Abstract
Background
Biomedical named-entity recognition (BioNER) is widely modeled with conditional random fields (CRF) by regarding it as a sequence labeling problem. The CRF-based methods yield structured outputs of labels by imposing connectivity between the labels. Recent studies for BioNER have reported state-of-the-art performance by combining deep learning-based models (e.g., bidirectional Long Short-Term Memory) and CRF. The deep learning-based models in the CRF-based methods are dedicated to estimating individual labels, whereas the relationships between connected labels are described as static numbers; thereby, it is not allowed to timely reflect the context in generating the most plausible label-label transitions for a given input sentence. Regardless, correctly segmenting entity mentions in biomedical texts is challenging because the biomedical terms are often descriptive and long compared with general terms. Therefore, limiting the label-label transitions as static numbers is a bottleneck in the performance improvement of BioNER.
Results
We introduce DTranNER, a novel CRF-based framework incorporating a deep learning-based label-label transition model into BioNER. DTranNER uses two separate deep learning-based networks: Unary-Network and Pairwise-Network. The former is to model the input for determining individual labels, and the latter is to explore the context of the input for describing the label-label transitions. We performed experiments on five benchmark BioNER corpora. Compared with current state-of-the-art methods, DTranNER achieves the best F1-score of 84.56% beyond 84.40% on the BioCreative II gene mention (BC2GM) corpus, the best F1-score of 91.99% beyond 91.41% on the BioCreative IV chemical and drug (BC4CHEMD) corpus, the best F1-score of 94.16% beyond 93.44% on the chemical NER, the best F1-score of 87.22% beyond 86.56% on the disease NER of the BioCreative V chemical disease relation (BC5CDR) corpus, and a near-best F1-score of 88.62% on the NCBI-Disease corpus.
Conclusions
Our results indicate that the incorporation of the deep learning-based label-label transition model provides distinctive contextual clues to enhance BioNER over the static transition model. We demonstrate that the proposed framework enables the dynamic transition model to adaptively explore the contextual relations between adjacent labels in a fine-grained way. We expect that our study can be a stepping stone for further prosperity of biomedical literature mining.
Keywords: Bioinformatics, Data mining, Named entity recognition, Neural network
Introduction
Biomedical named-entity recognition (BioNER) automatically identifies specific mentions of interest such as chemicals, diseases, drugs, genes, DNAs, proteins, viruses etc. in biomedical literature. As the fundamental step for various downstream linguistic tasks, e.g., adverse drug event extraction [1], bacteria biotope task [2], drug-drug interaction [3], and protein-protein interaction detection [4], the performance of BioNER is crucial in the overall biomedical knowledge discovery process [2].
BioNER operates by predicting a class label for each token across biomedical literature. It is typically considered as a sequence labeling problem and is thus widely modeled by a first-order linear-chain conditional random field (CRF) [5, 6]. CRF yields chain-structured label sequences by collectively assessing possible label-label transition relations between words over the entire input sequence.
In recent years, deep learning (briefly, DL) has become prevalent across various machine learning-based natural language processing (NLP) tasks since neural network-based learning systems can effectively identify prominent features in a data-driven way, replacing task-specific feature engineering based on high-level domain knowledge [7, 8]. For NER tasks, recent methods [9–14] have reported state-of-the-art performance by introducing a bidirectional long short-term memory (BiLSTM) into CRF. Accordingly, the combination of BiLSTM and CRF has been widely considered as a standard architecture for various sequence labeling problems.
The combined models (i.e., BiLSTM-CRFs) for NER typically consist of two major components: a token-level BiLSTM and a real-valued transition matrix. The BiLSTM is dedicated to estimate the best-suited label on each token, while the transition matrix is solely responsible for describing the transition compatibility between all possible pairs of labels on neighboring tokens; in detail, the numerical score at the ith row and jth column of a transition matrix represents the transition compatibility from the ith label to the jth label. Note that the transition matrix is once established by being suited to the statistics of given training data via its parameter learning and is frozen afterward. As a result, the transition matrix cannot provide the contextualized compatibility for the relationship of neighboring labels in a fine-grained way.
Accordingly, we contend that solely relying on the static transition matrix is not enough to explain the ever-changing label-label transition relations in BioNER, since biomedical entities are frequently descriptive, long or even contain conjunctions [15], e.g., “normal thymic epithelial cells,” “peripheral sensor neuropathy,” and “central nervous system and cardiac toxicity.” As a result, the boundaries of entity-mentions in biomedical texts are often too ambiguous to accurately segment them. Therefore, we argue that exploiting contextual information to describe label-label transition relations is important to facilitate the accurate identification of biomedical entities. Recently, Lin et al. [16] studied that explicitly modeling relations between parts in a structured model is applicable to semantic image segmentation, whereas it has been rarely studied in recent DL-based NLP methods.
To this end, we propose a novel framework, called Dynamic Transition for NER (DTranNER), to incorporate a DL-based model, which adaptively identify label-label transition relations to further improve the accuracy of BioNER. Overall, DTranNER makes use of two separate DL-based models: Unary-Network and Pairwise-Network. The addition of Pairwise-Network makes it possible to assess the transition compatibility between adjacent labels by exploring the context of an input sentence. Meanwhile, as another DL-based model, Unary-Network is used for individual labeling as in previous works. After all, Unary-Network and Pairwise-Network are arranged to yield agreed label sequences via this novel framework.
Because DTranNER is orthogonal to a DL-based model, any type of DL-based models such as attention [17] or transformer [18] can be employed to play the role of Unary-Network or Pairwise-Network. In this study, we conduct experiments using a BiLSTM as the underlying DL networks since it has been widely adopted in various sequence labeling problems so far.
We evaluated DTranNER by comparing with current state-of-the-art NER methods on five benchmark BioNER corpora to investigate the effectiveness of the DL-based label-label transition model. The results show that DTranNER outperformed the existing best performer on four out of five corpora and showed comparable accuracy to the existing best performer on one remaining corpus, thereby demonstrating the excellent performance of DTranNER.
Background
Problem definition: biomedical named entity recognition (BioNER)
An instance of a BioNER corpus consists of an input token sequence x=x1,…,xN and its associated output label sequence y=y1,…,yN. We use the IOBES tagging scheme by which tokens are annotated with one of “I,” “O,” “B,” “E,” or “S” labels. In the case of an entity spanning over multiple tokens, “B” is tagged to the token to indicate the beginning of the entity, “I” stands for “Inside,” and “E” indicates the ending token of the entity. For the case of an entity of a single token, the “S” label is tagged to it. The “O” label stands for “Outside,” which means that the token is not part of any named entity. To indicate the type of entities, one of the type tags, such as “Chemical,” “Disease,” “Gene,” or “Protein,” is additionally concatenated to each IOBES tag.
Linear-chain conditional random field (CRF)
As a class of discriminative probabilistic graphical models, a linear-chain conditional random field (CRF) describes the joint probability P(y|x) of the entire structured labels y with respect to the structure of an undirected graph, given a set of inputs x. CRF is widely used in various sequence labeling problems as well as BioNER by imposing the first-order Markov property on the output sequence labeling. There are two types—unary and pairwise—of elementary feature functions to organize an output label sequence. The unary feature functions are dedicated to estimating the suitability of candidate labels at each individual position, whereas the pairwise feature functions are designed to assess possible pairwise labels on two connected positions. Summing up, when an input sequence x of length N is given, the conditional distribution P(y|x) is represented as a product of position-dependent unary and pairwise feature functions; thus, it is formulated as in the following equation:
| 1 |
where sk(yi,x,i) denotes a member of the unary feature functions (i.e., s∈S) at the position i, and t(yi−1,yi,x,i) indicates a member of the pairwise feature functions (i.e., t∈T) at two consecutive positions i-1 and i. Traditionally, the unary and pairwise feature functions are manually designed to facilitate accurate sequence labeling, and they are usually real-valued binary indicators representing either true or false. The weights (i.e., λs∈θs and λt∈θt) associated to the feature functions are trainable parameters. Z(x) is the partition function as a normalization constant over all possible label assignments.
Bidirectional long short-term memory (BiLSTM)
Long short-term memory (LSTM) [19] is a specific variant of recurrent neural networks to mitigate the problem of vanishing and exploding gradients in modeling long-term dependencies of a sequence. LSTM is suited for modeling sequential data with recurrent connections of hidden states H={h1,h2,…,hN} and have become ubiquitous in a wide range of NLP tasks. At every time step, LSTM yields a current hidden state and internally updates a current cell state based on and calculated in the previous time step.
Given that LSTM is limited to using past context in the forward direction, a bidirectional LSTM (BiLSTM) is employed to exploit future context as well as past context. BiLSTM processes an input sequence in both forward and backward directions with two separate LSTMs. That is, the hidden states from both directional LSTMs are concatenated to make final output vectors .
Merger of BiLSTM and CRF: BiLSTM-CRF
BiLSTM-CRF has been widely employed in recent neural network-based NER studies [9–14, 20, 21] for sequence labeling. The architecture of BiLSTM-CRF is typically comprised of four layers: a token-embedding layer, a token-level BiLSTM layer, a binding layer, and a CRF layer. We denote an input token sequence of length N by x={x1,⋯,xN} and the corresponding output label sequence by y={y1,⋯,yN}. First, the token-embedding layer encodes input tokens into its fixed-dimensional vectors as e1,e2,…,eN. Next, the BiLSTM layer takes the token-embedding vectors as the inputs to generate the hidden-state vectors h1,h2,…,hN. Before being fed to the CRF layer, the hidden-state vectors are transformed to the score vectors U1,U2,…,UN with L-dimensionality, where L denotes the number of labels, via the binding layer so as to match the number of labels. The score vector contains the confidence values for possible labels on its corresponding token position. Namely, the stack from the token-embedding layer to the binding layer can be considered to play the role of the unary feature functions (i.e., s∈S) in Eq. 1. Besides, a real-valued transition matrix, denoted as A, accounts for all the label-label transition relations; it is likewise regarded to play the role of the pairwise feature functions (i.e., t∈T) in Eq. 1. Eventually, BiLSTM-CRF calculates the likelihood for a label sequence y given an input token sequence x via the following equation:
| 2 |
where Ui(yi) denotes the unary score for assigning the label yi on the ith token, Ai,j corresponds to the real-valued pairwise transition compatibility from ith label to jth label, and .
Related work
Recent state-of-the-art CRF-based NER studies [9–14, 20–22] have demonstrated the effectiveness of data-driven representation learning (i.e., DL) under CRF. We discuss several CRF-based methods for NER in terms of two kinds of feature functions: unary and pairwise feature functions. We also introduce BioBERT that showed the state-of-the-art performance in BioNER.
Lample et al. [9] proposed to bring BiLSTM into CRF for NER in general news domain. The model uses two BiLSTMs: one for token-level representation learning and the another for character-level representation learning. The BiLSTMs work as unary feature functions, whereas a static transition matrix comes in for pairwise feature functions. Afterward, Habibi et al. [10] adopted the model of Lample et al. [9] for BioNER.
Luo et al. [22] adopted BiLSTM-CRF for NER in chemistry domain and applied an attention mechanism to leverage document-level context information. They employ abbreviation embeddings using a specific external library to handle abbreviations that frequently appear in chemical entities’ naming. Their model also relies on a static matrix to retrieve all the label-label transition relations in CRF.
Dang et al. [12] developed D3NER to utilize various linguistic information under BiLSTM-CRF. D3NER creates a token embedding by aggregating several embeddings: a pre-trained word embedding, an abbreviation embedding, a POS embedding, and a character-level token embedding. Similarly, a transition matrix solely plays the role of pairwise feature functions.
Wang et al. [11] introduced a multi-task learning framework for BioNER. They trained a model using several biomedical corpora together to overcome a limited amount of annotated biomedical corpora. Their model also adopts BiLSTM-CRF with a transition matrix.
Yoon et al. [14] proposed aggregation of multiple expert models. They named it CollaboNet, where each expert model is mapped to a BiLSTM-CRF and is trained with each distinct corpus. Likewise, each BiLSTM-CRF has a transition matrix, corresponding to pairwise feature functions.
Peters et al. [13] introduced ELMo as a pre-trained model. ELMo provides contextualized word embeddings for various downstream tasks. They also trained the ELMo-enhanced BiLSTM-CRF for NER.
Lee et al. [23] released BioBERT by training BERT [24] for the use in the Bioinformatics domain. Similarly to ELMo, as a pre-trained model, BioBERT provides contextualized word embeddings and thus can be applied to downstream tasks. BioBERT achieved the state-of-the-art performance in several BioNER corpora.
DTranNER: architecture and method
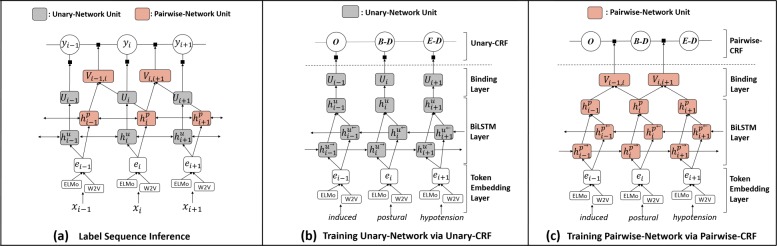
In this section, we present the proposed framework DTranNER as shown in Fig. 1. For parameter learning, the components (i.e., Unary-Network and Pairwise-Network) of DTranNER are systematically trained via two separate CRFs (i.e., Unary-CRF and Pairwise-CRF). Once trained, Unary-Network and Pairwise-Network are combined into a CRF for BioNER label sequence prediction. First of all, we describe how to build the token embeddings in our models. Although DTranNER is not limited to a specific DL architecture in the places of the underlying networks, from now on, we evaluate our framework using BiLSTM, which has been typically adopted in a majority of NER studies.
Fig. 1.
The overall architectures of the proposed framework DTranNER. a As a CRF-based framework, DTranNER is comprised of two separate, underlying deep learning-based networks: Unary-Network and Pairwise-Network are arranged to yield agreed label sequences in the prediction stage. The underlying DL-based networks of DTranNER are trained via two separate CRFs: Unary-CRF and Pairwise-CRF. b The architecture of Unary-CRF. It is dedicated to train Unary-Network. c The architecture of Pairwise-CRF. It is also committed to train Pairwise-Network. A token embedding layer is shared by Unary-Network and Pairwise-Network. A token-embedding is built upon by concatenating its traditional word embedding (denoted as “W2V”) and its contextualized token embedding (denoted as “ELMo”)
Token-embedding layer
Given a sequence of N tokens (x1,x2,..., xN), they are converted token-by-token into a series of fixed-dimensional vectors (e1,e2,..., eN) via the token-embedding layer. Each token embedding is designed to encode several linguistic information of the corresponding token in the sentence. Each token embedding is thus built up by concatenating the traditional context-independent token embedding and its contextualized token embedding. These token embeddings are subsequently fed to Unary-Network and Pairwise-Network as the inputs. We do not consider additional character-level token embeddings unlike several models [9–12, 14, 20, 21], because ELMo [13] as our contextualized token embedding provider basically includes a character-level CNN model.
Context-independent token embedding
We use the pre-trained token vectors, Wiki-PubMed-PMC, created by Pyysalo et al. [25] to initialize the traditional token-embedding vectors. The pre-trained token vectors were made up by being trained on three different datasets: the abstracts of the PubMed database, the full-text articles of the PubMed Central (PMC) database, and the texts of a recent Wikipedia dump. It is available at [26]. We replace every out-of-vocabulary (OOV) token with a special <UNK> vector.
Contextualized token embedding
We employ ELMo [13] for the contextualized token embeddings. Unlike context-independent token embeddings based on GloVe [27] or Word2Vec [28], ELMo creates context-dependent token embeddings by reconsidering the syntax and semantics of each token under its sentence-level context. In particular, we adopt the in-domain ELMo model pre-trained on the PubMed corpus, which is available at [29].
Unary-Network
As shown in Fig. 1b, Unary-Network takes token embeddings as inputs, put them into its own BiLSTM layer to extract task-specific contextual information in an ordered token-level sequence, and finally produces the L-dimensional score vectors as many as the number of tokens via its binding layer. The binding layer consists of two linear transformations with an activation function and a skip connection between them. That is, the binding layer is formulated as follows:
| 3 |
where Ui denotes the L-dimensional score vector exhibiting the suitability over all possible labels on the ith token, is the i-th hidden state from the BiLSTM layer, and are trainable weight matrices, and and are the bias vectors. Here, projects the d-dimensional vector obtained by both the feed-forward network and the skip connection to the L-dimensional output vector. We use an ELU as the activation function σ(·). As will be explained in the following section, Unary-Network is trained via the purpose-built CRF (i.e., Unary-CRF) for the parameter learning.
Pairwise-Network
Pairwise-Network aims to extract contextual information related to pairwise labeling. This design explains why two consecutive hidden state vectors of the BiLSTM are involved in describing an edge connection in the CRF layer as shown in Fig. 1c. Pairwise-Network therefore generates L2-dimensional score vectors to match the number of possible label pairs on two tokens. We employ a bilinear model-based method [30] to exploit interactive features of two neighboring hidden state vectors. This method approximates a classical three-dimensional tensor with three two-dimensional tensors, significantly reducing the number of parameters. It is shown in the following equation:
| 4 |
where fi−1,i denotes the m-dimensional vector via the bilinear model of two neighboring hidden state vectors (i.e., and ) of the underlying BiLSTM layer; , and are trainable matrices; and ∘ denotes Hadamard product (i.e., element-wise product of two vectors). The binding layer has a skip connection as in Unary-Network. It is thus formulated as the following equation:
| 5 |
where denotes the score vector indicating the confidence values over all label combinations on the neighboring (i−1)th and ith tokens, and are trainable weight matrices, and are the bias terms, and σ(·) is an ELU activation. Similarly to Unary-Network, Pairwise-Network is also trained via the purpose-built CRF (i.e., Pairwise-CRF) for the parameter learning.
Model training
Here, we explain how to train DTranNER. In order to facilitate the parameter learning of the two underlying networks (i.e., Unary-Network and Pairwise-Network), we establish two separate linear-chain CRFs, which are referred as Unary-CRF (Fig. 1b) and Pairwise-CRF (Fig. 1c), by allocating the two types of DL-based networks (i.e., BiLSTMs in our case) to the two purpose-built CRFs, respectively. The reason is that, when both Unary-Network and Pairwise-Network coexist in a single CRF, as Smith et al. [31] and Sutton et al. [32] claimed that the existence of a few indicative features can swamp the parameter learning of other weaker features, either one of the two networks starts to hold a dominant position, causing the other network to deviate from its optimal parameter learning. Our solution enables each network to notice own prediction error during the parameter learning. We explain in detail the effect of our training strategy in the Additional file 1.
In this study, note that each of Unary- and Pairwise-CRFs is a sufficient label sequence predictor or learner; in the sense, the conditional likelihood Pu of Unary-CRF is formulated as in Eq. 2, and the conditional likelihood Pp of Pairwise-CRF given the input sequence x with the length N is formulated as the following equation:
| 6 |
where is the normalization constant.
Rather than individually training multiple CRFs offline as in [31, 32], Unary-CRF and Pairwise-CRF are jointly trained in our training strategy by maximizing their product—i.e., —of the two likelihoods of Unary-CRF and Pairwise-CRF. By equivalently converting the objective function into the negative log likelihood, the optimization problem is written as the following equation:
| 7 |
where xe and ye denote the eth training sentence example and its ground-truth label sequence, and θu and θp denote the model parameters of Unary-CRF and Pairwise-CRF respectively.
Prediction
We explain the detail on how to infer label sequences with the trained DTranNER. Once trained via the two separate CRFs, Unary-Network and Pairwise-Network are arranged into a CRF to yield an agreed label sequence in the prediction stage. Note that Unary-Network and Pairwise-Network have distinct focuses derived by different roles, leading to learn their own specific representations. We combine them by multiplying them as a product of models [33]. More specifically, all the components obtained through the aforementioned training process—Unary-Network, Pairwise-Network, and the transition matrix—are organized in a CRF, as shown in Fig. 1a. The combined model is formulated in terms of the probability for a label sequence y given an input sequence x via the following equation:
| 8 |
As a result, we obtain the most likely label sequence using the Viterbi decoding.
Experimental setup
Datasets
We conducted our experiments with five BioNER benchmark corpora: BC2GM, BC4CHEMD, BC5CDR-chemical, BC5CDR-disease, and NCBI-Disease, which are commonly used in the existing literature [11, 12, 14, 23].
Table 1 shows the overall description of the five benchmark BioNER corpora. They are publicly available and can be downloaded from [34]. The BioCreative II Gene Mention (BC2GM) task corpus [35] consists of 20,128 sentences from biomedical publication abstracts and is annotated for mentions of the names of proteins, genes, and related entities. The BioCreative IV Chemical and Drug (BC4CHEMD) task corpus [36] contains the annotations for chemical and drug mentions in 10,000 biomedical abstracts. The BioCreative V Chemical Disease Relation (BC5CDR) corpus [37] is composed of mentions of chemicals and diseases that appeared in 1,500 PubMed articles. The NCBI-Disease corpus (NCBI-Disease) [38] is composed of 793 PubMed abstracts annotated for disease mentions. The aforementioned corpora cover four major biomedical entity types: gene, protein, chemical, and disease.
Table 1.
BioNER corpora in experiments
| Datasets | Number of Sentences | Entity Types | Entity Counts | Max Entity Length | Average Entity Length |
|---|---|---|---|---|---|
| BC2GM [35] | 20128 | Gene/Protein | 24583 | 26 tokens | 2.44 tokens |
| BC4CHEMD [36] | 87682 | Chemical/Drug | 84310 | 137 tokens | 2.19 tokens |
| BC5CDR-Chemical [37] | 13935 | Chemical/Drug | 15935 | 56 tokens | 1.33 tokens |
| BC5CDR-Disease [37] | 13935 | Disease | 12852 | 19 tokens | 1.65 tokens |
| NCBI-Disease [38] | 7284 | Disease | 6881 | 22 tokens | 2.21 tokens |
Training setup
In model training, we added L2 regularization penalty to the loss (i.e., Eq. 7) with the decay factor of 1×10−5. The Glorot uniform initializer of Glorot and Bengio [39] is used for initializing our weight matrices, and the biases are initialized with 0. All the activation functions are ELU (exponential linear unit). We set the minibatch size of model training to ten examples across all experiments. Our models are differentiable; thereby, the CRF and its underlying neural networks can be jointly trained end-to-end by backpropagation. We use the Adam optimizer of [40] with the learning rate of 0.001. In the training process, we renormalize all gradients whenever the L2 norm of the gradients exceeds 5 in every minibatch update. We applied layer normalization [41] to the outputs of the token embedding layer, and also applied weight normalization [42] to all the weight matrices of the binding layers of Unary-Network and Pairwise-Network. We used Dropout [43] with keep probability 0.5 in both the binding layers. We established our models within at most 50 epochs for all the corpora.
Evaluation metrics
We evaluated all the methods using the precision, recall, and F1 score on the test sets of all corpora. We defined each predicted entity as correct if and only if both the entity type and the boundary were exactly matched to the ground-truth annotation. We used the python version of the evaluation script designed for CoNLL-2000 Benchmark Task, which can be downloaded from [44]. To get reliable results, we repeated every test five times with different random initialization and report the arithmetic mean.
Results
Overall performance comparison
We compared DTranNER with five state-of-the-art methods: (1) Att-BiLSTM-CRF [22], (2) D3NER [12], (3) Collabonet [14], (4) the multi-task learning-based model of Wang et al. [11], and (5) BioBERT [23]. Note that all the models except BioBERT employ a CRF as their top layer and rely on a static transition matrix. The performance values in terms of the precision, recall, and F1-score over all the corpora are presented in Table 2. DTranNER outperformed the current state-of-the-art models on four of five corpora—BC2GM, BC4CHEMD, BC5CDR-Disease, and BC5CDR-Chemical—in terms of F1 scores.
Table 2.
Performance values in terms of the precision (%), recall (%) and F1-score (%) for the state-of-the-art methods and the proposed model DTranNER
| Corpus | BC2GM | BC4CHEMD | BC5CDR-Chemical | BC5CDR-Disease | NCBI-Disease | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Att-BiLSTM-CRF (2017) | - | - | - | 92.29 | 90.01 | 91.14 | 93.49 | 91.68 | 92.57 | - | - | - | - | - | - |
| D3NER (2018) | - | - | - | - | - | - | 93.73 | 92.56 | 93.14 | 83.98 | 85.40 | 84.68 | 85.03 | 83.80 | 84.41 |
| Collabonet (2018) | 80.49 | 78.99 | 79.73 | 90.78 | 87.01 | 88.85 | 94.26 | 92.38 | 93.31 | 85.61 | 82.61 | 84.08 | 85.48 | 87.27 | 86.36 |
| Wang et al. (2018) | 82.10 | 79.42 | 80.74 | 91.30 | 87.53 | 89.37 | 93.56 | 92.48 | 93.03 | 84.14 | 85.76 | 84.95 | 85.86 | 86.42 | 86.14 |
| BioBERT (2019) | 85.16 | 83.65 | 84.40 | 92.23 | 90.61 | 91.41 | 93.27 | 93.61 | 93.44 | 85.86 | 87.27 | 86.56 | 89.04 | 89.69 | 89.36 |
| DTranNER | 84.21 | 84.84 | 84.56 | 91.94 | 92.04 | 91.99 | 94.28 | 94.04 | 94.16 | 86.75 | 87.70 | 87.22 | 88.21 | 89.04 | 88.62 |
Note: The highest performance in each corpus is highlighted in Bold. We quoted the published scores for the other models. For Wang et al. [11], we conducted additional experiments to obtain the performance scores for two corpora (i.e., BC5CDR-Chemical and BC5CDR-Disease) using the software on their open source repository [45]
DTranNER achieved a much higher F1 score with higher precision than the current best performer (94.16% vs. 93.44%) for BC5CDR-Chemical, where its NER process was confused owing to many abbreviations despite its shorter average entity length as shown in Table 1. Thus, the pairwise transition network of DTranNER is shown to be advantageous in discovering abbreviation-formed entities.
Ablation studies
We investigated the effectiveness of main components of our proposed method DTranNER through ablation studies.
Impact of unary- and pairwise-Networks
To investigate the contribution of Unary- and Pairwise-Networks to DTranNER, we trained experimental models by deactivating each component (i.e., either Unary-Network or Pairwise-Network) in turn from DTranNER and then measured the performance of the variant models on three benchmark corpora: BC5CDR-Chemical, BC5CDR-Disease, and NCBI-Disease. The results are shown in Table 3.
Table 3.
Impact of Unary-Network and Pairwise-Network in terms of the F1-score (%)
| Settings | BC5CDR-Chemical | BC5CDR-Disease | NCBI-Disease |
|---|---|---|---|
| Unary-CRF | 93.01 | 86.14 | 86.94 |
| Pairwise-CRF | 93.27 | 86.05 | 86.71 |
| Unary+Pairwise ensemble | 93.25 | 86.78 | 87.09 |
| DTranNER | 94.16 | 87.22 | 88.62 |
Note: “Unary-CRF” denotes a variant model excluding Pairwise-Network from DTranNER, “Pairwise-CRF” denotes a variant model excluding Unary-Network from DTranNER, and “Unary+Pairwise ensemble” is an ensemble model of “Unary-CRF” and “Pairwise-CRF.” In the ensemble model, “Unary-CRF” and “Pairwise-CRF” were independently trained, and they voted over the sequence predictions by their prediction scores
The removal of either Unary-Network or Pairwise-Network from DTranNER caused the overall performance degradation in all the corpora by up to 1.91 percent points. That is, this ablation study presents that the performance achievement of DTranNER is attributed to not only an individual component but also the mutual collaboration of Unary-Network and Pairwise-Network. The relative importance between the two networks was not very clear.
We also compared DTranNER with an ensemble model of Unary-CRF and Pairwise-CRF, denoted as “Unary+Pairwise ensemble,” which were separately trained. The sequence prediction of the ensemble model was decided by voting with their sequence output scores. As shown in Table 3, the performance improvement of the ensemble model was marginal in BC5CDR-Chemical and NCBI-Disease. More important, the ensemble model was much worse than DTranNER in all corpora. This result indicates that yielding agreed label sequences between the two networks, which have separate views, as in DTranNER is more effective than their ensemble via simple voting.
Impact of separate BiLSTM layers of Unary- and Pairwise networks
Unary-Network and Pairwise-Network have an independent underlying layer which learns its role-specific representations. We investigate the impact of the separate underlying layers in the peer networks. For this purpose, we additionally built a variant model of DTranNER, denoted as “DTranNER-shared,” that forced Unary-Network and Pairwise-Network to share the parameters of their BiLSTM layers. As shown in Table 4 for the comparison result, it turned out that Unary-Network and Pairwise-Network benefit from the exclusive underlying layer.
Table 4.
Impact of separate BiLSTM layers in terms of the F1-score (%)
| Settings | BC2GM | BC5CDR-Chemical | BC5CDR-Disease | NCBI-Disease |
|---|---|---|---|---|
| DTranNER-shared | 83.69 | 93.57 | 86.75 | 88.01 |
| DTranNER | 84.56 | 94.16 | 87.22 | 88.62 |
Note: “DTranNER-shared” is a variant model that shares the BiLSTM layer in “Unary-Network” and “Pairwise-Network.”
Embedding layer
We here investigate the impact of each element in the token embedding layer of DTranNER. For this purpose, we built two variants of DTranNER: (1) a model (denoted as “W2V”) whose token embedding consists of only 200-dimensional pre-trained token embedding [26] and (2) another model (denoted as “ELMo”) whose token embedding is solely comprised of 1024-dimensional ELMo embedding, which is obtained from the ELMo model [29] pre-trained on the PubMed corpus. The comparison results are presented in Table 5. The context-dependent token embeddings via the ELMo model bring significant performance improvement on the four benchmark corpora, especially on NCBI-Disease. Nevertheless, the best performance is consistently achieved by the combination of the context-dependent ELMo embedding and the traditional context-independent embedding.
Table 5.
Impact of each component in the token embedding composition in terms of the F1-score (%)
| Settings | BC2GM | BC5CDR-Chemical | BC5CDR-Disease | NCBI-Disease |
|---|---|---|---|---|
| W2V | 82.03 | 92.64 | 85.17 | 84.88 |
| ELMo | 83.41 | 93.78 | 86.76 | 88.27 |
| ELMo + W2V(=DTranNER) | 84.56 | 94.16 | 87.22 | 88.62 |
Note: “W2V” is a variant model of DTranNER whose embedding layer uses only traditional context-independent token vectors (i.e., Wiki-PubMed-PMC [25]), “ELMo” is another variant model of DTranNER whose embedding layer uses only ELMo, and “ELMo + W2V” is equivalent to DTranNER
Case studies
To demonstrate the advantage of the DL-based label-label transition model, which is the main feature of DTranNER, we compared several example outcomes yielded by DTranNER and Unary-CRF as shown in Table 6. Note that Unary-CRF is not equipped with this main feature. In addition, the label sequence predictions of DTranNER in Table 6 coincide with the ground-truth annotations.
Table 6.
Case study of the label sequence prediction performed by DTranNER and Unary-CRF
| Diseases/Chemicals | ||
| Case 1 | Unary-CRF | to enable diagnosis of ureteric stones or obstruction in patients with HIV infection who receive indinavir theraphy |
| DTranNER | to enable diagnosis of ureteric stones or obstruction in patients with HIV infection who receive indinavir theraphy | |
| Case 2 | Unary-CRF | The present study was designed to investigate whether nociceptin / orphanin FQ and nocistatin could modulate |
| impairment of learning and memory induced by scopolamine | ||
| DTranNER | The present study was designed to investigate whether nociceptin / orphanin FQ and nocistatin could modulate | |
| impairment of learning and memory induced by scopolamine | ||
| Case 3 | Unary-CRF | We report the case of a female patient with rheumatoid arthritis who developed acute cytolytic hepatitis due to meloxicam |
| DTranNER | We report the case of a female patient with rheumatoid arthritis who developed acute cytolytic hepatitis due to meloxicam | |
| Case 4 | Unary-CRF | Reduced nicotinamide adenine dinucleotide phosphate - diaphorase (NADPH - d) histochemistry was also employed to |
| DTranNER | Reduced nicotinamide adenine dinucleotide phosphate - diaphorase (NADPH - d) histochemistry was also employed to | |
| Genes/Proteins | ||
| Case 5 | Unary-CRF | The MIC90 of ABK against coagulase type IV strains was rather high, 12.5 micrograms/ml |
| DTranNER | The MIC90 of ABK against coagulase type IV strains was rather high, 12.5 micrograms/ml | |
| Case 6 | Unary-CRF | subtle differences between individual subunits that lead to species - specific properties of RNA polymerase I transcription |
| DTranNER | subtle differences between individual subunits that lead to species - specific properties of RNA polymerase I transcription | |
| Case 7 | Unary-CRF | The S. typhimurium aspartyl / asparaginyl beta - hydroxylase homologue (designated lpxO) was cloned into |
| DTranNER | The S. typhimurium aspartyl / asparaginyl beta - hydroxylase homologue (designated lpxO) was cloned into | |
Note: Unary-CRF is the purpose-built model excluding Pairwise-Network from DTranNER. The named entities inferred by each model are underlined in sentences
For Case 1, Unary-CRF failed to detect one of the boundaries of the disease-type entity “ureteric stones or obstruction” because of the intervention of the inner conjunction “or,” while DTranNER precisely determined both boundaries. For Case 2, Unary-CRF failed to identify the chemical-type entities enumerated via the conjunctions “/” and “and,” whereas DTranNER exactly identified all the separate terms. For Case 3, Unary-CRF failed to determine the left boundary of the single-token entity “hepatitis” by mistakenly regarding “acute” and “cytolytic” as its constituent elements, whereas DTranNER exactly distinguished them from this entity by understanding the contextual relations. For Case 4, DTranNER correctly identified the two entities, where the latter is the abbreviation of the former, but Unary-CRF failed. For Case 5, Unary-CRF ignored the gene-type entity “coagulase type IV” by mistakenly regarding “type” and “IV” as generic terms, whereas DTranNER correctly identified it by reflecting the contextual correlations between its constituent elements. For Case 6, DTranNER correctly identified both boundaries of the gene-type entity “RNA polymerase I” by exploiting the contextual clues on the consecutive pairs, 〈“polymerase” and “I” 〉 and 〈“I” and “transcription” 〉, though “I” solely looks ambiguous; in contrast, Unary-CRF failed to determine the right boundary because it classified “I” as a generic term. For Case 7, DTranNER correctly extracted the lengthy entity by grasping the correlation between the neighboring tokens (i.e., “hydroxylase” and “homologue”), whereas Unary-CRF failed to handle this lengthy entity.
Summing up, DTranNER successfully supports various cases which would be very difficult without the contextual information, and these cases indeed show the benefit of DTranNER for BioNER.
Conclusion
In this paper, we proposed a novel framework for BioNER, for which we call DTranNER. The main novelty lies in that DTranNER learns the label-label transition relations with deep learning in consideration of the context in an input sequence. DTranNER possesses two separate DL-based networks: Unary-Network and Pairwise-Network; the former focuses on individual labeling, while the latter is dedicated to assess the transition suitability between labels. Once established via our training strategy, these networks are integrated into the CRF of DTranNER to yield agreed label sequences in the prediction step. In other words, DTranNER creates the synergy leveraging different knowledge obtained from the two underlying DL-based networks. As a result, DTranNER outperformed the best existing model in terms of the F1-score on four of five popular benchmark corpora. We are extending DTranNER to utilize unlabeled biomedical data. This extension is meaningful in several aspects: (1) building a more-generalized model using a wide range of biomedical literature, (2) rapidly incorporating up-to-date biomedical literature by skipping time-consuming annotation, and (3) reducing annotation cost.
Supplementary information
Additional file 1 Model training strategy in the proposed framework. The effect of the proposed model training strategy is not shown in the main manuscript. The learning curves during model training demonstrate the reason why the training strategy facilitates the parameter learning of the underlying deep learning-based models under our proposed framework.
Acknowledgements
Not applicable.
Abbreviations
- BiLSTM
Bidirectional long short-term memory
- BioNER
Biomedical named entity recognition
- CNN
Convolutional neural network
- CRF
Conditional random field
- DL
Deep learning
- NER
Named entity recognition
- NLP
Natural language processing
- POS
Part-of-speech
Authors’ contributions
SK and JG designed the study. SK implemented the model, performed experiments and analyses. SK drafted the manuscript. JG revised it. All authors have read and approved the final version of this manuscript. All data generated or analysed during this study are included in this published article (and its Additional file 1).
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (No. 2017R1E1A1A01075927).
Availability of data and materials
The code for our models and instructions for the use can be found on GitHub https://github.com/kaist-dmlab/BioNER. The datasets used for performance evaluation and analysis during the current study are available in the MTL-Bioinformatics-2016 repository, https://github.com/cambridgeltl/MTL-Bioinformatics-2016.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
S. K. Hong, Email: skhong831@kaist.ac.kr
Jae-Gil Lee, Email: jaegil@kaist.ac.kr.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12859-020-3393-1.
References
- 1.Gurulingappa H, Mateen-Rajpu A, Toldo L. Extraction of potential adverse drug events from medical case reports. J Biomed Semant. 2012;3(1):15. doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossy Robert, Jourde Julien, Manine Alain-Pierre, Veber Philippe, Alphonse Erick, van de Guchte Maarten, Bessi.res Philippe, N.dellec Claire. BioNLP Shared Task - The Bacteria Track. BMC Bioinformatics. 2012;13(Suppl 11):S3. doi: 10.1186/1471-2105-13-S11-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Chen Y, Liu F, Luo F, Tian G, Li X. Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data. BMC Bioinformatics. 2017;18(1):18. doi: 10.1186/s12859-016-1415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43(D1):447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In: Proceedings of the 18th International Conference on Machine Learning. ACM: 2001. p. 282–9. http://portal.acm.org/citation.cfm?id=655813.
- 6.Sutton C, McCallum A. An introduction to conditional random fields. Found Trends® Mach Learn. 2012;4(4):267–373. doi: 10.1561/2200000013. [DOI] [Google Scholar]
- 7.Settles B. Biomedical named entity recognition using conditional random fields and rich feature sets. In: Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications: 2004. p. 104–7. 10.3115/1567594.1567618.
- 8.Leaman R, Wei C-H, Lu Z. tmchem: a high performance approach for chemical named entity recognition and normalization. J Cheminformatics. 2015;7(1):3. doi: 10.1186/1758-2946-7-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies: 2016. p. 260–70. 10.18653/v1/n16-1030.
- 10.Habibi M, Weber L, Neves M, Wiegandt DL, Leser U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics. 2017;33(14):37–48. doi: 10.1093/bioinformatics/btx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Xuan, Zhang Yu, Ren Xiang, Zhang Yuhao, Zitnik Marinka, Shang Jingbo, Langlotz Curtis, Han Jiawei. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics. 2018;35(10):1745–1752. doi: 10.1093/bioinformatics/bty869. [DOI] [PubMed] [Google Scholar]
- 12.Dang TH, Le H. -Q., Nguyen TM, Vu ST. D3NER: biomedical named entity recognition using CRF-biLSTM improved with fine-tuned embeddings of various linguistic information. Bioinformatics. 2018;34(20):3539–46. doi: 10.1093/bioinformatics/bty356. [DOI] [PubMed] [Google Scholar]
- 13.Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L. Deep contextualized word representations. In: Proc. of NAACL. Association for Computational Linguistics (ACL): 2018. https://www.aclweb.org/anthology/N18-1202/.
- 14.Yoon W, So CH, Lee1 J, Kang J. Collabonet: collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinformatics. 2019; 20(10):249. [DOI] [PMC free article] [PubMed]
- 15.Zhou G, Zhang J, Su J, Shen D, Tan C. Recognizing names in biomedical texts: a machine learning approach. Bioinformatics. 2004;20(7):1178–90. doi: 10.1093/bioinformatics/bth060. [DOI] [PubMed] [Google Scholar]
- 16.Lin G, Shen C, van den Hengel A, Reid I. Exploring context with deep structured models for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2018;40(6):1352–66. doi: 10.1109/TPAMI.2017.2708714. [DOI] [PubMed] [Google Scholar]
- 17.Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate: 2014. https://arxiv.org/abs/1409.0473.
- 18.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. In: Advances in Neural Information Processing Systems. Neural Information Processing Systems Foundation: 2017. p. 5998–6008. http://papers.nips.cc/paper/7181-attention-is-all-you-need.pdf.
- 19.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–80. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 20.Chiu JP, Nichols E. Named entity recognition with bidirectional LSTM-CNNs. Trans Assoc Comput Linguist. 2016;4:357–70. doi: 10.1162/tacl_a_00104. [DOI] [Google Scholar]
- 21.Ma X, Hovy E. End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics: 2016. p. 1064–74. 10.18653/v1/p16-1101.
- 22.Luo L, Yang Z, Yang P, Zhang Y, Wang L, Lin H, Wang J. An attention-based BiLSTM-CRF approach to document-level chemical named entity recognition. Bioinformatics. 2017;34(8):1381–8. doi: 10.1093/bioinformatics/btx761. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. Biobert: a pre-trained biomedical language representation model for biomedical text mining. arXiv preprint. 2019. arXiv:1901.08746. [DOI] [PMC free article] [PubMed]
- 24.Devlin J, Chang M. -W., Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint. 2018. arXiv:1810.04805.
- 25.Pyysalo S, Ginter F, Moen H, Salakoski T, Ananiadou S. Distributional semantics resources for biomedical text processing. In: Proceedings of the Fifth International Symposium on Languages in Biology and Medicine. Database Center for Life Science: 2013. p. 39–44. https://pdfs.semanticscholar.org/e2f2/8568031e1902d4f8ee818261f0f2c20de6dd.pdf.
- 26.Sampo Pyysalo FilipGinter Hans Moen. Word vectors for biomedical natural language processing. 2013. http://evexdb.org/pmresources/vec-space-models/. Accessed 22 Aug 2019.
- 27.Pennington J, Socher R, Manning CD. Glove: Global vectors for word representation. In: Empirical Methods in Natural Language Processing (EMNLP): 2014. p. 1532–43. http://www.aclweb.org/anthology/D14-1162. https://doi.org/10.3115/v1/d14-1162.
- 28.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems. Neural Information Processing Systems Foundation: 2013. p. 3111–9. http://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf.
- 29.Matthew E. Peters. PubMed-based ELMo Model. 2018. https://s3-us-west-2.amazonaws.com/allennlp/models/elmo/contributed/pubmed/elmo_2x4096_512_2048cnn_ 2xhighway_weights_PubMed_only.hdf5. Accessed 22 Aug 2019.
- 30.Kim J-H, On KW, Lim W, Kim J, Ha J-W, Zhang B-T. Hadamard Product for Low-rank Bilinear Pooling. In: The 5th International Conference on Learning Representations. International Conference on Learning Representations (ICLR): 2017. https://arxiv.org/abs/1610.04325.
- 31.Smith A, Cohn T, Osborne M. Logarithmic opinion pools for conditional random fields. In: Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics. Association for Computational Linguistics: 2005. p. 18–25. 10.3115/1219840.1219843.
- 32.Sutton C, Sindelar M, McCallum A. Reducing weight undertraining in structured discriminative learning. In: Proceedings of the Main Conference on Human Language Technology Conference of the North American Chapter of the Association of Computational Linguistics. Association for Computational Linguistics: 2006. p. 89–95. 10.3115/1220835.1220847.
- 33.Hinton GE. Training products of experts by minimizing contrastive divergence. Neural Comput. 2002;14(8):1771–800. doi: 10.1162/089976602760128018. [DOI] [PubMed] [Google Scholar]
- 34.Gamal Crichton SampoPyysalo Billy Chiu and Anna Korhonen. MTL-Bioinformatics-2016. 2016. https://github.com/cambridgeltl/MTL-Bioinformatics-2016. Accessed 22 Aug 2019.
- 35.Smith L, Tanabe LK, nee Ando RJ, Kuo C-J, Chung I-F, Hsu C-N, Lin Y-S, Klinger R, Friedrich CM, Ganchev K, et al. Overview of biocreative ii gene mention recognition. Genome Biol. 2008;9(2):2. doi: 10.1186/gb-2008-9-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krallinger M, Rabal O, Leitner F, Vazquez M, Salgado D, Lu Z, Leaman R, Lu Y, Ji D, Lowe DM, et al. The chemdner corpus of chemicals and drugs and its annotation principles. J Cheminformatics. 2015;7(1):2. doi: 10.1186/1758-2946-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Sun Y, Johnson R, Sciaky D, Wei C. -H., Leaman R, Davis AP, Mattingly CJ, Wiegers TC, Lu Z. Annotating chemicals, diseases, and their interactions in biomedical literature. In: Proceedings of the Fifth BioCreative Challenge Evaluation Workshop: 2015. p. 173–82. https://pdfs.semanticscholar.org/eb1c/c140b14d0a8f5f789ba26e5e497a9776dd7e.pdf.
- 38.Doğan RI, Leaman R, Lu Z. Ncbi disease corpus: a resource for disease name recognition and concept normalization. J Biomed Informa. 2014;47:1–10. doi: 10.1016/j.jbi.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. International Conference on Artificial Intelligence and Statistics (AISTATS): 2010. p. 249–56. http://proceedings.mlr.press/v9/glorot10a/glorot10a.pdf?hc_location=ufi.
- 40.Kingma DP, Ba J. Adam: A method for stochastic optimization. In: Proceedings of the 3rd International Conference on Learning Representations. International Conference on Learning Representations (ICLR): 2015. https://arxiv.org/abs/1412.6980.
- 41.Lei Ba J, Kiros JR, Hinton GE. Layer normalization. arXiv preprint. 2016. arXiv:1607.06450.
- 42.Salimans T, Kingma DP. Weight normalization: A simple reparameterization to accelerate training of deep neural networks. In: Advances in Neural Information Processing Systems. Neural Information Processing Systems Foundation: 2016. p. 901–9. http://papers.nips.cc/paper/ 6113-weight-normalization-a-simple-reparameterization-to-accelerate-training-of-deep-neural-networks.
- 43.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–58. [Google Scholar]
- 44.Sampo Pyysalo. Python version of the evaluation script from CoNLL 2000. 2016. https://github.com/spyysalo/conlleval.py. Accessed 22 Aug 2019.
- 45.Wang Xuan, Zhang Yu, Ren Xiang, Zhang Yuhao, Zitnik Marinka, Shang Jingbo, Langlotz Curtis, Han Jiawei. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics. 2018;35(10):1745–1752. doi: 10.1093/bioinformatics/bty869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Model training strategy in the proposed framework. The effect of the proposed model training strategy is not shown in the main manuscript. The learning curves during model training demonstrate the reason why the training strategy facilitates the parameter learning of the underlying deep learning-based models under our proposed framework.
Data Availability Statement
The code for our models and instructions for the use can be found on GitHub https://github.com/kaist-dmlab/BioNER. The datasets used for performance evaluation and analysis during the current study are available in the MTL-Bioinformatics-2016 repository, https://github.com/cambridgeltl/MTL-Bioinformatics-2016.