Abstract
Background
The emergence of Vancomycin resistant enterococci (VRE) poses a major public health problem since it was first reported. Although the rising rates of VRE infections are being reported elsewhere in the worldwide; there is limited national pooled data in Ethiopia. Therefore, this study was aimed to estimate the pooled prevalence of VRE and antimicrobial resistance profiles of enterococci in Ethiopia.
Methods
Literature search was done at PubMed, EMBASE, Google scholar, African Journals online (AJOL) and Addis Ababa University repository following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline. Both published and unpublished studies reporting the prevalence of VRE until June 30, 2019 were included. Data were extracted using Microsoft Excel and copied to Comprehensive Meta-analysis (CMA 2.0) for analysis. Pooled estimate of VRE was computed using the random effects model and the 95% CIs. The level of heterogeneity was assessed using Cochran’s Q and I2 tests. Publication bias was checked by visual inspection of funnel plots and Begg’s and/or Egger’s test.
Results
Twenty studies fulfilled the eligibility criteria and found with relevant data. A total of 831 enterococci and 71 VRE isolates were included in the analysis. The pooled prevalence of VRE was 14.8% (95% CI; 8.7–24.3; I2 = 74.05%; P < 0.001). Compared to vancomycin resistance, enterococci had higher rate of resistance to Penicillin (60.7%), Amoxicillin (56.5%), Doxycycline (55.1%) and Tetracycline (53.7%). Relatively low rate of resistance was found for Daptomycin and Linezolid with a pooled estimate of 3.2% (95% CI, 0.5–19.7%) and 9.9% (95% CI, 2.8–29.0%); respectively. The overall pooled multidrug resistance (MDR) rate of enterococci was 60.0% (95% CI, 42.9–75.0%).
Conclusion
The prevalence of VRE and drug resistant enterococci are on the rise in Ethiopia. Enterococcal isolates showed resistance to one or more of the commonly prescribed drugs in different or the same drug lines. Multidrug resistant (MDR) enterococci were also found. Although the rates were low, the emergence of resistance to Daptomycin and Linezolid is an alarm for searching new ways for the treatment and control of VRE infections. Adherence to antimicrobial stewardship, comprehensive testing and ongoing monitoring of VRE infections in the health care settings are required.
Keywords: Enterococcus, Vancomycin resistance, Systematic review, Ethiopia
Background
Today, antimicrobial resistance (AMR) is one of the most important public health problem in the world and continues to challenge treatment especially in bacteria [1]. Widespread use and misuse of antibiotics is thought to increase the prevalence and emergence of resistance bacterial strains. As a growing problem; AMR complicates the treatment of bacterial infections leading to increased mortality, morbidity and healthcare related costs. The emergence of Vancomycin resistant enterococci (VRE) poses a major public health problem since it was first reported. VRE are among the most common resistant pathogens frequently causing healthcare associated infections and a growing concern for health care professionals [1–4].
Enterococci are gram-positive bacterial flora of the intestinal tract of humans, animals and birds [5–7]. Despite their commensal characteristics, they cause serious nosocomial infections in humans including urinary tract, bloodstream infections and endocarditis [8]. They are “tough bugs” that can survive in/and on the environment for long periods and became one of the main nosocomial pathogens. Enterococci are also able to form biofilms that contribute to the virulence, resistance to antibiotics and phagocytosis making their eradication extremely difficult [9, 10].
Enterococci become resistant to a variety of antimicrobials through intrinsic and acquired mechanisms. Isolates of E. gallinarum and E. flavescens develop an inherent, low-level resistance to Vancomycin [11]. Enterococci readily accumulate mutations and exogenous genes that confer additional resistance. They develop resistance to vancomycin by exchange of genetic material among themselves and/or with another genera [12]. The enterococci may acquire resistance through van associated genetic elements (vanA, vanB, vanD, vanE, vanG, vanL); of which vanA and vanB are the most prevalent genotypes in clinical isolates [11, 13, 14]. The vanA and vanB gene clusters are most commonly found in E. faecium and increasingly reported throughout the world [12, 15]. Other transposable elements are also reported to be involved in the spread of antimicrobial resistance [16].
Vancomycin was considered as one of the last lines of treatment against multidrug resistant organisms including ampicillin resistant enterococci and methicillin-resistant Staphylococcus aureus (MRSA) [8]. However, enterococci develop high level of resistance and the incidences of VRE infections among hospitalized patients has increased rapidly [9, 13, 17]. Infections due to VRE have been also reported to be associated with longer hospital stays, increased mortality and higher healthcare costs than infections with vancomycin susceptible enterococci [15, 18–20].
Enterococcal infections are now getting attention due to their ability to develop resistance to multiple antimicrobial agents which probably explain their large part of isolation in nosocomial infections [21, 22]. The two species (E. faecalis and E. faecium) are responsible for majority of the infections in humans. They are also constituting a reservoir for antibiotic resistance among the gut enterococci [23]. In 2017, the World Health Organization (WHO) has published the priority lists of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Hence; Vancomycin resistant E. faecium was categorized as high priority pathogens for which new and effective treatments are need [24]. Reports are also emerging on the development of resistance to Daptomycin and Linezolid which are being used to treat Vancomycin resistant enterococcal infections [14]; this could explain the challenging nature of these bacteria in the current medicine and as well as to the future. Other studies reported the continuous increase of VRE causing nosocomial infections [25].
In Ethiopia; different reports showed that antimicrobials are widely misused by health care providers, unskilled practitioners, animal husbandry operators and drug users. Antimicrobial misuse is one of the major driver and contributor of the emergence and survival of resistance strains. To prevent and contain the spread of drug resistance, the Ethiopian Public Health Institute (EPHI) established AMR surveillance centers and identified national priority surveillance pathogens in 2017 [4]. A previous systematic review has also reported the growing challenges of antibacterial drug resistance in Ethiopia [26]; but VRE were included neither in the national priority surveillance pathogens nor in previous systematic reviews. Although the rising rates of VRE infections are being reported elsewhere in the worldwide; there is limited national pooled data in Ethiopia. Therefore; this study was aimed at summarizing the findings of local studies to estimate the pooled prevalence of VRE and antimicrobial resistance profiles of enterococci in Ethiopia.
Methods
Search strategy
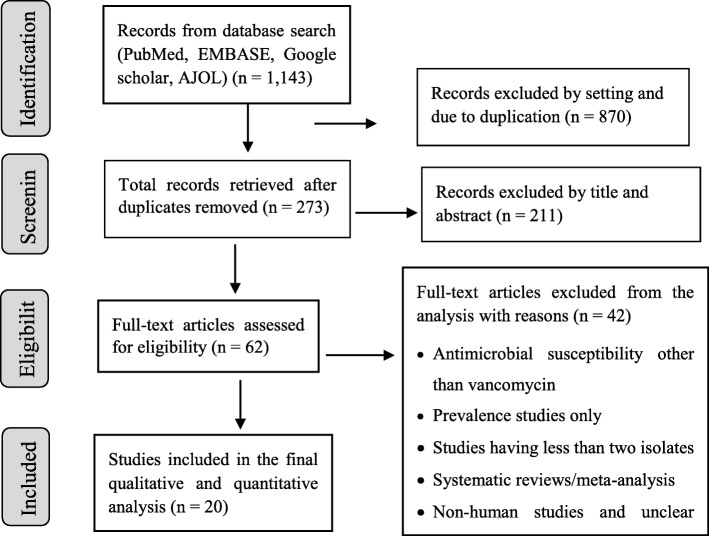
A comprehensive search was conducted at PubMed, EMBASE, Google scholar and African journals online (AJOL). To include unpublished studies (theses, dissertations); the repository of Addis Ababa University was searched. Reference lists of included studies were also sought. The database search was done following the PRISMA guideline/checklists [27] (Fig. 1). The PubMed was searched using MeSH terms and Boolean operators. The search string in PubMed was: ((((((((Enteroccoc*) OR Enterococcus faecalis) OR Enterococcus faecium) OR E. faecalis OR E. faecium AND Vancomycin resistan*) OR antibiotic resistan*) OR antimicrobial resistan*) OR drug resistan*) OR VRE) AND Ethiopia)))))))). Search results were combined in to EndNote X6 (Clarivate Analytics USA) and duplicates were removed. Studies published/reported up to June 30, 2019 and fulfilled the eligibility criteria (Table 1) were included.
Fig. 1.
PRISMA flow chart of study selection
Table 1.
Eligibility criteria
| Inclusion criteria | Exclusion criteria |
|---|---|
|
• Study settings: conducted in Ethiopian on any settings • Study subjects/population: humans • Study design: any study reported the prevalence of VRE or numbers of VRE and total enterococci isolates • Sample size: studies isolated not less than two enterococci • Language: published/reported in English • Type of study: peer-reviewed, full text available before June 30, 2019 |
• Studies on antimicrobial susceptibility tests other than vancomycin (studies that did not include VRE) • Prevalence studies only • Studies having less than two isolates • Studies not reporting enterococcal isolates separately (no population denominator) • Reviews, comments and duplications • Studies on non-human subjects |
Quality assessment
The quality of included studies was assessed by the Joanna Briggs Institute (JBI) critical appraisal checklist for prevalence data [28] (additional file 1); which contains nine sections. The assessment was done independently by two authors (AM and TA). Studies were included in the analysis if consensus was reached among the two reviewers. The quality of the 20 included studies is given in (additional file 2).
Data extraction
After studies were identified based on the predefined eligibility criteria; author name with year of publication, study period, region of study, study design, sample size, study population, types of specimens, antimicrobial susceptibility testing (AST) methods, number of isolates (both the total and vancomycin resistant enterococci), types of isolated species and history of publication were extracted using Microsoft Excel 2013 data collection sheet especially designed for this study. Resistance profiles of enterococci to other antimicrobials were also extracted and the study level proportions were pooled. The data extraction was done independently by two authors (AM and TA).
Data analysis
Whenever studies were not reporting the prevalence of VRE, it was calculated by dividing the numbers of VRE isolates to the total numbers of tested enterococcal isolates and multiplying by 100. Studies reporting a zero number of VRE isolates were imputed to 0.5 as a continuity correction to be include in the meta-analysis [29]. Subgroup analyses were done by the study region, study period, publication history, AST and types of specimens used to isolate enterococci.
Acknowledging the presence of heterogeneity in observational studies conducted in diverse settings, the random effects model was used in determining the pooled prevalence of VRE as well as resistance to other antimicrobials. Heterogeneity was evaluated by the Cochran’s Q-test and I2 statistics. Funnel plots were drawn to see the presence of publication bias and the Begg’s rank correlation and Egger’s regression tests were used to quantify the degree of publication bias. P-values < 0.05 in any of the Begg’s rank correlation and Egger’s regression tests were indicative of significant publication bias. In asymmetrical funnel plots, the Trim-and-Fill method was applied to include missing studies and estimate adjusted effect sizes. Sensitivity analysis in a leave-one-out approach was done to see the stability of the pooled prevalence of VRE and to explore the potential source of heterogeneity between studies. Data were analyzed using CMA version 2.0 for windows and used to generate forest and funnel plots.
Results
Study selection
The results of database search and process of study selection is shown in the flow chart below (Fig. 1). The search returned 1143 records; of which 62 studies were subjected for full text review for inclusion against the eligibility criteria. Finally, 42 studies were excluded and only 20 were included in our analysis.
Characteristics of included studies
All of the 20 studies included in this review were cross-sectional by design. Most of the studies were reported from Amhara region (n = 8) [30–37] and Addis Ababa (n = 7) [38–44]. The remaining studies were from Oromia (n = 4) [45–48] and Southern nations (n = 1) [23]. Studies were not available from administrative regions of Tigray, Afar, Dire Dawa, Harari, Somali, Gambela and Benishangul-Gumuz. Nineteen studies were conducted in hospital settings. Among the 6017 study participants included, 831 enterococci were isolated and tested with a variety of antimicrobials; of which 71 isolates were resistant to vancomycin. Stool, urine, blood and swab specimens were used to isolate enterococci. The highest numbers of enterococcal and VRE isolates were identified from stool followed by multi-site specimens.
Seventeen studies used disc diffusion and three studies employed dilution/minimum inhibitory concentration (MIC) as antimicrobial susceptibility testing (AST) method to determine Vancomycin resistance. Resistance to antimicrobial agents by either methods was defined based on the performance standards for antimicrobial susceptibility testing guidelines prepared by Clinical and Laboratory Standards Institute (CLSI, various editions). The prevalence of VRE ranged from 1.8% in Jimma to 60% in Addis Ababa. Species level enterococci were reported by four studies [23, 39, 47, 48] and E. faecalis and E. faecium were the most frequently isolated species. Six of the included studies were unpublished and 14 were published between 2013 and 2019. Details of the characteristics of the included studies is summarized in (Table 2) below.
Table 2.
Lists and characteristics of included studies
| Author, publication year | Study period | Study area/ region | Study design | Study subjects | Sample size | Prevalence of enterococci, N (%) | Type of specimen | AST method | Prevalence of VRE, N (%) | Types of isolates (species) | Publication history |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abamecha, 2015 [48] | January to July 2013 | Jima University Specialized Hospital, Oromia | CS | Hospitalized patients | 150 | 114 (76.00) | Stool, rectal swabs | Disc diffusion, MIC for VRE | 2 (1.8) | E.faecium, E.faecalis, E.gallinarum, E.casseliflavus, E.durans | Published |
| Abebe, 2014 [37] | July to September 2013 | University of Gondar Teaching Hospital, Amhara | CS | HIV positive and HIV negative clients | 226 | 201 (88.94) | Stool | Disc diffusion | 11 (5.5) | Not identified to species level | Published |
| Agegne, 2018 [36] | February to May 2017 | West Amhara Hospitals, Amhara | CS | HIV patients on ART | 349 | 220 (63.04) | Stool | Disc diffusion | 17 (7.7) | Not identified to species level | Published |
| Ali, 2018 [35] | February to May, 2017 | Dessie Referral Hospital, Amhara | CS | HIV positive and HIV negative clients | 300 | 112 (37.33) | Stool | Disc diffusion | 7 (6.3) | Not identified to species level | Published |
| Ayelign, 2018 [33] | February to June 2015 | University of Gondar Hospital, Amhara | CS | Pediatric patients | 310 | 3 (0.97) | Urine | Disc diffusion | 1 (33.3) | Not identified to species level | Published |
| Birri, 2013 [23] | Not reported | Dilla town, SNNPR | CS | Healthy infants aged 3 to 26 weeks | 28 | 53 (189.29)a | Stool | Dilution/MIC | 1 (1.9) | E.faecium, E.faecalis, E.avium, E.canintestini, E.maldoratus, E.raffinosus, E.gallinarum | Published |
| Eshetu, 2017 [44] | April to September 2016 | Tikur Anbessa Specialized Hospital, Addis Ababa | CS | Blood stream infection suspects | 422 | 5 (1.18) | Blood | Disc diffusion | 3 (60.0) | Not identified to species level | Unpublished |
| Fentie, 2018 [32] | February to April 2017 | University of Gondar teaching Hospital, Amhara | CS | Cancer patients | 216 | 2 (0.93) | Blood, urine, wound swab, ear discharge | Disc diffusion | 1 (50.0) | Not identified to species level | Published |
| Ferede, 2018 [41] | April to May 2016 |
Black Lion/Tikur Anbesa Specialized Hospital, Addis Ababa |
CS | Patients suspected for UTI, wound infection, septicemia, endocarditis, meningitis | 422 | 15 (3.55) | Blood, urine, body fluid, CSF, Pus | Disc diffusion | 1 (6.7) | Not identified to species level | Published |
| Gebrish, 2019 [47] | February to March 2016 | Jimma University Specialized Hospital, Oromia | CS | Hospitalized pediatric patients | 52 | 12 (23.08) | Stool, rectal swabs | Disc diffusion | 1 (8.3) | E.faecalis, E.faecalis, E.gallinarum | Published |
| Jemal, 2017 [34] | July to December 2016 | Felege Hiwot Referral Hospital, Amhara | CS | HIV patients on ART | 384 | 4 (1.04) | Blood | Disc diffusion | 0.5b (10.0) | Not identified to species level | Unpublished |
| Lega, 2015 [42] | April to July 2015 |
Yekatit 12 Hospital Medical College, Addis Ababa |
CS | Diabetic patients | 246 | 2 (0.81) | Urine | Disc diffusion | 1 (50.0) | Not identified to species level | Unpublished |
| Mitiku, 2018 [40] | September 2017 to June 2018 | Tikur Anbesa Specialized Hospital, Addis Ababa | CS | Under 5 children with febrile illness | 340 | 11 (3.24) | Blood | Disc diffusion | 0.5b (4.2) | Not identified to species level | Unpublished |
| Mohammed, 2017 [31] | March to May, 2014 |
University of Gondar Referral Hospital, Amhara |
CS | Patients with wound infections | 137 | 2 (1.46) | Wound swab | Disc diffusion | 1 (50.0) | Not identified to species level | Published |
| Molalign, 2016 [39] | September 2015 to May 2016 | Arsho Advanced Medical laboratory, Addis Ababa | CS | UTI patients | 712 | 15 (2.11) | Urine | Dilution/MIC | 7 (46.7) |
E.faecalis, E.gallinarum |
Unpublished |
| Sorsa, 2019 [46] | April 2016 to May 2017 | Asella teaching and referral hospital, Oromia | CS | Neonates with sepsis | 303 | 6 (1.98) | Blood | Disc diffusion | 1 (16.7) | Not identified to species level | Published |
| Teklehaymanot, 2016 | July to September 2015 | Tikur Anbessa Specialized Hospital, Addis Ababa | CS | Patients suspected for body fluid pathogens | 384 | 2 (0.52) | CSF, ascites, pleural fluid, synovial fluid | Disc diffusion | 0.5 b (16.7) | Not identified to species level | Unpublished |
| Toru, 2018 [45] | April to September 2016 |
Jimma University Specialized Hospital, Oromia |
CS | Pediatric patients (< 15 years) | 403 | 22 (5.46) |
Urine, blood, swabs, closed abscess, body fluids, CSF |
Disc diffusion | 5 (22.7) | Not identified to species level | Published |
| Woldemariam, 2019 [38] | April to July 2015 | St. Paul Specialized Hospital Millennium Medical College, Addis Ababa | CS | Adult diabetic patients | 248 | 6 (2.42) | Urine | Disc diffusion | 1 (16.7) | Not identified to species level | Published |
| Yilema, 2017 [30] | February to May 2014 | University of Gondar Teaching Hospital, Amhara | CS | Patients requiring culture and AST | 385 | 24 (6.23) |
Urine, blood, wound swabs, ear discharge, ascites, abscess |
Disc diffusion | 10 (41.7) | Not identified to species level | Published |
AST Antimicrobial Susceptibility testing, ART Antiretroviral Therapy, CS Cross-sectional, CSF Cerebrospinal Fluid, VRE Vancomycin resistant enterococci, MIC Minimum Inhibitory Concentration, SNNPR Southern Nations, Nationalities and Peoples Region
a: multiple enterococcal species were isolated from a single infant; b: 0.5 was added as a continuity correction to include the study in the analysis
Pooled prevalence of VRE
The pooled prevalence of VRE was estimated at 14.8% (95% CI; 8.7–24.3%; I2 = 74.05%; P < 0.001) (Fig. 2). Significant heterogeneity (Q = 73.21; I2 = 74.05%; P < 0.001) was observed in the estimation of overall pooled result. But, the sensitivity analysis revealed that no single study significantly influenced the heterogeneity and pooled prevalence of VRE. The pooled prevalence of VRE in the sensitivity analysis ranged from 13.2 to 16.7% which lies within the 95% CI bounds of the overall pooled estimate. The presence of publication bias was observed from the drawn asymmetric funnel plot (Fig. 3a). The Trim-and-Fill method was then applied to include the “missing” studies from the analysis. The asymmetric studies were trimmed to locate the unbiased effect and fills the plot by re-inserting the trimmed studies as well as their imputed counterparts. Accordingly, one study was missed and fall at the left side of the pooled estimate (Fig. 3b). In the Trim-and-Fill method, the adjusted estimate of VRE was 13.5% (95% CI; 7.8–22.2%); almost similar with the original pooled estimate. The Egger’s regression (intercept = 0.91; 95% CI; − 0.75 – 2.57; p = 0.263) and Begg’s rank test (p = 0.381) did not suggest significant publication bias.
Fig. 2.
Forest plot showing the pooled prevalence of VRE in Ethiopia
Fig. 3.
Funnel plot showing publication bias; before (a) and after (b) the Trim-and-Fill method is applied
Subgroup prevalence of VRE
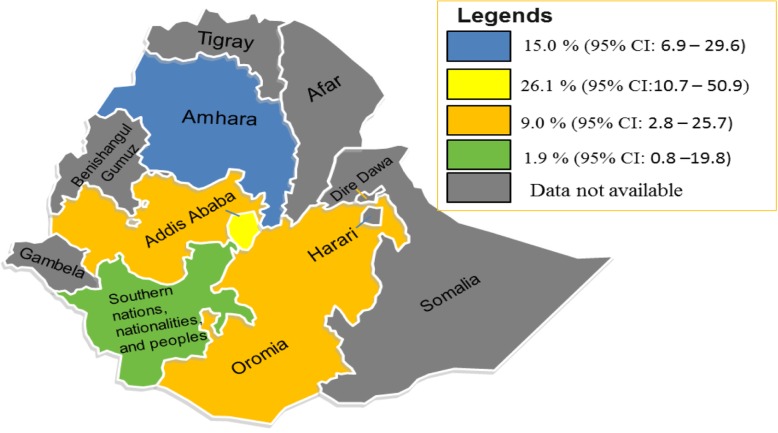
The prevalence of VRE was computed by region, type of antimicrobial testing (AST) method, study period, types of specimen used to isolate enterococci, and publication history. The prevalence of VRE by region was 26.1% (95% CI: 10.7–50.9%; I2 = 41.65%; P = 0.113) in Addis Ababa, 15.0% (95% CI: 6.9–29.6%; I2 = 79.39%; P < 0.001) in Amhara, 9.0% (95% CI: 2.8–25.7%; I2 = 71.49%; P = 0.015) in Oromia and 1.9% (95% CI: 0.1–23.1%) in Southern nations, nationalities and peoples region (SNNPR) (Table 3, Fig. 4). The prevalence of VRE pooled from studies conducted in the period before 2015 was 16.5% and that of the post-2015 was 16.3%; which indicates unchanged trend of VRE infections in Ethiopia. On the other hand, the pooled prevalence of VRE from studies which used disc diffusion to determine AST was 16.9% and it was 7.9% when AST was measured by dilution/minimum inhibitory concentrations (MICs). Relatively; high rates of VRE were isolated from urine (37.3%) and blood (22.0%) specimens. Use of multisite specimens did not increase the isolation rate of enterococci. Unpublished studies reported high rate of VRE than published studies (31.9% Vs. 11.3%; respectively) (Table 3).
Table 3.
Pooled prevalence of VRE by subgroups
| Subgroups | Numbers of studies | No of enterococci isolates tested, N | Pooled prevalence of VRE, N (%) | 95% CI | I2 | P-value |
|---|---|---|---|---|---|---|
| Region | ||||||
| Addis Ababa | 7 | 56 | 13 (26.1) | 10.7–50.9 | 41.65 | 0.113 |
| Amhara | 8 | 568 | 38 (15.0) | 6.9–29.6 | 79.39 | < 0.001 |
| Oromia | 4 | 154 | 19 (9.0) | 2.8–25.7 | 71.49 | 0. 015 |
| SNNPR | 1 | 53 | 1 (1.9) | 0.8–19.8 | – | – |
| Study perioda | ||||||
| Before/in 2015 | 8 | 354 | 27 (16.5) | 6.5–31.5 | 81.09 | < 0.001 |
| After 2015 | 11 | 424 | 43 (16.3) | 7.6–31.3 | 69.20 | < 0.001 |
| AST method | ||||||
| Disc diffusion | 17 | 649 | 61 (16.9) | 9.3–28.9 | 66.89 | < 0.001 |
| Dilution/MIC | 3 | 182 | 10 (7.9) | 1.9–27.6 | 91.88 | < 0.001 |
| Type of specimen | ||||||
| Stool | 5 | 598 | 37 (5.9) | 2.8–11.7 | 0.00 | 0.629 |
| Urine | 4 | 26 | 10 (37.3) | 15.8–63.3 | 0.00 | 0.665 |
| Blood | 4 | 26 | 4 (22.0) | 6.9–51.9 | 45.06 | 0.141 |
| Wound swab | 1 | 2 | 1 (50.0) | – | – | – |
| Multi-siteb | 6 | 179 | 19 (16.8) | 8.0–31.9 | 77.87 | < 0.001 |
| Publication history | ||||||
| Published | 14 | 792 | 60 (11.3) | 6.4–19.2 | 72.86 | < 0.001 |
| Unpublished | 6 | 39 | 11 (31.9) | 12.9–59.7 | 25.82 | 0.241 |
a One study did not report its study period; b Studies used more than one type of specimen to isolate enterococci; MIC: Minimum Inhibitory Concentration
Fig. 4.
Map showing regional distribution of VRE in Ethiopia; Map adapted from en.Wikipedia.org
Antimicrobial resistant enterococci
The resistance profile of enterococci was also pooled for antimicrobials other than Vancomycin. Resistance rates were pooled if at least two studies reported on a specific bacterium-antibiotic combinations. High level of resistance was observed to all classes of tested antimicrobials except to Daptomycin and Linezolid. The pooled resistance rate of enterococci to Daptomycin was 3.2% (95% CI; 0.5–19.7%) and that of Linezolid was 9.9% (95% CI; 2.8–29.0%). The pooled resistance rate to other antimicrobials was 60.7% (95% CI; 39.2–78.3%) to Penicillin, 56.5% (95% CI; 49.6–63.2%) to Amoxicillin, 53.7% (95% CI; 35.8–70.7%) to Tetracycline, 55.1% (95% CI; 22.2–84.9%) to Doxycycline, and 49.6% (95% CI; 36.5–62.7%) to Erythromycin. Studies reporting resistance to three or more antimicrobials were also pooled to estimate the prevalence of multidrug resistant (MDR) enterococci in Ethiopia. Hence; the overall prevalence of MDR enterococci was 63.0% (95% CI; 48.6–75.4%; I2 = 90.27%; P < 0.001) (Table 4).
Table 4.
Pooled resistance profile of enterococcal isolates in Ethiopia
| Antibiotics | No of studies | No of enterococci isolates tested, N | Pooled resistance N, (%) | 95% CI | I2 (%) | P-value |
|---|---|---|---|---|---|---|
| Amoxicillin | 2 | 203 | 115 (56.5) | 49.6–63.2 | 0.00 | 0.382 |
| Amox-clavulanate | 2 | 225 | 71 (45.3) | 13.9–80.9 | 92.37 | < 0.001 |
| Ampicillin | 16 | 807 | 344 (44.5) | 29.2–61.0 | 90.83 | < 0.001 |
| Chloramphenicol | 12 | 777 | 188 (32.9) | 20.8–47.8 | 87.24 | < 0.001 |
| Ceftriaxone | 2 | 8 | 4 (50.0) | 20.0–80.0 | 0.00 | > 0.05 |
| Ciprofloxacin | 17 | 765 | 266 (36.5) | 27.0–47.3 | 75.30 | < 0.001 |
| Clindamycin | 4 | 224 | 59 (26.9) | 21.5–33.2 | 0.00 | 0.478 |
| Daptomycin | 2 | 29 | 0.5 (3.2)a | 0.5–19.7 | 0.00 | 0.974 |
| Doxycycline | 3 | 254 | 85 (55.1) | 22.2–84.0 | 90.21 | < 0.001 |
| Erythromycin | 14 | 780 | 374 (49.6) | 36.5–62.7 | 86.19 | < 0.001 |
| Gentamycin | 10 | 533 | 248 (37.7) | 22.2–56.1 | 88.86 | < 0.001 |
| Linezolid | 2 | 30 | 2 (9.9) | 2.8–29.0 | 0.00 | 0.336 |
| Nitrofurantoin | 9 | 404 | 117 (31.5) | 23.4–41.0 | 38.76 | 0.110 |
| Norfloxacin | 5 | 350 | 100 (39.9) | 18.6–66.6 | 90.21 | < 0.001 |
| Penicillin | 8 | 343 | 181 (60.7) | 39.9–78.3 | 86.63 | < 0.001 |
| Streptomycin | 3 | 179 | 74 (36.8) | 10.4–73.1 | 91.62 | < 0.001 |
| Tetracycline | 9 | 450 | 199 (53.7) | 35.8–70.7 | 86.95 | < 0.001 |
| SXT | 10 | 241 | 104 (39.1) | 21.48–59.6 | 45.58 | 0.088 |
| MDR - enterococci | 20 | 825 | 543 (60.0) | 42.9–75.0 | 90.27 | < 0.001 |
SXT Trimethoprim-Sulfamethoxazole, MDR Multidrug resistance
a Continuity correction (0.5) is added to the study
Discussion
Determining the prevalence of antibiotic resistance is an important step in the formulation of interventions to control emergence and transmission of resistant pathogens. In recent years, an increase in invasive VRE infections have been reported elsewhere in the worldwide [13, 17, 25, 49]. Although antimicrobial resistance surveillance centers were established and priority surveillance pathogens were identified to prevent the spread of drug resistance in Ethiopia, VRE were not included in the lists of priority pathogens. A previous systematic review [26] reporting the growing challenges of antibacterial resistance in Ethiopia had not assessed the burden of drug resistant enterococci. The prevalence of VRE has been reported by several studies in Ethiopia but a comprehensive review covering different parts of Ethiopia has not been conducted. This systematic review and meta-analysis was conducted to estimate the pooled prevalence of VRE and antimicrobial resistance profile of enterococci in Ethiopia.
Twenty studies reporting the prevalence and/or number of VRE isolates were included in this study. Majority (80%) of the included studies failed to report the isolated enterococci at species level and simply highlighted the corresponding antimicrobial resistance profile. This might be due to poor laboratory capacity to identify species of enterococci. This indirectly indicates the potential existence of drug resistant enterococci in health care settings in Ethiopia and possible spread to the communities unless appropriately maintained., Although there was considerable methodological difference between studies, they were pooled for the purpose of this review. Therefore; the pooled prevalence of VRE in Ethiopia was estimated at 14.8%. This estimate is comparable with reports from Iran (14, 18.75%) [50, 51].
On the other hand, our finding was lower than studies reported from North America (21%), Asia (24%) and Europe (20%) [52]. Another study from Iran reported high rate of VRE (48.9%) among hospitalized patients [53]. These differences might be related with study population that hospitalized and critically ill patients are more likely to acquire VRE [13, 54] than the largely non-hospitalized study populations pooled in our analysis. In addition, the study period may contribute for the high rate of isolation in these countries. The studies were also conducted in the 1990’s and 2000’s following the first reports of VRE [21, 22]; while all of the studies included in our analysis were done in the 2010’s where clinical use of Vancomycin was being discouraged [11].
In contrast, higher rates of VRE was observed in our study than reports from Singapore (9.3%) [55], Germany (9.8%) [49], Iran (9.4%) [56] and United Kingdom (9.2%) [57]. Different factors were identified as risk factors for acquiring VRE infections including previous hospitalization, patient transfer, urinary catheters, critical illnesses, underlying diseases, contact with VRE patients and inappropriate use of antibiotics [54, 55, 58, 59]; all of which could contribute for the high prevalence of VRE in Ethiopia. Generally, infections and colonization with VRE were reported to be associated with health care contacts [18]. This could be true in settings where infection control knowledge, attitudes and practices among healthcare workers is poor in Ethiopia [60]. High frequency of inappropriate use of antibiotics and empirical therapies by healthcare professionals was also reported in Eastern Ethiopia [61]. In addition, the antimicrobial susceptibility testing method was based chiefly on disc diffusion and resistance was defined following the CLSI guideline.
Regional prevalence of VRE was also estimated. The highest estimated prevalence was obtained from Addis Ababa (26.1%); almost two times higher than Amhara (15.0%) and three times higher than Oromia (9.0%). This regional difference might be attributed by different study settings (hospital set up), study period, study population, variation in antibiotic use, method of antimicrobial susceptibility testing and type of specimens used to isolate enterococci. Stool, urine and blood were the most common specimens from which VRE were isolated. This is not surprising because enterococci have been reported as the most common organisms isolated from intestinal tract, urinary tract and blood stream infections [5, 8, 15, 20, 48, 52, 57, 62].
Enterococci are not only resistant to Vancomycin but also to other commonly used antimicrobials including Penicillin, Amoxicillin, Doxycycline, Tetracycline, Erythromycin, Daptomycin, Linezolid and others (see Table 4 above). Multidrug resistant (MDR) enterococci were also observed that could pose a critical health problem in patients and health care settings in Ethiopia. As there is no specific recommendation for the antimicrobial prescription of VRE and a follow up surveillance is not conducted at different health care centers where the studies included in this review were conducted, the prevalence of VRE is expected to continuously increase. With these concerns in mind, there has been success stories in treating VRE infections with Daptomycin and Linezolid [62]. In our analysis however; resistance to Daptomycin and Linezolid was observed in about 3.2 and 9.9% of enterococcal isolates, respectively. Although it requires strong studies, our analysis indicated that these drugs may select vancomycin resistant strains in some potentially pathogenic enterococci through antibiotic selection pressure as they showed some sort of resistance to Daptomycin and Linezolid.
Strengths and limitations of the study
A comprehensive search with clear inclusion and exclusion criteria was used, examined commonly used specimens and methods of susceptibility testing, and included unpublished studies retrieved from Addis Ababa University repository. The Trim-and-Fill method was applied to asymmetric funnel plots to produce adjusted estimates. There were a number of limitations in the depth and breadth of data. First; inability to report pooled estimates of VRE at species level due to the paucity of included studies reporting enterococci at species level. Second; the definition of VRE was not consistent across studies and different AST methods were combined limiting comparability and strength of this analysis. Third; data was not available from 54.5% of the regions, outside health care setting and non-human studies were excluded that may be difficult to generalize the pooled results. Fourth; combing resistance results from different patients across different regions might pool out the peaks of resistance in some settings. Lastly; the study protocol was not registered at PROSPERO.
Conclusion
The prevalence of VRE and drug resistant enterococci are on the rise in Ethiopia. Enterococcal isolates showed resistance to one or more of the commonly prescribed drugs in different or the same drug lines. Multidrug resistant (MDR) enterococci were also found. Although the rates were low, the emergence of resistance to Daptomycin and Linezolid is an alarm for searching new ways for the treatment and control of VRE infections. This review provides data about the current burden of VRE in Ethiopia and showed gaps that would be addressed in future studies to maintain the spread of VRE infections. Adherence to antimicrobial stewardship, comprehensive testing and ongoing monitoring of VRE infections in the health care settings are required.
Supplementary information
Additional file 1. The Joanna Briggs Institute (JBI) Critical Appraisal Checklist for Studies Reporting Prevalence Data
Additional file 2. Quality of the included 20 studies evaluated by JBI critical appraisal checklists
Acknowledgements
Not applicable.
Abbreviations
- AMR
Antimicrobial resistance
- ART
Antiretroviral Therapy
- AST
Antimicrobial Susceptibility Testing
- CLSI
Clinical and Laboratory Standards Institute
- EPHI
Ethiopian Public Health Institute
- JBI
Joanna Biggs Institution
- MDR
Multi Drug resistance
- MIC
Minimum Inhibitory Concentration
- SXT
Trimethoprim-Sulfamethoxazole
- VRE
Vancomycin Resistant Enterococci
- WHO
World Health Organization
Authors’ contributions
AM: Conceived and designed the study; select and assess quality of studies; extracted and analyzed data; interpreted results; and drafted the manuscript. TA: select and assess quality of studies; extracted data and interpret results. CG: interpret results and review the manuscript. All authors read and approved the manuscript.
Funding
The authors declare that they did not receive any funding from any source.
Availability of data and materials
The datasets used and/or analyzed during the current study are included in the manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Addisu Melese, Email: addisum22@gmail.com.
Chalachew Genet, Email: chaliegenet@gmail.com.
Tesfaye Andualem, Email: tesfayeandu@gmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12879-020-4833-2.
References
- 1.Prestinaci Francesca, Pezzotti Patrizio, Pantosti Annalisa. Antimicrobial resistance: a global multifaceted phenomenon. Pathogens and Global Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall, S.B.L.B., Antibacterial resistance worldwide: causes, challenges and responses. Nature Medicine Supplement. 2004;10(12):122–29. [DOI] [PubMed]
- 3.Marianne Frieri KK, Boutin A. Antibiotic resistance. J Infection Public Health. 2017;10:369–378. doi: 10.1016/j.jiph.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Institute, E.P.H . Ethiopia antimicrobial resistance surveillance, annual report 2017/18. 2018. [Google Scholar]
- 5.Banla LI, Salzman NH, Kristich CJ. Colonization of the mammalian intestinal tract by enterococci. Curr Opin Microbiol. 2019;47:26–31. doi: 10.1016/j.mib.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yitbarek Getachew LH, Zakaria Z, Aziz SA. Genetic Variability of Vancomycin-Resistant Enterococcus faecium and Enterococcus faecalis Isolates from Humans, Chickens, and Pigs in Malaysia. Appl Environ Microbiol. 2013;79(15):4528–4533. doi: 10.1128/AEM.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bekele Hailu MA. Distribution of drug resistance among enterococci and Salmonella from poultry and cattle in Ethiopia. Trop Anim Health Prod. 2010;42:857–864. doi: 10.1007/s11250-009-9499-0. [DOI] [PubMed] [Google Scholar]
- 8.Ana L. Flores-Mireles, J.N.W., Michael Caparon and Scott J. Hultgren, Urinary tract infections - epidemiology, mechanisms of infection and treatment options. Nature Reviews Microbiol. 2015;13:269–84. [DOI] [PMC free article] [PubMed]
- 9.Yomna A. Hashem, H.M.A., Tamer M. Essam, Aymen S. Yassin & Ramy K. Aziz, Biofilm formation in enterococci - genotype-phenotype correlations and inhibition by vancomycin. Sci Rep. 2017;7(5733):1–12. [DOI] [PMC free article] [PubMed]
- 10.Wagner T, Joshi B, Janice J, Askarian F, Škalko-Basnet N, Hagestad OC, Mekhlif A, Wai SN, Hegstad K, Johannessen M. Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J Proteome. 2018;187:28–38. doi: 10.1016/j.jprot.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 11.G Werner, Coque TM, Hammerum AM, et al, Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance. 2008;13(47):1–11. [PubMed]
- 12.Nade’ge Bourgeois-Nicolaos, C.M., Nicole Mangeney, Marie-Jos ´e Butel & Florence Doucet-Populaire, Comparative study of vanA gene transfer from Enterococcus faecium to Enterococcus faecalis and to Enterococcus faecium in the intestineof mice. FEMS Microbiol Lett, 2005. 254(2006): p. 27–33. [DOI] [PubMed]
- 13.Adams Daniel J., Eberly Matthew D., Goudie Anthony, Nylund Cade M. Rising Vancomycin-Resistant Enterococcus Infections in Hospitalized Children in the United States. Hospital Pediatrics. 2016;6(7):404–411. doi: 10.1542/hpeds.2015-0196. [DOI] [PubMed] [Google Scholar]
- 14.Arias Cesar A., Murray Barbara E. The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CA DG, Zimmer SM, Klein M, Jernigan JA. Comparison of Mortality Associated with Vancomycin-Resistant and Vancomycin-Susceptible Enterococcal Bloodstream Infections: A Meta-analysis. Clin Infect Dis. 2005;41:327–333. doi: 10.1086/430909. [DOI] [PubMed] [Google Scholar]
- 16.Hegstad K., Mikalsen T., Coque T.M., Werner G., Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clinical Microbiology and Infection. 2010;16(6):541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 17.Schroder UC, et al. Detection of vancomycin resistances in enterococci within 3 (1/2) hours. Sci Rep. 2015;5:8217. doi: 10.1038/srep08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camins Bernard C., Farley Monica M., Jernigan John J., Ray Susan M., Steinberg James P., Blumberg Henry M. A Population-Based Investigation of Invasive Vancomycin-Resistant Enterococcus Infection in Metropolitan Atlanta, Georgia, and Predictors of Mortality. Infection Control & Hospital Epidemiology. 2007;28(8):983–991. doi: 10.1086/518971. [DOI] [PubMed] [Google Scholar]
- 19.Gearhart M. M.J., Rudich S, Thomas M, Wetzel D, Solomkin J, Hanaway MJ, Aranda-Michel J, weber F, Trumball L, bass M, Zavala E, Woodle ES, Buell JF, Consequences of vancomycin-resistant Enterococcus in liver transplant recipients: a matched control study. Clin Transpl. 2005;19:711–716. doi: 10.1111/j.1399-0012.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- 20.Cheah A.A.Y., Spelman T., Liew D., Peel T., Howden B.P., Spelman D., Grayson M.L., Nation R.L., Kong D.D.M. Enterococcal bacteraemia: factors influencing mortality, length of stay and costs of hospitalization. Clinical Microbiology and Infection. 2013;19(4):E181–E189. doi: 10.1111/1469-0691.12132. [DOI] [PubMed] [Google Scholar]
- 21.Rice Louis. Emergence of Vancomycin-Resistant Enterococci. Emerging Infectious Diseases. 2001;7(2):183–187. doi: 10.3201/eid0702.010205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray Barbara. Diversity among Multidrug-Resistant Enterococci. Emerging Infectious Diseases. 1998;4(1):37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birri DJ, et al. Bacteriocin production, antibiotic susceptibility and prevalence of haemolytic and gelatinase activity in faecal lactic acid bacteria isolated from healthy Ethiopian infants. Microb Ecol. 2013;65(2):504–516. doi: 10.1007/s00248-012-0134-7. [DOI] [PubMed] [Google Scholar]
- 24.WHO . Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. [Google Scholar]
- 25.Cornelius Remschmidt, C.S., Michael Behnke, Petra Gastmeier, Christine Geffers and Tobias Siegfried Kramer, Continuous increase of vancomycin resistance in enterococci causing nosocomial infections in Germany - 10 years of surveillance. Antimicrob Resist Infect Control. 2018;7(54). [DOI] [PMC free article] [PubMed]
- 26.Moges F, et al. The growing challenges of antibacterial drug resistance in Ethiopia. J Glob Antimicrob Resist. 2014;2(3):148–154. doi: 10.1016/j.jgar.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Moher David, Liberati Alessandro, Tetzlaff Jennifer, Altman Douglas G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Medicine. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munn Z, M.S., Lisy K, Riitano D, Tufanaru C., Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and incidence data. Int J Evid Based Healthc. 2015;13(3):1–7. [DOI] [PubMed]
- 29.Andrea C Tricco, C.H.N., Vladimir Gilca, Andrea Anonychuk, Ba’ Pham and Shirra Berliner, Canadian oncogenic human papillomavirus cervical infection prevalence: Systematic review and meta-analysis BMC Infectious Diseases. 2011;11(235). [DOI] [PMC free article] [PubMed]
- 30.Yilema A, et al. Isolation of enterococci, their antimicrobial susceptibility patterns and associated factors among patients attending at the University of Gondar Teaching Hospital. BMC Infect Dis. 2017;17(1):276. doi: 10.1186/s12879-017-2363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammed A, et al. Bacterial Isolates and Their Antimicrobial Susceptibility Patterns of Wound Infections among Inpatients and Outpatients Attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2017;2017:8953829. doi: 10.1155/2017/8953829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fentie A, et al. Bacterial profile, antibiotic resistance pattern and associated factors among cancer patients at University of Gondar Hospital, Northwest Ethiopia. Infect Drug Resist. 2018;11:2169–2178. doi: 10.2147/IDR.S183283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayelign B, et al. Bacterial isolates and their antimicrobial susceptibility patterns among pediatric patients with urinary tract infections. Turk J Urol. 2018;44(1):62–69. doi: 10.5152/tud.2017.33678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohabaw Jemal, A.B., Yesuf Adem, Bacterial bloodstream infections and their antimicrobial susceptiblity pattern among HIV/AIDS patients at Felege Hiwot Referral Hospital, Bahir Dar, Amahara regional state, Northwest Ethiopia. 2017.
- 35.Ali S, et al. Vancomycin-resistant enterococci and its associated risk factors among HIV-positive and -negative clients attending Dessie referral hospital, Northeast Ethiopia. Int J Microbiol. 2018;2018:4753460. doi: 10.1155/2018/4753460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agegne M, et al. Magnitude of Vancomycin-resistant enterococci (VRE) colonization among HIV-infected patients attending ART Clinic in West Amhara government hospitals. Int J Microbiol. 2018;2018:7510157. doi: 10.1155/2018/7510157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abebe W, et al. Prevalence of vancomycin resistant enterococci and associated risk factors among clients with and without HIV in Northwest Ethiopia: a cross-sectional study. BMC Public Health. 2014;14:185. doi: 10.1186/1471-2458-14-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woldemariam HK, et al. Common uropathogens and their antibiotic susceptibility pattern among diabetic patients. BMC Infect Dis. 2019;19(1):43. doi: 10.1186/s12879-018-3669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamirat Molalign AB. Spectrum and antibiotic susceptibility profile of bacteriuria isolated from patients attending arsho advanced medical laboratory with urinary tract infections by using VITEK 2 compact system, Addis Ababa, Ethiopia. 2016. [Google Scholar]
- 40.Mequanint Mitiku KD. Multi-drug resistant bacterial Isolates among septicemia suspected under five children in Tikur Anbesa specialized hospital, Addis Ababa Ethiopia. 2018. [Google Scholar]
- 41.Ferede ZT, et al. Prevalence and antimicrobial susceptibility pattern of Enterococcus species isolated from different clinical samples at black lion specialized teaching hospital, Addis Ababa, Ethiopia. BMC Res Notes. 2018;11(1):793. doi: 10.1186/s13104-018-3898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tebarek Lega IA, Desta K, Kebede T. Bacterial Uropathogens and their Drug Resistance Pattern in Diabetic Patients Attending Yekatit 12 Hospital Medical College, Addis Ababa, Ethiopia. 2015. [Google Scholar]
- 43.Frehiwot Teklehaymanot KD, Hailu M. Bacterial profiles and their antimicrobial susceptibility patterns from body fluids at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. 2016. [Google Scholar]
- 44.Seneshat Eshetu AB, Getachew T, Gizaw S. Bacterial profile and antimicrobial susceptibility pattern of blood culture isolates at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. 2017. [Google Scholar]
- 45.Toru M, et al. Prevalence and phenotypic characterization of Enterococcus species isolated from clinical samples of pediatric patients in Jimma University specialized hospital, south West Ethiopia. BMC Res Notes. 2018;11(1):281. doi: 10.1186/s13104-018-3382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorsa A, et al. Blood culture result profile and antimicrobial resistance pattern: a report from neonatal intensive care unit (NICU), Asella teaching and referral hospital, Asella, south East Ethiopia. Antimicrob Resist Infect Control. 2019;8:42. doi: 10.1186/s13756-019-0486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gebrish S., F.B., Asfaw T., Magnitude of Drug-resistant Enterococcus species from Intestinal Tracts of Hospitalized Pediatric Patients in Ethiopia. 2019.
- 48.Abamecha A, Wondafrash B, Abdissa A. Antimicrobial resistance profile of Enterococcus species isolated from intestinal tracts of hospitalized patients in Jimma, Ethiopia. BMC Res Notes. 2015;8:213. doi: 10.1186/s13104-015-1200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobias Siegfried Kramer , C.R., Sven Werner, Michael Behnke, Frank Schwab, Guido Werner, Petra Gastmeier and Rasmus Leistner, The importance of adjusting for enterococcus species when assessing the burden of vancomycin resistance: a cohort study including over 1000 cases of enterococcal bloodstream infections. Antimicrob Resist Infect Control. 2018;7(133). [DOI] [PMC free article] [PubMed]
- 50.Ali Jahansepas, M.A.R., Alka Hasani, Yaeghob Sharifi, Marjan Rahnamaye Farzami, Alireza Dolatyar, and Mohammad Aghazadeh, Molecular Epidemiology of Vancomycin–Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Clinical Specimens in the Northwest of Iran. Microb Drug Resist, 2018. 00(00). [DOI] [PubMed]
- 51.Abbas Moghimbeigi MM, Dousti M, Kiani F, Sayehmiri F, Sadeghifard N, Nazari A. Prevalence of vancomycin resistance among isolates of enterococci in Iran: a systematic review and meta-analysis. Adolesc Health Med Ther. 2018;9:177–188. doi: 10.2147/AHMT.S180489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michail Alevizakos, A.G., Dimitrios Nasioudis, Katerina Tori, Myrto Eleni Flokas, and Eleftherios Mylonakis, Colonization With Vancomycin-Resistant Enterococci and Risk for Bloodstream Infection Among Patients With Malignancy: A Systematic Review and Meta-Analysis open forum infectious diseases, 2016. [DOI] [PMC free article] [PubMed]
- 53.Leili Shokoohizadeh, A.M.M., Mohammad Reza Zali, Reza Ranjbar, Masoud Alebouyeh, Türkan Sakinc and Liaqat Ali, High frequency distribution of heterogeneous vancomycin resistant Enterococcous faecium (VREfm) in Iranian hospitals. Diagn Pathol. 2013;8(163). [DOI] [PMC free article] [PubMed]
- 54.Mazuski, J.E., Vancomycin-Resistant Enterococcus: Risk Factors, Surveillance, Infections, and Treatment. Surg Infect. 2008;9(6):567–71. [DOI] [PubMed]
- 55.Kok-Soong Yang Y-TF, Lee H-Y, Kurup A, Koh T-H, Koh D, Lim M-K. Predictors of Vancomycin-resistant Enterococcus (VRE) Carriage in the First Major VRE Outbreak in Singapore. Ann Acad Med Singap. 2007;36:379–383. [PubMed] [Google Scholar]
- 56.Emaneini M, Hosseinkhani F, Jabalameli F, Nasiri MJ, Dadashi M, Pouriran R, Beigverdi R. Prevalence of vancomycin-resistant Enterococcus in Iran - a systematic review and meta-analysis. Eur J ClinMicrobiol Infect Dis. 2016;35:1387–1392. doi: 10.1007/s10096-016-2702-0. [DOI] [PubMed] [Google Scholar]
- 57.Liam Toner NP, Aliyu SH, Dev H, Lawrentschuk N, Al-Hayek S. Vancomycin resistant enterococci in urine cultures: Antibiotic susceptibility trends over a decade at a tertiary hospital in the United Kingdom. ICUrology. 2016;57:129–134. doi: 10.4111/icu.2016.57.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.KUO-MING YEH, Siu LK, Chang JC, Chang FY., Vancomycin-Resistant Enterococcus (VRE) Carriage and Infection in Intensive Care Units. Microb Drug Resist. 2014;10(2):177–83. [DOI] [PubMed]
- 59.Melissa Barger, E.B., Sol Pena, Wendy Mack and Tse-Ling Fong, VRE in cirrhotic patients. BMC Infect Dis. 2019;19(711). [DOI] [PMC free article] [PubMed]
- 60.Tenna A, Stenehjem EA, Margoles L, Kacha E, Blumberg HM, Kempker RR. Infection Control Knowledge, Attitudes, and Practices among Healthcare Workers in Addis Ababa, Ethiopia. Infect Control Hosp Epidemiol. 2013;24(12):1289–1296. doi: 10.1086/673979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tafa B, Endale A, Bekele D. Paramedical staffs knowledge and attitudes towards antimicrobial resistance in Dire Dawa, Ethiopia: a cross sectional study. Ann Clin Microbiol Antimicrob. 2017;16(1):64. doi: 10.1186/s12941-017-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jennifer D. Twilla, Finch CK, Justin B. Usery, Michael S. Gelfand, Joanna Q. Hudson, and Joyce E. Broyles, Vancomycin-Resistant Enterococcus Bacteremia: An evaluation of Treatment with Linezolid or Daptomycin. J Hosp Med. 2012;7(3):243–8. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The Joanna Briggs Institute (JBI) Critical Appraisal Checklist for Studies Reporting Prevalence Data
Additional file 2. Quality of the included 20 studies evaluated by JBI critical appraisal checklists
Data Availability Statement
The datasets used and/or analyzed during the current study are included in the manuscript.