medica, Shanghai University of Traditional Chinese Medicine, Shanghai, China. The authors also thanks Professor Lihong Wu (Shanghai University of Traditional Chinese Medicine) and Jiuzhi Yuan (Shenyang Pharmaceutical University) for specimen authentication.
Abbreviations
- MSRHM
multi-species resourced herb medicine
- G. macrophylla
Gentiana macrophylla Pall
- G. straminea
Gentiana straminea Maxim
- G. crasicaulis
Gentiana crassicaulis Duthie ex Burk
- G. daurica
Gentiana dahurica Fisch
- ITS2
internal transcribed spacer 2
- UPLC-Q-TOF–MS
ultra performance liquid chromatography quadrupole time-of-flight mass spectrometry
- 1H-NMR
proton nuclear magnetic resonance spectrometer
- PCA
principal component analysis
- OPLS-DA
orthogonal partial least squares discrimination analysis
- ANOVA
one-way analysis of variance
- VIP
variable importance in the projection value
Authors’ contributions
All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript. Furthermore, each author certifies that this material or similar material has not been and will not be submitted to or published in any other publication. The specific contributions made by each author have been listed as follows: Zeyun Li: conception and design of study, acquisition of data and drafting the manuscript; Yue Du, Yongliang Yuan: acquisition of data, analysis and interpretation of data; Xiaojian Zhang, Xin Tian: revising the manuscript critically for important intellectual content; Zhengtao Wang: conception and design of study, analysis and interpretation of data. All authors read and approved the final manuscript.
Funding
The work was supported by National Natural Science Foundation of China (Grant No.: 81603287), China Postdoctoral Science Foundation (Grant No.: 2019M662555) and postdoctoral research grant in Henan Province (Grant No.: 1902004).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhengtao Wang, Email: ztwang@shutcm.edu.cn.
Xin Tian, Email: tianx@zzu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13020-020-0292-3.
References
- 1.Wang Y, Lou Z. Study of Radix Gentianae Macrophyllae. Chin Pharm J. 1987;22:153–159. [Google Scholar]
- 2.Nie A, Lin Z, Wang Y, Zhang B. Study on chemical constituents and pharmacological action of gentiana gentiana macrophylla. Chin Trad Herb Drugs. 2017;48:597–608. [Google Scholar]
- 3.China Pharmacopoeia National Pharmacopoeia Committee, Beijing.
- 4.Wagner H, Bauer R, Melchart D, Xiao PG, Staudinger A. Chromatographic fingerprint analysis of herbal medicines Volume III. 中草药(英文版). 2011. pp. 137–46.
- 5.Shi D, Zhang J, Sun L. Necessity of the research on equal quantity based on the quality investigation of three multi-origin pieces. China J Trad Chin Med Pharm. 2018;33:320–324. [Google Scholar]
- 6.Cao X, Wang Z. Simultaneous determination of four iridoid and secoiridoid glycosides and comparative analysis of Radix Gentianae Macrophyllae and their related substitutes by HPLC. Phytochem Anal. 2010;21:348–354. doi: 10.1002/pca.1206. [DOI] [PubMed] [Google Scholar]
- 7.Qi S, Shang P, Zhang Y, Jia N, Jiao H, Zhao W, et al. HPLC fingerprint and LC-TOF-MS analysis on extract from roots of Gentiana macrophylla. Chin Herbal Med. 2012;04:245–251. [Google Scholar]
- 8.Wang Y, Ahmad B, Duan B, Zeng R, Huang L. Chemical and genetic comparative analysis of Gentiana crassicaulis and Gentiana macrophylla. Chem Biodivers. 2016;13:776–781. doi: 10.1002/cbdv.201500247. [DOI] [PubMed] [Google Scholar]
- 9.Michel CI, Meyer RS, Taveras Y, Molina J. The nuclear internal transcribed spacer (ITS2) as a practical plant DNA barcode for herbal medicines. J Appl Res Med Arom Plants. 2016;3:94–100. [Google Scholar]
- 10.Luo Y, Ma P, Yao H, Xin T, Hu Y, Zheng S, et al. Identification of Gentianae Macrophyllae Radix using the ITS2 barcodes. Acta Pharmaceutica Sinica. 2012;47:1710–1717. [PubMed] [Google Scholar]
- 11.Freitas J, Alves EF, Silva L, Zocolo GJ, De EB, Gramosa N. Chemometric analysis of NMR and GC datasets for chemotype characterization of essential oils from different species of Ocimum. Talanta. 2018;180:329. doi: 10.1016/j.talanta.2017.12.053. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Yao H, Han J, Xin T, Pang X, Shi L, et al. Principles for molecular identification of traditional Chinese materia medica using DNA barcoding. China J Chin Materia Med. 2013;38:141–148. [PubMed] [Google Scholar]
- 13.Luo K, Chen SL, Chen KL, Song JY, Yao H, Ma XY, et al. Assessment of candidate plant DNA barcodes using the Rutaceae family. Sci China Life Sci. 2010;53:701–708. doi: 10.1007/s11427-010-4009-1. [DOI] [PubMed] [Google Scholar]
- 14.Rubingh CM, Bijlsma S, Derks EPPA, Bobeldijk I, Verheij ER, Kochhar S, et al. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics. 2006;2:53–61. doi: 10.1007/s11306-006-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Zhan G, Jin M, Zhang H, Dang J, Zhang Y, et al. Botany, traditional use, phytochemistry, pharmacology, quality control, and authentication of Radix Gentianae Macrophyllae—a traditional medicine: a review. Phytomedicine. 2018;46:142–163. doi: 10.1016/j.phymed.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Dong H, Bao Y, Chen X, Hai Y, Zeng R. Rapid analysis on chemical constituents from roots of Gentiana crasicaulis by ultra-high performance liquid chromatography coupled with hybrid quadrupole-orbitrap mass spectrometry. Chin Trad Herbal Drugs. 2016;47:3175–3180. [Google Scholar]
- 17.Yang X, Zhang K, Li Z, Wang Z. Targeted profiling qHNM for determination of Gentiopicroside and loganic acid in 4 species of Gentianae Macrophyllae Radix. Chin J Exp Trad Med Formulae. 2017;23:98–103. [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Ma X, Lu Y, Lv P, Lin L, Wang Z, et al. Contents of seven indicative constituents in seven kinds of Gentiana macrophylla by UPLC. Chin Trad Herb Drugs. 2016;47:1968–1973. [Google Scholar]
- 20.Cao X, Wang Z, Wang Z. Comparative analysis of contents of four iridoid glucosides in different organs of four species of Gentiana L. J Plant Resourc Environ. 2012;21:58–63. [Google Scholar]
- 21.Li Z, Welbeck E, Yang L, He C, Hu H, Song M, et al. A quantitative (1)H nuclear magnetic resonance (qHNMR) method for assessing the purity of iridoids and secoiridoids. Fitoterapia. 2015;100:187–194. doi: 10.1016/j.fitote.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Welbeck E, Wang R, Liu Q, Yang Y, Chou G, et al. A universal quantitative 1H nuclear magnetic resonance (qNMR) method for assessing the purity of dammarane-type ginsenosides. Phytochem Anal. 2015;26:8–14. doi: 10.1002/pca.2527. [DOI] [PubMed] [Google Scholar]
- 23.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 24.Dieterle F, Riefke B, Schlotterbeck G, Ross A, Senn H, Amberg A. NMR and MS methods for metabonomics. Methods Mol Biol. 2011;691:385–415. doi: 10.1007/978-1-60761-849-2_24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Drift of retention times, m/z and the RSD of peak areas of 5 selected characteristic features from QC samples during the analysis.
Additional file 2: Fig. S1. XIC chromatography of 28 identified constituents.
Additional file 3: Table S2. Part of the variable with VIP value > 1.0 from OPLS-DA analysis of G. crassicaulis and G. dahurica.
Additional file 4: TAble S3. P value of CV-ANOVA for OPLS-DA models based on MS or NMR analysis.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.