Abstract
Carriers of premutation CGG expansions in the fragile X mental retardation 1 (FMR1) gene are at higher risk of developing a late-onset neurodegenerative disorder named Fragile X-tremor ataxia syndrome (FXTAS). Given that mitochondrial dysfunction has been identified in fibroblasts, PBMC and brain samples from carriers as well as in animal models of the premutation and that mitochondria are at the center of intermediary metabolism, the aim of this study was to provide a complete view of the metabolic pattern by uncovering plasma metabolic perturbations in premutation carriers. To this end, metabolic profiles were evaluated in plasma from 23 premutation individuals and 16 age- and sex-matched controls. Among the affected pathways, mitochondrial dysfunction was associated with a Warburg-like shift with increases in lactate levels and altered Krebs’ intermediates, neurotransmitters, markers of neurodegeneration, and increases in oxidative stress-mediated damage to biomolecules. The number of CGG repeats correlated with a subset of plasma metabolites, which are implicated in mitochondrial disorders but also in other neurological diseases such as Parkinson’s, Alzheimer’s and Huntington’s diseases. For the first time, the identified pathways shed light on disease mechanisms contributing to morbidity of the premutation, with the potential of assessing metabolites in longitudinal studies as indicators of morbidity or disease progression, especially at the early pre-clinical stages.
Keywords: Fragile X, metabolomics, mitochondrial dysfunction, neurodegeneration, trinucleotide repeat disease
Summary statement
Our study defines the potential use of plasma metabolic profiling to monitor brain pathophysiology in carriers of a 50–200 CGG expansion (premutation) in the 5’UTR fragile X mental retardation 1 (FMR1) gene before the onset of the neurodegenerative disorder FXTAS.
Introduction
A modestly expanded CGG nucleotide repeats (55–200) in the 5’-UTR of the fragile X mental retardation gene 1 (FMR1 [1–3]) represents the genetic hallmark of premutation carriers. Originally, premutation carriers were thought to be free of phenotypic traits; however, reports regarding fragile X-associated primary ovarian insufficiency (FXPOI; [4]), followed by the discovery of fragile X-associated tremor ataxia syndrome (FXTAS; OMIM:300623) identified in adult carriers [5] discredited this notion. Premutation carriers may also suffer from psychological problems, visuospatial deficits, and immune dysregulation [2, 3, 6–8] and affected children are often diagnosed with ADHD, autism, anxiety and other psychopathologies [9, 10], At the cellular level, fibroblasts from premutation carriers (humans or animal models of the premutation) are generally accompanied by high FMR1 gene expression, normal or lower levels of FMRP and mitochondrial dysfunction [11–14].
Currently, premutation carriers include 1 million women and 320,000 men in the United States [15]. It is currently unknown which carriers of the premutation will develop FXTAS. Clinical diagnosis fails to identify those carriers before significant neurological symptoms are evident, thus there is an immediate need for early detection and effective drugs for the cure or the prevention of FXTAS development. An understanding of the molecular characteristics underlying the disease processes of the premutation is a prerequisite for providing therapeutic strategies.
To fulfill this immediate need, metabolic profiling performed on readily accessible body fluids, such as cerebrospinal fluid, serum, urine or saliva is one of the most important techniques that provides a complete view of the metabolic status and uncovering metabolic perturbations in pathways [16–18] for diagnosis of many diseases [19] including metabolic disorders [20], motor neuron disease [21], Parkinson’s [22] and Alzheimer’s [23] as well as chemical toxicity and aging [24–27]. To this end, plasma metabolomics was evaluated in premutation carriers and age- and sex- matched controls with the aim of providing a complete view of the status of intermediary metabolism with the potential of uncovering perturbations in metabolic pathways [17, 18] associated with the presence of the FMR1 premutation.
Materials and methods
Characteristics of the subjects enrolled in this study
The study was conducted at the MIND Institute and approved by the IRB ethics committee at UC Davis Medical Center. Exclusion criteria were refusal of the patient or his guardian, infection or malignancy. Blood samples were obtained by venipuncture with informed consent. Controls and carriers of the premutation were recruited through the Fragile X Treatment and Research Center at the MIND Institute at University of California, Davis, and who participated in our genotype-phenotype study of families with fragile X between the years 2013 and 2015. All blood draws were performed at the MIND Institute between the hours 8 and 10 am (fasting was not advised). No exercise prior to the blood draw was reported; however, it seems an unlikely event since the subjects are usually at the Clinic between 7–8 am on the day of the exam. Clinical evaluations with Dr. Hagerman and associates were performed after the blood draw. CGG repeat number in all individuals included in this study was evaluated by using Southern Blot and PCR analysis as previously described [28]. Participants were not excluded from this study if they were taking prescription medications. However, careful record of all prescription medications was obtained.
Subjects
The “control group” consisted of 16 individuals (female-to-male ratio = 1.3; mean age ± SD of 37 ± 13 y). The “premutation group” was constituted by 23 premutation carriers (female-to-male ratio = 1.1; mean age ± SD of 37 ± 19 y; Table 1). Four of the subjects in the premutation group were diagnosed with FXTAS utilizing the criteria by Jacquemont et al. [29], Of the subjects included in this study, 6 controls and 13 carriers were on multivitamins/probiotics or nutritional supplements, 1 control and 8 carriers were on antidepressants, 3 controls and 4 carriers on cyclooxygenase inhibitors, 2 controls and 2 carriers on hormone replacement therapy, 1 control and 2 carriers on antihistamines, 1 control and 1 carrier on nitric-oxide producing drugs. Other medications included hydroxymethylglutarylCoA reductase inhibitors (2 carriers), proton pump inhibitors (2 carriers), beta2 agonist (2 carriers), levodopa (1 carrier), alpha2A receptor agonist (1 carrier), alphal adrenergic blocker (1 control), ACE inhibitor (1 carrier), anticoagulant (1 control), barbiturate (1 carrier), beta-blocker (1 carrier), and inhibitor of monoamine transport (1 carrier). Although not significant, carriers were more likely to take vitamins and supplements than controls (56.5% vs. 37.5%; p = 0.059).
Table 1.
Demographics and clinical characteristics of plasma donors included in this study
| Clinical groups | Agea(y) | CGG repeat expansionb | Sexc | FXTAS Stage |
|---|---|---|---|---|
| C1 | 29.0 | 30 | M | 0 |
| C2 | 54.0 | 30 | M | 0 |
| C3 | 23.0 | 29, 30 | F | 0 |
| C4 | 50.5 | 21 | M | 0 |
| C5 | 24.0 | 30 | M | 0 |
| C6 | 41.2 | 43 | M | 0 |
| C7 | 28.8 | 20, 33 | F | 0 |
| C8 | 26.0 | 30 | M | 0 |
| C9 | 33.7 | 23, 30 | F | 0 |
| C10 | 54.0 | 25, 33 | F | 0 |
| C11 | 25.0 | 24, 33 | F | 0 |
| C12 | 45.0 | 22, 33 | F | 0 |
| C13 | 24.0 | 23, 35 | F | 0 |
| C14 | 26.3 | 30, 37 | F | 0 |
| C15 | 41.5 | 20 | M | 0 |
| C16 | 57.4 | 23, 30 | F | 0 |
| P1 | 46.3 | 61 | M | 0 |
| P2 | 9.7 | 31, 63 | F | 0 |
| P3 | 8.4 | 180 | M | 0 |
| P4 | 24.0 | 31, 93 | F | 0 |
| P5 | 19.7 | 177 | M | 0 |
| P6 | 55.6 | 104 | M | 0 |
| P7 | 49.3 | 31, 86 | F | 0 |
| P8 | 17.3 | 16, 67 | F | 0 |
| P9 | 45.3 | 69 | M | 0 |
| P10 | 49.9 | 20, 98 | F | 0 |
| P11 | 9.1 | 160 | M | 0 |
| P12 | 55.4 | 30, 69 | F | 0 |
| P13 | 53.0 | 16, 67 | F | 0 |
| P14 | 33.1 | 30, 137 | F | 0 |
| P15 | 43.2 | 30, 106 | F | 0 |
| P16 | 38.4 | 33, 60 | F | 0 |
| P17 | 8.4 | 180 | M | 0 |
| P18 | 24.0 | 30, 79 | F | 0 |
| P19 | 25.0 | 67 | M | 0 |
| P20 | 62.5 | 105 | M | 4 |
| P21 | 61.3 | 96 | M | 4 |
| P22 | 61.8 | 110–130 | M | 1 |
| P23 | 59.1 | 33, 107 | F | 3 |
C and P refer to controls and carriers of the premutation, respectively. The number following the letter identifies the subject from which the plasma samples were obtained.
FXTAS-affected carrier, stage not available
Mean age ± SD = 37 ± 13 y and 37 ± 19 y respectively for controls and premutation (p = 1.000).
Mean CGG expansion ± SD = 31 ± 5 and 103 ± 40 respectively for controls and carriers (p < 0.0001).
Female-to-male ratio =1.3 and 1.1 respectively for the control and premutation groups (Chi square p = 0.802).
Sample preparation for metabolomics
The protocol for plasma metabolomics was reported in detail before [30]. Plasma samples were extracted following the protocols published [31]. Samples were derivatized by methoxyamine hydrochloride in pyridine and subsequently by N-methyl-N-trimethylsilyltrifluoroacetamide for trimethylsilylation of acidic protons. Data were acquired according to [32]. Absolute spectra intensities were further processed by a filtering algorithm implemented in the metabolomics BinBase database. The BinBase algorithm used the settings: validity of chromatogram (<10 peaks with intensity >107 counts s−1), unbiased retention index marker detection (MS similarity>800, validity of intensity range for high m/z marker ions), retention index calculation by 5th order polynomial regression. Spectra were cut to 5% base peak abundance and matched to database entries from most to least abundant spectra using the following matching filters: retention index window ± 2,000 units (equivalent to about ±2s retention time), validation of unique ions and apex masses (unique ion must be included in apexing masses and present at >3% of base peak abundance), mass spectrum similarity must fit criteria dependent on peak purity and signal/noise ratios and a final isomer filter. Failed spectra were automatically entered as new database entries if S/N >25, purity <1.0 and presence in the biological study design class was >80%. Quantification was reported as peak height using the unique ion as default, unless a different quantification ion was manually set in the BinBase administration software BinView. A quantification report table was produced for all database entries that were positively detected in > 10% of the samples of a study design class (as defined in the miniX database) for unidentified metabolites. A subsequent post-processing module was employed to automatically replace missing values from the *.cdf files. Replaced values were labeled as ‘low confidence’ by color coding, and for each metabolite, the number of high-confidence peak detections was recorded as well as the ratio of the average height of replaced values to high-confidence peak detections. These ratios and numbers were used for manual curation of automatic report data sets to data sets released for submission. Metabolites were identified by matching the ion chromatographic retention index, accurate mass, and mass spectral fragmentation signatures with reference library entries created from authentic standard metabolites under the identical analytical procedure as the experimental samples.
Statistics
Post-hoc analysis to compute the achieved power given the actual sample size utilized in this study (alpha = 0.05) indicated that it was >0.999 when performed with these outcomes (G*Power, v.3.0. 10). Metabolite identification was performed through the use of several databases including PubChem Compound [33], KEGG [34] and HMDB [35–37] and the online chemical translation service [38]. Before analysis, raw data were filtered by the presence of metabolites in at least 80% of patients and all data were mean-centered and standardized. Identification of the characteristic metabolites with significance between clusters was performed using the PLS-Discriminant Analysis (PLS-DA) method implemented in XLSTAT (version 2016.04.32331). The importance of each metabolite in the PLS-DA was evaluated by variable importance in the projection (VIP) score. The VIP score positively reflects the metabolite’s influence on the classification, and metabolites with a score >0.8 were considered important in this study. Additionally, plasma metabolites along with their relative concentrations were also analyzed using a Metabolite Set Enrichment Analysis (MSEA). The MSEA was performed by using quantitative enrichment analysis (QEA). The relative concentrations of metabolites were analyzed using the metabolic pathway-associated metabolite set library and the enrichment analysis was performed by using the globaltest package [39]. The QEA was performed using a generalized linear model to estimate a Q-statistic for each metabolite set, which allows describing the correlation between compound concentration and disease. The results were summarized as the average Q statistics for each metabolite in the input set.
Results
a. Plasma metabolomics
Metabolomics was performed on plasma samples from carriers of the premutation and age- and sex-matched controls (Table 1). The average CGG repeats at the 5’UTR of FMR1 of the mutant allele in heterozygous carriers was 83 ± 19 (mean ± SD), whereas that of hemizygous carriers (males only) was significantly longer (123 ± 45; p = 0.021). In both cases, the CGG repeats were significantly longer than those observed in controls (31 ± 6, p = 5×10−5; Table 1).
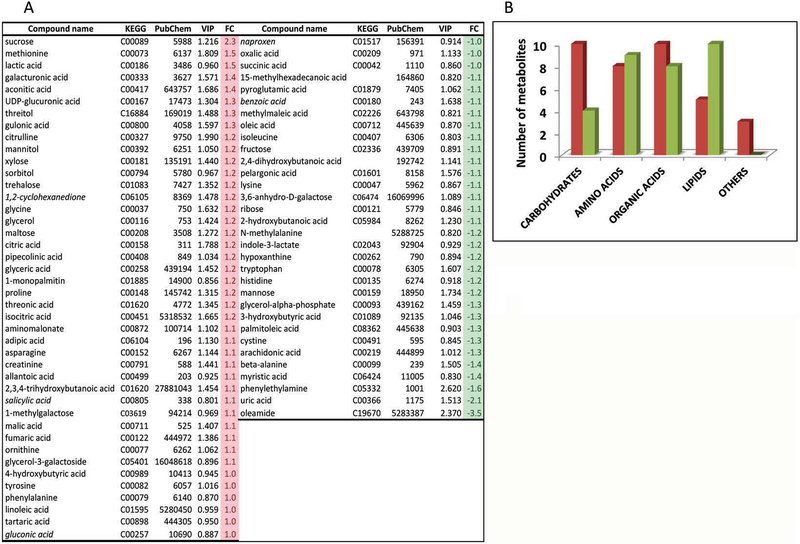
A total of 143 metabolites were identified in plasma samples from these subjects. The partial least squares-discriminant analysis (PLS-DA) was utilized to identify the performance of multiple metabolite biomarkers associated with premutation or, in other words, the predictive value of the class membership of subjects based on their metabolomic profile (Figure 1). Variable importance in projection (VIP) scores from the PLS-DA of all metabolites was calculated to evaluate those that contributed the most to the segregation of the diagnostic groups (VIP ≥ 0.8; Figure 1A). Considering that variables having a VIP score >1.0 are interpreted as being highly influential, values between 0.8 < VIP score < 1.0 indicate moderately influential variables, and VIP scores < 0.8 represent less important variables [40], the VIP cut-off value ≥ 0.8 (also used in other studies such as [41]) allowed the inclusion of not only potential biomarkers but also that of other metabolites with a relatively significant contribution in discriminating the groups (control vs premutation), as well as a significant number of metabolites needed for a global interpretation of the changes in the metabolic pathways. Using the above-mentioned cut-off, the number of metabolites discriminating the premutation carriers from controls was reduced to 64 (48% of all originally identified). Following the BRITE classification within the KEGG database, plasma metabolites with biological roles were divided into the following categories (Figure 1B): carbohydrates and related (n = 17); amino acids, derivatives and biogenic amines (n = 17); carboxylic acids (n = 18); lipids (n = 15); and others (n = 3). Of note, a significant increase in metabolites belonging to the “carbohydrate” category was observed in the premutation group, which mirrored the decline in those within the “lipid category” (Figure 1B).
Figure 1. Plasma metabolite levels in carriers of the premutation and matched-controls.
A. Partial least squares-discriminant analysis of the metabolites obtained with both diagnostic groups shown with VIP ≥ 0.8 and their corresponding fold change. In italics, xenobiotics. B. Number of plasma metabolites from carriers separated by their biological roles according to BRITE (KEGG database). Metabolites with biological roles were divided into the following categories: carbohydrates and related (n = 17); amino acids, derivatives and biogenic amines (n = 17); carboxylic acids (n = 18); lipids (n = 15); and others (n = 3). Red and green columns represent respectively the number of metabolites with higher and lower abundance in carriers than controls.
b. Pathway discovery analyses
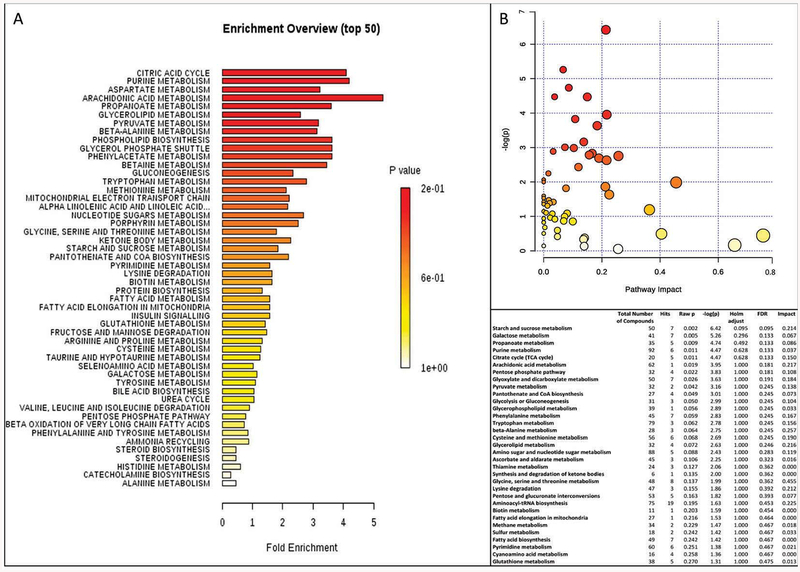
By performing PLS-DA or fold-change analyses, the potential to identify subtle, but consistent changes among a group of related compounds, could be undermined. To overcome this obstacle, plasma metabolites along with their relative concentrations were analyzed using a Metabolite Set Enrichment Analysis (MSEA). This approach identifies biologically meaningful patterns that are significantly enriched in quantitative metabolomic data. MSEA is a metabolomic version of the popular GSEA (Gene Set Enrichment Analysis) approach with its own collection of metabolite libraries analyzing directly a set of functionally related metabolites without the need to preselect compounds based on some arbitrary cut-off threshold. Metabolomic data from controls and carriers showed that the pathways significantly enriched were (in decreasing order of significance and with FDR < 0.05; Figure 2A) (i) citric acid cycle, (ii) the metabolism of the following compounds: purine, Asp, arachidonic acid, propanoate, glycerolipid, pyruvate, beta-alanine, and phospholipid biosynthesis, (iii) the glycerolphosphate shuttle (which serves to transport reducing equivalents from the cytosol to mitochondria), and (iv) the metabolism of phenylacetate and betaine.
Figure 2. Pathway over-representation analysis derived from plasma metabolomics.
A. Pathways overrepresented in premutation vs. controls were obtained by utilizing the Metabolite Set Enrichment Analysis (MSEA). B. Metabolites were analyzed using the Pathway Analysis module of MetaboAnalyst, which combines results from the pathway enrichment analysis with the pathway topology analysis to identify the most relevant pathways in the premutation. The table includes the data with p ≤ 0.27 associated with the pathway impact plot.
In parallel, we utilized the Pathway Analysis module of MetaboAnalyst, which combines results from the pathway enrichment analysis with the pathway topology analysis to identify the most relevant pathways in the premutation category. Pathway enrichment analysis usually refers to quantitative enrichment analysis directly using the compound concentration values, as compared to compound lists used by over-representation analysis. As indicated above, this analysis is more sensitive and has the potential to identify subtle but consistent changes among compounds involved in the same biological pathway. For this analysis, concentrations of each metabolite—normalized to averaged control values—were used as input values, Globaltest was the pathway enrichment analysis method and the node importance measure for topological analysis was the relative betweenness centrality (which measures the number of shortest paths going through the node for metabolite importance measure focusing on global network topology [42]). A graphical and detailed list of the pathways identified and their relative impact are shown in Figure 2B. The most significant ones (in decreasing order) were carbohydrate and lipid metabolism including galactose, propanoate, TCA, arachidonic acid, pentose phosphate pathway, pyruvate metabolism, panthotenate and glycolysis or gluconeogenesis.
Taken together, both analyses concurred on most of the pathways (TCA and fatty acid metabolism including glycerolphosphate shuttle) and extended some of the conclusions to other pathways (pentose phosphate pathway, gluconeogenesis).
b.1. Fatty acid and carbohydrate metabolism
Lower levels of plasma fatty acids (C9–C20) and derivatives were observed in carriers vs. controls (Figure 1A; Table 2). These findings along with elevated levels of adipic acid (a dicarboxylic acid), glycine and glycerol and lower levels of the ketone body 3-hydroxybutyrate resembled some of the features—although not as marked—of the nonketotic, hyperglycinemia with glyceric acidemia syndrome. This scenario may be interpreted as an increased lipolysis (higher glycerol) followed by increased hepatic fatty acid oxidation (decreased circulating FFA, preferentially outside mitochondrial fatty acid oxidation) to fuel the hepatic gluconeogenesis pathway. The fact that glycine levels were higher in carriers than controls is suggestive of a decreased activity of the mitochondrial glycine cleavage system. If this were the case, an imbalanced redox status could be occurring in mitochondria characterized by higher [NADH]/[NAD+] ratios. Higher [NADH]/[NAD+] ratios may inhibit NAD-dependent dehydrogenases such as pyruvate dehydrogenase complex (PDHC), alpha-ketoglutarate dehydrogenase (AKGDH) and isocitrate dehydrogenase (ICDH). The lower entry of pyruvate into the TCA via PDHC could result in two options: one, to form oxaloacetate via the anaplerotic reaction catalyzed by pyruvate carboxylase or, two, form lactate via lactate dehydrogenase (Figure 3).
Table 2:
Fatty acid levels and ratios in plasma of carriers and controls
| Outcome | LOG2 P/C ratio | p-value |
|---|---|---|
| Saturated Fatty Acids | ||
| pelargonic acid | −1.12 | 0.015 |
| lauric acid | −1.12 | 0.218 |
| myristic acid | −1.38 | 0.079 |
| palmitic acid | −1.06 | 0.332 |
| stearic acid | −1.02 | 0.395 |
| behenic acid | −1.10 | 0.225 |
| arachidic acid | −1.01 | 0.399 |
| Monounsaturated Fatty Acids | ||
| palmitoleic acid | −1.64 | 0.018 |
| oleic acid | −1.36 | 0.025 |
| Polyunsaturated Fatty Acids | ||
| n-3 PUFA | ||
| docosahexaenoic acid | 1.09 | 0.179 |
| n-6 PUFA | ||
| linoleic acid | 1.02 | 0.232 |
| arachidonic acid | −1.27 | 0.021 |
| Ratio of n-3/n-6 | −1.38 | 0.015 |
| Estimation of enzymatic activity | ||
| SCD index (Δ6D and Δ5D; 20:4 n-6/20:3 n-6) | −1.30 | 0.039 |
| SCD n-7 index (SCD-16; 16:1 n-7/16:0) | −1.46 | 0.017 |
| SCD n-9 index (SCD-18; 18:1 n-9/18:0) | −1.21 | 0.075 |
| SCD index (MUFA/SFA) | −1.38 | 0.015 |
| De novo lipogenesis index (16:0/18:2 n-6) | −1.12 | 0.106 |
| Elongation index (18:0/16:0) | 1.05 | 0.079 |
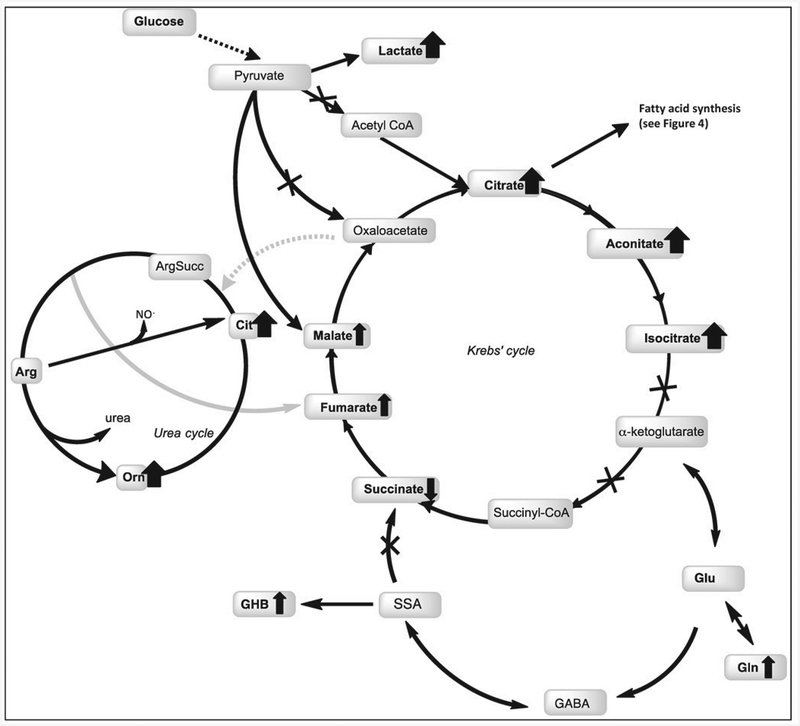
Figure 3. Overview of metabolites related to glycolysis and bioenergetics in the premutation.
Metabolites identified in this study are shown in bold. Bold wide arrows depict strong alterations (LOG2 ≥ ± 1.2; increase if up or decrease if down) whereas thinner ones indicate moderate fold changes (LOG2 ≥ ± 1.1). Gin in the production of GABA and alpha-ketoglutarate in the Krebs’ cycle is also shown. The lower entry of pyruvate into the TCA via PDHC could result in two options: 1) to form oxaloacetate via the anaplerotic reaction catalyzed bv pyruvate carboxylase or, 2) form lactate via lactate dehydrogenase. In support of the latter, several intermediates of the TCA cycle (citrate, aconitate, isocitrate), some TCA analogs (tartaric acid or 3-hydroxymalic acid), and lactate were found significantly higher in carriers suggesting a lower TCA activity. Furthermore, the higher ratio of Gln-to-Glu observed in plasma of premutations compared to controls suggested an increased flux from TCA to Gin while the higher levels of GHB suggested an increased synthesis of GABA from Glu. The higher observed ratio of GHB-to-succinate in carriers is indicative of a lower succinic semialdehyde dehydrogenase (SSADH) activity, probably due to either a direct oxidation of critical Cys (e.g., increased oxidative stress conditions within mitochondria) or genetics contributing to the permutation phenotype. Other abbreviations used: GABA, 4-aminobutyric acid; GHB: 4-hydroxybutyrate; SSA, succinic semialdehyde; Cit, citrulline.
In support of the latter option, several intermediates of the TCA cycle, especially those located within the first half of the cycle—namely citrate, aconitate, isocitrate—, some TCA analogs (such as tartaric acid or 3-hydroxymalic acid), and lactate were higher in carriers than controls (1.6 to 1.9-fold) suggesting a lower TCA activity (Figure 3). Furthermore, the higher ratio of Gln-to-Glu observed in plasma of premutations compared to controls (2.3-fold) suggested an increased flux from TCA to Gin while the higher 4-hydroxybutyrate levels (GHB, a GABA derivative) suggested an increased synthesis of GABA from Glu. Of note, the higher ratio of GHB-to-succinate in carriers than controls (2.2-fold; p = 0.035) is indicative of a lower succinic semialdehyde dehydrogenase (SSADH) activity, probably due to either a direct oxidation of critical Cys [43] (e.g., increased oxidative stress conditions within mitochondria) or genetics contributing to the premutation phenotype.
b.2. Fatty acids in prostaglandin pathways
To gain a deeper understanding of the fatty acid profile in carriers, we evaluated both the levels of free fatty acids between controls and carriers of the premutation and their ratios, which are predictive of the activation of prostaglandins of the series 2 vs. series 3.
The levels of pelargonic, palmitoleic, oleic and arachidonic acids were lower than controls (respectively −1.12, −1.64, −1.36, −1.47 with p = 0.015, 0.018, 0.025, 0.021; Figure 1A; Table 2) as well as the ratio of fatty acids of the n-3 series over the n-6 series (docosahexaenoic over that of linoleic and arachidonic acids; −1.38-fold; p = 0.015; Table 2). The levels of myristic acid followed the same trend as that of other fatty acids (−1.38 of controls) but without reaching a statistical significance (p = 0.079). Fatty acid ratios were used to estimate enzymatic activities of steps involved in fatty acid desaturation, elongation and de novo lipogenesis (Figure 4; Table 2). As such, the estimation of the activity of the stearoylCoA desaturase (SCD-1), rate-limiting enzyme in monounsaturated fatty acid biosynthesis, which converts palmitic to palmitoleic and stearic acid to oleic acid, was calculated as the SCD-16 (C16:l n-7/C16:0) and (C18:l n-9/C18:0) ratios as described by [44], The SCD-16 (C16:l n-7/C16:0) was significantly lower in carriers than controls (−1.46; p = 0.017;_Table 2) indicating that the synthesis of palmitoleic acid was lower in these subjects. Similarly, the SCD-18 (C18:l n-9/C18:0), which promotes the endogenous synthesis of oleic acid from stearic acid, followed a similar trend but without reaching statistical significance (−1.21; p = 0.075; Table 2). The monounsaturated to saturated fatty acids ratio [MUFA/SFA = (C18:l n-9 + C16:l n-7)/(C18:0 + C16:0); Table 2], also used as an index of the desaturase activity, confirmed the previous findings and it was significantly lower in carriers than controls (−1.38; p = 0.015; Table 2).
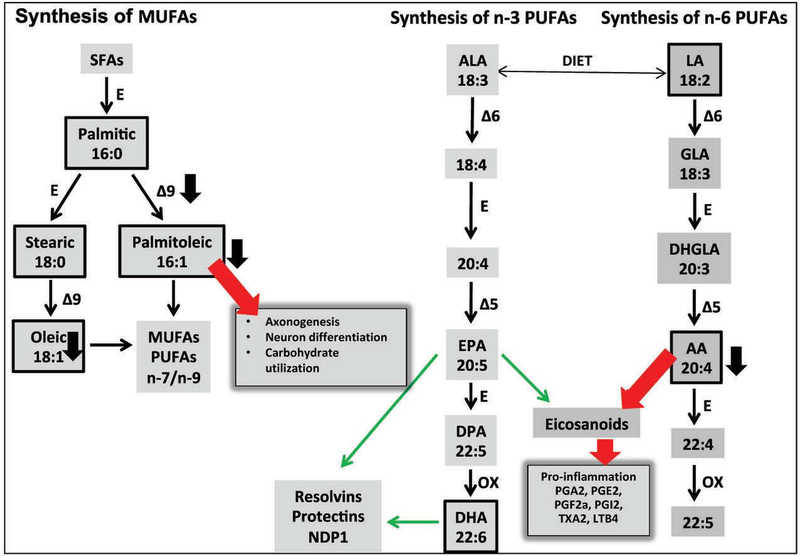
Figure 4. Overview of metabolites identified in the premutation in the context of the biosynthesis of fatty acids and eicosanoids.
Different fatty acid ratios were used to estimate enzymatic activities of steps involved in fatty acid desaturation, elongation and de novo lipogenesis. The ratio of arachidonic to linoleic acid ratio (C20:4 n-6/C18:2 n-6) was evaluated as an index of the activation of the pathway leading to the formation of prostaglandins of the 2 series starting from linoleic acid, via the formation of arachidonic acid. An increased pro-inflammatory status in carriers was inferred from the lower ratio of fatty acids of the n-3 series over that of the n-6 series, as prostaglandins generated via the Δ5–6 desaturase pathway are more pro-inflammatory than those originated from alpha-linolenic acid which generates prostaglandins of the 3 series (thick red arrows vs. thin green arrows). Boxed metabolites were identified by metabolomics. Black, thick arrows are based on the abundance indicated under Table 2. Abbreviations: ALA, alpha-linolenic acid; LA, linoleic acid; EPA, eicosapentaenoic acid; DPA, docosapentaenoic acid; DHA, docosahexaenoic acid; AA, arachidonic acid; DHGLA, dihomo-gamma-linolenic acid; MUFAs, monounsaturated fatty acids; PUFAS, polyunsaturated fatty acids; SFAs, saturated fatty acids; E, elongase; desaturases are indicated as delta followed by the bond affected; OX, peroxisomal beta-oxidation. Eicosanoids with anti-inflammatory properties (resolvins, protectins) are indicated with thin green arrows whereas pro-inflammatory (prostanglandins of the series 2 and LTB4) are indicated with thick red arrows.
The ratio of arachidonic to linoleic acid ratio (C20:4 n-6/C18:2 n-6) was evaluated as an index of the activation of the pathway leading to the formation of prostaglandins of the 2 series (PGA2, PGE2, PGF2α, PGI2, TXA2, LTB4; Figure 4) starting from linoleic acid, via the formation of arachidonic acid. This ratio (Δ6D and Δ5D in Table 2) was significantly lower in carriers than controls (−1.30; p = 0.039; Table 2) suggesting a lower activation of the n-6 long chain polyunsaturated fatty acids (i.e., Δ6D and Δ5D) or, more likely, an increased generation of prostaglandins of the series 2 from arachidonic acid which is not met by the demand.
b.3. Increased oxidative stress
Increased mitochondrial ROS production, elevated biomarkers of lipid and protein oxidative-nitrative damage, and decreased antioxidant defenses have been observed in lymphocytes, postmortem brain samples and fibroblasts from premutation individuals [11, 45, 46], Consistent with these findings, increased levels of metabolites derived from oxidative stress-mediated damage to proteins (aminomalonate) and carbohydrates (galacturonic acid) was also noted in the plasma of carriers. Lower levels of the amino acid derivative beta-Ala can result from lower catabolism of carnosine (dipeptide of beta-Ala and His taken from diet or synthesized in muscle or brain by carnosine synthetases), anserine (dipeptide of beta-Ala and MethylHis), dihydrouracyl or coenzyme A. Carnosine has been claimed to serve as a quencher of lipid peroxidation products such as 4-hydroxy-2-nonenal and malondialdehyde, to preclude glycation of proteins [47, 48] and to prevent neuronal cell death [49, 50], The fact that lower levels of both beta-Ala (−1.4-fold) and His (−1.2-fold) were observed in plasma from carriers compared to controls (Figure 1) might suggest an increased crosslinking of carnosine with peroxidation or oxidation products in an attempt to control increases in oxidative stress.
Plasma sorbitol and threonic acid (likely derived from glycated proteins) levels were significantly increased in plasma from premutation carriers (1.2-fold for both; Figure 1). While sorbitol has been linked to intestinal dysfunction [51], increases in both sorbitol and threonic acid concentrations in carriers are more consistent with increases in oxidative stress [52] as a result of a hyperactivation of the polyol metabolic pathway.
Damaged proteins are rapidly catabolized and the excess of nitrogen is disposed as urea. Indeed, the levels of citrulline and ornithine were higher (1.2-fold and 1.1-fold respectively; Figure 1) in plasma from carriers than controls, which may indicate an increased activity of the urea cycle.
In terms of antioxidant defenses, the higher levels of xylose (1.2-fold; Figure 1), erythritol (1.3-fold; Figure 1), and threitol (1.3-fold; Figure 1), probably to increase the availability of NADPH for antioxidant defenses, inferred higher glucose flux through the pentose phosphate pathway. However, the lower levels of both reduced and oxidized Cys (−1.3-fold, Figure 1), 3-hydroxybutyrate (−1.3-fold; Figure 1), and higher levels of Met (1.5-fold; Figure 1) were suggestive of a shift of homocysteine towards the transmethylation pathway (increased Met) from the trans-sulfuration one (to form Cys, 2-hydroxybutyrate). This shift may have an impact on glutathione metabolism, for Cys is a required building block for the synthesis of this antioxidant molecule.
A lower turnover of RNA (including mRNA) and DNA is supported by the lower plasma levels of hypoxanthine (−1.2-fold; Figure 1), allantoic (oxidation product of uric acid) and uric acids (purine metabolism; 1.1- and −2.1-fold respectively; Figure 1), beta-Ala (dihydrouracil; −1.4-fold; Figure 1) and aminoisobutyric acid (pyrimidine catabolism; −1.2-fold). These lower levels may suggest a decreased repair capacity of nucleic acids [53, 54], decreased transcription rate (decreased protein synthesis), and/or lower rate of cell division.
c. Correlation with CGG repeats expansion
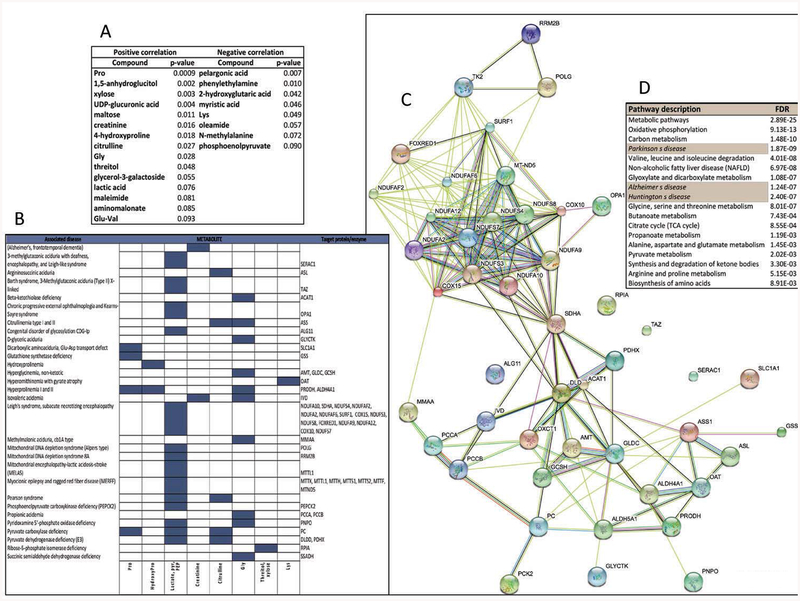
Considering that a number of mitochondrial outcomes (i.e., protein expression of ATPB, MnSOD and cytochrome c oxidase, subunit IV), as well as oxidative stress markers (nitrated ATPB) correlated with the CGG expansion in fibroblasts from carriers aged 41 to 81 years [11], and that the CGG expansion has also been shown to directly correlate with a disrupted/fragmented mitochondrial network [14], we tested whether plasma metabolite levels correlated with the triplet nucleotide expansion. From 143 metabolites, only 23 correlated significantly with CGG repeats, with 15 of them showing a direct correlation (Figure 5A; Supplementary Material, “Linear Regression” tab). Among these 15, five were amino acids or derivatives (Pro, Gly, hydroxyproline, citrulline and Glu-Val), seven were carbohydrates and derivatives (1,5-anhydroglucitol, xylose, UDP-glucuronic acid, maltose, threitol, glycerol-3-galactoside and lactic acid), and the rest were of different origin (creatinine, maleimide, aminomalonate). Those that followed a reciprocal correlation with the CGG expansion were three fatty acids and derivatives (pelargonic and myristic acids and oleamide), two amino acids and derivatives (Lys, methyalalanine), and others (phenylethylamine, 2-hydroxyglutaric acid and phosphoenolpyruvate). Recently we have shown that a panel of four core serum metabolites (namely, phenylethylamine, oleamide, aconitate and isocitrate) can be used for sensitive and specific diagnosis of the premutation [55], and one of these ratios (i.e., oleamide/isocitrate) showed significant potential as a specific biomarker for FXTAS [55], Indeed this ratio showed a trend decreasing as the stages of FXTAS increased in severity (Supplementary Figure 1). Although not significant due to the low number of carriers/stage, further research needs to expand the pool of FXTAS-affected carriers to confirm these findings (Table 1).
Figure 5. Correlation of plasma metabolites with CGG repeats and identification of the associated biological, cellular and molecular pathways.
A. List of plasma metabolites that showed a linear correlation with CGG expansion with the corresponding p values. More detailed information on the linear regression analysis is presented in the Supplementary Material. B. Prediction of enzyme or protein targets for those metabolites shown under panel A, whose concentrations were either elevated or reduced in plasma of genetic disorders listed under OMIM, Rare Metabolic Disease, HMDB, and Metagene databases. Rows represent disorders and columns represent the metabolites. If known, the causative gene’s name is indicated on the right. C. Using the putative targets identified under panel B as input data, a protein-protein interaction network was built with STRING. The interactions included both direct (physical) and indirect (functional) associations; they stem from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other (primary) databases. Detailed information of the analysis based on gene ontology for molecular function, biological process, cellular component and pathways is shown in Supplementary Material. D. The network generated under Panel C was used to generate a pathway analysis with the KEGG database. On the table it is shown the pathway/disease name (left) and false discovery rate (right).
To derive a more refined understating of those metabolites correlated with the CGG expansion, we manually extracted metabolite data from the OMIM, Rare Metabolic Disease, HMDB, and Metagene databases for genetic diseases in which any of these metabolites matched the same trend of levels observed in the carriers (e.g., elevated lactic acid compared to controls). This manual curation process permitted a finer tuned resolution of problems arising in the validation data, for example, from differences in metabolite nomenclature. The subset of genetic diseases that fulfilled at least one of these laboratory parameters, were used to generate a list of potential enzyme or protein targets (Figure 5B). A quick glance at the identified diseases revealed that several of them are considered mitochondrial disorders (Leigh’s, MELAS< MERFF, Alpers’, Pearson) suggesting that the plasma metabolites that correlated with CGG expansion are indeed associated with mitochondrial dysfunction. The above identified potential protein targets (Figure 5B) were used to build a protein-protein interaction network (Figure 5C), and these results were used for analyzing data in terms of their gene ontology (Figure 5D and Supplementary Material). In support of the previous conclusions, the most significant cellular component was identified as mitochondria, followed by cytosol, and myelin sheath (Supplementary Material, “Cellular compartment” tab). Along with this finding, molecular function and biological process gene ontology analyses revealed oxidoreductases and mitochondria-associated processes as the most significant ones (Supplementary Material, “Molecular function” and “Biological process” tabs). Pathway analysis performed by using the KEGG database, indicated not only the relevance of mitochondrial processes (oxidative phosphorylation, one-carbon metabolism or folate, branched-chain amino acids catabolism, Gly-Ser-Thr metabolism, Kreb’s cycle, Ala-Asp-Glu metabolism, ketone bodies metabolism) but also the connection to other neurological diseases such as Parkinson’s, Alzheimer’s, and another triplet nucleotide repeat disease (Huntington’s disease).
d. Xenobiotics of pharmaceutical or dietary origin
Current metabolomics approaches, in addition to measuring metabolites originated from endogenous cellular metabolism, also detect those derived exogenously from drugs, food, and cosmetics. Presumptive diet- or treatment-related findings were also noted for carriers as well as controls. Examples include, but were not limited to, food component/plant origin and pharmaceutical-related metabolites such as salicylate, naproxen, benzoic acid, quinic acid, gluconic acid, 1,2-cyclohexanedione, and glycolic acid. From the variety of medications that both groups were receiving (see Methods), carriers were more likely to be taking antidepressants than controls (30.4% vs. 6.25%; p < 0.0001); among them, the most common class was the selective serotonin reuptake inhibitors (SSRI; n = 6, including one control). However, no difference in metabolite levels was observed in our study consistent with the lack of differences in PUFA levels between medicated and unmedicated subjects reported by others [56, 57], No unmedicated subjects were present in any group, but we did not find that the metabolomics data correlated with the presence of the iatrogenic compounds indicated above or with antidepressants, antipsychotics, or mood stabilizers.
Discussion
The significant similarity of affected pathways based on changes in plasma and CSF metabolites and canonical pathways observed for Alzheimer’s and Parkinson’s diseases [58] supports the notion that plasma could be used to depict closely biochemical fingerprints of brain changes in carriers of the premutation. In this study, by using a non-targeted plasma metabolomics screening approach in a clinically well-characterized cohort of subjects, several pathways were identified as being altered in carriers of the premutation. Although it could be argued that plasma levels of metabolites do not represent those in brain, unesterified fatty acids readily cross the blood–brain barrier into the brain [59] representing the major peripheral form that mirrors PUFA metabolism in the brain. Additionally, it has been reported that decreases in RBC membrane PUFAs from subjects with schizophrenia correlates with the degree of demyelination in brain white matter [60].
Among the pathways affected inferred by the plasma metabolomics data, mitochondrial dysfunction was associated with a Warburg-like shift (confirming our previous study performed with lymphocytes from premutation individuals [46]) with increases in lactate levels, altered TCA intermediates and analogs including Glu and GHB, and a pro-inflammatory state as supported by the lipid profile as well as the increases in oxidative stress-mediated damage to carbohydrates and proteins.
Biological implications of the affected pathways in the context of the premutation
Consistent with our previous findings obtained with human carriers and animal models of the premutation, the current study also identified multiple pathways related to energy metabolism and, more specifically, to mitochondrial function that were already significantly affected in the premutation cohort [11–13, 61–64], Furthermore, specific markers that were elevated or reduced in plasma of carriers and that correlated with CGG repeats, identified not only mitochondrial pathways but also dysfunctional pathways shared by other neurological diseases such as Parkinson’s.
Evidence for mitochondrial dysfunction resulted from the higher levels of a dicarboxylic acid (adipate) and lactate (Table 3), both associated with mitochondrial respiratory chain disorders. Increases in TCA intermediates have also been observed in some mitochondrial diseases [65], generally associated with the formation of analogs of TCA intermediates (e.g., tartaric acid, methylmaleic acid or citraconic acid). These TCA analogs are known inhibitors of fumarase [66], which catalyzes the conversion of fumarate to malate. Indeed, fumarate concentrations were higher in premutation than controls (Figure 1A) suggesting that excess of fumarate may inhibit AKG-dependent prolylhydroxylases. This process would stabilize HIF-1α under normoxic conditions resulting in a constitutive activation of this factor and downstream genes, including glycolytic ones providing a mechanism for the metabolomic differences.
Table 3:
Affected biochemical pathways in premutation and their association to clinical manifestations
| Outcome measured | Phenotype or symptom |
|---|---|
| Increased adipate and lactate levels | Mitochondrial respiratory chain |
| disorders/deficits | |
| Increased TCA intermediates | Mitochondrial respiratory chain |
| disorders/deficits | |
| Decreased overall fatty acids levels | Depression |
| Decreased levels of oleic and arachidonic acids | Parkinson’s, Alzheimer’s |
| Decreased levels of n-3 and n-6 FA | Learning and memory function deficits |
| Decreased levels of n-3 over n-6 FA | Increased inflammation |
| Decreased levels of palmitoleic acid | Decreased axogenesis, neuronal |
| differentiation, carbohydrate utilization | |
| Increased mitochondria-derived oxidative stress | Anxiety-related disorders |
| Altered Glu, GHB | Anxiety-related disorders |
| Increased levels of sorbitol | Mood disorders |
| Mitochondrial dysfunction |
In Italics are clinical outcomes reported in premutation individuals.
Besides putative energy deficits, which may accompany mitochondria dysfunction, neurotransmission perturbations could also result from this scenario. Indeed, the altered plasma levels of Glu and GHB may signal an imbalance in neurotransmission, which is a hallmark of anxiety disorders [67] (Table 3). This is relevant considering that carriers affected with FXTAS have higher incidence of anxiety/mood disorders, and daughters of men with FXTAS have a higher prevalence of neurological symptoms including anxiety when compared with non-carrier female controls [1, 68, 69].
Lower plasma levels of FFA in carriers may reflect (a) the higher incidence of depression in carriers as lower levels of FFA have been reported in post-mortem brain samples from subjects affected with unipolar and bipolar depression [70], (b) neurodegeneration as lower levels of oleic and arachidonic acids have been reported in frontal cortex from subjects with Alzheimer’s disease [71] and with Parkinson’s disease [72], respectively (Table 3).
The lower ratio of fatty acids of the n-3 series over that of the n-6 series is suggestive of learning and memory issues in carriers vs. controls (Table 3) as it has been shown that n-3 PUFAs foster neuronal activity [73] counteracting memory deficits in aged mice [74], enhance the expression of mitochondrial ATPase (whereas its deficiency led to deficient glucose transport in cortex; [75]), increase the expression of Glu transporters 1 and 2 influencing neurotransmission [76], ameliorate spatial memory in rats by increasing the expression of subtypes of endocannabinoid receptors [77], increase the expression of transcription factors involved in learning and memory [78], and improve brain function and decreased levels of tau phosphorylation in a mouse model of Alzheimer’s with enhanced endogenous production of n-3 PUFA [79].
The significant decrease in plasma of palmitoleic acid in carriers of the premutation vs. controls along with the lower estimated enzymatic activity of SCD1 deserves further discussion considering the role of these factors in axonogenesis, neuron differentiation, and carbohydrate utilization (Table 3). Axonogenesis requires the de novo synthesis of monounsaturated fatty acids based on the evidence that brain-derived neurotrophic factor (BDNF) promotes both axonogenesis during brain development [80] while selectively increasing intracellular levels of palmitoleic acid and SCD1 [81], and that SCD-1 is highly expressed in axotomized neurons of the regenerating facial and hypoglossal nucleus [82]. More recently, a role for palmitoleic acid as an insulin-sensitizing lipokine has been proposed (although still controversial), suggesting that lower levels of this fatty acid may limit carbohydrate utilization. This is an important observation taking in consideration that type II diabetes mellitus is associated with an increased risk of cognitive dysfunction and dementia in Alzheimer’s disease [83].
An increased pro-inflammatory status in carriers was inferred from the lower ratio of fatty acids of the n-3 series over that of the n-6 series in which prostaglandins generated via the Δ5–6 desaturase pathway (or prostaglandins of the 2 series) are more pro-inflammatory than those originated from alpha-linolenic acid which generates prostaglandins of the 3 series [84] (Table 3 and Figure 4, thick red arrows vs. thin green arrows). In this regard, PGE2 in neural injury in Alzheimer’s disease is well documented, and includes modulation of protein-lipid interactions, trans-membrane and trans-synaptic signaling [85]. Moreover, PGE2 levels in CSF have been identified as one of the key pathways linked to Alzheimer’s disease severity [58]: PGE2 is higher in patients with mild memory impairment, but lower in those with more advanced Alzheimer’s disease [86].
While a pro-inflammatory state is usually accompanied by increases in free radicals, other pathways also related to oxidative stress were found affected in carriers, confirming findings recently obtained in fibroblasts from premutation individuals, in which increased mitochondrial ROS production was accompanied by increased mtDNA deletions and increased biomarkers of lipid and protein oxidative-nitrative damage [45], as well as in post-mortem brain samples [11], in which increase in oxidative/nitrative stress damage was preceded by mitochondrial dysfunction. Two metabolites identified in plasma belonging to the polyol pathway, a minor metabolic pathway of glucose running parallel to glycolysis, suggested a hyperactivation of this pathway, which leads to the formation of reactive aldehydes and biomolecule damage as evidenced by several metabolites in plasma (e.g., aminomalonate). Of interest, high levels of sorbitol have been reported in the CSF of subjects affected with mood disorder [87] and in cases with mitochondrial dysfunction [88] (Table 3).
Untargeted plasma metabolomics in the context of the premutation and other neurological disorders
We have recently shown that an untargeted serum metabolomic profiling approach, combined with sequential metabolite ratio analysis, has the potential to discriminate specific plasma biomarkers in FXTAS-free and FXTAS-affected carriers [55]. In particular, our results demonstrated that a panel of four core serum metabolites (phenylethylamine, oleamide, aconitate and isocitrate) could be used for sensitive and specific diagnosis of the premutation with and without FXTAS, and one of these ratios (oleamide/isocitrate) as a specific biomarker of FXTAS [55]. Here, we showed a statistically significant correlation between CGG expansion and 23 metabolites, some of which are associated with mitochondrial dysfunction (e.g., increased lactic acid) and two of them were among the biomarkers identified for carriers (phenylethylamine and oleamide) [55]. Taking together, these studies confirm previous evidence of mitochondrial deficits in human samples from carriers as well as in a number of cellular and animal models of the premutation [11–14, 46].
The advantage of using untargeted metabolomics over traditional targeted assays relates to the detection of several hundred analytes, which could not be detected in clinical biochemical genetics laboratory even when using the full repertoire of tests available. Therefore, plasma metabolomic analysis may be an attractive alternative when the carrier’s phenotype shows unusual presentation of symptoms and the clinician is considering ordering a combination of biochemical tests. One effective option would be to start with a metabolomic testing followed by endophenotype specific testing for disorders that might be missed by metabolomic analysis. Through this approach metabolomics analysis may expedite additional diagnosis while allowing for the possibility to identify sources of phenotypic heterogeneity within the broad umbrella of the FMR1 premutation. It is worth reiterating that all specimens were collected while the carriers—as well as controls—were on clinical management designed to alleviate their clinical phenotype. Interestingly, the fact that most subjects were on multiple medications did not impact our ability to identify significant differences between groups. This could be partially explained by the overlap of the medications or supplements (see Methods), the fact that the medication did not significantly affect the metabolomics data or that the medication was not optimized for the intended treatment.
Strength of the study is in utilizing metabolomics to detect changes in a broad variety of metabolites that reflect the complexity of metabolic networks altered in carriers. The accuracy of our findings was enhanced by the selection of carriers that closely match the control group on demographic factors. Future studies will need to assess the progression of changes in the identified pathways to shed light on the mechanisms contributing to the morbidity or as indicators of disease progression, especially at the early pre-clinical stages while considering the effect of sex, X-inactivation ratio, CGG repeats as well as FMR1 mRNA expression, and age on the morbidity of the disorder [89–92].
Supplementary Material
Acknowledgements
We wish to thank all subjects that participated in this study.
Funding information
This study was funded by National Institutes for Health (ES12691 and HD036071), Simons Foundation (#271406) and by the MIND Institute Intellectual and Developmental Disabilities Research Center (U54 HD079125).
Abbreviations list
- AKGDH
alpha-ketoglutarate dehydrogenase
- FFA
free fatty acids
- FXTAS
fragile X-associated tremor ataxia syndrome
- FMR1
fragile X mental retardation 1 gene
- FMRP
fragile X mental retardation protein
- GHB
4-hydroxybutyrate
- GSEA
gene set enrichment analysis
- ICDH
isocitrate dehydrogenase
- MSEA
metabolite set enrichment analysis
- PDHC
pyruvate dehydrogenase complex
- PLS-DA
partial least squares-discriminant analysis
- QEA
quantitative enrichment analysis
- SSADH
succinic semialdehyde dehydrogenase
- TCA
tricarboxylic acid cycle
- VIP
variable importance in projection
Footnotes
Declaration of interest
The authors have no conflicts of interest to disclose. R. Hagerman has received funding from Novartis, Roche/Genentech, Alcobra, Zynerba, and Neuren for treatment trials in fragile X syndrome, autism and Down syndrome. She has also consulted with Novartis and Roche/Genentech regarding treatment for fragile X syndrome. The other authors have no financial disclosures relevant to this article.
REFERENCES
- 1.Kogan CS, Turk J, Hagerman RJ and Cornish KM (2008) Impact of the Fragile X mental retardation 1 (FMR1) gene premutation on neuropsychiatric functioning in adult males without fragile X-associated Tremor/Ataxia syndrome: a controlled study. Am. J. Med. Genet. B Neuropsychiatr. Genet 147B: 859–872 doi 10.1002/ajmg.b.30685 [DOI] [PubMed] [Google Scholar]
- 2.Tassone F, Greco CM, Hunsaker MR, Seritan AL, Berman RF, Gane LW, Jacquemont S, Basuta K, Jin LW, Hagerman PJ and Hagerman RJ (2012) Neuropathological, clinical and molecular pathology in female fragile X premutation carriers with and without FXTAS. Genes Brain Behav. 11: 577–585 doi 10.1111/j.1601-183X.2012.00779.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battistella G, Niederhauser J, Fornari E, Hippolyte L, Gronchi Perrin A, Lesca G, Forzano F, Hagmann P, Vingerhoets FJ, Draganski B, Maeder P and Jacquemont S (2013) Brain structure in asymptomatic FMR1 premutation carriers at risk for fragile X-associated tremor/ataxia syndrome. Neurobiol. Aging 34: 1700–1707 doi 10.1016/j.neurobiolaging.2012.12.001 [DOI] [PubMed] [Google Scholar]
- 4.Cronister A, Schreiner R, Wittenberger M, Amiri K, Harris K and Hagerman RJ (1991) Heterozygous fragile X female: historical, physical, cognitive, and cytogenetic features. Am. J. Med. Genet 38: 269–274 [DOI] [PubMed] [Google Scholar]
- 5.Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B and Hagerman PJ (2001) Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 57: 127–130 [DOI] [PubMed] [Google Scholar]
- 6.Winarni TI, Chonchaiya W, Sumekar TA, Ashwood P, Morales GM, Tassone F, Nguyen DV, Faradz SM, Van de Water J, Cook K, Hamlin A, Mu Y, Hagerman PJ and Hagerman RJ (2012) Immune-mediated disorders among women carriers of fragile X premutation alleles. Am. J. Med. Genet. A 158A: 2473–2481 doi 10.1002/ajmg.a.35569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong LM, Goodrich-Hunsaker NJ, McLennan Y, Tassone F, Harvey D, Rivera SM and Simon TJ (2012) Young adult male carriers of the fragile X premutation exhibit genetically modulated impairments in visuospatial tasks controlled for psychomotor speed. J. Neurodev. Disord 4: 26 doi 10.1186/1866-1955-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagerman R and Hagerman P (2013) Advances in clinical and molecular understanding of the FMR1 premutation and fragile X-associated tremor/ataxia syndrome. Lancet Neurol. 12: 786–798 doi 10.1016/S1474-4422(13)70125-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chonchaiya W, Tardif T, Mai X, Xu L, Li M, Kaciroti N, Kileny PR, Shao J and Lozoff B (2013) Developmental trends in auditory processing can provide early predictions of language acquisition in young infants. Dev. Sci 16: 159–172 doi 10.1111/desc.12012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farzin F, Perry H, Hessl D, Loesch D, Cohen J, Bacalman S, Gane L, Tassone F, Hagerman P and Hagerman R (2006) Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J. Dev. Behav. Pediatr 27: S137–144 [DOI] [PubMed] [Google Scholar]
- 11.Ross-Inta C, Omanska-Klusek A, Wong S, Barrow C, Garcia-Arocena D, Iwahashi C, Berry-Kravis E, Hagerman RJ, Hagerman PJ and Giulivi C (2010) Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochem. J 429: 545–552 doi BJ20091960 [pii] 10.1042/BJ20091960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napoli E, Ross-Inta C, Wong S, Omanska-Klusek A, Barrow C, Iwahashi C, Garcia-Arocena D, Sakaguchi D, Berry-Kravis E, Hagerman R, Hagerman PJ and Giulivi C (2011) Altered zinc transport disrupts mitochondrial protein processing/import in fragile X-associated tremor/ataxia syndrome. Hum. Mol. Genet 20: 3079–3092 doi 10.1093/hmg/ddr211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Napoli E, Ross-Inta C, Song G, Wong S, Hagerman R, Gane LW, Smilowitz JT, Tassone F and Giulivi C (2016) Premutation in the Fragile X Mental Retardation 1 (FMR1) Gene Affects Maternal Zn-milk and Perinatal Brain Bioenergetics and Scaffolding. Front. Neurosci 10: 159 doi 10.3389/fnins.2016.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napoli E, Song G, Wong S, Hagerman R and Giulivi C (2016) Altered Bioenergetics in Primary Dermal Fibroblasts from Adult Carriers of the FMR1 Premutation Before the Onset of the Neurodegenerative Disease Fragile X-Associated Tremor/Ataxia Syndrome. Cerebellum: 1–13 doi 10.1007/s12311-016-0779-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seltzer MM, Baker MW, Hong J, Maenner M, Greenberg J and Mandel D (2012) Prevalence of CGG expansions of the FMR1 gene in a US population-based sample. Am. J. Med. Genet. B Neuropsychiatr. Genet 159B: 589–597 doi 10.1002/ajmg.b.32065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner E, Heilier JF, Ducruix C, Ezan E, Junot C and Tabet JC (2008) Mass spectrometry for the identification of the discriminating signals from metabolomics: current status and future trends. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871: 143–163 doi 10.1016/j.jchromb.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 17.Werner E, Croixmarie V, Umbdenstock T, Ezan E, Chaminade P, Tabet JC and Junot C (2008) Mass spectrometry-based metabolomics: accelerating the characterization of discriminating signals by combining statistical correlations and ultrahigh resolution. Anal. Chem 80: 4918–4932 doi 10.1021/ac800094p [DOI] [PubMed] [Google Scholar]
- 18.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG and Kell DB (2004) Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22: 245–252 doi 10.1016/j.tibtech.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 19.German JB, Hammock BD and Watkins SM (2005) Metabolomics: building on a century of biochemistry to guide human health. Metabolomics. 1: 3–9 doi 10.1007/s11306-005-1102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Kong H, Guan Y, Yang J, Gu J, Yang S and Xu G (2005) Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Anal. Chem 77: 4108–4116 doi 10.1021/ac0481001 [DOI] [PubMed] [Google Scholar]
- 21.Rozen S, Cudkowicz ME, Bogdanov M, Matson WR, Kristal BS, Beecher C, Harrison S, Vouros P, Flarakos J, Vigneau-Callahan K, Matson TD, Newhall KM, Beal MF, Brown RH Jr. and Kaddurah-Daouk R (2005) Metabolomic analysis and signatures in motor neuron disease. Metabolomics. 1: 101–108 doi 10.1007/s11306-005-4810-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherzer CR, Eklund AC, Morse LJ, Liao Z, Locascio JJ, Fefer D, Schwarzschild MA, Schlossmacher MG, Hauser MA, Vance JM, Sudarsky LR, Standaert DG, Growdon JH, Jensen RV and Gullans SR (2007) Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. U. S. A 104: 955–960 doi 10.1073/pnas.0610204104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barba I, Fernandez-Montesinos R, Garcia-Dorado D and Pozo D (2008) Alzheimer’s disease beyond the genomic era: nuclear magnetic resonance (NMR) spectroscopy-based metabolomics. J. Cell. Mol. Med 12: 1477–1485 doi 10.1111/j.1582-4934.2008.00385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudonck KJ, Mitchell MW, Nemet L, Keresztes L, Nyska A, Shinar D and Rosenstock M (2009) Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol. Pathol 37: 280–292 doi 10.1177/0192623309332992 [DOI] [PubMed] [Google Scholar]
- 25.Boudonck KJ, Rose DJ, Karoly ED, Lee DP, Lawton KA and Lapinskas PJ (2009) Metabolomics for early detection of drug-induced kidney injury: review of the current status. Bioanalysis. 1: 1645–1663 doi 10.4155/bio.09.142 [DOI] [PubMed] [Google Scholar]
- 26.Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA and Milburn MV (2008) Analysis of the adult human plasma metabolome. Pharmacogenomics. 9: 383–397 doi 10.2217/14622416.9.4.383 [DOI] [PubMed] [Google Scholar]
- 27.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C and Chinnaiyan AM (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 457: 910–914 doi 10.1038/nature07762 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 28.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE and Hagerman PJ (2000) Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 66: 6–15 doi 10.1086/302720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacquemont S, Hagerman RJ, Leehey M, Grigsby J, Zhang L, Brunberg JA, Greco C, Des Portes V, Jardini T, Levine R, Berry-Kravis E, Brown WT, Schaeffer S, Kissel J, Tassone F and Hagerman PJ (2003) Fragile X premutation tremor/ataxia syndrome: molecular, clinical, andneuroimaging correlates. Am. J. Hum. Genet 72: 869–878 doi 10.1086/374321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Napoli E, Tassone F, Wong S, Angkustsiri K, Simon TJ, Song G and Giulivi C (2015) Mitochondrial Citrate Transporter-dependent Metabolic Signature in the 22qll.2 Deletion Syndrome. J. Biol. Chem 290: 23240–23253 doi 10.1074/jbc.M115.672360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL and Adams SH (2010) Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One. 5: el5234 doi 10.1371/journal.pone.0015234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, Moon S and Nikolau B (2008) Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 53: 691–704 doi 10.1111/j.1365-313X.2007.03387.x [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J and Bryant SH (2016) PubChem Substance and Compound databases. Nucleic Acids Res. 44: D1202–1213 doi 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Kawashima M, Furumichi M and Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44: D457–462 doi 10.1093/nar/gkvl070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R and Scalbert A (2013) HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 41: D801–807 doi 10.1093/nar/gksl065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ and Forsythe I (2009) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37: D603–610 doi 10.1093/nar/gkn810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ and Querengesser L (2007) HMDB: the Human Metabolome Database. Nucleic Acids Res. 35: D521–526 doi 10.1093/nar/gk1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wohlgemuth G, Haldiya PK, Willighagen E, Kind T and Fiehn O (2010) The Chemical Translation Service—a web-based tool to improve standardization of metabolomic reports. Bioinformatics. 26: 2647–2648 doi 10.1093/bioinformatics/btq476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goeman JJ, van de Geer SA, de Kort F and van Houwelingen HC (2004) A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 20: 93–99 [DOI] [PubMed] [Google Scholar]
- 40.Eriksson L, J. E, Kettaneh-Wold N and Wold S (2001) Multi- and Megavariate Data Analysis Principles and Applications. Umetrics Academy, Umeå, Sweden [Google Scholar]
- 41.Ahmed SSSJ (2013) Systems biology in unruptured intracranial aneurysm: a metabolomics study in serum for the detection of biomarkers. Metabolomics. 10: 52–62 doi 10.1007/s11306-013-0551-8 [DOI] [Google Scholar]
- 42.Aittokallio T and Schwikowski B (2006) Graph-based methods for analysing networks in cell biology. Brief. Bioinform 7: 243–255 doi 10.1093/bib/bbl022 [DOI] [PubMed] [Google Scholar]
- 43.Kim YG, Lee S, Kwon OS, Park SY, Lee SJ, Park BJ and Kim KJ (2009) Redox-switch modulation of human SSADH by dynamic catalytic loop. EMBO J. 28: 959–968 doi 10.1038/emboj.2009.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ntambi JM and Miyazaki M (2004) Regulation of stearoyl-CoA desaturases and role in metabolism. Prog. Lipid Res. 43: 91–104 [DOI] [PubMed] [Google Scholar]
- 45.Song G, Napoli E, Wong S, Hagerman R, Liu S, Tassone F and Giulivi C (2016) Altered redox mitochondrial biology in the neurodegenerative disorder fragile X-tremor/ataxia syndrome: use of antioxidants in precision medicine. Mol Med. 22 doi 10.2119/molmed.2016.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Napoli E, Song G, Schneider A, Hagerman R, Eldeeb MA, Azarang A, Tassone F and Giulivi C (2016) Warburg effect linked to cognitive-executive deficits in FMR1 premutation. FASEB J. doi 10.1096/fj.201600315R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guiotto A, Ruzza P, Babizhayev MA and Calderan A (2007) Malondialdehyde scavenging and aldose-derived Schiff bases’ transglycation properties of synthetic histidyl-hydrazide carnosine analogs. Bioorg. Med. Chem 15: 6158–6163 doi 10.1016/j.bmc.2007.06.029 [DOI] [PubMed] [Google Scholar]
- 48.Reddy VP, Garrett MR, Perry G and Smith MA (2005) Carnosine: a versatile antioxidant and antiglycating agent. Sci. Aging Knowledge Environ. 2005: pel2 doi 10.1126/sageke.2005.18.pe12 [DOI] [PubMed] [Google Scholar]
- 49.Baek SH, Noh AR, Kim KA, Akram M, Shin YJ, Kim ES, Yu SW, Majid A and Bae ON (2014) Modulation of mitochondrial function and autophagy mediates carnosine neuroprotection against ischemic brain damage. Stroke. 45: 2438–2443 doi 10.1161/STROKEAHA.114.005183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulebyakin K, Karpova L, Lakonsteva E, Krasavin M and Boldyrev A (2012) Carnosine protects neurons against oxidative stress and modulates the time profile of MAPK cascade signaling. Amino Acids. 43: 91–96 doi 10.1007/s00726-011-1135-4 [DOI] [PubMed] [Google Scholar]
- 51.Cavanna A, Molino G, Ballare M, Torchio M, Fracchia M, Avagnina P and Bircher J (1987) Non-invasive evaluation of portal-systemic shunting in man by D-sorbitol bioavailability. J. Hepatol 5: 154–161 [DOI] [PubMed] [Google Scholar]
- 52.Johansen JS, Harris AK, Rychly DJ and Ergul A (2005) Oxidative stress and the use of antioxidants in diabetes: linking basic science to clinical practice. Cardiovasc. Diabetol 4: 5 doi 10.1186/1475-2840-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pang B, McFaline JL, Burgis NE, Dong M, Taghizadeh K, Sullivan MR, Elmquist CE, Cunningham RP and Dedon PC (2012) Defects in purine nucleotide metabolism lead to substantial incorporation of xanthine and hypoxanthine into DNA and RNA. Proc. Natl. Acad. Sci. U. S. A 109: 2319–2324 doi 10.1073/pnas.1118455109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bass BL (2002) RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 71: 817–846 doi 10.1146/annurev.biochem.71.110601.135501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giulivi C, Napoli E, Tassone F, Halmai J and Hagerman R (2016) Plasma biomarkers for monitoring brain pathophysiology in FMR1 premutation carriers. Front. Mol. Neurosci In press doi 10.3389/fnmol.2016.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saunders EF, Reider A, Singh G, Gelenberg AJ and Rapoport SI (2015) Low unesterified:esterified eicosapentaenoic acid (EPA) plasma concentration ratio is associated with bipolar disorder episodes, and omega-3 plasma concentrations are altered by treatment. Bipolar Disord. 17: 729–742 doi 10.1111/bdi.12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu CC, Huang SY, Su KP, Lu ML, Huang MC, Chen CC and Shen WW (2003) Polyunsaturated fatty acid deficit in patients with bipolar mania. Eur. Neuropsychopharmacol 13: 99–103 [DOI] [PubMed] [Google Scholar]
- 58.Trushina E, Dutta T, Persson XM, Mielke MM and Petersen RC (2013) Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS One. 8: e63644 doi 10.1371/journal.pone.0063644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouellet M, Emond V, Chen CT, Julien C, Bourasset F, Oddo S, LaFerla F, Bazinet RP and Calon F (2009) Diffusion of docosahexaenoic and eicosapentaenoic acids through the blood-brain barrier: An in situ cerebral perfusion study. Neurochem. Int 55: 476–482 doi 10.1016/j.neuint.2009.04.018 [DOI] [PubMed] [Google Scholar]
- 60.Peters BD, Duran M, Vlieger EJ, Majoie CB, den Heeten GJ, Linszen DH and de Haan L (2009) Polyunsaturated fatty acids and brain white matter anisotropy in recent-onset schizophrenia: a preliminary study. Prostaglandins Leukot. Essent. Fatty Acids. 81: 61–63 doi 10.1016/j.plefa.2009.04.007 [DOI] [PubMed] [Google Scholar]
- 61.Napoli E, Song G, Wong S, Hagerman R and Giulivi C (2016) Altered Bioenergetics in Primary Dermal Fibroblasts from Adult Carriers of the FMR1 Premutation Before the Onset of the Neurodegenerative Disease Fragile X-Associated Tremor/Ataxia Syndrome. Cerebellum doi 10.1007/s12311-016-0779-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conca Dioguardi C, Uslu B, Haynes M, Kurus M, Gul M, Miao DQ, De Santis L, Ferrari M, Bellone S, Santin A, Giulivi C, Hoffman G, Usdin K and Johnson J (2016) Granulosa cell and oocyte mitochondrial abnormalities in a mouse model of fragile X primary ovarian insufficiency. Mol. Hum. Reprod doi 10.1093/molehr/gaw023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaplan ES, Cao Z, Hulsizer S, Tassone F, Berman RF, Hagerman PJ and Pessah IN (2012) Early mitochondrial abnormalities in hippocampal neurons cultured from Fmrl pre-mutation mouse model. J Neurochem. 123: 613–621 doi 10.1111/j.1471-4159.2012.07936.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conca Dioguardi C, Uslu B, Haynes M, Kurus M, Gul M, Miao D-Q, De Santis L, Ferrari M, Bellone S, Santin A, Giulivi C, Hoffman G, Usdin K and Johnson J (2016) Granulosa cell and oocyte mitochondrial abnormalities in a mouse model of fragile X primary ovarian insufficiency. Mol. Hum. Reprod doi 10.1093/molehr/gaw023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crippa BL, Leon E, Calhoun A, Lowichik A, Pasquali M and Longo N (2015) Biochemical abnormalities in Pearson syndrome. Am. J. Med. Genet. A 167A: 621–628 doi 10.1002/ajmg.a.36939 [DOI] [PubMed] [Google Scholar]
- 66.van Vugt-Lussenburg BM, van der Weel L, Hagen WR and Hagedoorn PL (2013) Biochemical similarities and differences between the catalytic [4Fe-4S] cluster containing fumarases FumA and FumB from Escherichia coli. PLoS One. 8: e55549 doi 10.1371/journal.pone.0055549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNaughton N (1997) Cognitive dysfunction resulting from hippocampal hyperactivity--a possible cause of anxiety disorder? Pharmacol. Biochem. Behav 56: 603–611 [DOI] [PubMed] [Google Scholar]
- 68.Bourgeois JA, Cogswell JB, Hessl D, Zhang L, Ono MY, Tassone F, Farzin F, Brunberg JA, Grigsby J and Hagerman RJ (2007) Cognitive, anxiety and mood disorders in the fragile X-associated tremor/ataxia syndrome. Gen. Hosp. Psychiatry 29: 349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chonchaiya W, Nguyen DV, Au J, Campos L, Berry-Kravis EM, Lohse K, Mu Y, Utari A, Hervey C, Wang L, Sorensen P, Cook K, Gane L, Tassone F and Hagerman RJ (2010) Clinical involvement in daughters of men with fragile X-associated tremor ataxia syndrome. Clin. Genet 78: 38–46 doi 10.1111/j.1399-0004.2010.01448.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conklin SM, Runyan CA, Leonard S, Reddy RD, Muldoon MF and Yao JK (2010) Age-related changes of n-3 and n-6 polyunsaturated fatty acids in the anterior cingulate cortex of individuals with major depressive disorder. Prostaglandins Leukot. Essent. Fatty Acids. 82: 111–119 doi 10.1016/j.plefa.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin V, Fabelo N, Santpere G, Puig B, Marin R, Ferrer I and Diaz M (2010) Lipid alterations in lipid rafts from Alzheimer’s disease human brain cortex. J. Alzheimers Dis. 19: 489–502 doi 10.3233/JAD-2010-1242 [DOI] [PubMed] [Google Scholar]
- 72.Fabelo N, Martin V, Santpere G, Marin R, Torrent L, Ferrer I and Diaz M (2011) Severe alterations in lipid composition of frontal cortex lipid rafts from Parkinson’s disease and incidental Parkinson’s disease. Mol. Med 17: 1107–1118 doi 10.2119/molmed.2011.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robson LG, Dyall S, Sidloff D and Michael-Titus AT (2010) Omega-3 polyunsaturated fatty acids increase the neurite outgrowth of rat sensory neurones throughout development and in aged animals. Neurobiol. Aging 31: 678–687 doi 10.1016/j.neurobiolaging.2008.05.027 [DOI] [PubMed] [Google Scholar]
- 74.Labrousse VF, Nadjar A, Joffre C, Costes L, Aubert A, Gregoire S, Bretillon L and Laye S (2012) Short-term long chain omega3 diet protects from neuroinflammatory processes and memory impairment in aged mice. PLoS One. 7: e36861 doi 10.1371/journal.pone.0036861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harbeby E, Jouin M, Alessandri JM, Lallemand MS, Linard A, Lavialle M, Huertas A, Cunnane SC and Guesnet P (2012) n-3 PUFA status affects expression of genes involved in neuroenergetics differently in the fronto-parietal cortex compared to the CA1 area of the hippocampus: effect of rest and neuronal activation in the rat. Prostaglandins Leukot. Essent. Fatty Acids. 86: 211–220 doi 10.1016/j.plefa.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 76.Blondeau N, Nguemeni C, Debruyne DN, Piens M, Wu X, Pan H, Hu X, Gandin C, Lipsky RH, Plumier JC, Marini AM and Heurteaux C (2009) Subchronic alpha-linolenic acid treatment enhances brain plasticity and exerts an antidepressant effect: a versatile potential therapy for stroke. Neuropsychopharmacol. 34: 2548–2559 doi 10.1038/npp.2009.84 [DOI] [PubMed] [Google Scholar]
- 77.Pan JP, Zhang HQ, Wei W, Guo YF, Na X, Cao XH and Liu LJ (2011) Some subtypes of endocannabinoid/endovanilloid receptors mediate docosahexaenoic acid-induced enhanced spatial memory in rats. Brain Res. 1412: 18–27 doi 10.1016/j.brainres.2011.07.015 [DOI] [PubMed] [Google Scholar]
- 78.Dyall SC, Michael GJ and Michael-Titus AT (2010) Omega-3 fatty acids reverse age-related decreases in nuclear receptors and increase neurogenesis in old rats. J. Neurosci. Res 88: 2091–2102 doi 10.1002/jnr.22390 [DOI] [PubMed] [Google Scholar]
- 79.Lebbadi M, Julien C, Phivilay A, Tremblay C, Emond V, Kang JX and Calon F (2011) Endogenous conversion of omega-6 into omega-3 fatty acids improves neuropathology in an animal model of Alzheimer’s disease. J. Alzheimers Dis. 27: 853–869 doi 10.3233/JAD-2011-111010 [DOI] [PubMed] [Google Scholar]
- 80.Cohen-Cory S and Fraser SE (1995) Effects of brain-derived neurotrophic factor on optic axon branching and remodelling in vivo. Nature. 378: 192–196 doi 10.1038/378192a0 [DOI] [PubMed] [Google Scholar]
- 81.Suzuki S, Hongli Q, Okada A, Kasama T, Ohta K, Warita K, Tanaka K, Miki T and Takeuchi Y (2012) BDNF-dependent accumulation of palmitoleic acid in CNS neurons. Cell. Mol. Neurobiol 32: 1367–1373 doi 10.1007/s10571-012-9863-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Breuer S, Pech K, Buss A, Spitzer C, Ozols J, Hoi EM, Heussen N, Noth J, Schwaiger FW and Schmitt AB (2004) Regulation of stearoyl-CoA desaturase-1 after central and peripheral nerve lesions. BMC Neurosci. 5: 15 doi 10.1186/1471-2202-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biessels GJ, Kappelle LJ and Utrecht Diabetic Encephalopathy Study, G. (2005) Increased risk of Alzheimer’s disease in Type II diabetes: insulin resistance of the brain or insulin-induced amyloidpathology? Biochem. Soc. Trans 33: 1041–1044 doi 10.1042/BST20051041 [DOI] [PubMed] [Google Scholar]
- 84.Das UN (2005) A defect in the activity of Delta6 and Delta5 desaturases may be a factor predisposing to the development of insulin resistance syndrome. Prostaglandins Leukot. Essent. Fatty Acids. 72: 343–350 doi 10.1016/j.plefa.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 85.Bazan NG, Colangelo V and Lukiw WJ (2002) Prostaglandins and other lipid mediators in Alzheimer’s disease. Prostagl. Other LipidMediat. 68–69: 197–210 [DOI] [PubMed] [Google Scholar]
- 86.Combrinck M, Williams J, De Berardinis MA, Warden D, Puopolo M, Smith AD and Minghetti L (2006) Levels of CSF prostaglandin E2, cognitive decline, and survival in Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 77: 85–88 doi 10.1136/jnnp.2005.063131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Regenold WT, Kling MA and Hauser P (2000) Elevated sorbitol concentration in the cerebrospinal fluid of patients with mood disorders. Psychoneuroendocrinol. 25: 593–606 [DOI] [PubMed] [Google Scholar]
- 88.Regenold WT, Phatak P, Makley MJ, Stone RD and Kling MA (2008) Cerebrospinal fluid evidence of increased extra-mitochondrial glucose metabolism implicates mitochondrial dysfunction in multiple sclerosis disease progression. J. Neurol. Sci 275: 106–112 doi 10.1016/j.jns.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alvarez-Mora MI, Rodriguez-Revenga L, Feliu A, Badenas C, Madrigal I and Mila M (2016) Skewed X Inactivation in Women Carrying the FMR1 Premutation and Its Relation with Fragile-X-Associated Tremor/Ataxia Syndrome. Neurodegener. Dis 16: 290–292 doi 10.1159/000441566 [DOI] [PubMed] [Google Scholar]
- 90.Berry-Kravis E, Potanos K, Weinberg D, Zhou L and Goetz CG (2005) Fragile X-associated tremor/ataxia syndrome in sisters related to X-inactivation. Ann. Neurol 57: 144–147 doi 10.1002/ana.20360 [DOI] [PubMed] [Google Scholar]
- 91.Al-Hinti JT, Nagan N and Harik SI (2007) Fragile X premutation in a woman with cognitive impairment, tremor, and history of premature ovarian failure. Alzheimer. Dis. Assoc. Disord 21: 262–264 doi 10.1097/WAD.0b013e31811ec130 [DOI] [PubMed] [Google Scholar]
- 92.Rodriguez-Revenga L, Pagonabarraga J, Gomez-Anson B, Lopez-Mourelo O, Madrigal I, Xuncla M, Kulisevsky J and Mila M (2010) Motor and mental dysfunction in mother-daughter transmitted FXTAS. Neurol. 75: 1370–1376 doi 10.1212/WNL.0b013e3181f73660 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.