Dear Editor,
CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated genes) surveillance complexes are RNA-based adaptive immune systems employed by prokaryotes against invading nucleic acids from bacteriophages and plasmids.1,2 The CRISPR-derived RNAs (crRNAs) guide the Cas effector complex to target and degrade the invading nucleic acids. Recently, bioinformatics analyses have revealed the presence of CRISPR–Cas loci in bacterial Tn7-like transposons, thereby implicating a functional relationship between RNA-guided DNA targeting and transposition, with the latter representing a new role unrelated to host defense.3 Support for this concept has emerged from recent functional studies on type I-F and type V-K effectors involved in sequence-specific DNA transposition,4,5 thereby significantly broadening the potential biological applications of CRISPR–Cas technology. To complement the available functional studies, our efforts have focused on structural studies of the Vibrio cholerae Tn6677 multi-subunit type I-F CascadecrRNA–TniQ complex, whereby transposition subunit TniQ initiates DNA transposition with the eventual help of other transposition-associated proteins TnsA, TnsB and TnsC in the gene cluster (Fig. 1a).
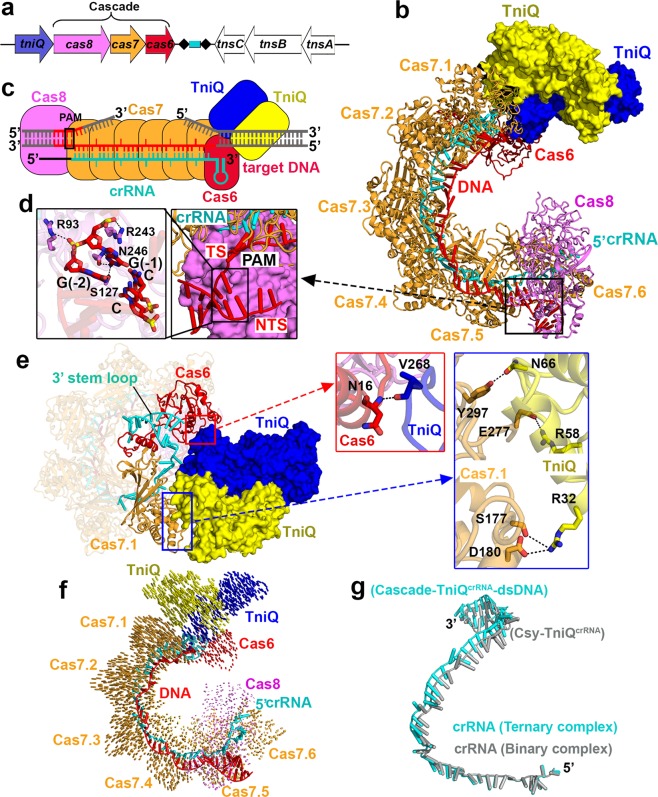
Fig. 1. Cryo-EM structures of type I-F CascadecrRNA–TniQ complexes.
a The cas and transposition-related genes in the Vibrio cholerae Tn6677 type I-F CRISPR–transposon system. b, c Schematic (c) and ribbon (b) representations of cryo-EM structure of CascadecrRNA–TniQ–dsDNA ternary complex. d PAM recognition by Cas8. Detailed interactions are shown in the expanded panels. e Interactions between the individual transposition protein TniQ and Cas6 and Cas7.1. Inset panels pointed by arrows provide detailed interactions. f Structure comparison between CascadecrRNA–TniQ binary complex and CascadecrRNA–TniQ–dsDNA ternary complex based on alignment of the Cas8 subunit. Vector length correlates with the domain movement scale. g Superposition of crRNA in the binary (in silver) and ternary (in cyan) complexes.
Here we report cryo-EM structures of a V. cholerae type I-F CascadecrRNA in complex with transposition subunit TniQ before (binary CascadecrRNA–TniQ) and after (ternary CascadecrRNA–TniQ–dsDNA) target double-stranded DNA (dsDNA) binding at an average resolution of 2.9 Å and 3.2 Å, respectively (Supplementary information, Figs. S1 and 2). Cas6 and TniQ can be readily traced in the 2.9 Å structure of the binary complex, thereby providing insights into how the three Cas subunits (Cas8, Cas7, and Cas6) of the multi-subunit type I-F Cascade are assembled in an intertwined helical topology with crRNA and TniQ.
The binary complex reveals a 1:6:1 Cas8:Cas7:Cas6 subunit stoichiometry similar to that reported for type I-F CascadecrRNA complexes,6–8 but contains a head-to-tail aligned TinQ dimer, whose individual monomers are bound to Cas7.1 and Cas6, so as to facilitate the first step of DNA transposition (Supplementary information, Fig. S3a). The complex is assembled with a 60-nucleotide (nt) crRNA processed by the CRISPR-specific endoribonuclease Cas6 from long precursor CRISPR transcripts (pre-crRNA), which contains 28-nt repeat sequences separated by 32-nt plasmid- or phage-derived spacer sequences. The partially palindromic repeat sequence leads to a stable stem-loop structure that could be recognized and cleaved by Cas6 (Supplementary information, Fig. S3b). Cas6 binds to the 3′ stem-loop of crRNA, while forming multiple polar interactions with Cas7.1 (Supplementary information, Fig. S3b, left insert). It has been shown that the trimming of pre-crRNA into crRNA is essential for both complex assembly9 and DNA transposition, given that the H29A mutant located in the pre-crRNA cleavage pocket shows no DNA transposition activity.5 Following Cas6-mediated maturation of crRNA, Cas proteins are assembled with crRNA, with the 5′ end recognized by Cas8 and the 3′ stem-loop held by Cas6, which is kinked by Cas7 thumb motifs in a periodic “5+1” pattern (Supplementary information, Fig. S3c).
Our CascadecrRNA–TniQ–dsDNA ternary complex contains a G–G/C–C PAM (protospacer adjacent motif) sequence (Fig. 1b), which is required for binding of the target sequence, allowing CRISPR–Cas systems to discriminate between self and non-self.10 The target sequence contains 12-base pair (bp) of PAM-proximal duplex DNA and 51-bp of PAM-distal duplex DNA (Fig. 1c; Supplementary information, Fig. S4). We could observe only 5-bp of PAM-proximal duplex DNA with 2-nt overhang at the 3′ end of the non-target strand (NTS), and a traceable 32-nt target DNA strand complementary and paired to 32-nt spacer RNA, but no clear density for the PAM-distal duplex segment, which is adjacent to bound TniQ. The traceable parts of the crRNA–dsDNA are colored, while non-traceable parts are shown in grey (Fig. 1c; Supplementary information, Fig. S4). The G–G/C–C PAM is specifically recognized by Ser127 and Asn246 in Cas8 (Fig. 1d). Arg243 forms a wedge and stacks with PAM G (–1), facilitating the formation of crRNA–target DNA heteroduplex and displacing the NTS, thus leading to the onset of R-loop formation.
It has been proposed that the transposition protein TniQ serves as an important connection between sequence-specific DNA targeting by the CascadecrRNA complex and DNA transposition by the accompanying transposase subunits TnsA, TnsB, and TnsC.5 In line with this concept, we observe that TniQ forms a head-to-tail dimer whose individual monomers simultaneously bind to Cas6 and Cas7.1 of the CascadecrRNA complex (Fig. 1b, c), with clear density observed at the interface between TniQ and both Cas6 and Cas7.1. One TniQ interacts with Cas6 via a main chain polar interaction between Val268 and Asn16, whereas the other TniQ interacts with Cas7.1 by multiple polar interactions between side chains over a larger interface (Fig. 1e, inset). We also determined the crystal structure of apo-TniQ at 2.1 Å resolution, demonstrating that TniQ forms a head-to-tail dimer with a dimeric interface of 1931 Å2 (Supplementary information, Fig. S5a). The dimer result was confirmed by SEC-MALS in solution (Supplementary information, Fig. S5b). Superposition of the TniQ dimer in the cryo-EM structure of ternary CascadecrRNA–TniQ–dsDNA complex (in color) with the crystal structure of the apo-form (in grey) reveals minimal conformational changes with an RMSD of 1.19 Å; the loop interacting with Cas6 becomes ordered and the helix interacting with Cas7.1 undergoes slight movement (circled in green, Supplementary information, Fig. S5c).
Notably, the binary and DNA-bound ternary CascadecrRNA–TniQ complexes superpose with an RMSD of 2.33 Å. Minimal conformational changes are observed on ternary complex formation in the presence of bound target DNA, reflecting slight opening of the entire complex (Fig. 1f) and crRNA (Fig. 1g), especially within the TniQ-bound end.
Our studies, and a parallel contribution11 provide insights into DNA targeting by the CascadecrRNA–TniQ complex, which represents an essential initial step in crRNA-guided DNA integration. The studies also provide structural insights into the potential use of RNA-guided Tn7-like transposons for genome editing. Further studies are required to investigate how the CascadecrRNA–TniQ complex recruits other transposon-associated proteins (TnsA, TnsB, and TnsC) to facilitate DNA transposition.
The atomic coordinates and EM maps have been deposited into the Protein Data Bank with the accession numbers 6V9P, 6V9Q, and 6VBW, and the EM Data Bank with the accession numbers EMD-21126 and EMD-21146.
Supplementary information
Supplementary information, Figures and Tables
Acknowledgements
We thank Chongyuan Wang for advice and discussion on cryo-EM aspects of the research and Hui Yang for sharing her crystal structure of apo-TniQ prior to publication. X-ray diffraction studies were based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on 24-ID-C beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. This research was supported by funds from the Geoffrey Beene Cancer Research Center and NIH GM129430 to D.J.P., and by Memorial Sloan-Kettering Cancer Center Core Grant (P30CA008748).
Author contributions
N.J. undertook all aspects of research from sample preparation and purification of Csm complexes, to cryo-EM grid preparation, data collection and processing, and structure refinement under the supervision of D.J.P. W.X. helped SEC-MALS analysis and sample preparation. In addition, M.D. and E.T.E. helped in cryo-EM data collection. N.J. and D.J.P. wrote the paper.
Competing interests
The authors declare no competing interests.
Contributor Information
Ning Jia, Email: jian@mskcc.org.
Dinshaw J. Patel, Email: pateld@mskcc.org
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0272-2.
References
- 1.Marraffini LA, Sontheimer EJ. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters JE, Makarova KS, Shmakov S, Koonin EV. Proc. Natl Acad. Sci. USA. 2017;114:E7358–E7366. doi: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strecker J, et al. Science. 2019;365:48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH. Nature. 2019;571:219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 6.Rollins MF, et al. Mol. Cell. 2019;74:132–142. doi: 10.1016/j.molcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo TW, et al. Cell. 2017;171:414–426. doi: 10.1016/j.cell.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury S, et al. Cell. 2017;169:47–57. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haurwitz RE, Sternberg SH, Doudna JA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah SA, Erdmann S, Mojica FJ, Garrett RA. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halpin-Healy, T. S., Klompe, S. E., Sternberg, S. H. & Fernández, I. S. Nature. 577, 271–274 (2020). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figures and Tables