Abstract
Background:
Genetic risk factors play an important role in the pathogenesis of familial intracranial aneurysm (FIA), however, the molecular mechanisms remain largely unknown. We investigate potential FIA-causing genetic variants by rare variant interrogation and a family-based genomics approach in a large family with an extensive multigenerational pedigree with FIA.
Method:
Exome sequencing (ES) was performed in a likely dominant family with IA disease. Variants were analyzed by an in-house developed pipeline and prioritized using various filtering strategies, including population frequency, variant type, and predicted variant pathogenicity. Sanger sequencing was also performed to evaluate the segregation of the variants with the phenotype.
Results:
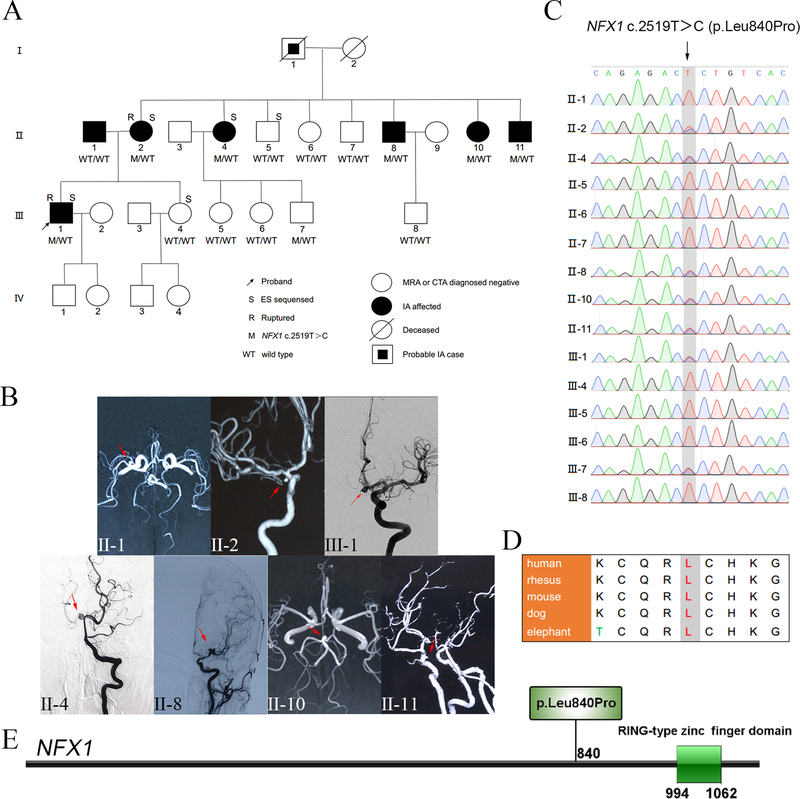
Based on the ES data obtained from five individuals from a family with 7/21 living members affected with IA, a total of 14 variants were prioritized as candidate variants. Familial segregation analysis revealed that NFX1 c.2519T>C (p.Leu840Pro) segregated in accordance with Mendelian expectations with the phenotype within the family; i.e. all IA-affected cases and absent from all unaffected members of the second-generation. This missense variant is absent from public databases (1000genome, ExAC, gnomAD, ESP5400), and has damaging predictions by bioinformatics tools (Gerp++ score =5.88, CADD score =16.43, MutationTaster score =1, LRT score =0). In addition, 840Leu in NFX1 is robustly conserved in mammals and maps in a region before the RING-type zinc finger domain.
Conclusion:
NFX1 c.2519T>C (p.Leu840Pro) likely contributes to the pathogenesis of FIA.
Keywords: familial intracranial aneurysm, exome sequencing, genetics, NFX1
Introduction
Intracranial aneurysm (IA [MIM: 105800]) is a complex disorder characterized by dilation or ballooning of the cerebral artery. IA affects 3.2% of the general population, with a mean age-of-onset of 50 years 1. IAs are generally asymptomatic, but the rupture of an IA may result in life-threatening subarachnoid hemorrhage (SAH, incidence rate in IA cases=0.7% 2), which could lead to death in half of the patients within 1 month 3. The pathogenesis of IA remains enigmatic. The wall of IAs is often characterized by lack of elastic laminas, leading to the collagen fibers being exposed to more mechanical load 4,5. Besides the known risk factors (hypertension, cigarette smoking, and alcohol consumption 6,7), mounting evidence suggests that genetic risk factors play an important role in the pathogenesis of IAs; IA may be considered as a complex trait and understanding potential gene X environmental (G X E) interactions might possibly elucidate modifiable risk. It is known that first degree relatives of patients with the disorder have up to seven times greater risk than the general population, and about 10% of patients with aneurysmal SAH have first or second-degree relatives with unruptured intracranial aneurysms 8. Korja et al. have reported that the estimated heritability for aneurysmal SAH is 41% in the Nordic Twin Cohort, suggesting that there is a moderate role for genetic factors in the etiology of SAH 9.
Of note, IAs are observed in a subset of families with dominant polycystic kidney disease (ADPKD) due to pathogenic variants in PKD1/2, Ehlers-Danlos syndrome IV caused by mutations in collagen type 3, Loeys–Dietz syndrome associated with variants in the transforming growth factor beta (TGFβ) signaling pathway genes, most frequently TGFBR1 and TGFBR2, and Marfan syndrome caused by pathogenic variants in FBN1 10. These latter Mendelizing disease traits are all syndromic examples with IA as an associated endophenotype, but clearly support an underlying genetic etiology for the pathobiology of IAs.
Both strategies of common and rare variant identification have been used in detecting disease-associated or disease-causing genetic factors potentially contributing to IAs. Genome-wide association studies (GWAS) have been applied extensively in sporadic IAs which have focused on the role of common variants that may have a minor effect on disease risk 11–16. In addition, replicated associations have identified some susceptibility loci for IAs including 4q31.23 (EDNRA), 8q12.1 (SOX17), 9p21.3 (CDKN2A/CDKN2B/CDKN2BAS), 10q24.32 (CNNM2), 12q22, 13q13.1(KL/STARD13), 18q11.2 (RBBP8), and 20p12.1 17–19. But although the locus association is robust to replicate, the actual potential gene involved at distinct locus is less clear. Moreover, these loci can only explain a small fraction (~5%) of the population-attributable risk for IAs 17,20. Thus, the genes contributing to the genetic predisposition of IAs is largely unknown. The detection of rare variants which are expected to have a larger effect size on disease risk is potentially one approach to unravel disease biology and the pathogenesis of the disorder.
Exome sequencing (ES) has emerged as a robust technology for identifying coding variation at the genome-wide level and enabling researchers to identify causative rare variants predisposing to disease. Broad application of ES has led to a better understanding of the genetic architecture of predisposition to some Mendelian diseases such as intracranial vertebral-basilar artery dissecting aneurysm (IVAD) 21, neurogenetic disorders 22,23, and brain arteriovenous malformations (BAVM) 24,25, as well as familial intracranial aneurysms (FIAs), in recent years. With the approach of ES, variants in ADAMTS15 26, TMEM132B 27, THSD1 28, RNF213 29, ANGPTL6 30 and LOXL2 31 were associated with FIAs. These recent advances provide new insights into the genetics of IA and demonstrate the usefulness of pedigree analysis and family-based ES to explore pathogenesis underlying IA formation.
In the present study, a large four-generation family with multiple cases of IA presenting an autosomal dominant (AD) likely inheritance pattern was ascertained and studied. By ES of selected family member samples and further segregation analysis using Sanger methodology, we identified a novel missense variant in Nuclear Transcription Factor X-box Binding 1 (NFX1 [MIM: 603255]) as the causative mutation in the family.
Materials and methods
Family Recruitment
We enrolled a family of Chinese Han ethnicity (Figure 1A) with 7/21 living members affected by IA. Participants were reviewed by two experienced neuroradiologists independently to validate the diagnosis of IAs by radiology imaging of the cerebrovascular system (MRA/DSA) and to rule out IAs resulting from syndromic disorders such as autosomal dominant polycystic kidney disease (ADPKD, MIM: 173900), Ehlers-Danlos syndrome type IV (MIM: 130050), Loeys–Dietz syndrome (MIM: 610192), Marfan syndrome (MIM: 154700), or brain arteriovenous malformations (BAVM, [MIM: 108010]) by physical examination and medical records review (past medical history, ultrasound, biochemical examination, X-ray). Peripheral blood samples were obtained from available family members.
Figure 1.
Family pedigree, clinical images, and segregation results (A-C). Protein location and evolutionary conservation of the NFX1 c.2519T>C variant (D-E). (A) Pedigree and NFX1 genotype segregation in the family. WT/WT represents both wild type alleles, i.e. bi-allelic, while M/WT designates NFX1 c.2519T>C pathogenic variant as heterozygous allele. (B) MRA/DSA images of the presenting family members IA; i.e. clinically affected cases. Red arrows indicate the site of IAs. (C) Validation of the NFX1 c.2519T>C variant via Sanger sequencing. (D) Protein sequence alignments indicate that 840Leu in NFX1 is robustly conserved in mammals. (E) NFX1 p.Leu840Pro mapping, located in a region where no domain structure has been delineated before the RING-type zinc finger domain of NFX1 (http://ibs.biocuckoo.org/).
This research was approved by the ethics committee of Beijing Tiantan Hospital under 2016YFC1300800. Informed written consent was obtained from all of the participants.
Genomic DNA Preparation and ES
Genomic DNA for each individual was extracted from peripheral blood lymphocytes using a standard phenol-chloroform method. Exome sequencing (ES) was performed on three IA cases and two phenotypically normal members from the family (Figure 1A, marked with “S”). DNA samples were prepared in Illumina libraries and then underwent whole-exome capture with the SureSelect Human All Exon V6+UTR r2 core design (91 Mb, Agilent), followed by sequencing on the Illumina HiSeq 4000 platform with 150-bp paired-end reads mode.
Variant-calling and Annotation
All reads were mapped to the human reference sequence (hg 19) using BWA-MEM (version 0.7.12). Picard (version 2.5.0, http://picard.sourceforge.net) and SAMtools (version 0.1.19) were used to mark duplicate reads and process the alignment file. Genome Analysis Toolkit (GATK version 3.4.0) was then used to refine the alignments by performing local indel realignment and subsequent base quality recalibration. Single-nucleotide variants (SNVs) and insertions/deletions (indels) were called with the Haplotype Caller of the GATK. Filtering of variant quality was performed by variant quality score recalibration (VQSR) measurement using GATK’s recommended parameters incorporating 892 in-house exome data (available upon request). Retained variants were annotated by the in-house ‘PUMCH’ annotation pipeline 32 which applied ANNOVAR (Annotation of Genetic Variants), VEP (Variant Effect Predictor) and additional annotation tools and clinical databases. Computational prediction tools (SIFT 33, Polyphen-2 34, MutationTaster 35, LRT 36, Gerp++ 37 and CADD 38) were used to predict the conservation and pathogenicity of candidate variants. All variants were compared against publicly available databases such as the 1000 Genomes Project (http://internationalgenome.org/), the Exome variant server, NHLBI GO Exome Sequencing Project (ESP, http://evs.gs.washington.edu/EVS/), the Exome Aggregation Consortium database (ExAC, http://exac.broadinstitute.org/), and Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/).
Manual Review and Prioritization
From the set of variants passed from quality control (QC) (Step 1) performed by VQSR measurement using GATK’s recommended parameters, we retained novel variants using Exome Aggregation Consortium_East Asian (ExAC_EAS) population (Step 2). Then all protein-altering or splice region variants were retained (Step 3). Variants segregated with disease, which present in the three affected individuals and absent in the phenotypically normal participants sent for ES were retained for further analysis (Step 4). Then, predicted deleterious (truncating or CADD (phred-like) score≥10) variants were list as the candidate variants (Step 5). These variants were manually reviewed by visual inspection of sequence reads using the Integrative Genomics Viewer (IGV) 39.
Variant Confirmation and Familial Segregation Analysis
To validate the variants identified by ES with an orthogonal sequencing technology, and evaluate the segregation of the variants with the phenotype, we sequenced all the available family members using direct Sanger sequencing. Variants segregated fully with definite IA phenotype in the family (Step 6), and absent from the second-generation unaffected members (Step 7) were considered as the final etiologic variant. Polymerase chain reaction (PCR) was performed with the primers summarized in Table S1, and products were purified with Axygen-AP-GX-50 Toolkit and sequenced on an ABI Prism 3730 Avant DNA sequencer (Applied Biosystems).
Results
Clinical Information
The proband (individual III-1; Figure 1A) was diagnosed after a subarachnoid hemorrhage (SAH) at age 31y, with a ruptured anterior communicating artery aneurysm (Figure 1B). In the following years, his aunt (Ⅱ−4) and his mother (Ⅱ−2) were sequentially stricken by SAH at age 52y and 63y, notably older than the proband’s age at diagnosis. Because of the significant recurrence of ruptured IAs in this family (III-1, II-2, II-4), a systematic cerebral artery screening was performed among all available relatives by digital subtraction angiography (DSA)/ magnetic resonance angiography (MRA). The proband’s another aunt (II-10) and two uncles (II-8, II-11) were observed to carry IAs (Figure 1B) without any clinical symptoms. His grandfather (I-1) died in his 60s after an episode suggestive of aneurysmal SAH. In combination with his unaffected sister (III-4), all findings consist with Mendelian expectation for AD likely IA disease trait in this family. Interestingly, the proband’s father (Ⅱ−1), without a familial history of IA, was also found to be carrying an IA in the right middle cerebral artery (Figure 1B). Characteristics and aneurysm images of IA cases are shown in Table 1 and Figure 1B independently. Demographic information of the other members is shown in Table S2. Three affected individuals (II-2, II-4, III-1) and two unaffected relatives (II-5, III-4) were selected for ES.
Table 1.
Clinical Characteristics of patients with intracranial aneurysm.
| Ⅱ−1 | Ⅱ−2 | Ⅱ−4 | Ⅱ−8 | Ⅱ−10 | Ⅱ−11 | Ⅲ−1 | |
|---|---|---|---|---|---|---|---|
| Age at diagnosis or last evaluation /sex | 68/M | 63/F | 52/F | 49/M | 46/F | 41/M | 31/M |
| Presentation | Asymptomatic | SAH | SAH | Asymptomatic | Asymptomatic | Asymptomatic | SAH |
| IA Location | MCA M4 | ICA C7 | BA terminus | MCA M4 | BA terminus | ICA C4 | ACoA |
| Size (mm) | 3×3 | 4×4 | 5×5 | 5×5 | 3×3 | 3×3 | 4×4 |
| Smoking | N | N | N | Y | N | Y | Y |
| Alcohol drinking | Y | N | N | Y | N | Y | Y |
| Hypertension | N | Y | Y | N | N | N | N |
| Diabetes | N | N | N | N | N | N | N |
| Hyperlipidemia | N | N | N | N | N | N | N |
M, man; F, female; MCA, middle cerebral artery; ICA, internal carotid artery; BA, basilar artery; ACoA, anterior communicating artery; Y, yes; N, no
Exome Sequencing Identified 14 Candidate Variants
ES on DNA isolated from blood of the selected five individuals generated approximately 10 billion bases for each individual with an average depth-of-coverage of 99.73×. And 96.35% (95.85 % to 96.65 %) of target exon regions were covered by at least 20× (Table S3). After alignment and a series of quality control procedures, 384102 single nucleotide variants (SNVs) and 45898 insertions/deletions (indels) were identified. We primarily focused on the novel, heterozygous variants in the coding region predicted by conceptual translation to affect protein-coding sequences. After filtering against reference from public databases (ExAC_EAS), 1589 non-synonymous SNVs and 478 indels were retained (Table 2, Step 3). Among them, 14 SNVs and 3 indels co-segregated with the IA phenotype among the 5 family members sent for ES (Table 2, Step 4). Taking into consideration of the pathogenicity, 14 variants were prioritized as candidate variants (Table 3). The rare variant filtering steps and results are illustrated in Table 2.
Table 2.
Exome-Variant Filtration Steps and Results of the Pedigree
| Filtration Steps | SNVs | indels |
|---|---|---|
| Step 1): QC passed variants a | 384102 | 45898 |
| Step 2): novel variants b | 204610 | 33009 |
| Step 3): Protein-altering c or splice region variants | 1589 | 478 |
| Step 4): Variants segregated with disease d | 14 | 3 |
| Step 5): Variants predicted to be deleterious e | 11 | 3 |
| Candidate variants list | 11 | 3 |
| Step 6): familial segregation analysis f | 4 | 2 |
| Step 7): absent from the second-generation unaffected members | 1 | 0 |
| The final etiologic variant | NFX1 c.2519T>C | |
SNVs and indel variants filtered by variant quality score recalibration (VQSR) measurement using GATK’s recommended parameters;
Novel variants, absent from public databases (ExAC_EAS);
Nonsynonymous, stop-gain, frameshift, start-lost, stop-lost;
Variants present in the three affected individuals (II-2, II-4, III-1) and absent in the phenotypically normal participants (II-5, III-4) sent for ES;
Variants predicted to be deleterious with CADD≥10 or truncating variants;
Variants segregated fully with definite IA phenotype in the family.
Table 3.
Information of Candidate Variants
| Gene Symbol | Variant Type | Zygosity | Chr_Position | Ref mRNA | Amino Acid Change | Gerp++ Score | CADD Score | pLI Score | ExAC_EAS Count |
|---|---|---|---|---|---|---|---|---|---|
| EFHD1 | Missense | Het | 2_233498511 | NM_025202.3 | c.97G>A(p.Ala33Thr) | −1.08 | 11.56 | – | 0 |
| FNDC1 | Missense | Het | 6_159654771 | NM_032532.2 | c.3227A>G(p.Gln1076Arg) | −2.84 | 13.63 | – | 0 |
| IFNA2 | Missense | Het | 9_21385094 | NM_000605.3 | c.235C>T(p.Leu79Phe) | −0.19 | 14.89 | – | 0 |
| NFX1 | Missense | Het | 9_33351652 | NM_002504.4 | c.2519T>C(p.Leu840Pro) | 5.88 | 16.43 | – | 0 |
| MYOCD | Missense | Het | 17_12666834 | NM_153604.2 | c.2690C>T(p.Pro897Leu) | 6.08 | 18.70 | – | 0 |
| RHO | Missense | Het | 3_129247637 | NM_000539.3 | c.61C>T(p.Arg21Cys) | 4.59 | 26.2 | – | 0 |
| PLK3 | Missense | Het | 1_45271250 | NM_004073.2 | c.1841C>T(p.Thr614Ile) | 5.70 | 29.9 | – | 0 |
| LENG8 | Missense | Het | 19_54967909 | NM_052925.2 | c.1540G>T(p.Ala514Ser) | 3.07 | 21.5 | – | 0 |
| SERAC1 | Missense | Het | 6_158535967 | NM_032861.3 | c.1538C>T(p.Thr513Met) | 2.39 | 35 | – | 0 |
| HYDIN | Missense | Het | 16_71127814 | NM_001198542 | c.1433G>C(p.Arg478Pro) | 4.95 | 17.69 | – | 0 |
| KIAA1524 | Missense | Het | 3_108279578 | NM_020890.2 | c.1745A>T(p.Lys582Met) | 5.68 | 12.66 | – | 0 |
| ESYT3 | Frameshift | Het | 3_138153945 | NM_031913.3 | c.308delG(p.Gly103AlafsTer30) | 2.19 | 13.28 | 0 | 0 |
| TIGD7 | Frameshift | Het | 16_3350476 | NM_033208.3 | c.135_139delTAAAA(p.Lys47Ter) | 4.29 | 36 | 0 | 0 |
| HEATR4 | Deletion | Het | 14_73989704 | NM_203309.2 | c.151_153delTTC(p.Phe51del) | 0.45 | 12.04 | 0 | 0 |
| NOTCH3 | Missense | Het | 19_15297996 | NM_000435.2 | c.1760G>A(p.Arg587His) | 3.22 | 11.87 | – | 1 |
Missense Variant in NFX1 Co-segregating with the Disease
The 14 candidate variants were validated using Sanger sequencing in all fifteen individuals (7 cases, 8 controls) in the family. Co-segregation analysis identified a missense variant in NFX1 shared by all the affected IA cases and absent from the second-generation unaffected members. In the third-generation, NFX1 c.2519T>C was detected in only one 29 year old member (Ⅲ−7) who may not manifest IA at the time probably due to the late age of onset for IA (Figure 1C); these data suggested the causal role of NFX1 c.2519T>C in this family.
NFX1 encodes a nucleic acid-binding protein that interacts with the conserved X1 box cis-element, and is conserved through yeast, Drosophila, Caenorhabditis elegans, Arabidopsis, mice, and humans 40. Studies of NFX1 homologues demonstrate its importance in normal cell growth, function, and homeostasis across species40. In addition, 840Leu in NFX1 is robustly conserved in mammals (Figure 1D) indicating its evolution may have preserved function. NFX1 c.2519T>C (p.Leu840Pro) maps where no domain structure has been delineated in a region before the RING-type zinc finger domain (Figure 1E); variation to Pro may produce a kink in the protein secondary structure. This missense variant is absent from public databases (1000genome, ExAC, gnomAD, ESP5400), and has substantial damaging predictions by bioinformatics tools (Gerp++ score =5.88, CADD score =16.43, MutationTaster score =1, LRT score =0).
Identification of a Paternally Inherited Missense Variant in NOTCH3
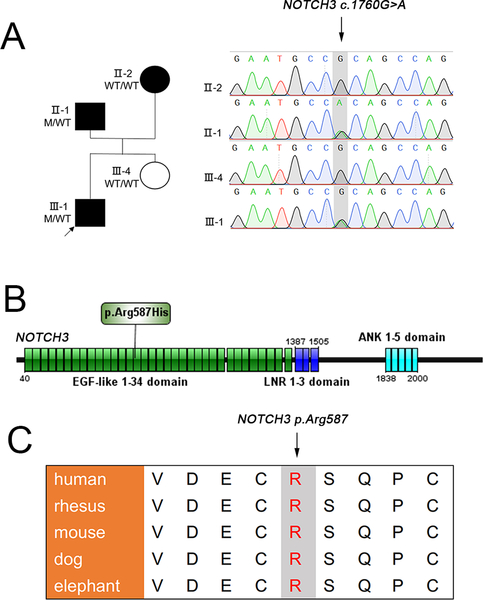
Since the father of the proband also developed an IA (individual Ⅱ−1; Figure 1B), we also analyzed paternally inherited rare variants in the proband. After filtering with the conditions of low minor allele frequency and inheritance modal, we found a deleterious missense variant (c.1760G>A) in NOTCH3 which was confirmed in his affected father while absent in his unaffected sister using Sanger method (Figure 2A). The NOTCH3 c.1760G>A (p.Arg587His) variant is located in the EGF-like 15 domain (Figure 2B). With the predictions by bioinformatics tools (Gerp++ score =3.22, CADD score =11.78, MutationTaster score =0.996), this missense variant is present in only one individual in ExAC_EAS. Meanwhile, 587Arg in NOTCH3 is also strongly conserved in mammals, including human, rhesus, mouse, dog and elephant (Figure 2C). However, we are unable to testify the participation of this variant in the pathogenesis of IA in the proband or his affected father due to the lack of genetic evidence.
Figure 2.
Pedigree of the proband’s family, and Sanger sequencing, protein location and evolutionary conservation of the NOTCH3 c.1760G>A variant (A-C). (A) Validation of NOTCH3 c.1760G>A variant via Sanger sequencing in the family. WT/WT represents both two wild type alleles, i.e. bi-allelic, while M/WT designates NOTCH3 c.1760G>A pathogenic variant as heterozygous allele. (B) NOTCH3 p.Arg587His is located in the EGF-like 15 domain (http://ibs.biocuckoo.org/). (C) Protein sequence alignments indicate that 587Arg in NOTCH3 is robustly conserved in mammals.
Discussion
In the present study, we identified one novel missense variant, c.2519T>C (p.Leu840Pro), in the 16th exon of Nuclear Transcription Factor X-box Binding 1 (NFX1), which was heterozygous in all six IA-affected members and only one out of the eight unaffected relatives in the pedigree. This variant is absent from public databases, and predicted to be deleterious by bioinformatics tools.
In addition, we found a deleterious missense variant c.1760G>A (p.Arg587His) in NOTCH3 which was detected in the proband (Ⅲ−1) and his affected father but not in his mother and sister. This variant leads to the same residue change c.1759C>T (p.Arg587Cys) enrolled in HGMD as causative for CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) 41, a vascular degenerative disease that is the most common form of hereditary stroke disorder, leading to dementia due to systemic vascular degeneration. While intracranial aneurysms are present in both the proband and father’s cerebral vascular imaging, there are no clinical manifestations of migraine, recurrent cerebral ischemia, emotional disturbances, and dementia, or any evidence of leukoencephalopathy to suggest a CADASIL diagnosis.
Concurrent variants in the two genes suggest the possibility of epistasis or mutational burden effects. The proband (Ⅲ−1) who is heterozygous for both of the deleterious variants had more severe IA characterized by an early age of onset of IA rupture. For example, the age of the proband on his IA rupture (31 years old) is much younger than that of the other two affected members on his maternal side of the family (52 and 63 years old). The impact of mutational burden on phenotypic expression and severity of disease has been described in families with peripheral neuropathy demonstrating intrafamilial phenotypic variability 42. Such second hit mutations in another gene that trigger the disease process in the region of the lesion also exist in several vascular diseases 28,43–45. Genetic heterogeneity, phenocopy, age dependent penetrance, and gene-environment interactions are all factors that make it complicated to identify the pathogenesis of IA. NFX1 has not been previously implicated in cerebrovascular disease. Therefore, this finding may represent a novel disease association for NFX1.
The major limitation of our study is the lack of investigation of the functional significance of the identified NFX1 and NOTCH3 variants. Our study design focused on rare, deleterious variants in potential novel IA-related genes. Intronic and regulatory region were not covered by ES, thus were unable to be studied.
Conclusions
In conclusion, NFX1 c.2519T>C (p.Leu840Pro) is likely contributing to the pathogenesis of FIA.
Supplementary Material
Acknowledgements
We thank the family who participated in this research. We also thank GeneSeeq Inc. for technical support of sequencing.
A) Funding Statement:
This work was supported by grants No. 2016YFC1300800 from the National Key Research and Development Plan of China, Nos. 81801156, 81801158, 81471167 & 81671139 from the National Natural Science Foundation of China, and No. 2018–4-1077 from the Special Research Project for Capital Health Development (All to Dr. Xinjian Yang); and grants Nos. 81822030 and 81501852 from the National Natural Science Foundation of China; 2016 Milstein Medical Asian American Partnership Foundation Fellowship Award in Translational Medicine, No. 7172175 from Beijing Natural Science Foundation and No. 2016-I2M-3–003 from the CAMS Initiative for Innovative Medicine, (All to Dr. Nan Wu). J.R.L was supported by the US National Institutes of Health, National Institute of Neurological Disorders and Stroke (NINDS R01 NS058529 and R35 NS105078), National Human Genome Research Institute/National Heart, Lung, and Blood Institute to the Baylor Hopkins Center o Mendelian Genomics (NHGRI/NHLBI UM1 HG006542). J.E.P. was supported by the National Human Genome Research Institute (NHGR K08 HG008986).
The funding sources had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
List of abbreviations:
- SAH
subarachnoid hemorrhage
- ES
exome sequencing
- IVAD
intracranial vertebral-basilar artery dissecting aneurysm
- BAVM
brain arteriovenous malformations
- AD
autosomal dominant
- NFX1
Nuclear Transcription Factor X-box Binding 1
- DSA
digital subtraction angiography
- MRA
magnetic resonance angiography
- SNV
single nucleotide variant
- GWAS
genome-wide association study
- indels
insertions/deletions
- ExAC_ EAS
Exome Aggregation Consortium_East Asian
- MAC
Minor allele count
- pLI
probability of loss of function (LoF) intolerance
- PCR
Polymerase chain reaction
- VSMCs
vascular smooth muscle cells
- DTAAD
descending thoracic aortic aneurysm and dissection
- CADASIL
cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
- CCM
cerebral cavernous malformation
Footnotes
B) Competing interests Statement:
The authors declare that they have no competing interests.
D) Data Sharing:
All exome data in this study are available upon request to the corresponding author.
Reference:
- 1.Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. The Lancet Neurology 2011;10:626–36. [DOI] [PubMed] [Google Scholar]
- 2.Rinkel GJ, Djibuti M, Algra A, van Gijn J. Prevalence and risk of rupture of intracranial aneurysms: a systematic review. Stroke 1998;29:251–6. [DOI] [PubMed] [Google Scholar]
- 3.Steiner T, Juvela S, Unterberg A, Jung C, Forsting M, Rinkel G, et al. European Stroke Organization guidelines for the management of intracranial aneurysms and subarachnoid haemorrhage. Cerebrovascular diseases 2013;35:93–112. [DOI] [PubMed] [Google Scholar]
- 4.Frosen J. Smooth muscle cells and the formation, degeneration, and rupture of saccular intracranial aneurysm wall--a review of current pathophysiological knowledge. Translational stroke research 2014;5:347–56. [DOI] [PubMed] [Google Scholar]
- 5.Frösen J PA, Paetau A, Kangasniemi M, Niemelä M, Hernesniemi J, Jääskeläinen J. Remodeling of saccular cerebral artery aneurysm wall is associated with rupture: histological analysis of 24 unruptured and 42 ruptured cases. Stroke 2004;35:2287–93. [DOI] [PubMed] [Google Scholar]
- 6.Teunissen LL, Rinkel GJ, Algra A, van Gijn J. Risk factors for subarachnoid hemorrhage: a systematic review. Stroke 1996;27:544–9. [DOI] [PubMed] [Google Scholar]
- 7.Ruigrok YM, Buskens E, Rinkel GJE. Attributable risk of common and rare determinants of subarachnoid hemorrhage. Stroke 2001;32:1173–5. [DOI] [PubMed] [Google Scholar]
- 8.Ruigrok YM, Rinkel GJ, Wijmenga C. Genetics of intracranial aneurysms. The Lancet Neurology 2005;4:179–89. [DOI] [PubMed] [Google Scholar]
- 9.M K,K S, P M, S Z, A S, A H, et al. Genetic epidemiology of spontaneous subarachnoid hemorrhage: Nordic Twin Study. Stroke 2010;41 2458–62. [DOI] [PubMed] [Google Scholar]
- 10.Hitchcock E, Gibson WT. A Review of the Genetics of Intracranial Berry Aneurysms and Implications for Genetic Counseling. Journal of genetic counseling 2017;26:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, Zu Fraunberg M, van Duijn CM, et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nature genetics 2008;40:1472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foroud T, Koller DL, Lai D, Sauerbeck L, Anderson C, Ko N, et al. Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke 2012;43:2846–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nature genetics 2010;42:420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foroud T, Lai D, Koller D, Van’t Hof F, Kurki MI, Anderson CS, et al. Genome-wide association study of intracranial aneurysm identifies a new association on chromosome 7. Stroke 2014;45:3194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama K, Narita A, Nakaoka H, Cui T, Takahashi T, Yasuno K, et al. Genome-wide association study to identify genetic variants present in Japanese patients harboring intracranial aneurysms. Journal of human genetics 2010;55:656–61. [DOI] [PubMed] [Google Scholar]
- 16.Low SK, Takahashi A Fau - Cha P-C, Cha Pc Fau - Zembutsu H, Zembutsu H Fau - Kamatani N, Kamatani N Fau - Kubo M, Kubo M Fau - Nakamura Y, et al. Genome-wide association study for intracranial aneurysm in the Japanese population identifies three candidate susceptible loci and a functional genetic variant at EDNRA. Hum Mol Genet 2012;21:2102–10. [DOI] [PubMed] [Google Scholar]
- 17.Yasuno K, Bakircioglu M, Low SK, Bilguvar K, Gaal E, Ruigrok YM, et al. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. P Natl Acad Sci USA 2011;108:19707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deka R, Koller DL, Lai D, Indugula SR, Sun G, Woo D, et al. The relationship between smoking and replicated sequence variants on chromosomes 8 and 9 with familial intracranial aneurysm. Stroke 2010;41:1132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashikata H, Liu W, Inoue K, Mineharu Y, Yamada S, Nanayakkara S, et al. Confirmation of an association of single-nucleotide polymorphism rs1333040 on 9p21 with familial and sporadic intracranial aneurysms in Japanese patients. Stroke 2010;41:1138–44. [DOI] [PubMed] [Google Scholar]
- 20.Tromp G, Weinsheimer S, Ronkainen A, Kuivaniemi H. Molecular basis and genetic predisposition to intracranial aneurysm. Annals of medicine 2014;46:597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Zhao S, Zhang Q, Yuan J, Liu J, Ding X, et al. Whole-exome sequencing reveals known and novel variants in a cohort of intracranial vertebral-basilar artery dissection (IVAD). Journal of human genetics 2018;63:1119–28. [DOI] [PubMed] [Google Scholar]
- 22.Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Akdemir ZC, et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron 2015;88:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiszniewski W, Gawlinski P, Gambin T, Bekiesinska-Figatowska M, Obersztyn E, Antczak-Marach D, et al. Comprehensive genomic analysis of patients with disorders of cerebral cortical development. European journal of human genetics : EJHG 2018;26:1121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Zhao S, Liu B, Zhang Q, Li Y, Liu J, et al. Perturbations of BMP/TGF-beta and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM). J Med Genet 2018;55:675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolaev SI, Fish JE, Radovanovic I. Somatic Activating KRAS Mutations in Arteriovenous Malformations of the Brain. N Engl J Med 2018;378:1561–2. [DOI] [PubMed] [Google Scholar]
- 26.Yan J, Hitomi T, Takenaka K, Kato M, Kobayashi H, Okuda H, et al. Genetic study of intracranial aneurysms. Stroke 2015;46:620–6. [DOI] [PubMed] [Google Scholar]
- 27.Farlow JL, Lin H, Sauerbeck L, Lai D, Koller DL, Pugh E, et al. Lessons learned from whole exome sequencing in multiplex families affected by a complex genetic disorder, intracranial aneurysm. PloS one 2015;10:e0121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santiago-Sim T, Fang X, Hennessy ML, Nalbach SV, DePalma SR, Lee MS, et al. THSD1 (Thrombospondin Type 1 Domain Containing Protein 1) Mutation in the Pathogenesis of Intracranial Aneurysm and Subarachnoid Hemorrhage. Stroke 2016;47:3005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou SR, Ambalavanan A, Rochefort D, Xie PX, Bourassa CV, Hince P, et al. RNF213 Is Associated with Intracranial Aneurysms in the French-Canadian Population. Am J Hum Genet 2016;99:1072–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourcier R, Le Scouarnec S, Bonnaud S, Karakachoff M, Bourcereau E, Heurtebise-Chretien S, et al. Rare Coding Variants in ANGPTL6 Are Associated with Familial Forms of Intracranial Aneurysm. Am J Hum Genet 2018;102:133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Li Z, Shi Y, Chen L, Tan H, Wang Z, et al. Exome Sequencing Identifies LOXL2 Mutation as a Cause of Familial Intracranial Aneurysm. World Neurosurg 2017;109:e812–e8. [DOI] [PubMed] [Google Scholar]
- 32.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature genetics 2011;43:309–15. [DOI] [PubMed] [Google Scholar]
- 33.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nature protocols 2016;11:1–9. [DOI] [PubMed] [Google Scholar]
- 34.Adzhubei Ia Fau - Schmidt S, Schmidt S Fau - Peshkin L, Peshkin L Fau - Ramensky VE, Ramensky Ve Fau - Gerasimova A, Gerasimova A Fau - Bork P, Bork P Fau - Kondrashov AS, et al. A method and server for predicting damaging missense mutations. Nature methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods 2014;11:361–2. [DOI] [PubMed] [Google Scholar]
- 36.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome research 2009;19:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. Plos Comput Biol 2010;6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics 2014;46:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson Jt Fau - Thorvaldsdottir H, Thorvaldsdottir H Fau - Winckler W, Winckler W Fau - Guttman M, Guttman M Fau - Lander ES, Lander Es Fau - Getz G, Getz G Fau - Mesirov JP, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA. NFX1–123 and human papillomavirus 16E6 increase Notch expression in keratinocytes. Journal of virology 2013;87:13741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim Y, Choi EJ, Choi CG, Kim G, Choi JH, Yoo HW, et al. Characteristics of CADASIL in Korea: a novel cysteine-sparing Notch3 mutation. Neurology 2006;66:1511–6. [DOI] [PubMed] [Google Scholar]
- 42.Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep 2015;12:1169–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atri D, Larrivee B, Eichmann A, Simons M. Endothelial signaling and the molecular basis of arteriovenous malformation. Cellular and molecular life sciences : CMLS 2014;71:867–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monkley SJ, Kostourou V, Spence L, Petrich B, Coleman S, Ginsberg MH, et al. Endothelial cell talin1 is essential for embryonic angiogenesis. Dev Biol 2011;349:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leblanc GG, Golanov E, Awad IA, Young WL, Biology of Vascular Malformations of the Brain NWC. Biology of vascular malformations of the brain. Stroke 2009;40:e694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.